Abstract
Clinical text is rich in acronyms and abbreviations, and they are highly ambiguous. As a pre-processing step before subsequent NLP analysis, we are developing and evaluating clinical abbreviation disambiguation methods. The evaluation of two sequential steps, the detection and the disambiguation of abbreviations, is reported here, for various types of clinical notes. For abbreviations detection, our result indicated the SPECIALIST Lexicon LRABR needed to be revised for better abbreviation detection. Our semi-supervised method using generated training data based on expanded form matching for 12 frequent abbreviations in our clinical notes reached over 90% accuracy in five-fold cross validation and unsupervised approach produced comparable results with the semi-supervised methods.
Introduction
Clinical notes are rich in acronyms and abbreviations. They allow for faster data entry, but are often ambiguous. According to Liu, 33.1% of UMLS (Unified Medical Language System) Metathesaurus abbreviations have multiple meanings,1 and we observed an even higher proportion of ambiguous abbreviations in clinical notes: 54.3%2. In these notes, the same abbreviation can have different meanings in different healthcare institutions, in different medical specialties, between different healthcare practitioners, and even in the same clinical note. For example, in the “History of Present Illness” section, in a sentence like “The PT was admitted for a left tibial closed spiroid fracture”, PT means Patient; but in the “Labs” section, in the sentence “His PT was initially measured at 21.s”, PT means Prothrombin Time; and in a “Discharge Instructions” section, in a sentence like “Follow-up with PT weekly for the next 2 months”, PT means Physical Therapy.
Moreover, ambiguous abbreviations often have different meanings in biomedical text (i.e., scientific publications) compared to clinical text. Natural Language Processing (NLP) applications developed for biomedical text have difficulties disambiguating abbreviations when used with clinical text. We would like an application that would understand “MD” as “Doctor of Medicine” in clinical text and “Mental Depression” in biomedical text.
Well aware of this issue, but also convinced by the quality of NLP applications developed for biomedical text such as MetaMap,3 we are developing an abbreviation disambiguation tool for clinical text in the context of the POET (Parsable Output Extracted from Text) project at the University of Utah. The evaluation of the tool, called ABRADe (ABbReviations and Acronyms Disambiguation) is presented here.
In this paper, we describe our methods for abbreviation detection and disambiguation, and their evaluation. We implemented a dictionary lookup module for abbreviations detection based on the SPECIALIST Lexicon LRABR resource. For abbreviation disambiguation, we used two approaches: a semi-supervised multi-class SVM classifier, and an unsupervised clustering method. The evaluation of both approaches is presented here.
Background
In the NLP domain, the correct interpretation of abbreviations has often been approached as a word sense disambiguation or text normalization problem.4,5,6 Several researchers have tried to resolve the ambiguity by using the proximity of the abbreviation with its expanded form, such as in “abbreviation (expanded form),” and this method showed good performance with biomedical text.7,8 However, this method does not work well with clinical notes. These notes are written for clinical experts and assume that these experts know the meaning of each abbreviation, and therefore it is extremely rare to see abbreviations with their expanded form.
Disambiguating abbreviations involves two main tasks: detecting abbreviations, and choosing the correct expanded form (i.e., meaning) for each abbreviation detected. Dictionary lookup and morphology-based matching have been used for the first task. For the second task, machine-learning approaches have been a popular choice. This task also requires more domain-specific knowledge, such as the UMLS SPECIALIST lexicon,9 a large biomedical and general English syntactic lexicon developed to provide the lexical information needed by various NLP applications developed by the National Library of Medicine. This lexicon includes syntactic, morphological, and orthographic information for each lexical item (word or term), and also includes the LRABR, a list of abbreviations and acronyms with their possible meanings. It lists 30,409 abbreviation-expanded form pairs (2009 release), with 18,335 unique abbreviations and 28,449 unique expanded forms.
For abbreviations detection, Liu et al.1 extracted 163,666 unique pairs of abbreviation and expanded form from 137,850 UMLS concept names with a subset of 16,855 UMLS abbreviations. They showed that 33.1% of abbreviations with six characters or less were ambiguous with an average of 2.28 expanded forms (i.e. meanings). They also found that the UMLS Metathesaurus included about 66% of the abbreviations in their clinical corpus.
Kim et al.2 used 30,409 abbreviation-expanded form pairs from LRABR, and found that 17.8% of LRABR abbreviations were ambiguous with 4.68 different expanded forms on average. Also, using simple dictionary lookup, they detected 166,905 abbreviations (of ≥ 2 characters) representing 1,593 different abbreviations in 10,000 clinical notes and showed that 53.3% of the abbreviations detected were ambiguous.
Xu et al.10 experimented with abbreviations detection without dictionary lookup and without an abbreviations database, using only general English terms and a medical lexicon. They detected abbreviations in 10 admission notes, using a decision tree classifier with morphological features and reached a precision of 91.4% and a recall of 80.3% for 411 abbreviations. They measured that the UMLS Metathesaurus covered 35% of the abbreviation expanded forms.
For abbreviation disambiguation, Pakhomov11 introduced a semi-supervised learning method based on Maximum Entropy to classify automatically generated training data based on local context. His method was based on the assumption that an abbreviation and one of its expanded forms were likely to have a similar distribution. He showed that there was a reasonable overlap between the contexts of an abbreviation and one of its expanded forms. His method showed over 89% cross validation accuracy on the generated training set.
Joshi et al.12 used fully supervised learning for acronym disambiguation in clinical notes with an accuracy around 90% with ten-fold cross validation. They annotated 7,738 instances of 16 ambiguous acronyms.
Methods
The detection of abbreviations starts with documents preprocessing. We use a sentence splitter adapted from openNLP and using the cTAKES13 trained model, and a simple tokenizer. The abbreviation annotator uses pattern matching and LRABR to annotate all abbreviations in our text. Each annotated abbreviation is then disambiguated as explained below. We also investigated the possibility to derive the abbreviation-expanded form pairs from the UMLS Metathesaurus, but chose the LRABR instead, for its superior coverage of abbreviations we could find in our corpus.
To evaluate the detection of abbreviations, we built a reference standard of 37 randomly selected clinical notes that were manually annotated for all abbreviations by two authors (SMM and JFH), with disagreement adjudication.
As mentioned above, we used two different approaches for abbreviations disambiguation: a semi-supervised and an unsupervised method. For the former, we built a training corpus of 9,963 (10,000 – 37) randomly selected clinical notes from the University of Utah Health Sciences Center, and created “synthetic” training cases using a method similar to the one proposed by Pakhomov11. We used the LRABR to detect all mentions of expanded forms (e.g., “cerebrovascular accident”) in 9,963 clinical notes, and then replaced the expanded form with the corresponding abbreviation (e.g., “CVA”). We then built feature vectors from these instances, with features that included the abbreviation itself, and the five preceding and following words (within the sentence) without any normalization. We pruned words that occurred less than five times in the training corpus.
Our multi-class SVM classifier used LIBLINEAR14 with a one-against-all strategy. LIBLINEAR is a linear classifier and can deal with large-scale problems very efficiently. We created one classifier containing every instance instead of making a different classifier for each abbreviation. The goal of this method was to demonstrate that words only in a small context window already allow for good performance using multi-class SVM classification. We used 5-fold cross validation on the training cases.
Our second approach was based on unsupervised hierarchical clustering. This method allowed grouping the expanded forms of each abbreviation based on the cosine similarity in the same feature words used for semi-supervised learning. We chose the single-link distance method15 to measure the distance between each cluster: the distance between the two nearest objects from two clusters, and used the clustering module available in Ling Pipe.16 We implemented this method hoping that if our clustering reached reasonable performance, or at least comparable with the semi-supervised method, we could then directly apply this clustering method to instances with abbreviations and not rely on the assumption that contexts around expanded forms will be similar to those of abbreviations. We generated additional training instances with real abbreviations, and then applied the clustering method with the same feature used for expanded forms. After clustering, each cluster was annotated manually. This method allowed us to compare the differences between training data generated from expanded forms (as described earlier) or generated from abbreviations.
Results
Abbreviations detection:
When comparing the reference with all abbreviations detected by our system, only satisfactory performance was observed, as shown in Table 1. Even partial matches (i.e., our system annotated only part of the abbreviation included in the reference standard) only reached an F1-measure of about 75%.
Table 1:
Abbreviation detection evaluation results.
| Recall | Precision | F1-measure | |
|---|---|---|---|
| Exact match | 63.71 | 68.32 | 65.94 |
| Partial match | 72.82 | 78.10 | 75.37 |
These results show the limitations of using a resource built for biomedical text (i.e. LRABR) with clinical text. Our system could only detect abbreviations found in LRABR. Table 2 lists the most frequent missed (i.e., false negative) and spurious (i.e., false positive) abbreviations with the number of instances in our 37 documents reference standard.
Table 2:
Top 10 false negative and false positive abbreviations
| False negatives | False positives | ||
|---|---|---|---|
| Abbreviation | Instances | Abbreviation | Instances |
| p.o. | 48 | He | 128 |
| b.i.d. | 27 | q | 50 |
| q.d. | 21 | b | 27 |
| MedQ | 15 | In | 26 |
| Mrs. | 13 | At | 24 |
| HEENT | 10 | K | 12 |
| mL | 8 | C | 9 |
| dL | 8 | S1 | 5 |
| MRN | 8 | Ms | 5 |
| q.h.s. | 7 | rehab | 3 |
Spurious abbreviations were listed in LRABR but not in the reference standard. For instance, “He” is mostly used as a pronoun in our notes, but is listed as an abbreviation in LRABR (e.g., Helium) and was detected by our system.
Semi-supervised abbreviations disambiguation:
In earlier research2, we presented the 20 most frequent ambiguous abbreviations in 10,000 notes of ten different types (Social Service Note IP, Rheumatology Clinic Note, Plastic Surgery Clinic Note, Operative Report, Obstetrics Gynecology Clinic Note, Hematology Oncology Clinic Note, Discharge Summary, Cardiology Clinic Note, Burn Clinic Note, and Admission H&P). For this classification task, we selected the 12 most frequent ambiguous abbreviations in our 9,963 training set notes, excluding abbreviations when one of their expanded form listed in LRABR corresponded to their meaning more than 90% of the times. When using abbreviation and expanded form pairs from LRABR to build synthetic training cases as explained in the Methods, we extracted 25,822 instances with very different distributions of each abbreviation as shown in Table 3.
Table 3:
Abbreviation instances in the training corpus and the number of different expanded forms for each abbreviation.
| Abbreviation | Instances in corpus | Different expanded forms in LRABR | Different expanded forms in corpus | Usage of LRABR (%) |
|---|---|---|---|---|
| PO | 9,030 | 10 | 4 | 40.00 |
| CA | 8,390 | 66 | 22 | 33.33 |
| AP | 3,893 | 47 | 21 | 44.68 |
| ER | 1,530 | 21 | 5 | 23.81 |
| RA | 1,121 | 27 | 14 | 51.85 |
| LV | 698 | 13 | 8 | 61.54 |
| IV | 392 | 16 | 6 | 37.50 |
| DVT | 310 | 2 | 2 | 100.00 |
| MD | 163 | 38 | 16 | 42.11 |
| PND | 134 | 9 | 3 | 33.33 |
| CVA | 113 | 3 | 3 | 100.00 |
| CT | 48 | 4 | 4 | 100.00 |
We then proceeded with abbreviation expanded forms classification with fivefold cross validation on the 9,963 documents training corpus. Table 4 shows the accuracy of LIBLINEAR’s classification for each abbreviation, followed by the percent coverage of the most and three common expanded forms. Table 5 shows the three most common expanded forms for each abbreviation. The LIBLINEAR accuracy was higher than just choosing the most common abbreviation expanded form, except with “DVT.”
Table 4:
Results of the semi-supervised method with five-fold cross validation on training instances
| Abbreviation | LIBLINEAR accuracy (%) | The most common (%) | 3 most common (%) |
|---|---|---|---|
| PO | 97.29 | 49.86 | 99.99 |
| CA | 89.58 | 58.95 | 89.60 |
| AP | 86.62 | 43.98 | 77.96 |
| ER | 96.21 | 65.23 | 96.27 |
| RA | 77.07 | 33.81 | 72.08 |
| LV | 90.69 | 80.95 | 96.86 |
| IV | 83.42 | 71.94 | 90.56 |
| DVT | 68.71 | 75.16 | 100.00 |
| MD | 46.01 | 39.26 | 67.48 |
| PND | 89.55 | 89.55 | 100.00 |
| CVA | 98.23 | 61.06 | 99.99 |
| CT | 68.75 | 60.42 | 93.76 |
| Micro-average | 91.09 | 53.83 | 91.42 |
| Macro-average | 82.68 | 60.85 | 90.38 |
Table 5:
Three most common expanded forms found in clinical documents.
| Abbreviation | Three most common expanded forms | ||
|---|---|---|---|
| PO | postoperative (49.86) | posterior (48.67) | perioperative (1.46) |
| CA | cancer (58.95) | coronary artery (17.12) | carcinoma (13.53) |
| AP | abdominal pain (43.98) | alkaline phosphatase (22.09) | appendectomy (11.89) |
| ER | emergency room (65.23) | external rotation (28.69) | extended release (2.35) |
| RA | right arm (33.81) | rheumatoid arthritis (28.19) | right atrial (10.08) |
| LV | left ventricular (80.95) | leucovorin (9.03) | left ventricle (6.88) |
| IV | invasive (71.94) | intraventricular (11.99) | initial visit (6.63) |
| DVT | deep venous thrombosis (75.16) | deep vein thrombosis (24.84) | |
| MD | major depression (39.26) | muscular dystrophy (14.72) | myelodysplasia (13.50) |
| PND | paroxysmal nocturnal dyspnea (89.55) | postnasal drip (9.70) | prenatal diagnosis (0.75) |
| CVA | cerebrovascular accident | costovertebral angle (38.05) | cardiovascular accident (0.88) |
| CT | (61.06) computed tomography (60.42) | connecticut (22.92) | computed tomographic (10.42) |
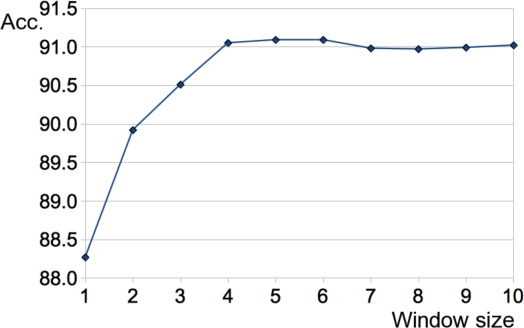
We also experimented with different sizes of context windows, ranging from one to ten words around the abbreviation, and observed a plateau after a size of 4 (Figure 1).
Figure 1.
Comparative result of different context window size
Unsupervised abbreviations clustering:
With the same features used for our semi-supervised classification (context window of size 5), we applied a clustering algorithm to our training instances, and got for example four clusters for “PO” and 22 for “CA.” Then, we were able to compare the clusters generated by LingPipe with training instances used for the semi-supervised method as reference standard. Table 6 shows the results of clustering by single-link with and a comparison with the accuracy of the semi-supervised method using LIBLINEAR. (Note: accuracy and F1-measure are equivalent when evaluating the semi-supervised method since the classifier must predict one expanded form and therefore produces no true negatives.) Recall and precision in clustering were calculated by comparing instances with the reference standard in each cluster, and counting them as true positives when placed in the cluster with the same manually assigned expanded form, false positives when having a different expanded form than the one assigned to the cluster, and false negatives when not found in the cluster with the same expanded form. For example, when examining the “per os” cluster, all instances of “per os” found in the cluster were true positives, all instances of other expanded forms found in the cluster were false positives, and all instances of “per os” not found in the cluster were false negatives.
Table 6:
Results of expanded form clustering for each abbreviation (Difference = LIBLINEAR – F1-measure)
| Abbreviation | Clustering | LIBLINEAR accuracy (%) | Difference | ||
|---|---|---|---|---|---|
| Recall | Precision | F1-measure | |||
| PO | 99.93 | 48.57 | 65.37 | 97.29 | 31.92 |
| CA | 99.64 | 39.77 | 56.85 | 89.58 | 32.73 |
| AP | 98.94 | 26.76 | 42.12 | 86.62 | 44.50 |
| ER | 99.42 | 50.88 | 67.31 | 96.21 | 28.90 |
| RA | 97.24 | 21.89 | 35.74 | 77.07 | 41.33 |
| LV | 98.89 | 67.45 | 80.20 | 90.69 | 10.49 |
| IV | 97.80 | 54.11 | 69.68 | 83.42 | 13.74 |
| DVT | 99.75 | 62.91 | 77.16 | 68.71 | −8.45 |
| MD | 82.33 | 20.70 | 33.08 | 46.01 | 12.93 |
| PND | 96.75 | 80.89 | 88.11 | 89.55 | 1.44 |
| CVA | 98.73 | 52.97 | 68.94 | 98.23 | 29.29 |
| CT | 90.56 | 44.48 | 59.66 | 68.75 | 9.09 |
| Micro Avg. | 99.33 | 42.17 | 58.45 | 91.09 | 32.64 |
| Macro Avg. | 96.67 | 47.62 | 62.02 | 82.68 | 20.66 |
The most different result was measured with “AP” (44.50%) and the smallest with “PND” (1.44%). While the unsupervised method results are usually lower than the semi-supervised method results (micro-average: 32.64%, macro-average: 20.66%), they were higher for “DVT”.
We applied the same method described above to generate new training instances, and extracted 24,065 instances containing one of 12 abbreviations instead of expanded forms. We then clustered them with the number of expanded form found in LRABR (for instance, 10 for “PO” was set as a cluster size, 66 for “CA,” etc.). Five words preceding and following the abbreviation were used to calculate cosine similarity. Clusters with less than five instances were removed (e.g., eight clusters for “PO” with less than 5 instances were removed).
A human expert (SMM) then reviewed the instances in each cluster and assigned the cluster to the most prevalent meaning (i.e., expanded form). A few clusters were merged when the human expert assigned them to the same expanded form. Table 7 includes information about each abbreviation’s clusters. The most frequent abbreviation in our corpus was “MD” with 7297 instances. In five abbreviations (“ER”, “LV”, “DVT”, “PND”, and “CT”), the assigned expanded forms were the same as the most common expanded form obtained with the semi-supervised method. However, abbreviations with numerous instances (such as, “MD”, “IV” and “PO”) had different expanded forms than with the semi-supervised method. We found out that “AP “and “ER” were sometimes used as initials of a reports author.
Table 7:
The results of expanded form clustering for each abbreviation from generated data based on abbreviation matching.
| Abbrev. | Instances | Number of cluster(s) | Nb. clusters after review | Most common expanded form(s) |
|---|---|---|---|---|
| PO | 1593 | 2 | 2 | per os (= orally), post office |
| CA | 775 | 1 | 1 | cancer antigen |
| AP | 893 | 3 | 3 | anteroposterior, alkaline phosphatase, initials of a reports author |
| ER | 913 | 2 | 2 | emergency room, initials of a reports author |
| RA | 590 | 1 | 1 | rheumatoid arthritis |
| LV | 588 | 2 | 1 | left ventricular |
| IV | 4595 | 1 | 1 | intravenous |
| DVT | 783 | 1 | 1 | deep vein thrombosis |
| MD | 7297 | 2 | 1 | Medicinae Doctor (= Doctor of Medicine) |
| PND | 395 | 1 | 1 | paroxysmal nocturnal dyspnea |
| CVA | 549 | 1 | 1 | costovertebral angle |
| CT | 5094 | 1 | 1 | computed tomography |
| All | 24065 | 18 | 16 |
Discussion
The abbreviation evaluation method we experimented with demonstrated that using a resource developed for biomedical text with clinical text did not allow for good performance. Adding domain- or institution-specific knowledge could increase recall significantly, and is the approach we plan to pursue, along with other improvements cited below. False positives could be reduced with some filtering heuristics or stop words (e.g., “He”, “In”, “At”). In this experiment, we detected abbreviations in a case sensitive way. It could therefore be possible to increase the recall by normalizing words to remove case information or punctuation in abbreviations. For instance, in the reference standard, “mL” is used for “milliliter” and “dl.” for “deciliter”. In LRABR, the abbreviations for these expanded forms are “ml” and “dl”. Detecting abbreviations with an edit distance method17 instead of strict string matching could also improve recall.
When evaluating our semi-supervised abbreviation disambiguation method, the accuracy measured with “DVT” was lower than simply using the most common expanded form because of the presence of two close synonyms in our corpus: “deep venous thrombosis” and “deep vein thrombosis”. Normalizing these terms could improve performance, and could be applied to other abbreviations like “left ventricular” and “left ventricle” for “LV,” or “computed tomography” and “computed tomographic” for “CT.” A terminological resource such as the UMLS Metathesaurus could also be used to link all synonyms (e.g., “deep venous thrombosis” and “deep vein thrombosis”) to one common concept.
The training cases we used relied on the “one sense per collocation” hypothesis proposed by Yarowsky18, that term (or abbreviations in our case) exhibit only one sense in a given collocation, in a given local context of surrounding words. A limitation here is that Yarowsky’s hypothesis could not always apply for abbreviations and their expanded forms. For instance, we found numerous “MD” in our corpus, used as an abbreviation of “Doctor of Medicine”, but almost never found the expanded form. Our method therefore failed to generate the training instances for “MD.” The distribution of abbreviation meanings was clearly different when manually annotating abbreviations (i.e., when building the reference standard), when searching for expanded forms and create the synthetic training cases (i.e., meanings; Table 5), and when assigning a meaning to clusters (Table 7), but the local context remained similar enough to provide consistent features for abbreviations meaning prediction (Table 4).
The unsupervised abbreviations clustering method gave us encouraging results, and has many opportunities for improvement such as a wider context window, using whole words in the sentence, using surrounding sentences as features etc. The cluster tree hierarchy of each abbreviation was very unbalanced, and most clusters had few instances and were removed. The main advantage of this method is its independence from the “one sense per collocation” hypothesis, that is could be used to detect several major expanded forms for each abbreviation.
Conclusion
Our abbreviations detection evaluation was based on a very useful resource, LRABR, but also showed its limitations to detect abbreviations in clinical notes when only using this resource. For better results, we will need more domain-specific knowledge, adding new abbreviations and expanded forms as found in clinical rather than biomedical text. Our semi-supervised abbreviation disambiguation method with 12 abbreviations reached over 90% accuracy with five-fold cross validation. We also implemented two preliminary experiments with unsupervised methods by calculating cosine similarity of context words.
Acknowledgments
NLM grants 5R21LM009967 and 3R21LM009967-01S1.
References
- 1.Liu H, Lussier YA, Friedman C. A study of abbreviations in the UMLS. Proc AMIA Symp. 2001:393–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim Y, Hurdle JF, Meystre SM. Acronyms and Abbreviations Ambiguity in Clinical Notes. AMIA Annu Symp Proc; 2010. [Google Scholar]
- 3.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Lesk M. Automatic sense disambiguation using machine readable dictionaries: how to tell a pine cone from an ice cream cone. Proceedings of the 5th annual international conference on Systems documentation; 1986. pp. 24–6. [Google Scholar]
- 5.Yarowsky D. Word-sense disambiguation using statistical models of Roget’s categories trained on large corpora. Proceedings of the 14th conference on Computational linguistics; 1992. pp. 23–8. [Google Scholar]
- 6.Sussna M. Word sense disambiguation for free-text indexing using a massive semantic network. Proceedings of the second international conference on Information and knowledge management; 1993. pp. 67–74. [Google Scholar]
- 7.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pac Symp Biocomput. 2003:451–62. [PubMed] [Google Scholar]
- 8.Chang JT, Schutze H, Altman RB. Creating an online dictionary of abbreviations from MEDLINE. J Am Med Inform Assoc. 2002 Nov-Dec;9(6):612–20. doi: 10.1197/jamia.M1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NLM UMLS SPECIALIST Lexicon. 2010. Available from: http://lexsrv3.nlm.nih.gov/SPECIALIST/index.html.
- 10.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes. Proc AMIA Symp. 2007:821–5. [PMC free article] [PubMed] [Google Scholar]
- 11.Pakhomov S. Semi-Supervised Maximum Entropy Based Approach to Acronym and Abbreviation Normalization in Medical Texts. Proceedings of 40th Annual Meeting of the ACL; 2002. pp. 160–7. [Google Scholar]
- 12.Joshi M, Pakhomov S, Pedersen T, Chute CG. A comparative study of supervised learning as applied to acronym expansion in clinical reports. Proc AMIA Symp. 2006:399–403. [PMC free article] [PubMed] [Google Scholar]
- 13.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan R, Chang K, Hsieh C, Wang X, Lin C. LIBLINEAR: A library for large linear classification. Journal of Machine Learning Research. 2008;9:1871–4. Software available at http://www.csie.ntu.edu.tw/~cjlin/liblinear. [Google Scholar]
- 15.Sibson R. SLINK: an optimally efficient algorithm for the single-link cluster method. Computer Journal. 1973;16:30–4. [Google Scholar]
- 16.Alias-i. LingPipe 2010. Available from: http://alias-i.com/lingpipe/.
- 17.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Physics Doklady. 1966;10(8):707–10. [Google Scholar]
- 18.Yarowsky D. Unsupervised Word Sense Disambiguation Rivaling Supervised Methods. ACL-95. Cambridge, MA: ACL; 1995. pp. 189–96. [Google Scholar]