Abstract
We performed an evaluation of the Unified Medical Language System (UMLS) in representing concepts derived from medical narrative documents from three domains: chest x-ray reports, discharge summaries and admission notes. We detected concepts in these documents by identifying noun phrases (NPs) and N-grams, including unigrams (single words), bigrams (word pairs) and trigrams (word triples). After removing NPs and N-grams that did not represent discrete clinical concepts, we processed the remaining with the UMLS MetaMap program. We manually reviewed the results of MetaMap processing to determine whether MetaMap found full, partial or no representation of the concept. For full representations, we determined whether post-coordination was required. Our results showed that a large portion of concepts found in clinical narrative documents are either unrepresented or poorly represented in the current version of the UMLS Metathesaurus and that post-coordination was often required in order to fully represent a concept.
Introduction
Because data in medical narrative documents is unstructured, it cannot be utilized in computerized applications, such as clinical decision support systems (CDSS). The need for automated processing and structuring of data in medical narrative documents was first recognized many years ago. In fact, research into medical Natural Language Processing (NLP) has been ongoing for nearly 40 years. Recent developments, including increased adoption of Electronic Health Records (EHRs) and the parallel growth of medical narrative data in digital form have made physician’s notes more available to computational processing than ever before. This, coupled with the fact that an estimated 50% of the total data in a patient’s medical record is in the form of medical narrative physician notes1 has increased the need for medical NLP systems that can extract information from this rapidly expanding data source.
A number of medical NLP systems have been described in the literature throughout the last 40 years2–5. One of the earliest reports of the potential use of NLP in a medical application was in 1978 by Collen6 where NLP techniques were proposed to aid in the difficulties of computers to acquire medical history data from patients directly. Other significant medical NLP advancements over the years include the development of the Medical Language Extraction and Encoding System (MedLEE), by Carol Friedman et al2 at Columbia University in the mid 1990s. In 2001, Aronson3 described the UMLS MetaMap program, produced by the National Library of Medicine (NLM). MetaMap is a highly configurable NLP program that maps biomedical text to concepts in the UMLS. In 2008 Elkin et al5 described the Multi-threaded Clinical Vocabulary Server (MCVS). Developed at the Mayo Clinic over several years, the MCVS parses free text reports into coded SNOMED CT reference terminology.
Two recent developments are likely to shape the future of medical NLP and could help speed medical NLP system development. First, in 2006 the first Informatics for Integrating Biology to the Bedside (i2b2) NLP Challenge was held7. Designated the smoking challenge, teams of NLP developers were tasked with creating systems that could accurately determine a patients smoking status based on medical text records. The i2b2 NLP challenge has become a yearly event with ever-increasing participation of teams of NLP developers7–9. The second significant development in the medical NLP community has been the release of open-source NLP toolkits, some specifically tailored to medical NLP. In April 2009, biomedical informatics researchers at Mayo Clinic and IBM launched a Web site for the newly founded Open Health Natural Language Processing (OHNLP) Consortium10. As part of the launch, Mayo Clinic and IBM released their clinical NLP technologies into the public domain. The site will allow researchers and developers working on NLP systems worldwide to contribute code and further develop the systems. Mayo Clinic and IBM jointly developed an NLP system for extracting information from more than 25 million free-text clinical notes based on IBM’s open-source Unstructured Information Management Architecture (UIMA)11. Mayo’s open-source solution, the clinical Text Analysis and Knowledge Extraction System (cTAKES)10, focuses on processing patient-centric clinical notes. IBM’s medKAT systems (medical Knowledge Analysis Tool) is a UIMA-based, modular and flexible system that uses advanced NLP techniques to extract structured information from unstructured data sources, such as pathology reports, clinical notes, discharge summaries and medical literature10. IBM’s Watson Deep QA system, which recently competed successfully on the American TV quiz show, Jeopardy!, was built using the UIMA system12. Another recent open-source NLP project is the General Architecture for Text Engineering (GATE) developed at the University of Sheffield13. Redesigned and re-released in 2002, GATE includes a free open source API, framework and graphical development environment to develop NLP systems.
It is clear that significant research and progress has been made in the field of medical NLP and in the technology necessary to identify and extract pertinent concepts from natural language medical reports. What is less clear is if we have the tools and terminologies necessary to adequately represent and structure these extracted concepts. In the broadest sense, for medical NLP systems to be the useful and effective they must perform two tasks:
Identify and extract all the relevant and pertinent concepts from medical narrative documents.
Map those concepts to a controlled medical terminology.
The ability to perform these two tasks is important in evaluating medical NLP systems. A primary goal of medical NLP is to take unstructured data (from free text reports) and structure it so it is then amenable to processing by computer applications such as CDSS and research queries. A medical NLP system that accurately identifies and extracts relevant concepts from text but poorly maps these concepts to a controlled medical terminology is not very useful. Likewise, an NLP system that effectively maps all extracted concepts to a controlled medical terminology, but can only identify and extract a small subset of the total concepts present in the document, is also not very useful.
The success of medical NLP systems to map extracted concepts to a controlled medical vocabulary depends on how well the concepts are represented by the terminology as well as the complexity of that representation. This is an important dependency since several medical NLP systems use a controlled terminology to define the universe of possible concepts they can identify3, 5. By default, such NLP systems can only identify concepts that are represented in the controlled medical terminology; a concept in the text not represented in the controlled medical terminology will be ignored by the system.
Given its importance in medical NLP, it seems appropriate to evaluate how well clinical concepts in medical corpuses are represented in controlled medical terminologies. Representation of concepts in a controlled medical terminology can be measured with two metrics: coverage and complexity. We define coverage here as the percentage of total corpus concepts that are represented by the terminology. Complexity refers to how complex it is to map the corpus concept to its controlled medical terminology representation. Mapping a clinical concept to it’s representation in a controlled medical terminology can be either simple or complex. The simplest mapping occurs when the clinical concept is fully represented in the vocabulary using pre-coordination – meaning the concept is fully represented using a single terminology entry. For example, the concept of ‘lower lobe pneumonia’ could map to a single coded entry in the terminology - ‘lower lobe pneumonia’. More complex mapping occurs when post-coordination is required – meaning that two or more terminology entries are needed to fully represent the concept. For example, the concept ‘lower lobe pneumonia’ could be represented in the terminology by entries such as ‘lower lobe’ + ‘pneumonia’ or ‘lower’ + ‘lobe’ + ‘pneumonia’.
To assess how well a controlled medical terminology represents the concepts contained in a corpus we need to first know what the possible concepts for a given corpus are. Unfortunately, this knowledge is often lacking especially in the clinical document domain. For example, do we know what concepts are possible in a corpus of radiology reports, and how frequently they occur?
The vast majority of medical NLP systems use the Unified Medical Language System (UMLS) Metathesaurus14 as the controlled medical terminology to represent and provide structure for corpus derived concepts. The most researched medical NLP system - MedLEE - includes UMLS codes (that have been mapped to its internal lexicon) in its XML output15. Most medical NLP systems in fact use only a small subset of the entire UMLS Metathesaurus - usually only SNOMED-CT (SCT) since it is the most clinically-rich and comprehensive clinically relevant vocabulary in the UMLS Metathesaurus. Using a single vocabulary decreases the need for disambiguation of matching terms and increases NLP processing speed. The UMLS Metathesaurus contains 158 highly variable vocabularies and not all have equal clinical relevance. Studies have shown that NLP system performance suffers when they attempt to map corpus derived concepts using all UMLS Metathesaurus vocabularies due to ambiguity of the mappings16.
Is SCT (or the UMLS in general) an appropriate controlled medical terminology for adequately representing corpus derived clinical concepts? Does the UMLS provide comprehensive representation of concepts that occur with high frequency in medical narrative documents? And if so, how complex is that representation? We attempt to answer these questions with our research.
Background
A few previous studies have evaluated the coverage of the UMLS Metathesaurus in representing clinical concepts in specific medical domains17, 18. In 2001, Friedman et al evaluated the UMLS as a source of lexical knowledge in 158 reports from two medical domains (chest x-rays and discharge summaries) using MedLEE18. They used a subset of the UMLS which they created using several filtering methods. They found that, compared to MedLEE’s internal lexicon, the UMLS lexicon did not perform as well at representing clinical concepts.
There is some evidence that the complexity of representation of clinical concepts in the UMLS is high. In 2007, Andrews et al19 compared the results of three professional coding services in mapping 319 clinical research question/answer sets to SCT terms. They found that only half of the time one companies’ codings could be even partially related to another companies codings, primarily via subsumption. And incredibly, the percentage of time the codings were equivalent between companies was extremely low. Coding equivalence between companies was only 9%, 4%, 34% for companies A to B, A to C and B to C respectively19. In 2006, Chaing et al20 measured agreement among three physicians using two SCT terminology browsers to encode 242 concepts from five ophthalmology case presentations published in a clinical journal. Inter-coder reliability, based on exact coding match by each physician, was only 44% using one browser and 53% using the other. In both studies, the authors conclude that the reliability of SCT coding is imperfect, and is in part a result of the terminology itself, the lack of a model for articulating rules of use for the terminology, as well as the absence of a model that formalizes SCT’s semantic structure in a manner more reflective of clinical use cases. Fung et al21 analyzed how well the UMLS represented concepts found in the narrow domain of problem lists. They found 1,134 (8%) of the terms in problem lists could not be mapped to the UMLS. A significant proportion of these could be derived from existing terms by the addition of modifiers, and they recommend the adoption of a consistent set of rules for combining concepts. Others have proposed the establishment of mapping rules to lessen complexity and achieve more consistent mappings22.
The above mentioned studies evaluated the coverage and/or mapping complexity of the UMLS Metathesaurus using small, domain specific corpuses and/or a limited number of clinical concept targets. In this research, we investigate the coverage and complexity of the UMLS in representing a much larger set of clinical concepts derived from medical narrative documents in three clinical domains.
Methods
All documents analyzed in this study are part of the Indiana Network for Patient Care (INPC), an operational health information exchange (HIE) that has been operating for nearly 12 years23. The INPC is one of the nation’s largest and longest tenured HIEs, containing over 30 million text reports representing over 11 million patients. It is maintained by the Regenstrief Institute24.
We randomly selected clinical narrative medical documents sent to the INPC during the interval from January 1, 2010 to June 30, 2010. We collected 3000 chest x-ray reports, 1000 discharge summaries and 1000 admission notes (history and physicals) from this period. All documents were in the Health Level Seven (HL7) message format, and were the identical messages sent to the INPC by medical transcription services. The classes of messages were identified by Logical Observation Identifiers Names and Codes (LOINC®)25. LOINC codes are assigned and inserted into the messages by the HIE upon receipt and local test code to LOINC code mappings are maintained by Regenstrief personnel.
To identify concepts in these documents, we identified noun phrases (NPs) as well as N-grams, including unigrams (single words), bigrams (word pairs) and trigrams (word triples). For each corpus, we also recorded the frequencies of NPs and N-grams at the corpus level. To identify NPs, we used an enhanced and modified version of the Stanford Parser26. The Stanford parser is a statistical parser trained on nonmedical documents (The Penn Treebank, WSJ section) and has been widely used in biomedical named entity recognition and relationship extraction27, 28. However, because it has been trained on nonmedical documents, certain report formatting and many biomedical and clinical terms likely to be found in medical report narratives were rarely encountered by the parser in training. Therefore, to improve the performance of the parser, we performed a number of enhancements. We added a preprocessing algorithm that improved tokenization and normalized word case. For example in some parts of these documents, such as section headings, words consist of all upper case characters. Since the parser uses lexical features such as word case during it’s processing, this text format can cause parser errors. We also performed a word frequency analysis of the documents we collected and adjusted the parser frequencies of some commonly used words when they were substantially different from those in our document set. Lastly, we augmented the parser lexicon by adding biomedical terms from the UMLS Specialist Lexicon and mapped the POS tags of these terms to standard Stanford Parser POS tags.
To identify N-grams, we used the sentence parser and word tokenizer that is part of the Regenstrief Extractor (REX) NLP software system. REX is a rule-based NLP software program that has successfully identified and extracted concepts from medical and other documents29–32. REX preprocesses documents in a number of ways including removing control characters, HTML tags, etc. and normalizes text by removing punctuation and converting text to lower case after sentence delimiting is performed. REX also calculated document level statistics including sentence number and word counts. Results from the Stanford parser and REX were output into a tab-delimited text file which was then imported into a MySQL database to aid in analysis.
We realize that the methods described above are inadequate to identify all concepts in medical documents. Likewise, we are aware that not all extracted NPs and N-grams will represent concepts. Our goal in this research is not to evaluate the accuracy of the Stanford Parser or our N-gram algorithm in identifying NPs or concepts. Nor is our goal to exhaustively identify every possible concept in the documents. Rather, our goal is to use these methods to identify a subset of likely concepts that occur repeatedly in order to evaluate the adequacy of the UMLS to represent them.
We used a number of methods to exclude from our analysis NPs and N-grams that did not represent discrete clinical concepts. We performed manual review of the NPs and N-grams to remove those that had negligible semantic meaning. For example, trigrams such as ‘and is otherwise’ and ‘be the only’ were removed. We also removed all single word NPs and excluded all two word NPs, unigrams and bigrams with stop words (common words that occur frequently in text) Longer multi-word NPs and trigrams with stop words were only included if the stop word did not begin or end the phrase. For example, we removed from our analysis bigrams such as “a pneumonia” and “lung the” and trigrams such as “a nonfluent aphasia”. We semi-automated the review process by using automated lexical analysis of the NPs and N-grams to isolate likely filtering candidates. We also automatically removed all NPs and N-grams that contained any numeric data (such as “#4”) using regular expressions. We used the Medical De-identification System (MeDS)33 to remove all NPs and N-grams that contained patient or provider identifying information such as “dr smith” and “January”. Finally, we performed a final manual review of all remaining NPs and N-grams to detect omission errors committed by the automated methods described above.
After the review, we processed all remaining NPs and N-grams with the UMLS MetaMap program 2010 Release3, 34. MetaMap is a NLP program that identifies UMLS concepts in text and returns them in a ranked list using a five step process that includes identifying simple NPs, generating variants of each phrase, finding matched phrases, assigning scores to matched phrases by comparing them with the input and composing mappings. MetaMap has been used in a number of applications35, 36. We set MetaMap parameters to allow it to search all UMLS sources for correct mappings and to return only what it considered to be the best mapping. We used no minimum mapping score threshold. We ran MetaMap using a strict model, which filters out terms with complex syntactic structure and is the MetaMap model recommended by NLM when performing semantic NLP processing37. We also set MetaMap parameters to return all source(s) and Semantic Types of the matching UMLS term(s). All other parameters remained in their default state. All NPs and N-grams were input into the MetaMap program as plain text strings.
We manually reviewed the results of MetaMap processing of NPs and N-grams. Because this was not an evaluation of MetaMap accuracy, when MetaMap found a matching UMLS map, we assumed the mapping was accurate and was the best possible mapping; we did not manually search the UMLS for alternative or better mappings. For each NP and N-gram we determined, through manual review of MetaMap output, whether MetaMap found a full representation or a partial representation of the concept. For full representations, we also determined whether post-coordination was required by counting the number of UMLS concepts (CUIs) MetaMap used in mapping the concept. For NPs and N-grams with no MetaMap mapping, we manually searched the UMLS to attempt to find a mapping. We used the UMLS Terminology Services Metathesaurus Browser (UMLS Browser)38 to perform this manual search.
To assess the coverage of SCT for representing corpus derived concepts, we performed an analysis of the frequency with which mapped terms identified by MetaMap were part of the SCT vocabulary. Because we set MetaMap to output all UMLS sources for all mapped terms, we performed this analysis by querying MetaMap output for the UMLS source code of ‘SNOMEDCT’. We performed this analysis on both complete and partial MetaMap matches.
Results
Our document collection consisted of a total of 5000 documents from three medical domains: 3000 chest x-ray reports, 1000 discharge summaries, and 1000 admission notes (history and physicals). Corpus level statistics are shown in Table 1.
Table 1.
Corpus level statistics.
| Corpus | Documents | Total sentences | Total words | Average words per report |
|---|---|---|---|---|
| Chest x-ray reports | 3000 | 34,378 | 331,919 | 111 |
| Discharge summaries | 1000 | 52,347 | 485,358 | 485 |
| Admission notes | 1000 | 105,856 | 874,655 | 875 |
We identified and extracted a total of 315,957 NPs and 1,009,063 N-grams from the three corpuses. Table 2 shows details of the NPs and N-grams extracted for each corpus.
Table 2.
NPs and N-grams identified for the three corpuses.
| Corpus | NPs | Unique NPs | Unigrams | Unique Unigrams | Bigrams | Unique Bigrams | Trigrams | Unique Trigrams |
|---|---|---|---|---|---|---|---|---|
| Chest x-ray reports | 43,775 | 5,975 (14%) | 76,485 | 2,073 (3%) | 41,300 | 8,363 (20%) | 34,669 | 13,724 (40%) |
| Discharge summaries | 93,919 | 19.790 (21%) | 135,203 | 8,549 (6%) | 72,866 | 28,592 (40%) | 71,513 | 41,369 (58%) |
| Admission notes | 178,263 | 21,181 (12%) | 243,377 | 9,137 (4%) | 140,899 | 32,027 (23%) | 136,796 | 47,480 (35%) |
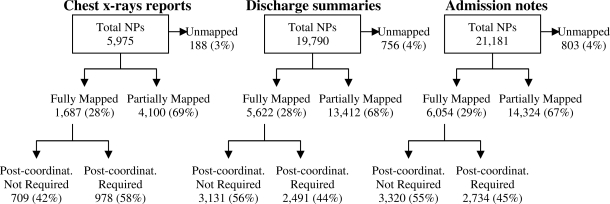
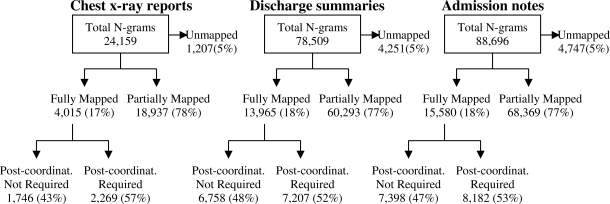
Figure 1 shows the results of MetaMap processing of the unique NPs for each corpus, while Figure 2 displays the results of MetaMap processing for the unique N-Grams.
Figure 1.
Results of MetaMap processing of unique noun phrases (NPs). Post-coordinat. = Post-coordination mapping.
Figure 2.
Results of MetaMap processing of unique N-grams. Post-coordinat. = Post-coordination mapping.
As shown in Figures 1 and 2, the majority of corpus derived concepts were at least partially mapped by MetaMap. For all corpuses, 5% or fewer concepts were completely unmapped. Nearly 28% of unique NPs were fully mapped while approximately 18% of the unique N-grams were fully mapped. Post-coordination was required slightly less often for fully mapped NPs (49%) compared to N-grams (54%); overall, post-coordination was required for 52 % of the fully mapped concepts.
MetaMap identified UMLS pre-coordinated concepts that represented fairly complex clinical concepts. Examples of these are shown in Table 3.
Table 3.
Examples of pre-coordinated UMLS representations of corpus derived concepts found by MetaMap.
| Corpus derived concept | UMLS representation |
|---|---|
| acute interstitial pneumonia | C1279945:Acute interstitial pneumonia |
| lymph node enlargement | C0497156:Enlargement of lymph nodes |
| adult onset diabetes | C0011860:Diabetes Mellitus Non-Insulin-Dependent |
| secondary malignant neoplasm | C0751623:Second Primary Cancers |
| random blood sugar | C1261429:Random blood glucose level result |
We analyzed the NPs and N-grams where MetaMap found no or only partial matching UMLS terms and we focused on the most frequently occurring corpus derived concepts. We empirically discovered five main reasons why MetaMap failed to completely map a corpus derived concept to a UMLS term. Below we define these reasons and provide estimates as to how often each resulted in non-mappings (in parentheses):
Specificity (45%) - The corpus derived concept was present in the UMLS, but in a more specific context than what was in the corpus.
Synonymy (28%) - The corpus derived concept was a synonym of an existing UMLS concept, but the synonymy relationship was missing from the UMLS.
MetaMap error (7%) - The corpus derived concept was a concept in the UMLS but MetaMap failed to map it.
Concept ambiguity (5%) - The corpus derived concept (or a part of it) was mapped to an incorrect UMLS term due to word sense ambiguity.
Concept missing (15%) - The corpus derived concept (or a part of it) was missing representation in the UMLS.
Due to space constraints, we will discuss only a few illustrative examples of each of these.
Specificity
For most corpus derived concepts where MetaMap found no or only a partial match, the UMLS had a variant of the concept but in a more specific form. For example, the trigram demonstrates no change was found 17 times in the chest x-ray report corpus. A typical sentence in the corpus with these terms is ‘Comparison made to prior study demonstrates no change’. MetaMap found no matching UMLS terms for this phrase, and a manual search for this term using the UMLS Browser revealed that the general concept ‘demonstrates’- meaning to ‘provide evidence for’ appears to be poorly represented. More specific UMLS terms such as ‘demonstrates flexibility’ and ‘demonstrates knowledge of medication management’ exist, but the singular concept ‘demonstrates’ is missing. An empirical review of a general radiology corpus revealed that this mental concept – where one entity provides evidence for another entity - is fairly common in this domain.
Another example in this category was the concept postprocedural which occurred 20 times in the discharge summary corpus. Typical phrases with this concept included ‘Postprocedural renal dialysis status’ and ‘Postprocedural aortocoronary bypass’. MetaMap found no mapping for this concept and a manual search of the UMLS for postprocedural revealed only more specific terms such as ‘C0341310:Postprocedural steatorrhea’ and ‘C2349671:Postprocedural fever’.
The concept atraumatic occurred 240 times in the admission note corpus. Typically, the term was used to mean no evidence of trauma in describing the results of an examination of the head such as ‘HEAD Atraumatic normocephalic’ but it was also used in describing extraocular eye movements, the scalp and the throat. A search of the UMLS for the term returned only these concepts:
C0186314: Reduction of atraumatic hip dislocation
C2717980: Dental Atraumatic Restorative Treatment
C2036876: atraumatic multidirectional subluxation of left shoulder
C2036878: atraumatic multidirectional subluxation of right shoulder
C0186315: Reduction of atraumatic hip dislocation with general anesthesia
Again, the concept of atraumatic by itself as a modifier for describing the results of an examination of a body part or region appears missing from the UMLS.
Synonymy
A fairly frequent concept found in chest x-ray reports is peribronchial cuffing which occurred 335 times in our chest x-ray corpus. Peribronchial cuffing, also referred to as peribronchial thickening or bronchial wall thickening, is a radiographic sign which occurs when excess fluid or mucus buildup in the small airway passages of the lung causes localized patches of atelectasis (lung collapse). MetaMap mapped this term only to ‘C0180207:Cuffs [Medical Device]’. A manual search of the UMLS for peribronchial cuffing returned no results. However, searching the UMLS for the synonymous term bronchial wall thickening returned ‘C1868833:Bronchial wall thickening [Disease or Syndrome]’. The UMLS lacks the knowledge that peribronchial cuffing is synonymous with bronchial wall thickening. It is likely that this phrase is not a local variant used only at our institution. A Google query for peribronchial cuffing returned 48,600 results, the Wikipedia had an entry for it, and PubMed returned 30 abstracts containing it.
MetaMap error
The concept increased bronchovascular markings was found 32 times in the chest x-ray report corpus. MetaMap found no matching UMLS term for this concept despite the presence of ‘C2073518:x-ray of chest: increased bronchovascular markings’ in the MEDCIN vocabulary.
Unnecessary post-coordination mapping was also due to apparent MetaMap errors. For example, the phrase left pleural effusion (which occurred 42 times in the chest x-ray report corpus) was mapped to ‘left [C1552822]’ and ‘pleural effusion disorder [C0032227]’ by MetaMap despite the presence of the UMLS concept ‘left-sided pleural effusion [C2063367]’. It is unclear why MetaMap ranked the mapping requiring post-coordination first; this behavior was not seen in other mappings. The UMLS concept ‘left-sided pleural effusion [C2063367]’ occurs only in a single source (MEDCIN). While we placed no source restrictions in running MetaMap, perhaps there are some internal source restrictions within the MetaMap strict model.
Concept ambiguity
The concept lungs are clear occurred 202 times in the chest x-ray report corpus. In this context, clinicians use this phrase to indicate that the lungs are free of consolidation, infiltrate or nodules. Sometimes it is used alone as in ‘Lungs are clear.’ and sometimes it is used to indicate what the lung is clear of as in ‘The lungs are clear of focal infiltrate’. MetaMap mapped this concept to ‘C0024109:Lungs’ and ‘C0522503:Clear (Translucent)’. It did not recognize that in this context ‘clear’ meant ‘without abnormality’ and not translucent. However, the UMLS does have the concept of ‘free of’ (‘C0332296: Free of (attribute)’) but no vocabulary has the synonymous term ‘clear of’.
Concept missing
Approximately 15% of the non-matched or partially matched concepts were the result of missing representation in the UMLS. A number of fairly frequent corpus derived concepts were not represented in the UMLS. For example, the concept organomegaly occurred 64 times in the admission note corpus. This term is frequently used by clinicians in order to expedite description of a physical exam and typically means no general organ enlargement on abdominal examination (instead of listing the abdominal organs individually such as no hepatomegaly, no splenomegaly, etc.) The UMLS does not include this concept and does not have a representation of ‘no general organ enlargement’, although it does have concepts that represent enlargement of the organs individually (I.e. hepatomegaly).
Another corpus derived concept that is missing from the UMLS is grossly which occurred in 201 chest xray reports. It is used by clinicians as an adjective to mean glaringly or flagrantly obvious as in ‘osseous structures are grossly intact, and ‘the lungs are grossly clear’. MetaMap found no matches for this concept, and a manual search of the UMLS revealed the only match to this term was ‘C0332441:Normal tissue morphology’.
The concept activity as tolerated occurred in 28 discharge summaries. It is used by clinicians to advise patients that they may resume normal activities after discharge and is typically part of the discharge instructions. MetaMap mapped this concept only to ‘C0441655:Activities’. A manual search of the UMLS found no adequate representation of this concept.
Some concepts, while not completely absent from the UMLS, are minimally represented. For example, the concept of silhouette is a frequently occurring concept in the chest x-ray report corpus. When used in radiology reports, it generally means the interface between a light and dark area on the film. It occurred 601 times in the chest-ray report corpus in various forms including ‘cardiac silhouette remains borderline enlarged’ and ‘hazy opacity silhouetting the left heart border’. MetaMap found no matches for this term, and a manual search of the UMLS revealed a single concept – ‘C0507134:Cardiac shadow viewed radiologically’ found only in two UMLS sources - Foundational Model of Anatomy Ontology (FMA) and the University of Washington Digital Anatomist (UWDA). This concept is missing from clinically relevant UMLS sources including SNOMED-CT and MESH. The concept advised to followup occurred in 28 discharge summaries. It is used by clinicians to document that they have advised the patient when or whom they should see for post-discharge care as in ‘advised to followup with his primary care physician, and ‘he was advised to followup in 2 weeks’. MetaMap found no matches for this concept and a manual search of the UMLS revealed ‘C0589120:Follow-up status’, ‘C1704685:Follow-Up Report’ and ‘C0016441:Follow-Up Studies’ but no concept representing a physician advising a patient regarding followup care.
Of the 238,260 corpus derived concepts mapped at least partially by MetaMap, 50,649 (21%) did not contain SCT as a UMLS source. This included 5,687, 21,562, and 23,400 concepts from the chest x-ray report, discharge summary, and admission note corpuses respectively. In other words, had we used only SCT as our source vocabulary, 50,649 concepts would have received no mapping by MetaMap.
Discussion
The aim of this study was to evaluate the use of the UMLS in representing concepts derived from clinical documents in three domains. We found that a large portion of concepts found in clinical narrative documents are either unrepresented or poorly represented in the current version of the UMLS Metathesaurus despite its incredibly diverse and comprehensive content. The problem of non-representation of clinical concepts is greater when examining specific UMLS vocabularies. Concepts occurring with high frequency in some corpuses, such as ‘pulmonary infiltrate’ in chest x-ray reports, are missing representation in SCT - the most comprehensive clinical content vocabulary within the UMLS. We hope these results encourage discussion on the need for more directed efforts to improve representation of corpus derived clinical concepts in the UMLS and its use for medical NLP.
Let’s assume for the moment that at some point in the near future, with further advances in NLP technology, faster processing speeds, and further expansion and development of complex knowledge bases, a medical NLP system will be developed that will identify and extract concepts from medical documents with near perfect accuracy. When such a system is developed, we will need a controlled medical terminology that is capable of effectively representing these concepts in standard form. While the UMLS is clearly the most comprehensive controlled medical terminology we have, it appears to need targeted modifications and enhancements before it can fully represent all clinical concepts in medical documents.
Use of the UMLS as a controlled medical terminology to represent and structure corpus derived concepts is becoming increasing complex and difficult due in part to the ever increasing size of the UMLS Metathesaurus. In 2003, the UMLS Metathesaurus contained 900 thousand concepts, and 2 million terms from 100 source vocabularies. Today it contains 2.3 million concepts, and 8.5 million terms from 158 source vocabularies14. The NLM acknowledges that as the size of the UMLS Metathesaurus increases, so does the problem of term ambiguity and the need for more effective word sense disambiguation (WSD)39. The larger size of the UMLS also increases medical NLP system processing time. One potential solution to address increased UMLS size is to filter the UMLS Metathesaurus for specific biomedical and clinical domains. Clearly, a significant number of the 8.5 million terms in the UMLS Metathesaurus are of little value for medical NLP purposes and are likely never to appear in biomedical and/or clinical text. And for the terms likely to appear in clinical documents, an even smaller number are likely to appear in clinical text within a given medical domain, such as discharge summaries. The NLM has recently begun to explore the creation of specific ‘content views’ of the UMLS Metathesaurus. In 2008, the Lister Hill NLP Content View (LNCV) project was initiated with the goal being to construct maintainable NLP content views of the Metathesaurus consisting of a biomedical literature view and multiple clinical views40. This work is ongoing and so far only the development and testing of methods to create content views for a subset of MEDLINE citations has been reported, but construction of multiple clinical content views are planned37. However, a prerequisite for construction of clinical content views is a thorough understanding of the target medical domain including knowledge about what concepts are important and how frequently they occur. Others have also recognized the difficulties caused by the ever-increasing size of the UMLS Metathesaurus and its effects on NLP precision and practicality. In 2010, Xu et al27 performed research to analyze the frequency and syntactic distribution of Metathesaurus terms in 18 million MEDLINE abstracts and to create a filtered UMLS Metathesaurus based on the MEDLINE analysis. They found that the resulting MEDLINE-filtered UMLS Metathesaurus contained just over 500k terms - an 87% decrease from its original size.
The size, efficiency, ease of use, and concept management and maintenance of a controlled terminology is related in part to its conceptual model. A terminology with many pre-coordinated terms is easier to use and results in more consistent and less complex concept mapping, but tends to be larger, less efficient, and harder to manage. A terminology with few pre-coordinated terms is smaller and more efficient, but can result in less consistent and more complex concept mapping. A terminology that allows for post-coordination, such as SCT, affords the greatest flexibility and expressiveness in creating new or complex concepts, but while use of post-coordination to represent concepts can be relatively straightforward and intuitive, at times it can be complex. When post-coordination is used to represent concepts, guidelines should be in place to ensure its correct use. As recommended by Rosenbloom et al.41 this includes restricting post-coordination to only meaningful concepts, preventing creation of duplicate concepts, and providing detailed instructions on how to use post-coordination within the terminologies data model. In our study, the vast majority of full or partially mapped clinical concepts required post-coordination for representation, which is similar to the findings in other studies19. This need for post-coordination and the complex mapping that results has been a reason for poor agreement among human coders in mapping concepts from text to SCT19, 20. Increasing the number of pre-coordinated terms in the UMLS that represent clinical concepts would help to lessen mapping complexity and help decrease coding variability.
There are several limitations to our study. Not all concepts in clinical documents are identified by our NP and N-gram extraction methods, so we clearly did not analyze the entire universe of concepts in these documents. Another limitation relates to our use of MetaMap. We used MetaMap to provide us with mappings, which we manually reviewed for the purposes of determining the coverage and mapping complexity of clinical concept representation within the UMLS. When MetaMap found a matching UMLS map, we assumed the mapping was accurate and was the best possible mapping; we did not search the UMLS for alternative or better mappings. Because we did not investigate for MetaMap errors, we therefore may have over-estimated the representation of clinical concepts in the UMLS. The manual review of MetaMap output and the UMLS search for unmapped concepts was performed by a single biomedical informatician who is also a physician (JF). It is possible another reviewer may have disagreed with the mapping analysis. We used only the UMLS Terminology Services Metathesaurus Browser (UMLS Browser) to perform manual UMLS searches of unmapped concepts. It is possible we would have found additional matching UMLS concepts had we used alternative UMLS search engines or browsers.
Conclusion
A large portion of frequently occurring concepts found in clinical narrative documents are either unrepresented or poorly represented in the current version of the UMLS Metathesaurus. Although it is the most comprehensive controlled medical terminology, the UMLS appears to require modifications and enhancements before it can fully represent all clinical concepts in medical documents.
References
- 1.de Lusignan S, van Weel C. The use of routinely collected computer data for research in primary care: opportunities and challenges. Family Practice. 2006 Apr;23(2):253–63. doi: 10.1093/fampra/cmi106. 2006. [DOI] [PubMed] [Google Scholar]
- 2.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994 Mar-Apr;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Aronsky D, Kasworm E, Jacobson JA, Haug PJ, Dean NC. Electronic screening of dictated reports to identify patients with do-not-resuscitate status. J Am Med Inform Assoc. 2004 Sep-Oct;11(5):403–9. doi: 10.1197/jamia.M1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elkin PL, Froehling D, Wahner-Roedler D, et al. NLP-based identification of pneumonia cases from free-text radiological reports. AMIA Annu Symp Proc; 2008. pp. 172–6. [PMC free article] [PubMed] [Google Scholar]
- 6.Collen MF. Patient data acquisition. Med Instrum. 1978 Jul-Aug;12(4):222–5. [PubMed] [Google Scholar]
- 7.Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner O. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc. 2009 Jul-Aug;16(4):561–70. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzuner O, Solti I, Xia F, Cadag E. Community annotation experiment for ground truth generation for the i2b2 medication challenge. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):519–23. doi: 10.1136/jamia.2010.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Open Health Natural Language Processing (OHNLP) Consortium Web site Available at: https://cabig-kcnci.nih.gov/Vocab/KC/index.php/OHNLP_Documentation_and_Downloads Accessed March 13, 2011. [Google Scholar]
- 11.Apache UIMA Web site. Available at: http://uima.apache.org/ Accessed January 13, 2011.
- 12.Ferrucci D, Brown E, Chu-Carroll J, et al. Building Watson: An Overview of the DeepQA Project. AI Magazine. 2010;31(3):59–79. [Google Scholar]
- 13.General Architecture for Text Engineering (GATE) Web site. Available at: http://gate.ac.uk/sale/gate-flyer/2009/gate-solution-profile-2-page.pdf Accessed March 13, 2011.
- 14.UMLS Web site. Available at: http://www.nlm.nih.gov/research/umls/ Accessed March 1, 2011.
- 15.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004 Sep-Oct;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leroy G, Rindflesch TC. Effects of information and machine learning algorithms on word sense disambiguation with small datasets. Int J Med Inform. 2005 Aug;74(7–8):573–85. doi: 10.1016/j.ijmedinf.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Humphreys BL, McCray AT, Cheh ML. Evaluating the coverage of controlled health data terminologies: report on the results of the NLM/AHCPR large scale vocabulary test. J Am Med Inform Assoc. 1997 Nov-Dec;4(6):484–500. doi: 10.1136/jamia.1997.0040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman C, Liu H, Shagina L, Johnson S, Hripcsak G. Evaluating the UMLS as a source of lexical knowledge for medical language processing. Proc AMIA Symp. 2001:189–93. [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews JE, Richesson RL, Krischer J. Variation of SNOMED CT coding of clinical research concepts among coding experts. J Am Med Inform Assoc. 2007 Jul-Aug;14(4):497–506. doi: 10.1197/jamia.M2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang MF, Hwang JC, Yu AC, Casper DS, Cimino JJ, Starren JB. Reliability of SNOMED-CT coding by three physicians using two terminology browsers. AMIA Annu Symp Proc; 2006. pp. 131–5. [PMC free article] [PubMed] [Google Scholar]
- 21.Fung KW, McDonald C, Srinivasan S. The UMLS-CORE project: a study of the problem list terminologies used in large healthcare institutions. J Am Med Inform Assoc. 2010 Nov 1;17(6):675–80. doi: 10.1136/jamia.2010.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wade G, Rosenbloom ST. Experiences mapping a legacy interface terminology to SNOMED CT. BMC Med Inform Decis Mak. 2008;8(Suppl 1):S3. doi: 10.1186/1472-6947-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald CJ, Overhage JM, Barnes M, et al. The Indiana network for patient care: a working local health information infrastructure. An example of a working infrastructure collaboration that links data from five health systems and hundreds of millions of entries. Health Aff (Millwood) 2005 Sep-Oct;24(5):1214–20. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 24.Regenstrief Institute Web site. Available at: http://www.regenstrief.org/ Accessed March 1, 2011.
- 25.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003 Apr;49(4):624–33. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 26.Klein D, Manning CD. Accurate Unlexicalized Parsing. Proc of the 41st Meeting of the Association for Computational Linguistics; 2003. pp. 423–30. [Google Scholar]
- 27.Xu R, Supekar K, Morgan A, Das A, Garber A. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. AMIA Annu Symp Proc; 2008. pp. 820–4. [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe HJ, Huang Y, Regula DP. Using a statistical natural language Parser augmented with the UMLS specialist lexicon to assign SNOMED CT codes to anatomic sites and pathologic diagnoses in full text pathology reports. AMIA Annu Symp Proc. 2009;2009:386–90. [PMC free article] [PubMed] [Google Scholar]
- 29.Friedlin J, McDonald CJ. A natural language processing system to extract and code concepts relating to congestive heart failure from chest radiology reports. AMIA Annu Symp Proc; 2006. pp. 269–73. [PMC free article] [PubMed] [Google Scholar]
- 30.Friedlin J, Grannis S, Overhage JM. Using natural language processing to improve accuracy of automated notifiable disease reporting. AMIA Annu Symp Proc; 2008. pp. 207–11. [PMC free article] [PubMed] [Google Scholar]
- 31.Were MC, Gorbachev S, Cadwallader J, et al. Natural language processing to extract follow-up provider information from hospital discharge summaries. AMIA Annu Symp Proc. 2010;2010:872–6. [PMC free article] [PubMed] [Google Scholar]
- 32.Friedlin J, Overhage M, Al-Haddad MA, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. AMIA Annu Symp Proc. 2010;2010:237–41. [PMC free article] [PubMed] [Google Scholar]
- 33.Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc. 2008 Sep-Oct;15(5):601–10. doi: 10.1197/jamia.M2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ. The NLM Indexing Initiative’s Medical Text Indexer. Stud Health Technol Inform. 2004;107(1):268–72. [PubMed] [Google Scholar]
- 35.Srinivasan S, Rindflesch TC, Hole WT, Aronson AR, Mork JG. Finding UMLS Metathesaurus concepts in MEDLINE. Proc AMIA Symp. 2002:727–31. [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan PF, Aronson AR. Towards linking patients and clinical information: detecting UMLS concepts in e-mail. J Biomed Inform. 2003 Aug-Oct;36(4–5):334–41. doi: 10.1016/j.jbi.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Demner-Fushman D, Mork JG, Shooshan SE, Aronson AR. UMLS content views appropriate for NLP processing of the biomedical literature vs. clinical text. Journal of Biomedical Informatics. 2010;43(4):587–94. doi: 10.1016/j.jbi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UMLS Terminology Services Web site . Available at: https://uts.nlm.nih.gov/home.html Accessed January 13, 2011. [Google Scholar]
- 39.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010 May 1;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aronson AR, Mork JG, Neveol A, Shooshan SE, Demner-Fushman D. Methodology for creating UMLS content views appropriate for biomedical natural language processing. AMIA Annu Symp Proc; 2008. pp. 21–5. [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenbloom ST, Miller RA, Johnson KB, Elkin PL, Brown SH. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc. 2006 May-Jun;13(3):277–88. doi: 10.1197/jamia.M1957. [DOI] [PMC free article] [PubMed] [Google Scholar]