Abstract
The interoperability specifications for electronic laboratory reporting specify the use of HL7, LOINC, SNOMED CT and UCUM. We explored the degree to which health care transactions comply with these standards by evaluating laboratory data captured in a health information exchange to support automated detection of public health notifiable diseases. We studied the NCD’s ability to detect and report Lead, Influenza and MRSA. We found that due to incomplete LOINC mapping, alternate approaches such as keyword searches within local test names and codes could identify additional potentially reportable messages. We also found that non-adherence to HL7 messaging standards and inconsistently recorded laboratory results require the use of complex systems with complementary NLP techniques to accurately report notifiable conditions. We conclude that the incomplete adoption of and adherence to specified standards poses challenges to deploying processes that utilize real-world data for secondary purposes.
Introduction
The development and promotion of standards is essential to the functionality of electronic health record systems especially in terms of interoperability and automated tools to support clinical practice1,2,3. Prompted by the Office of the National Coordinator (ONC), the Health Information Technology Standards Panel (HITSP) has harmonized and specified standards for several interoperability use cases4.
One such use case that relies on these interoperability specifications is the electronic health records laboratory results reporting5 (IS01), which identifies the use of HL7 v2.5.16, LOINC7 (Logical Observations Identifiers and Codes), SNOMED CT8 (Systemized Nomenclature of Medicine – Clinical Terms) and UCUM9 (Unified Code for Units of Measure). In spite of these attempts to clarify standards’ use, there continues to be inconsistent and incomplete adoption of agreed upon standards for laboratory reporting that makes it difficult to design and implement automated systems that rely on these data to enable so-called secondary use processes10. In order to assess and improve system functionality, we must understand the degree to which healthcare data complies with existing standards. In this paper we analyze the information characteristics of health information exchange clinical transactions that are leveraged by an automated Notifiable Condition Detector (NCD) system11,12.
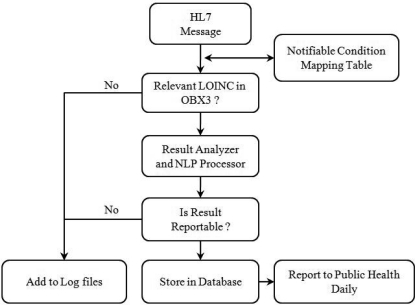
The NCD is open-source technology that leverages messaging and terminology standards such as HL7 and LOINC. It uses the rich inflow of clinical results into an operational regional health information exchange, the Indiana Network for Patient Care13 (INPC), to process over 300,000 messages daily from hundreds of sources and report 105 notifiable conditions to public health and other health care departments. Each inbound HL7 2.x message, containing laboratory results from INPC participants, is first evaluated for the presence of a relevant LOINC in its observation identifier field (OBX3). This is done through a Notifiable Condition Mapping Table14 (NCMT), which contains a list of all notifiable conditions (diseases) and the corresponding LOINCs that could potentially be used to evaluate and test for those conditions. Thus, each condition is associated with one or more LOINCs and each LOINC is associated with one or more conditions. If no LOINCs from the NCMT are found in the OBX3 field of an HL7 message then it is considered non-reportable and is not further processed. If the message contains a relevant LOINC then it is considered as potentially reportable and is processed by a complex set of algorithms that first identify the result as numeric, discrete or free-text type and then determine whether the message is reportable. Key in this determination is an NLP rule-based system called REX15 (Regenstrief Extraction tool) that uses regular expressions to detect the presence and context of keywords. All reportable messages are categorized and stored in a database that is forwarded to public health and infection control departments on a daily basis. All non-reportable messages are stored in log files. Thus the NCD performs two broad functions, it first screens inbound messages to detect those that are potentially reportable and then it processes these to determine the messages that are actually reported.
We chose to evaluate the NCD’s ability to accurately detect and report three conditions: Lead, which is usually reported as a numeric result; Influenza, which is usually reported as a discrete result (positive, negative); and Methicillin Resistant Staphylococcus Aureus (MRSA), which is usually reported as a free-text culture report. We also determined the data characteristics of these messages attributable to non-adherence of HITSP specifications.
Methods
Our study was conducted over one month from 01/03/2011 until 02/02/2011. We queried the NCD database to retrieve all messages reported by the system (positives) for Lead, Influenza and MRSA. We manually reviewed these to determine the true positives and false positives. We then obtained a set of relevant LOINCs associated with these three conditions using the most current version of our NCMT. We queried the NCD log files to retrieve all messages that contained any LOINCs from this set in their OBX3 fields, thus obtaining messages that were detected by the system as potentially reportable for these three conditions but not reported (negatives). We retrieved over 173,000 messages for MRSA alone since many of the LOINCs associated with this condition are also used to test and evaluate many other conditions. In order to minimize this number while retaining true potentially reportable MRSA messages, we only included those messages that had the words “mrsa” or “aureus” in any part of the message. Since the NCD does not report any messages that have an incomplete or preliminary flag in their result status field (OBX11) we excluded such messages. We manually reviewed the remaining messages to determine the true negatives and false negatives.
Incomplete mapping leads to false negatives. The NCD receives messages from many sources that have different local to LOINC mapping. We observed there were messages that contained valid LOINCs but are unmapped to a notifiable condition in our NCMT. Also, there were messages that lacked local to LOINC mapping but still contained notifiable conditions. These messages were not detected by the NCD as potentially reportable and hence were not further processed (negatives). We queried the log files for such messages by using keyword searches within their OBX3 fields. For Lead we used the keyword “lead”, for Influenza we used the keyword “influenza” and for MRSA we initially used the keywords “cul”, “org” or “isolate”. Here again, we retrieved over 183,000 messages for MRSA alone since the keywords used for MRSA were also associated with many other conditions. We only included those MRSA messages that also had the words “mrsa” or “aureus” in any part of the message. We manually reviewed the remaining messages to determine the true negatives and false negatives.
We also observed that there were potentially reportable messages that neither contained relevant LOINCs nor our keywords in their OBX3 fields but surprisingly did contain relevant LOINCs in their observation request field (OBR4). Since the NCD was not designed to look for relevant LOINCs in this field, all such messages failed to be detected as well and were not processed further (negatives). We queried the log files to retrieve these messages. Since we retrieved over 47,000 messages for MRSA alone, we only considered those messages that had the words “mrsa” or “aureus”. We manually evaluated all remaining messages to determine true negatives and false negatives.
We explored the characteristics of the actual results’ within messages that were detected and processed by the NCD by determining the number of messages that used SNOMED CT encoded results and for numeric and discrete results the number of messages that had units and reference range specified in their OBX6 and OBX7 fields respectively. We also determined the percentage of numeric and discrete results that were conflated with other types of information such as specimen type, when or where the test was performed, who was notified about the result, the reference range, units and comments on interpreting the results.
Results
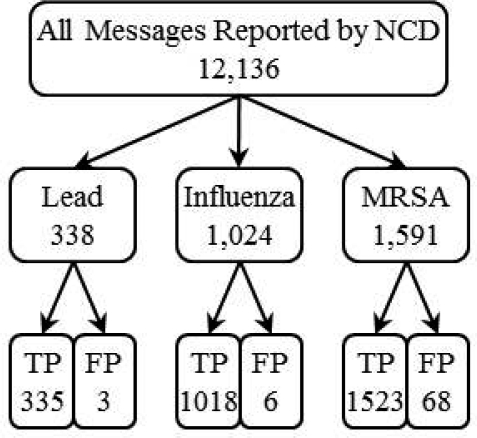
The NCD reported 12,136 messages for 50 notifiable conditions, over the one month period of our study, from the 9,364,136 incoming messages it processed. There were 2,953 reported messages for Lead, Influenza and MRSA and after manually reviewing these, we found 77 false positives. Individual counts are depicted in Figure 2.
Figure 2:
Number and accuracy of messages reported by the NCD for Lead, Influenza and MRSA
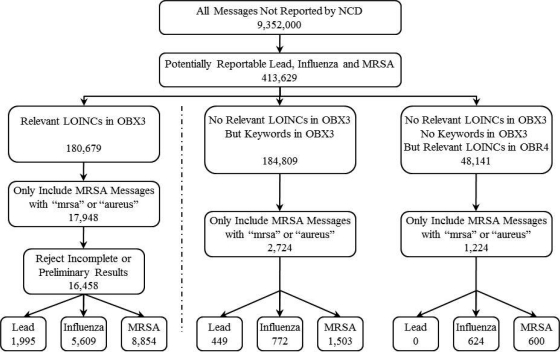
Of the remaining 9,352,000 messages not reported by the NCD, there were 16,458 messages detected as potentially reportable and 3,948 messages that the system failed to detect as potentially reportable for Lead, Influenza and MRSA because there were no relevant LOINCs in their OBX3 fields. Individual counts for each are depicted in Figure 3.
Figure 3:
Algorithm for retrieving potentially reportable messages from the log files.
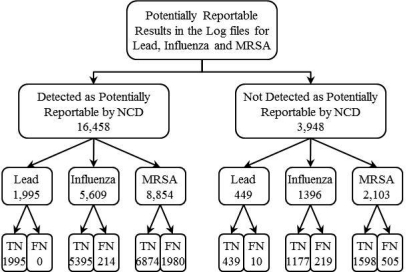
Of the 16,458 messages detected as potentially reportable by the NCD but not reported, we found 2,194 false negatives. Of the 3,948 messages that the NCD failed to detect as potentially reportable, we found 734 reportable messages. Individual counts for each are depicted in Figure 4.
Figure 4:
Manual review of all potentially reportable messages retrieved from the log files.
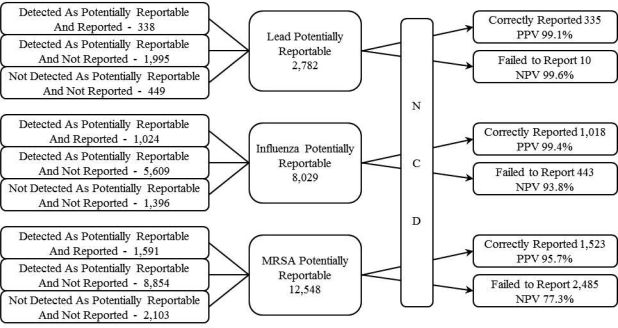
The NCD received 23,359 potentially reportable messages for Lead, Influenza and MRSA, of which it correctly reported 2,876 and failed to report 2,928 messages. Individual counts are depicted in Figure 5. Overall, the system’s sensitivity for reporting Lead, Influenza and MRSA was 97.1%, 70.2% and 38.0% respectively and its specificity was 99.9%, 99.9% and 99.2% respectively. The combined sensitivity, specificity, PPV and NPV for reporting all three conditions was 49.6%, 99.6%, 97.4% and 85.7% respectively.
Figure 5:
Total number of messages processed by the NCD for each condition and the results of that processing.
Of the 16,458 messages recognized by the system as potentially reportable through relevant LOINCs in OBX3, only 12,981 (79%) could have also been identified through our keyword searches of local test codes and names contained within OBX3 fields (81% for Lead, 70% for Influenza and 84% for MRSA).
We examined the additional 2,724 potentially reportable messages that were obtained by searching for keywords in OBX3. We discovered 18 potentially reportable LOINCs, 7 for Influenza and 11 for MRSA, that were missing from the NCMT.
We observed that the actual value of the lab results contained within the observation value fields (OBX5) of messages were often littered with other data. We found that 153 (7%) of numeric lab results and 3,753 (57%) of discrete lab results contained such additional (misplaced) data. We also found that 1,074 (46%) of the numeric lab results did not have a reference range in their reference range field (OBX7) and 589 (25%) did not have units specified in their units field (OBX6).
None of the messages contained SNOMED CT encoded results. We found 483 (4%) potentially reportable MRSA messages did not have any explicit mention of the words “mrsa” or “methicillin” in any part of the message and could only be identified as MRSA by the results of susceptibility testing.
Discussion
One of the first steps in detecting notifiable conditions is to accurately and consistently identify the subset of potentially reportable messages from all daily inbound messages from multiple different sources. The NCD system heavily relies on the NCMT and the presence of relevant LOINCs within messages to accomplish this. Our study showed that 17% of potentially reportable messages were missed because the NCD only searched OBX3 fields for relevant LOINCs. Some of the false negatives were due to the fact that sending facilities were using idiosyncratic local terms that had not been mapped to LOINC whereas the others were due to an incomplete mapping within NCMT. Indeed we were able to discover 18 LOINCs previously unassociated with Influenza or MRSA in the NCMT. One solution is to be more liberal with our existing mappings. For example there are 85 LOINCs currently associated with Influenza in the NCMT. Using the Regenstrief LOINC Mapping Assistant16 (RELMA) ver. 5.0, we were able to find a total of 316 LOINCs that could be used in the evaluation of Influenza. However, even if all 316 LOINCs are mapped to Influenza in the NCMT, there still needs to be a process for integrating newly created LOINCs either automatically or through periodic review14.
Identifying potentially reportable messages that do not contain any local code to LOINC mappings is more challenging. One potential approach is to use keyword searches for local codes or local test names within targeted fields such as OBX3. With our limited selection of keywords we were able to identify 2,724 (14%) additional potentially reportable messages. However, we also showed that relying on keywords alone is not sufficient as we were able to identify only 79% of the potentially reportable messages determined through relevant LOINCs in OBX3. For the keywords search approach to be successful, methods will be needed to systematically identify, validate and update keywords on a regular ongoing basis.
We found that at times, the relevant LOINCS are in the OBR4 field and not in OBX3. The NCD currently only examines OBX3 for relevant LOINCS. We discovered 1,224 additional messages that neither had relevant or mapped LOINCs nor our keywords within their OBX3 fields but surprisingly did have relevant LOINCs within their OBR4 fields. It is difficult to design systems to anticipate such nuances in data but this also highlights the importance of using multiple methods to identify all potentially reportable messages as completely as possible. We plan on adding this functionality to the NCD in the future.
Messages correctly identified as potentially reportable are processed by the result analyzers and NLP tools (REX) within the NCD. Our study shows that these subsystems failed to correctly report 13% (2,194) of the messages they processed. For Lead, the system was able to report all messages without any false negatives in spite of the absence of reference ranges in 46% of the messages. The NCD was able to overcome these non-standard lead results by retrieving pre-established reference ranges from the LOINC database. Although this is an example of adapting the system to real-world data shortcomings, it may have introduced some inaccuracy as we observed that different labs occasionally specified different reference ranges for the same LOINC.
For Influenza, which is predominantly reported as a discrete result (eg. positive, negative, detected, not detected) there were only 214 (4%) false negatives even when 57% of the results where conflated with additional and misplaced data. A typical example of this is an OBX5 field reported as “Detected∼Reference range: Not Detected∼Unit: not reported”. The system failed to report this message because it mistakenly paired the “Not” in the reference range with the result “Detected”. This error could be avoided if HL7 messaging standards were strictly adhered to by placing the reference range “Not Detected” in the OBX7 field, the units “not reported” in the OBX6 field and only reporting the actual observation value “Detected” in the OBX5 field.
For MRSA, which is usually reported as free-text results, the NCD failed to report 1,980 messages. Since LOINCs associated with MRSA are non-specific, REX processes free-text reports by first identifying the relevant notifiable condition and then its context though rules and regular expressions established from prior training sets. Some errors were caused by new patterns of keywords and punctuation not previously seen in the training sets that could be addressed by periodically retraining REX with new data sets. Another solution would be to use a combination of different NLP techniques in tandem. Both solutions require periodic labor intense evaluations to monitor performance. The use of SNOMED CT Lab Test Findings and SNOMED CT Organisms tables in reporting lab results would circumvent the majority of current limitations in NLP tools. We plan to conduct an error analysis of the algorithms, rules and regular expressions that are used by the NCD in detecting and reporting MRSA messages.
Our study has some limitations. We evaluated the NCD’s ability to detect and report only Lead, Influenza and MRSA. However, these three conditions represented the spectrum of numeric, discrete and free-text result types generally used to report other conditions as well. They also accounted for 21% of all messages reported by the NCD. Another limitation is that all messages were analyzed by a single reviewer.
Conclusions
Despite the broad availability of interoperability specifications, there is pervasive non-adherence to HL7 messaging standards, incomplete LOINC mapping, inconsistently reported results and no adoption of SNOMED CT standards when reporting electronic laboratory results. These barriers must be overcome in order to efficiently deploy systems that utilize real-world data for secondary use. Ideally this is accomplished by source systems adopting and strictly adhering to specified standards. However, our experience shows that this is an ineffective approach. Alternatively, complex systems are required that compensate for sub-standard data by using robust techniques for local code to LOINC mapping and multiple NLP algorithms. Such systems also require periodic evaluation to ascertain accuracy.
Figure 1:
Simplified architecture of the Notifiable Condition Detector
Acknowledgments
This work was performed at the Regenstrief Institute, Indianapolis, IN, and was supported in part by grant 5T 15 LM007117-14 from the National Library of Medicine. This study is also supported in part by the grant of the Indiana Center of Excellence in Public Health Informatics (1P01HK000077-01). We gratefully acknowledge the contribution of Rico Merriwether MBA, PMP; Regenstrief Institute, Project Manager.
References
- 1.Hammond WE. The making and adoption of health data standards. Health Aff (Millwood) 2005 Sep-Oct;24(5):1205–13. doi: 10.1377/hlthaff.24.5.1205. [DOI] [PubMed] [Google Scholar]
- 2.Brailer DJ. Interoperability: the key to the future health care system. Health Aff (Millwood) 2005. Jan-Jun. Suppl Web Exclusives: W5-19–W5-21. [DOI] [PubMed]
- 3.Kuperman GJ, Blair JS, Franck RA, Devaraj S, Low AF, NHIN Trial Implementations Core Services Content Working Group Developing data content specifications for the nationwide health information network trial implementations. J Am Med Inform Assoc. 2010 Jan-Feb;17(1):6–12. doi: 10.1197/jamia.M3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Health Information Technology Standards Panel Harmonization Framework. Available at: http://www.hitsp.org/harmonization.aspx. Accessed Mar 6, 2011.
- 5.Health Information Technology Standards Panel Laboratory Results Reporting. Available at: http://www.hitsp.org/InteroperabilitySet_Details.aspx?MasterIS=true&InteroperabilityId=44&PrefixAlpha=1&APrefix=IS&PrefixNumeric=01. Accessed Mar 6, 2011.
- 6.HL7’s Version 2.x Messaging Standards. Available at: http://www.hl7.org/implement/standards/v2messages.cfm. Accessed Mar 6, 2011.
- 7.McDonald Clem, Huff Stan, Mercer Kathy, Hernandez Jo Anna, Vreeman Daniel J. Logical Observation Identifiers Names and Codes (LOINC) Users' Guide. 2010. Dec, Available at http://loinc.org. Accessed Mar 6, 2011.
- 8.Systemized Nomenclature of Medicine – Clinical Terms (SNOMED CT). Available at http://www.ihtsdo.org/snomed-ct/. Accessed Mar 6, 2011.
- 9.Schadow G, McDonald CJ, Suico JG, Fohring U, Tolxdorff T. Units of measure in clinical information systems. J Am Med Inform Assoc. 1999;6(2):151–62. doi: 10.1136/jamia.1999.0060151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, Detmer DE. Expert Panel. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. J Am Med Inform Assoc. 2007 Jan-Feb;14(1):1–9. doi: 10.1197/jamia.M2273. Epub 2006 Oct 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overhage JM, Suico J, McDonald CJ. Electronic laboratory reporting: barriers, solutions and findings. J Public Health Manag Pract. 2001 Nov;7(6):60–6. doi: 10.1097/00124784-200107060-00007. [DOI] [PubMed] [Google Scholar]
- 12.Grannis S, Overhage JM, Friedlin J. Architectural and operational components of a real world operational automated notifiable condition processor. 2008 PHIN Annual Conference; August 2008. [Google Scholar]
- 13.McDonald CJ, Overhage JM, Barnes M, Schadow G, Blevins L, Dexter PR, Mamlin B, INPC Management Committee The Indiana network for patient care: a working local health information infrastructure. An example of a working infrastructure collaboration that links data from five health systems and hundreds of millions of entries. Health Aff (Millwood) 2005 Sep-Oct;24(5):1214–20. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 14.Grannis S, Vreeman D. A vision of the journey ahead: using public health notifiable condition mapping to illustrate the need to maintain value sets. AMIA Annu Symp Proc; 2010 Nov 13; 2010. pp. 261–5. [PMC free article] [PubMed] [Google Scholar]
- 15.Friedlin J, Grannis S, Overhage JM. Using natural language processing to improve accuracy of automated notifiable disease reporting. AMIA Annu Symp Proc; 2008 Nov 6; pp. 207–11. [PMC free article] [PubMed] [Google Scholar]
- 16.Regenstrief LOINC Mapping Assistant (RELMA) Users’ Guide. Available at http://loinc.org. Accessed Mar 6, 2011.