Abstract
This paper describes the design of the NeuroLOG middleware data management layer, which provides a platform to share heterogeneous and distributed neuroimaging data using a federated approach. The semantics of shared information is captured through a multi-layer application ontology and a derived Federated Schema used to align the heterogeneous database schemata from different legacy repositories. The system also provides a facility to translate the relational data into a semantic representation that can be queried using a semantic search engine thus enabling the exploitation of knowledge embedded in the ontology. This work shows the relevance of the distributed approach for neurosciences data management. Although more complex than a centralized approach, it is also more realistic when considering the federation of large data sets, and open strong perspectives to implement multi-centric neurosciences studies.
Introduction
The importance of suitable computer infrastructures supporting data sharing in life science is now largely recognized and promoted by institutional organizations in Europe1 as well as in the US2. It appears as a key factor for the success of translational research and medicine in the coming years3. However in spite of significant efforts during the last decade such infrastructures are still in their infancy and clearly not commonly used in research centres due to several technical, regulatory, ethical or cultural factors. This paper reports on the NeuroLOG project (2007–2010)*, supported by the French National Agency for Research, which addressed some of the technical and organisational issues that currently hamper the sharing of data and processing resources in the context of neuroimaging research. The paper is focused on the design and deployment of the ontology-based federated architecture of NeuroLOG, which aims at federating heterogeneous neuroimaging data sources and delivering advanced data querying capability. It shows the relevance of a decentralized federative approach, especially when dealing with large scale data sources. The paper is organized as follows. First, open questions and state of the art solutions proposed for sharing neuroimaging data for research purposes are presented. Our approach and the distributed architecture design proposed are then described. Section “Results” reports on the deployment of our NeuroLOG platform to federate five neuroimaging research centres located in France and provides real cases of data queries and retrieval. Our methodological and implementation choices are then discussed and finally the major features of our approach are stressed in section “Conclusion”.
Background
Roughly, the sharing of neuroimaging data can be envisaged along two approaches. The centralized approach consists in gathering in a single repository all data that the collaborative centres wish to share. This approach was implemented for instance in the NIH MRI study of normal brain development4, or in ADNI (Alzheimer’s Disease Neuroimaging Initiative), a large scale multi-centre US project for investigating Alzheimer’s disease. The main advantage of the centralized model is its technical simplicity. The main drawback is that each data provider should organize the data to share based on a predefined common schema. An alternative is the federated approach that offers more flexibility in terms of local database organization, and allows providers to keep control on their data. This approach relies on a mediation layer5 that interfaces to site legacy resources and provides a unified view of distributed and heterogeneous data to the end users. This unified view interestingly can be designed in reference to an ontological model. The advantages of federated systems over centralized ones are discussed in depth in Aishish et al.’s paper6. The Biomedical Informatics Research Network (BIRN)7 initiative provided pioneering work in data integration based on a federated approach, with an appealing proof of concept of ontology-based mediation applied to neuroscience research8. BIRN is a major cooperative action launched in 2001 with two main goals: (1) to setup an infrastructure to share data and compute resources, and (2) to support applications towards a better understanding of brain structure and functioning, “from cell to behaviour”. During the first phase of the project (2001–2008) significant effort was devoted to ontology development, leading to the BIRNLex ontology9 (in fact rather a lexicon than an ontology), available via the NCBO BioPortal. In parallel, a technical infrastructure was deployed based on the Storage Resource Broker (SRB), a system for virtualizing files in large federated systems10. The data organization relies mainly on the HID schema (Human Imaging Database), and uses the XNAT (Extensible Neuroimaging Archive)11 and the XCEDE (XML-based Clinical Experiment Data Exchange Schema) schemata. The BIRN federated system includes 11 HIDs.
In Europe, similar work was carried out in the context of the @NeurIST project, a four-year European Integrated Project launched in 2006, which “aimed at supporting the research and treatment of cerebral aneurysms by bringing together heterogeneous data, computing and complex processing services”. This project was not limited to research and considered care delivery issues for instance in identifying patients with high risk of rupture and defining personalised design of endovascular devices. From a technical standpoint, @NeurIST12 implemented a service-oriented infrastructure “offering generic services for data access, data staging, semantic mediation and Grid computing”. Data services virtualize heterogeneous data as Web services, and rely mainly on OGSA-DAI and OGSA-DQP. The project developed an ontology13 relying on the Descriptive Ontology for Language and Cognitive Engineering (DOLCE) foundational ontology14.
Similarly to BIRN and @NeurIST, the NeuroLOG project adopted a federated approach. The major difference with these two projects is the multi-layer domain ontology, OntoNeuroLOG, built to hide the data heterogeneity to the final user.
Methods
General approach
NeuroLOG delivers to the users the view of a single virtual data repository that integrates the data available from the different federated sites. This includes images acquired, used and/or produced in different research studies, stored as regular files, and metadata stored in relational databases. Metadata concern various types of data: 1) Images, to describe the contents of images (datasets), the conditions of image acquisition (e.g. MR equipment and MR sequences), the subjects involved (patients or healthy volunteers), or image processing (e.g. bias correction or segmentation) used to produce some images; 2) Contexts in which image acquisitions take place, namely imaging examinations and multi-centre studies in which patients are enrolled; 3) Scores obtained using tests and questionnaires to assess patients’ neurological states and cognitive performances. The local organization of data and metadata may differ among the different sites as well as the jargon used to express them (although the use of standards like DICOM tends to reduce the variability of the terms used). In particular, the relational schema may differ based on each site’s specific research domains. A key feature of the federated system is its ability to build up an integrated view of all data that hides the underlying heterogeneity, while preserving sites autonomy through a weak coupling. The federated view is built dynamically using mediation mechanisms provided by the mediator used (DataFederator, from SAP) and mappings between each site database schema and the common schema referred to as the “Federated Schema” (FS). The FS provides the “pivot language” used to align the heterogeneous data. In order to be formally grounded and as explicit as possible this schema is defined based on an application ontology, providing common semantics to the shared data.
In NeuroLOG, the common schema is not derived from the various schemata to be integrated, but built independently. The FS was defined as one of the by-products of the OntoNeuroLOG ontology, assumed to cover a broader scope than the one restricted to the data to integrate.
Design of the ontology
OntoNeuroLOG reuses preliminary works carried out in the context of the NeuroBase project (OntoNeuroBase ontology15). It was built as a modular multi-layer application ontology, grounded on DOLCE14 and a number of companion formal and core domain ontologies, mostly developed by Kassel and collaborators.
DOLCE, like other formal ontologies (ex: BFO, GFO), provides a set of – very – abstract concepts (e.g. physical object, event, quality) and relations (e.g. whole-parts, constitution). The development of an application ontology such as OntoNeuroLOG requires to extend this formal level (independent of any domain) for a practical use. For example, we need to account of plural entities such as collections of entities16 or to describe the function of entities (natural or artificial)17. For OntoNeuroLOG, we thus extended the formal level of DOLCE by adding several modules including a formal ontology of artifacts18. Based on the latter, we defined core ontologies of two domains of artifacts: an ontology of digital images, further specialized in the field of neuroimaging19; and an ontology of instruments used to assess the neurological state and cognitive performances of subjects20. This module was extended to describe general tests such as the Mini Mental State and more specialized ones like the EDSS used for patients suffering from Multiple Sclerosis (these assessment instruments are considered in their own rights as domains corresponding to classes of (local) instruments administered in centers). A list of the main modules developed is presented in Table 1.
Table 1.
Some domains covered by the ontological modules composing OntoNeuroLOG
| Major Formal and Core Ontologies | Major Domain ontologies |
|---|---|
| Particular (i.e. DOLCE) | Study |
| Action | Examination and Subject |
| Artefact | Neuroimaging Dataset |
| Participant role | Medical image expresssion |
| Capacity | Medical image file |
| Discourse, Message, and Discourse act | Medical image format |
| Number, Scalar quale, and Unit of measure | Dataset processing |
| Inscription, Expression, Conceptualization | Dataset acquisition |
| Language and Computer language | MR protocol |
| Computer language expression | MR sequence |
| Assessment-Instrument | Specific Assessment-Instruments (MMS, EDSS, etc.) |
Concerning the process of construction of our application ontology, we followed the OntoSpec21 method where two stages are distinguished. The design step is based on a semantically rich and semi-informal specification of the ontology for finely characterizing concepts and documenting the modeling choices. The implementation step allows encoding knowledge in an operational language, typically a dialect of OWL. During this second step, the level of expressiveness (e.g. OWL-Lite versus OWL-DL) and the representation entities to be considered (for example an object versus a terminal data) depend on the reasoning needs and on the application context (i.e. the reasoner to be used or the expected response time). In NeuroLOG we chose OWL-Lite increased by rules, a language interpreted by the semantic search engine CORESE22. A significant subset of the ontology was manually translated into a relational FS to be used as a common view to query the heterogeneous databases schemata.
Data Management Layer architecture
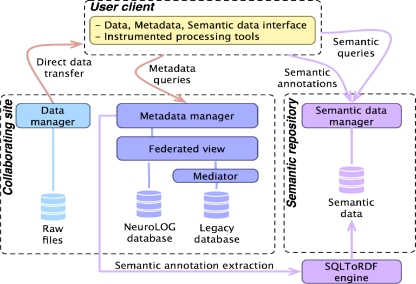
Custom legacy environments covering neuroimaging data storage, processing resources, databases and applications have been developed within various neuroscience sites to address their research challenges. Collaboration between several sites requires transcending laboratories boundaries and overcoming the heterogeneity of their respective legacy environments. The NeuroLOG Data Management Layer (DML) illustrated in Figure 1 addresses both the heterogeneity and the distribution of legacy neuroscience data to enhance collaborative activities by providing a seamless and secure access to distributed data (security issues are beyond the scope of this paper, refer to Gaignard et al. 23 for further details).
Figure 1:
Data Management Layer (DML) architecture
From the end-user point of view the DML provides a unified view over three data repositories: (i) a single virtual relational repository (Metadata manager) based on the Federated Schema derived from the NeuroLOG ontology; (ii) a centralized semantic repository (Semantic data manager) allowing for rich semantic-aware queries; and (iii) a flat file catalog (Data manager) enabling the retrieval of raw image files indexed through their associated metadata.
Preserving the autonomy of participating sites imposes that the NeuroLOG middleware have no control whatsoever on the legacy systems federated: legacy databases are operated by site administrators and thus must not be altered by the federation. To do so, the DML is granted a read-only access to the legacy databases. In addition, the DML provides its own database, the NeuroLOG Database, compliant with the FS, that stores new data produced when processing and analyzing source data (the data processing capability of the NeuroLOG system is not detailed in this paper). NeuroLOG relies on the DataFederator mediation and federation tool to integrate both the legacy and new metadata, making it possible to seamlessly share newly produced data as part of the overall federated view.
Besides, the DML does not build up a virtual file system by aggregating several file systems all together in a tree-like structure, which would not make much sense for the end-user. Instead, the Metadata manager provides different search modes to easily browse the federated metadata, which is the entry point that shall ultimately lead the user to actual image files.
Finally, the centralized semantic repository is populated through an automated and periodic generation of semantic annotations by the METAMorphoses translation tool24 consuming relational metadata published by collaborating sites, and by the operation of the platform by end-users. Semantic annotations are produced at runtime, enabling for the reinsertion of valuable processing results metadata. Implementing RDFS, and most of OWL-Lite entailments, the CORESE semantic engine22 allows to exploit the NeuroLOG ontology through SPARQL semantic queries over annotations originating from distributed and heterogeneous data sources.
Mediation layer
The Mediation Layer dynamically aligns the legacy data schema to the FS by means of mappings that describe how to convert legacy database columns and tables into their equivalent entities in the Federated Schema. The mappings description is an iterative process that involves not only technical staff but also sites’ domain experts (who participated in the definition of the legacy schema), and ontology experts who shall guarantee that semantics is preserved all along the process. Some mappings are rather simple in case the source and target columns bear the same semantics with typically a one-to-one correspondence in the domain of values (e.g. converting {false, true} into {0, 1}). Other cases require more complex manipulations when the source and target semantics do not exactly match, or when data representation is different, e.g. an n-to-n relation may be implemented as a multi-valued column or using an intermediate relation table. The accuracy of the mappings is critical as it may challenge the global coherence of the federated view. Several options were considered to handle complex mappings on a case-by-case basis: (i) no satisfying mapping can be figured out, then the data must be excluded from the federated view: better have less data than erroneous one; (ii) the legacy schema lacks expressivity, a schema evolution may be considered; or (iii) the ontology lacks some semantic details that are of interest for the whole federation, then its update is considered.
Another issue challenges the global coherence of the federated view: some logical entity may be duplicated in legacy databases of several sites resulting in multiple concrete representations. Typically sites participating in a multi-centre study may have their own representation of the study entity in their legacy database. The DML addresses this issue by setting up a master-slave tagging mechanism that helps identify entities referring to the same logical entity. When queried for that logical entity, the DML will filter slave replicas out of the results, letting only the single master replica be returned as part of the results (see25 for further details).
Results
Platform deployment
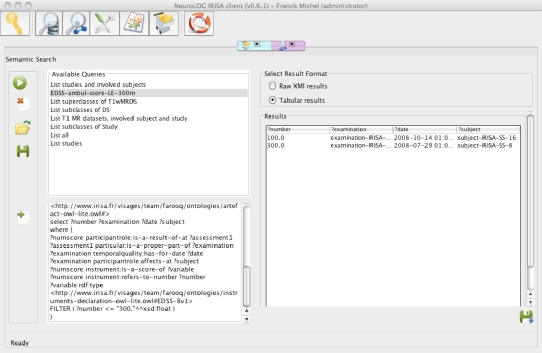
A platform was deployed federating the data of five centres located at IRISA in Rennes, Pitié-Salpêtrière Hospital in Paris, GIN in Grenoble, INRIA and I3S in Sophia-Antipolis. The shared data files are mostly MR images (T1-weighted, T2-weighted, FLAIR and Diffusion-weighted images). The platform currently hosts about 500 image datasets from 77 subjects or patients with various neurological disorders: Multiple sclerosis, stroke, tumor, and Alzheimer’s disease, and enrolled in 12 different studies that were carried out at the different sites. It also contains patient’s clinical and neuropsychological scores, as well as other contextual data. Figure 2 illustrates the navigation in the metadata to select datasets, based on criteria related to the concerned subject or Study. Figure 3 shows an example of a semantic query to retrieve subjects whose EDSS score is above a cut-off value denoting a high-level of disability, which can then be correlated with observations in MR images in a clinico-radiological approach.
Figure 2.
User interface of the client application: selection criteria can be defined; the selected entities, e.g. datasets, can then be put in the cart, for subsequent downloading or processing.
Figure 3.
Example of semantic query: the left part of the window shows the SPARQL query entered by the user, and the right part shows in tabular form the response to this query.
Discussion
Federated approaches are obviously more complex to setup than centralized ones, since they require tackling the heterogeneity of the distributed data. In the case of small health care facilities contributing to multi-centric clinical research studies, it is probably more efficient that they send their data to a centralized repository rather than installing and operating their own local repository. However, for large collaborating centres, there is no doubt that each will prefer to keep control on its own data and organize it as it feels optimum in terms of functional needs, quality management constraints, confidentiality, etc., thus requiring a federated sharing infrastructure rather than a centralized one. Morever, although one may admit that in a relatively focused domain of research (such as AD research for example) the actors may accept to rely on a single repository for data storage, it seems nevertheless highly desirable that the data may also be accessed / compared / mined in a multidisciplinary framework. It is generally admitted that such binding of experimental observations from different domains and contexts (e.g. other diseases, animal models) is a key feature for significant breakthrough in translational medicine in general26 and neuroinformatics in particular27. Ontology-based federated approaches seem to us to be the best approach for enabling such multidisciplinary data sharing.
The NeuroLOG project demonstrated the feasibility of our ontology-based federated approach to aggregate pre-existing distributed neuroimaging data. This includes the development of a suitable application ontology and its use in a mediation layer to integrate diverse data such as MR images, study context, related imaging protocols, as well as neuropsychological and neuroclinical scores from various existing distributed repositories.
Not all the design objectives could be met. In particular, the domain covered by the ontology was not as broad as expected initially. For example, it was not possible to us to include Region of Interest (ROI) annotations, an interesting topic we had studied in previous work19, and which would have allowed segmented images to be annotated with information on anatomy and pathology. One of the reasons why this could not be achieved was that no tools were available to extract relevant subsets from large ontologies such as the Foundational Model of Anatomy, something that is now possible with tools like vSPARQL28.
Experimentations performed using the current version of the platform exhibited some limitations inherent to our approach. For instance, we experienced that the complete autonomy of sites could lead to disturbance of the platform’s operation, since the latter relies on tightly coupled relational database technology. Indeed, in a wide-area distributed environment, network and computing service interruptions happen on a regular basis and all functionalities delivered within the platform should be as little sensitive as possible to the disconnection or failure of the resources. As the size of the federated system increases, it becomes critical to provide automatic reconfiguration capabilities, in order to dynamically isolate the platform components that are no longer accessible. Another limitation concerned the extension of the ontology. In practice we realized that upgrading the Federated Schema required significant work to update the mappings of the databases or even to re-import source data.
Clearly, in the long term, the ontology, which is the cornerstone of our approach, should be extended. To develop ontologies that meet our needs, i.e. covering many domains and showing a great heterogeneity, a modular and multi-level of abstraction approach seems unavoidable. It allows the same general modeling principles to be applied to all domains, facilitating even more the maintenance of the ontology. Nevertheless, a fundamental question remains: the one of the availability of sufficiently complete formal ontologies and of core ontologies covering enough domains so that developers of application ontologies would just have to model concerned domains (at the level of medical disciplines) without having to cope with the formal level that is complex to master. De facto, the field of formal ontologies is still in its childhood, as evidenced by the lack of consensus on how to define and introduce in these formal ontologies abstract concepts such as collection, process, capacity, function, component, etc. The absence of such consensus reduces capabilities of developers to reuse existing ontologies, such as those published on the NCBO BioPortal. To address this situation, in the NeuroLOG project, we have defined (rather than reused) most of the modules composing OntoNeuroLOG, developing ontologies in the domains of images, image acquisition protocols, and image processing. This is a major achievement and this can now be used by the community, and hopefully facilitate the implementation of similar projects. It covers a number of important domains, however it is still incomplete especially regarding to anatomical annotation of the images, using e.g. the Foundational Model of Anatomy, as stated earlier.
Finally, assessing the added value provided by such ontology-based mediation (compared to an approach relying on relational databases) is another important issue. Recall that an ontology is a domain model which is semantically rich, compared to a relational schema (at least when an expressive ontology definition language is used to specify it). The stakes are hence twofold: i) the query evaluation system must provide the reasoning capabilities needed to exploit this semantic richness, and ii) built ontologies have to exploit the expressiveness provided. On the first point, we have used in the NeuroLOG project the search engine CORESE that interprets ontologies specified in OWL-Lite increased by rules: this is a quite poor expressive language but, in the short term, we intend to use a search engine which exploits more semantics including algebraic properties of relations, as shown in29. On the second point: today OntoNeuroLOG fully exploits multiple inheritance (as illustrated by our taxonomy of datasets) and in the short term we plan to integrate an ontology of anatomical structures (e.g. a suitable subset of FMA) for which more expressiveness will be needed to account for the semantics of the relations. Finally, let us emphasize another added value of the ontological approach: the ontology, once built, can be completed by expert decision rules to offer to the end-users added-value services such as an aid for diagnosis (e.g. as demonstrated by Tu et al. 30 in the case of autism).
Conclusion
We presented the principles and the architecture of the NeuroLOG middleware, and its deployment to federate five neuroimaging data repositories located in Paris, Rennes, Grenoble and Sophia Antipolis. In this work, the key features are the development of the application ontology called OntoNeuroLOG and its use to align the heterogeneous data to a common model. The project is pursued to refine the development of the middleware, and to extend and exploit this platform for two target research applications where a multi-centre pluridisciplinary approach is essential: 1) neurodegenerative diseases (Alzheimer’s disease) and 2) epilepsy (surgical treatment of drug-resistive epilepsy). The technical enhancements that are envisaged concern primarily the semantic search engine suitable to integrate both semantic and relational repositories, and providing the required mediation interface to cope with schema heterogeneity.
Acknowledgments
This project was funded by the French National Agency for Research under contract number ANR-06-TLOG-024. The authors warmly thank all the contributors to the NeuroLOG project as well as the clinical colleagues providing the image data exploited in the testbed (Pr Gilles Edan and Dr Jean-Christophe Ferré in Rennes, Pr Jean-François Lebas in Grenoble, clinicians of the neuroradiology and neuropsychology departments of the Pitié Salpêtrière hospital).
Footnotes
References
- 1.2007. OECD principles and guidelines for access to research data from public funding. Organisation for Economic Co-operation and Development.
- 2.Walport M, Brest P, et al. Sharing research data to improve public health. Lancet. 2011 Feb 12;377(9765):537–9. doi: 10.1016/S0140-6736(10)62234-9. [DOI] [PubMed] [Google Scholar]
- 3.Burgun A, Bodenreider O. Accessing and integrating data and knowledge for biomedical research. Yearb Med Inform. 2008:91–101. [PMC free article] [PubMed] [Google Scholar]
- 4.Evans AC. The NIH MRI study of normal brain development. Brain Development Cooperative Group Neuroimage. 2006 Mar;30(1):184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 5.Wiederhold G. Mediators in the Architecture of Future Information Systems. IEEE Computer. 1992;25(3):38–49. [Google Scholar]
- 6.Ashish N, Ambite JL, Muslea M, Turner JA. Neuroscience Data Integration through Mediation: An (F)BIRN Case Study. Front Neuroinformatics. 2010;4:1–11. doi: 10.3389/fninf.2010.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biomedical Informatics Research Network BIRN web site, available from: http://www.birncommunity.org/
- 8.Martone ME, Gupta A, Ellisman MH. E-neuroscience: challenges and triumphs in integrating distributed data from molecules to brains. Nat Neurosci. 2004;7(5):467–72. doi: 10.1038/nn1229. [DOI] [PubMed] [Google Scholar]
- 9.Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, Larson SD, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex Vocabularies: Building Comprehensive Ontologies for Neuroscience. Neuroinformatics. 2008;6(3):175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keator DB, Grethe JS, Marcus D, Ozyurt B, Gadde S, Murphy S, Pieper S, Greve D, Notestine R, Bockholt HJ, Papadopoulos P. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN) IEEE Trans Inf Technol Biomed. 2008;12:162–172. doi: 10.1109/TITB.2008.917893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit (XNAT): An informatics platform for managing, exploring, and sharing neuroimaging data. NeuroInformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 12.Benkner S, Arbona A, Berti G, Chiarini A, Dunlop R, Engelbrecht G, Frangi AF, Friedrich CM, Hanser S, Hasselmeyer P, Hose RD, Iavindrasana J, Köhler M, Iacono LL, Lonsdale G, Meyer R, Moore B, Rajasekaran H, Summers PE, Wöhrer A, Wood S. @neurIST: infrastructure for advanced disease management through integration of heterogeneous data, computing, and complex processing services. IEEE Trans Inf Technol Biomed. 2010 Nov;14(6):1365–77. doi: 10.1109/TITB.2010.2049268. [DOI] [PubMed] [Google Scholar]
- 13.Boeker M, Stenzhorn H, Kumpf K, Bijlenga P, Schulz S, Hanser S. The @neurIST ontology of intracranial aneurysms: providing terminological services for an integrated IT infrastructure. AMIA Annu Symp Proc; 2007 Oct 11; pp. 56–60. [PMC free article] [PubMed] [Google Scholar]
- 14.Masolo C, Borgo S, Gangemi A, Guarino N, Oltramari A, Schneider L. The WonderWeb Library of Foundational Ontologies and the DOLCE ontology. 2003. WonderWeb Deliverable D18, Final Report, vr. 1.0,
- 15.Temal L, Lando P, Gibaud B, Dojat M, Kassel G, Lapujade A. OntoNeuroBase: a multi-layered application ontology in neuroimaging. Second Workshop: Formal Ontologies Meet Industry (FOMI 2006); Trento (Italy). 2006. [Google Scholar]
- 16.Wood Z, Galton A. A taxonomy of collective phenomena. Applied Ontology. 2009;4(3–4):267–292. [Google Scholar]
- 17.Borgo S, Carrara M, Garbacz P, Vermaas PE. Formalization of Functions within the DOLCE Ontology. In: Horvath I, Mandorli F, Rusak Z, editors. Proceedings of the Eighth International Symposium on Tools and Methods of Competitive Engineering TMCE 2010, Vol. 1, Tools and Methods of Competitive Engineering (Delft University of Technology); Ancona, Italy: Apr 12–16, pp. 113–126. [Google Scholar]
- 18.Kassel G. A formal ontology of artefacts. Applied Ontology. 2010;5(3–4):223–246. [Google Scholar]
- 19.Temal L, Dojat M, Kassel G, Gibaud B. Towards an Ontology for Sharing Medical Images and Regions of Interest in Neuroimaging. Journal of Biomedical Informatics. 2008;41:766–778. doi: 10.1016/j.jbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Batrancourt B, Dojat M, Gibaud B, Kassel G. A core ontology of instruments used for neurological, behavioral and cognitive assessments. In: Galton A, Mizoguchi R, editors. Proceedings of the Sixth International Conference on formal Ontology in Information Systems (FOIS 2010); Toronto (Ca): IOS Press; May, 2010. pp. 185–198. [Google Scholar]
- 21.Kassel G. Integration of the DOLCE top-level ontology into the OntoSpec methodology. 2005. LaRIA Research Report 2005-08, available at: http://hal.ccsd.cnrs.fr/ccsd-00012203.
- 22.Corby O, Dieng-Kuntz R, Faron-Zucker C. Querying the Semantic Web with Corese Search Engine. Proceedings of the 15th ECAI/PAIS; Valencia, Spain. 2004. [Google Scholar]
- 23.Gaignard A, Montagnat J. HealthGrid’09. IOS Press; Jun, 2009. A distributed security policy for neuroradiological data sharing; pp. 257–262. [PubMed] [Google Scholar]
- 24.Svihla M, Jelínek I. Benchmarking RDF Production Tools. Proceedings of 18th International Conference on Database and Expert Systems Applications - DEXA; Heidelberg: Springer; 2007. pp. s. 700–710. [Google Scholar]
- 25.Michel F, Gaignard A, Ahmad F, Barillot C, Batrancourt B, Dojat M, Gibaud B, Girard P, Godard D, Kassel G, Lingrand D, Malandain G, Montagnat J, Pélégrini-Issac M, Pennec X, Rojas Balderrama J, Wali B. HealthGrid’10 (HG’10) IOS Press; Paris, France: Jun, 2010. Grid-wide neuroimaging data federation in the context of the NeuroLOG project; pp. 112–123. [PMC free article] [PubMed] [Google Scholar]
- 26.Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, Doherty D, Forsberg K, Gao Y, Kashyap V, Kinoshita J, Luciano J, Marshall MS, Ogbuji C, Rees J, Stephens S, Wong GT, Wu E, Zaccagnini D, Hongsermeier T, Neumann E, Herman I, Cheung KH. Advancing translational research with the Semantic Web. BMC Bioinformatics. 2007;8(Suppl 3):S2. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akil H, Martone ME, Van Essen DC. Challenges and opportunities in mining neuroscience data. Science. 2011;331:709–711. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw M, Detwiler LT, Noy N, Brinkley J, Suciu D. vSPARQL: A view definition language for the semantic web. Journal of Biomedical Informatics. 2011;44:102–117. doi: 10.1016/j.jbi.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner JA, Mejino JL, Brinkley JF, Detwiler LT, Lee HJ, Martone ME, Rubin DL. Application of neuroanatomical ontologies for neuroimaging data annotation. Front Neuroinformatics. 2010;4(10):1–12. doi: 10.3389/fninf.2010.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu SW, Tennakoon L, O’Connor M, Shankar R, Das A. Using an integrated ontology and information model for querying and reasoning about phenotypes: The case of autism. AMIA Annu Symp Proc; 2008. Nov 6, pp. 727–31. [PMC free article] [PubMed] [Google Scholar]