Abstract
LOINC codes are seeing increased use in many organizations. In this study, we examined the barriers to semantic interoperability that still exist in electronic data exchange of laboratory results even when LOINC codes are being used as the observation identifiers. We analyzed semantic interoperability of laboratory data exchanged using LOINC codes in three large institutions. To simplify the analytic process, we divided the laboratory data into quantitative and non-quantitative tests. The analysis revealed many inconsistencies even when LOINC codes are used to exchange laboratory data. For quantitative tests, the most frequent problems were inconsistencies in the use of units of measure: variations in the strings used to represent units (unrecognized synonyms), use of units that result in different magnitudes of the numeric quantity, and missing units of measure. For non-quantitative tests, the most frequent problems were acronyms/synonyms, different classes of elements in enumerated lists, and the use of free text. Our findings highlight the limitations of interoperability in current laboratory reporting.
Introduction
Logical Observation Identifiers Names and Codes (LOINC®) was developed in 1994 to provide a universal vocabulary for reporting laboratory and clinical observations1–3. We and others have previously evaluated LOINC usage by analyzing coverage, consistency and the correctness of LOINC usage among 3 large institutions4–6. One goal of LOINC is to facilitate the aggregation of laboratory data collected from different institutions to support research other kinds of secondary use of clinical data6. Our working definition of interoperability is that data from different institutions are interoperable if they are mutually substitutable, that is, if the data from one institution can be used in the patient care and decision support programs of the second institution without the need to translate or convert the data. For example, “body weight 80 kg” and “body weight 176.4 lb” are not semantically interoperable because the units of measure would need to be converted before these two representations would be mutually substitutable. In our previous study5, when observing the same laboratory tests (having the same LOINC codes) among different institutions, we discovered that laboratory data contained many heterogeneous data formats. For example, laboratory data for “Patient Body Weight (LOINC: 29463-7)” among three institutions could be “80 kg”, “160 lb” or “158 lbs”. Another example is for reporting “Aspergillus fumigatus 2 Ab (LOINC: 29334-0)”, the report value could be “negative”, ”NEG”, “positive” or “POS” among three institutions. This implies that even with proper LOINC use, aggregating data across different institutions will require the conversion of different units of measure and creating synonym tables. Therefore, we investigated the common problems that occur when aggregating data that has been represented using LOINC codes. We collected laboratory data reported using LOINC codes from three large institutions for our investigations. Our goal was to understand:
How many heterogeneous data formats are represented when combining tests based on LOINC codes?
What kinds of efforts are needed for converting the heterogeneous data formats so that data can be aggregated?
Background
1). LOINC in Action
Currently, LOINC is used for reporting laboratory data in many health organizations, including major laboratories (e.g. ARUP, Quest and LabCorp), large healthcare providers (e.g. Indiana Network for Patient Care and Intermountain Healthcare) and insurance companies (e.g. United Healthcare)3. LOINC is also used in many clinical applications, including reporting public health data7, retrieving laboratory data for clinical studies6, providing standardized terminology in supporting sharable clinical decision support logic8 and reporting adverse events9.
2). Previous Evaluations of LOINC
Terminological Systems (TSs) are not perfect, but can be improved through cycles of scrutiny to detect errors and omissions and then correction. The early stages of developing TSs focused on building them based on functional, structural, and policy perspectives10, 11. As TSs are now in use in many applications, investigators have begun to evaluate TSs based on practical issues12–14. In some ways LOINC development has followed a similar path. At first we mainly focused on discussing LOINC design philosophy1, 2. As LOINC has become more widely used, we are now more interested in evaluating its performance in real applications4–6, 15. We have evaluated LOINC usage among three large institutions based on examining LOINC coverage4 and correctness of LOINC mapping5.
3). The Design of LOINC
The Entity-attribute-value (EAV) triplet is a common design used in representing clinical data16. For example, a body weight measurement would be represented conceptually as “Observation (entity) has Test name = Body weight (attribute); value =84.7 kg (value)”. LOINC codes are designed to be used as observation identifiers in Health Level Seven (HL7) messages. Here are two sets of examples of the actual syntax of HL7 Version 2.X OBX (observation/result) segments:
Example 1:
OBX|1|NM|29463-7^Body Weight^LN|1|84.7|kg |||”
OBX|1|NM|29463-7^Body Weight^LN|1|156|lb |||”
OBX|1|NM|29463-7^Body Weight^LN|1|166|lbs |||”
Example 2:
OBX|1|NM| 29334-0^ Aspergillus fumigatus 2 Ab^LN|1|Pos| |||”
OBX|1|NM| 29334-0^ Aspergillus fumigatus 2 Ab^LN|1|Positive| |||”
In example 1, the LOINC code “29463-7” represents the test, “Body Weight”, and the “84.7 kg”, “156 lb” and “166 lbs” are the values of “Quantitative” measurements. In the example 2, the LOINC code “29334-0” represents the test, “Aspergillus fumigatus 2 Ab”, and “Pos” and “Positive” are “Ordinal” measurement values. LOINC codes are defined using a six-axis model: “component”, “property”, “timing”, “system”, “scale”, and “method”. The scale portion of the LOINC name specifies whether the measurement is “quantitative”, “ordinal”, “nominal”, “narrative” etc. (Table 1)1, 2, 17.
Table 1.
Examples and descriptions for each type of measurement in “Scale” axis.
| Scale Type | Abbreviation | Descriptions |
|---|---|---|
| Quantitative | Qn | This is for reporting continuous numeric scale. Valid values are “–7.4”, “0.125”,”<10”,”1–10”,”1:256” |
| Ordinal | Ord | Ordered categorical responses, e.g, “1+”,”positive”, ”negative”, “reactive”, “indeterminate” |
| Nominal | Nom | Nominal or categorical response that do not have a natural ordering, e.g. “name of bacteria”, “yellow”, ”clear”, “bloody” |
| Narrative | Nar | Text narrative, e.g. description of microscopic part of a surgical papule test. |
| Quantitative or ordinal | OrdQn | Test can be reported as either “Qrd” or “Qn”. LOINC committee discourages the use of this type. |
4). Current work to standardize laboratory data
With the development of EHRs, transferring clinical data among physicians, laboratories, healthcare organizations, and clinical researchers has become a complicated task where users often have to deal with many different data formats. To address this issue, the California HealthCare Foundation (CHCF) started the EHR-lab Interoperability and Connectivity Specification (ELINCS) in 200518. ELINCS was developed to provide for standardized formatting and coding of electronic messages used to exchange data between clinical laboratories and EHR systems. In 2007, HL7 approved the ELINCS standard for transmitting laboratory data, and it has since been evaluated in pilot implementations 19. Also as an attempt to reduce the variability of data, the International Standards Organization (ISO) developed the ISO/IEC 11179 standard as a framework for consistent data representation20. To conform to ISO/IEC 11179 standard, a data element should contain a data element concept (DEC) and one value domain. The value domain consists of a set of permissible values, which is an expression of a value meaning allowed in a specific value domain20. The ISO/IEC 11179 standard has been widely adopted for developing common data elements (CDEs) in cancer research21.
5). Extensional definitions (EDs) to characterize laboratory data
A systematic method can be used to characterize laboratory data by grouping tests having the same local codes together and describing their usage in the system. A test can be characterized by how frequently it is done, its mean value, the standard deviation of the value, its associated unit of measure, and the frequencies of coded values that it has. These test profiles, which are called EDs, reflect the meaning of tests in the system (Table 2). The approach of generating EDs has been applied to automatically map local laboratory tests from 3 institutions22 and to verify the correctness of the LOINC mappings5.
Table 2.
The example of extensional definitions (EDs).
| Extensional attribute | Example | Value of information for meaning and mapping |
|---|---|---|
| Local description | “Creatinine, 24 hr urine”, ”Sodium urine” | Provides a human readable meaning for the test |
| Mean | 1.46,137 | Mean - the average value for quantitative tests. Provides information for reviewing quantitative tests. |
| Standard deviation | 0.54, 7.02 | Standard deviation - a measure of the physiologic consistency of the values of Quantitative tests |
| Units of measure | g/24 h, mmol/L, mg/dl | Units of measure - provides scale information. Sometimes it also provides time information (e.g. g/24h implies 24 hour) |
| Coded variables | Yellow (45), Negative (1345), Rare (697), 1+(143) | After grouping the same tests in each institution by their local codes, we calculated the frequency of each coded variable. |
| Frequency | 50, 184 | Frequency - implies whether tests are frequent (e.g. biochemistry tests) or rare (e.g. allergen test) |
Methods
Data sources and scope
This study is based on two LOINC evaluation studies4, 5. After sending out invitations, three institutions agreed to join in this research. They were: 1. Associated Regional and University Pathologists, ARUP Laboratories (Salt Lake City, UT) 2. Intermountain Healthcare, Intermountain (Salt Lake City, UT) 3. Regenstrief Institute, Inc. (Indianapolis, IN). ARUP Laboratory is a national clinical and anatomical pathology reference laboratory, which is owned and operated by the Pathology Department of the University Utah. Intermountain Healthcare is a not-for-profit health care provider organization, which consists of many hospitals located in major cities in Utah. Regenstrief Institute, Inc., is an informatics and healthcare research organization, which operates a regional health information exchange in central Indiana called the Indiana Network for Patient Care (INPC)23 that includes data from more than a hundred source facilities and thirteen health systems. Regenstrief Institute is located on the campus of the Indiana University School of Medicine in Indianapolis. In our previous study, the Regenstrief dataset was retrieved from five institutions, which share similar resources on their laboratory systems. To avoid selection bias, we only used data collected from the largest institution. With IRB approval, de-identified patient data for the year of 2007 as reported by general laboratory systems for each institution were selected for this study.
Generate extensional definitions (EDs)
The raw patient data were stored in the source institutions with various formats, e.g. HL7 messages or flat files. First, we customized the individual interface for each institution to transform raw data into standardized comma separated value (CSV) files. In CSV files, we loaded the following data elements:
Local code: The internal test identifier. We can use these codes to group the same tests together for generating EDs.
Local description: The local test name, which suggests the meaning of test.
Numeric value: The numeric result of the test, which was used for calculating mean value and standard deviation of value.
Units of measure: We kept raw presentations of units of measure, without any normalization process of text string.
Coded variables: The results of non-quantitative tests, e.g. (positive, pos, 1+, rare, yellow, light pink)
LOINC mappings: The LOINC code for this test (mapped by the source institution), which we use for grouping the same tests among different institutions.
Then, we developed a parsing program written in JAVA and Python to process CSV files to generate EDs (Table 2). To avoid transferring the patient data out from its original institution, we distributed pre-installed parsing programs to each institution and asked collaborators to process the de-identified patient data within the virtual machines. Only processed statistical results were sent back to us for analysis.
Analysis
There are two ways to count numbers of tests. One is to calculate the number of unique concept and another is to calculate the total frequency of each concept. For example, there are 10 “Body weight” tests in the system. If we count the unique concept of “Body weight”, it is one. But if we count the volume of “Body weight” tests, it is ten. In the laboratory system, the distributions of tests are highly skewed24 and counting concepts by their volume provides the most information about the true usage of the test in the system.
After receiving the EDs for all tests at the three institutions, we:
Calculated the numbers of LOINC codes for each different “Scale” by counting their unique concept and their total volume
Calculated numbers of UOM by counting their unique concept and their total volume
Calculated the number of variables by counting their unique concept and their total volume
Manual Review
In order to characterize the differences in presentation of the laboratory among three institutions, we conducted a more detailed manual review on a subset of the data. We grouped tests from the three source institutions by LOINC codes, and then selected a one-tenth sample. To aid in the analysis we divided tests into classes of quantitative and non-quantitative. In on our last study5, we sampled one-tenth of the LOINC mappings for manual review for correctness of the LOINC mappings. The LOINC mapping errors identified in that prior review were excluded from the current study. We also excluded LOINC concepts that only existed in a single institution. We then reviewed the result values and characterized the differences in presentation by identifying common patterns. A given test could be assigned to more than one taxonomy, because it could contain more than two heterogeneous presentations.
Results
After receiving the data from all three institutions, there were 4,876 local laboratory tests, which were mapped to 3,078 unique LOINC codes. Among these 4,876 tests, the frequency and percentage of each scale type was determined as shown in Table 3. The most frequent categories were “Qn” and “Ord”. The tests in “Narrative” category were relative small, and consisted of unstructured information and were not considered further in the analysis. The distribution of LOINC ‘Class’ was shown in Table 4.
Table 3.
The frequency and percentage of each scale.
| ARUP | Intermountain | Regenstrief | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Scale | A | B | C | A | B | C | A | B | C |
| Qn | 1473 | 77% | 35% | 859 | 68% | 81% | 1194 | 71% | 95% |
| Ord | 323 | 17% | 46% | 233 | 18% | 6% | 397 | 23% | 3% |
| Nominal | 96 | 5% | 17% | 102 | 8% | 1% | 80 | 5% | 2% |
| OrdQn | 21 | 1% | 2% | 66 | 5% | 2% | 12 | 1% | <1% |
| Narrative | 4 | <1% | 1% | 7 | 1% | <1% | 9 | 1% | <1% |
| Total | 1917 | 1267 | 1692 | ||||||
A: Number of unique LOINC codes, B: Percentage of each LOINC code by counting unique LOINC codes, C: Percentage of each LOINC code by counting their total volume
Table 4.
The distributions of LOINC ‘Class’.
| ARUP | Intermountain | Regenstrief | |||
|---|---|---|---|---|---|
| LOINC class | Percentage | LOINC class | Percentage | LOINC class | Percentage |
| CHEM | 42.87% | HEM/BC | 40.04% | CHEM | 61.58% |
| MICRO | 15.00% | CHEM | 36.65% | HEM/BC | 29.78% |
| HEM/BC | 13.41% | MICRO | 5.11% | UA | 4.50% |
| SERO | 5.34% | UA | 4.97% | MICRO | 1.55% |
| DRUG/TOX | 4.64% | ABXBACT | 2.41% | SPEC | 0.87% |
| ALLERGY | 3.77% | SPEC | 2.36% | DRUG/TOX | 0.50% |
| SPEC | 3.74% | CLIN | 2.18% | BLDBK | 0.31% |
| COAG | 2.46% | COAG | 1.47% | COAG | 0.26% |
| UA | 1.54% | BLDBK | 1.35% | SERO | 0.17% |
| OB.US | 1.23% | DRUG/TOX | 1.02% | CELLMARK | 0.17% |
The ‘CHEM’, ‘HEM/BC’,’MICRO’ are the most frequently used LOINC classes.
Characterization of the different presentations of laboratory data
The frequency distribution of UOM was highly skewed. In Intermountain and Regenstrief, about 10 UOM account more than 85% of all test volume. The examples of the top 10 UOM in each institution are listed (Table 5). The total unique UOM used in ARUP, Intermountain and Regenstrief were 103, 80 and 105 respectively.
Table 5.
Example of the top 15 UOM in three institutions.
| ARUP | Intermountain | Regenstrief | |||
|---|---|---|---|---|---|
| UOM | Percentage | UOM | Percentage | UOM | Percentage |
| mg/dL | 12% | mg/dL | 18% | mg/dL | 25% |
| % | 10% | % | 16% | mmol/L | 23% |
| ng/mL | 7% | mmol/L | 16% | % | 11% |
| g/dL | 5% | 10^3/uL | 9% | k/cumm | 9% |
| mmol/L | 5% | g/dL | 6% | GM/dL | 8% |
| IV | 5% | U/L | 5% | Units/L | 6% |
| K/uL | 5% | K/uL | 5% | million/cumm | 5% |
| kU/L | 4% | fL | 5% | fL | 2% |
| pg/mL | 3% | /100 WBC@s | 3% | pg | 2% |
| U/L | 3% | 10^6/uL | 3% | mL/min/1.73m2 | 2% |
| Total number of unique units of measure | |||||
| 103 | 80 | 105 | |||
The percentage is the sum of the total test volume.
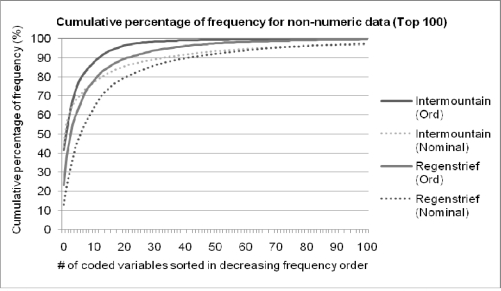
We calculated the frequency of each coded variable by summing their whole volume. The frequency distribution of non-quantitative tests was also highly skewed (Figure 1). Among three non-quantitative groups (Ord, Nominal and QnOrd), the “Nominal” category contains the most varied formats and frequent “See note” “See Description” and “HIDE” among three institutions. The examples of frequently used coded variables are shown in Table 6.
Figure 1.
The cumulative percentage of frequencies for non-quantitative tests (Ord and Nominal). The percentage was summed for the total test volume.
Table 6.
Example of coded variables used in ACnc, Nominal and OrdQn category, which were summed to their total volume.
| Ord | Nominal | OrdQn | ||||||
|---|---|---|---|---|---|---|---|---|
| A | I | R | A | I | R | A | I | R |
| NEG | NEG | Negative^Nega tive | SEE NOTE | HIDE | CLEAR^Clear | NONE | SS | Syn-S |
| NEGATIVE | HIDE,*, | Normal^Normal | NO | NORMAL | YELLOW^Yellow | SEE NOTE | R | Syn-R |
| NOT APPL | PLTOK | NEG^Negative | NEGATIVE | (null) | 310215^Escherichia coli | HIGH | I | space |
| DETECTED | P1,*, | Few (1+)^Few (1+) | WHITE | CCS | 310784^Staphylococcus aureus | POSS | NONE | >=8 |
| POSITIVE | (null) | neg^Negative | CLEAR | SOK | See description^See description | NEGATIVE | DEL | Susceptible |
| NONE DET | NR | CX7NEG^Negative | US | XT | LTYELL^Light Yellow | <0.1 | <=0.25 | <=8 |
| SEE NOTE | P2,*, | Neg^Negative | LNMP | COMP AT | SLCLDY^Slightly Cloudy | NDO | Resistant | |
| NON REAC | NDE | NR^Negative | NOT APPL | DEL | 760835^Pseudomon as aeruginosa | <=8 | >=256 | |
| 1+ | OBSER,*, | RARE^Rare | HISPANIC | LRCA | Normal^Normal | <=0.06 | Ceftazidime | |
| NORMAL | TRACE,*, | Positive^Positive | IFE DONE | ;BL | No fungal elements seen. | HIDE | ||
They are sorted in decreasing frequency. A: ARUP, I: Intermoutain, R:Regenstrief. “P1” means “one plus” and is a synonym for “1+”,
II. Comparing the different presentations of laboratory data
A one-tenth sample of the 3,078 unique LOINC codes contained 479 laboratory tests and 308 unique LOINC codes (Table 7). After removing the mapping errors, there were 445 tests. Only tests appearing in more than one institution were selected for review, which left 229 tests containing 92 LOINC concepts. The most frequent reasons for differences in quantitative test presentation were “missing UOM” and the “Synonymous units” (Table 8). The most frequent reasons for differences in presentation among non-quantitative tests were “Acronym/Synonym” and “Different enumeration” (Table 9).
Table 7.
The number of tests and LOINC codes in the data sets.
| Number of tests | Number of LOINC | |
|---|---|---|
| Total number of tests | 4876 | 3078 |
| After one-tenth sampling | 479 | 308 |
| After removing error mapping | 445 | 293 |
| Appearing in more than one institution (quantitative + non-quantitative) | 229 (186+43) | 92 (75+17) |
Table 8.
Comparison of the different presentations of UOM as extracted from three institutions.
| Category Type | Count | Examples | |
|---|---|---|---|
| Institution A | Institution B | ||
| Synonym | 16 | mg/24hr | mg/d |
| 10^3/uL | K/uL | ||
| mCg/mL | ug/ml | ||
| Different magnitude | 3 | 3.98 mol/L | 100 umol/L |
| 23.5 mg/dL | 43.2 ug/ml | ||
| 84.7 kg | 166 lb | ||
| Exact match | 38 | mg/dl | mg/dl |
| Missing UOM | 19 | ||
| Number of LOINC | 75 | ||
Table 9.
Comparison of the different presentations of non-quantitative test results.
| Category Type | Count | Examples | |
|---|---|---|---|
| Institution A | Institution B | ||
| Acronym/Synonym | 8 | NEG | Negative |
| SLCLDY | Slight Cloudy | ||
| CLDY | Cloudy | ||
| Different enumeration lists | 12 | Light Pink, Pale Pink, Slightly Yellow | Pink, Yellow |
| BLDY, CLEAR, TURB, SLCLDY, CLDY | Turbid, Clear, Hazy, Cloudy | ||
| 1+,2+,3+ | Rare, Moderate, Many | ||
| Free text | 3 | 1+ (few) Acid fast bacilli in concentrated smear | Rare Acid fast bacilli seen .br Results called to and read back by Dr xxx |
| Perfect match | 2 | Positive, Negative | Positive, Negative |
| # of LOINC | 17 | ||
The “Acronym/Synonym” and “Different enumeration lists” were the most frequent reasons for inconsistent presentations. These examples were extracted from three institutions.
Discussion
Having a universal observation identifier (e.g. a LOINC code) to aggregate laboratory test data is necessary but not sufficient for full semantic interoperability. Current LOINC codes could cover 99% of the volume of laboratory tests in daily operation in Intermountain and Refenstrief4. Yet, this analysis shows that laboratory data have significant variation in their delivered units of measure for quantitative tests and for the answers of non-quantitative tests. Further work (e.g. to define guidelines for units of measure and coded values for laboratory data) for solving those problems is needed. Currently, users combining existing laboratory data might encounter the following issues.
Heterogeneous formats of quantitative tests
1) Missing UOM: Quantitative tests without a UOM is not meaningful;25 to know the accurate meaning of quantitative tests, users need to know the UOM. The quantitative tests reported in the HL7 OBX segment should always specify UOM. Although not available for this analysis, one problematic messaging pattern we have observed in our experience (and a potential reason why the UOM was missing from our data) is sending the UOM in the NTE segment of the HL7 message.
2) Lack of a standardized code for UOM: 21% (16/75) of the quantitative LOINCs from our sample had variations of synonymous UOM, thus highlighting the potential value of adopting a standardized UOM representation. One standardized UOM developed by Regenstrief is “The Unified Code for Units of Measure (UCUM)”25. Other standards are ISO 2955, ANSI X3.501, “ISO+” developed by HL7 and ASTM 1238, and the European Standard ENV 124356.
3) There is a need for converting UOM: Inevitability we have to face varied formats of UOM in existing systems, which might have different magnitudes, e.g. we need to convert “lb” to “kg”. One approach to solve this problem is to create conversion programs for UOM based on the “dimension” of the measurement. The base system of dimensions consist of length, time, mass, charge, temperature, luminous intensity and angle. For example, “mg/dL” could be represented by “L-3M”. UOM within the same dimension can be converted algorithmically. The UCUM project has developed an open-source Java implementation for UOM conversion25.
Heterogeneous formats of non-quantitative tests
1) Lack of standardized terminology: There are substantial number of synonyms and acronyms used in reporting non-quantitative tests. This variation creates a large burden for those attempting to aggregate data, because it forces users to create mapping tables for all of the synonyms and acronyms appearing as result values. We also observed both “neg” and “negative” used for values of the same test within a single institution, but this type of inconsistency is more commonly seen between different tests and institutions.
2) Lack of standardized enumeration lists (value sets) for reporting encoded data: For example, e.g. reporting urine color as “yellow” in one institution, while another institution could have more fine-grain descriptions, e.g. “light yellow”, “yellow” and “dark yellow”. Lack of standardized value sets will hinder data integration. The best way to solve this is to specify the standardized value sets for reporting laboratory data, but for existing systems, a possible solution is to use an ontological approach for grouping different granularities of information under common parents.
3) Lack of permissible value: It is common to observe the same laboratory test containing a quantitative measurement (<1:16) in some instances, and a non-quantitative measurement (negative) in other instances. This happens because of a typical laboratory practice. When measuring the existence or quantity of a “Drug”, “Bacteria” or “Antibody” in a sample, users first derive the quantitative measurement from the machine, then compare the measured value to a cut-off value to interpret the quantitative test as being positive or negative. It would be best if both the measured value and the interpretation were sent from the laboratory.
4) Lack of standardized models for reporting complicated data: Sometimes value sets are not sophisticated enough for reporting complicated data, such as reporting genetic tests. For example, reporting “ALPHA-1-ANTITRYPSIN PHENOTYPE’, the report value could be “M1M1”, “M1M2”,”MM”,”M3M3”,”M1Z” or “M1S”. The phenotypes of “ALPHA-1-ANTITRYPSIN” have different alleles variants “M”, “S”, and “Z”, and “M” variants could be divided into six M subtypes. The report value, e.g. “M1M2” implies a model of “types of variants (M, Z, or S)” and “subtype of variants (M1,M2,…,M6)”. Standardized models for reporting complicated data are needed to help clinical applications consistently represent the results of complex tests.
5) Lack of standardized strategies for sending some result information: Result reporters often find convenient (but less than optimal) ways of dealing with the complexity of sending both numeric and interpretive data by sending the true test result (and perhaps some additional interpretive or “boilerplate” information) as NTE segments in the HL7 message. We saw evidence of this practice by phrases like “see note” or “see description” appearing in the OBX-5 (observation value) field of the message. Storing results in different strategies, e.g. storing information in NTE segments instead of OBX-5 segment, would hinder data integration.
Limitations
This study only collected data from three institutions. These institutions also provided their laboratory test names for creating the initial set of LOINC codes. Because these three institutions have better knowledge and resources for using LOINC than typical institutions, we cannot necessarily extrapolate our results to other institutions. Another limitation was that we did not compare the consistency of the use of UOM and coded values within the individual institutions. This dataset was collected in 2007 and some newer laboratory tests, e.g. genetic tests, were fewer at that time. For those newer tests, a more recent dataset is needed.
Conclusion
Greater interoperability could be achieved if national standards bodies or the LOINC Committee provided more guidance on best practices in coding of laboratory results. Some possible suggests are: 1) For numeric data: When UOM are appropriate for a given test, reporting UOM should be required and, standard UCUM codes should be used. 2) For enumerated lists, standardized terms and codes should be developed and use in reporting should be required. The differences in enumerated lists could be resolved by creating an ontology for combining different enumerated lists. 3) For complicated data (e.g. genetic tests results), standardized models or patterns for results reporting are needed. It is reasonable to predict that genomic tests will be increase in frequency and will become important tests in clinical decision support systems. We need to standardize how to report these kinds of complex data. Healthcare providers, should be aware of the issues related to coding of laboratory results and adopt best practices in their daily operations. LOINC use in their production databases can provide valuable information for improving LOINC design. Finally, users should profile their LOINC usage periodically to monitor the quality of TSs practice.
References
- 1.Forrey AW, McDonald CJ, DeMoor G, Huff SM, Leavelle D, Leland D, et al. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem. 1996 Jan;42(1):81–90. [PubMed] [Google Scholar]
- 2.Huff SM, Rocha RA, McDonald CJ, De Moor GJ, Fiers T, Bidgood WD, Jr, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc. 1998 May-Jun;5(3):276–92. doi: 10.1136/jamia.1998.0050276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003 Apr;49(4):624–33. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 4.Lin MC, Vreeman DJ, McDonald CJ, Huff SM. A Characterization of Local LOINC Mapping for Laboratory Tests in Three Large Institutions. Methods Inf Med. Aug 20;49(5) doi: 10.3414/ME09-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin MC, Vreeman DJ, McDonald CJ, Huff SM. Correctness of Voluntary LOINC Mapping for Laboratory Tests in Three Large Institutions. AMIA Annu Symp Proc; 2010. pp. 447–51. [PMC free article] [PubMed] [Google Scholar]
- 6.Baorto DM, Cimino JJ, Parvin CA, Kahn MG. Combining laboratory data sets from multiple institutions using the logical observation identifier names and codes (LOINC) Int J Med Inform. 1998 Jul;51(1):29–37. doi: 10.1016/s1386-5056(98)00089-6. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention National Electronic Disease Surveillance System (NEDSS). Centers for Disease Control and Prevention. Available from: http://www.cdc.gov/NEDSS/.
- 8.Greenes RA. Clinical decision support : the road ahead. Boston: Elsevier Academic Press; 2007. [Google Scholar]
- 9.Brandt CA, Lu CC, Nadkarni PM. Automating identification of adverse events related to abnormal lab results using standard vocabularies. AMIA Annu Symp Proc; 2005. p. 903. [PMC free article] [PubMed] [Google Scholar]
- 10.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998 Nov;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 11.2002. ISO/TC215. Health informatics -- Controlled health terminology -- Structure and high-level indicators. Report NO.:17117.
- 12.Andrews JE, Richesson RL, Krischer J. Variation of SNOMED CT coding of clinical research concepts among coding experts. J Am Med Inform Assoc. 2007 Jul-Aug;14(4):497–506. doi: 10.1197/jamia.M2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenreider O, Mitchell JA, McCray AT. Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. Proc AMIA Symp; 2002. pp. 61–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 15.Dugas M, Thun S, Frankewitsch T, Heitmann KU. LOINC codes for hospital information systems documents: a case study. J Am Med Inform Assoc. 2009 May-Jun;16(3):400–3. doi: 10.1197/jamia.M2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadkarni PM, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P. Organization of heterogeneous scientific data using the EAV/CR representation. J Am Med Inform Assoc. 1999 Nov-Dec;6(6):478–93. doi: 10.1136/jamia.1999.0060478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LOINC Committee . Indianapolis, IN: 2011. LOINC Manual. Available from: http://loinc.org/downloads/files/LOINCManual.pdf. [Google Scholar]
- 18.The California HealthCare Foundation (CHCF) Indianapolis, IN: 2011. ELINCS: The Lab Data Standard for Electronic Health Records. Available from: http://elincs.chcf.org/. [Google Scholar]
- 19.Raths D. ELINCS gets adopted. HL 7 is taking over ELINCS to ensure lab data and EHRs continue towards interoperability. Healthc Inform. 2007 Mar;24(3):20, 4. [PubMed] [Google Scholar]
- 20.ISO/IEC 11179, Information Technology -- Metadata registries (MDR). Indianapolis, IN 2011; Available from: http://metadata-standards.org/11179/.
- 21.Covitz PA, Hartel F, Schaefer C, De Coronado S, Fragoso G, Sahni H, et al. caCORE: a common infrastructure for cancer informatics. Bioinformatics. 2003 Dec 12;19(18):2404–12. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- 22.Zollo KA, Huff SM. Automated mapping of observation codes using extensional definitions. J Am Med Inform Assoc. 2000 Nov-Dec;7(6):586–92. doi: 10.1136/jamia.2000.0070586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald CJ, Overhage JM, Barnes M, Schadow G, Blevins L, Dexter PR, et al. The Indiana network for patient care: a working local health information infrastructure. An example of a working infrastructure collaboration that links data from five health systems and hundreds of millions of entries. Health Aff (Millwood) 2005 Sep-Oct;24(5):1214–20. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 24.Vreeman DJ, Finnell JT, Overhage JM. A rationale for parsimonious laboratory term mapping by frequency. AMIA Annu Symp Proc; 2007. pp. 771–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Schadow G, McDonald CJ, Suico JG, Fohring U, Tolxdorff T. Units of measure in clinical information systems. J Am Med Inform Assoc. 1999 Mar-Apr;6(2):151–62. doi: 10.1136/jamia.1999.0060151. [DOI] [PMC free article] [PubMed] [Google Scholar]