Abstract
Identification of a cohort of patients with specific diseases is an important step for clinical research that is based on electronic health records (EHRs). Informatics approaches combining structured EHR data, such as billing records, with narrative text data have demonstrated utility for such tasks. This paper describes an algorithm combining machine learning and natural language processing to detect patients with colorectal cancer (CRC) from entire EHRs at Vanderbilt University Hospital. We developed a general case detection method that consists of two steps: 1) extraction of positive CRC concepts from all clinical notes (document-level concept identification); and 2) determination of CRC cases using aggregated information from both clinical narratives and structured billing data (patient-level case determination). For each step, we compared performance of rule-based and machine-learning-based approaches. Using a manually reviewed data set containing 300 possible CRC patients (150 for training and 150 for testing), we showed that our method achieved F-measures of 0.996 for document level concept identification, and 0.93 for patient level case detection.
INTRODUCTION
Electronic health records (EHRs) contain a longitudinal record of patient health, disease, and response to treatment useful for epidemiologic, clinical, genomic, and informatics research. Notable recent examples include those explored by the electronic Medical Records and Genomics (eMERGE) network [1]. However, accurate identification of cohorts of patients having specific diseases or receiving certain treatments from EHR can be challenging. As much of the detailed patient information is embedded in clinical narratives, a traditional method for case detection is to conduct manual chart review by physicians. However, it is a very costly and time-consuming task to manually collect, find, and abstract all clinical documents of possible patients.
Recent advances in natural language processing (NLP) have offered automated methods to extract information from free text, including clinical narratives. Over the last three decades, a number of clinical NLP systems have been developed, including some earlier systems such as Medical Language Processing (MLP) system from the Linguistic String Project (LSP) [2, 3], MedLEE (Medical Language Extraction and Encoding System) [4–6], and SymText/MPlus [7–9], as well as more recent open source systems such as cTAKES [10] and HiTEX [11]. Some systems, such as MetaMap [12] and KnowledgeMap [13], focus on extracting concepts from biomedical text including biomedical literature and clinical notes. After that, contextual information of those concepts such as assertion status (i.e., is a medical problem present or negated?) can be recognized through additional algorithms or programs, such as NegEx [14] for negation status, and ConText [15] for other broad contextual information.
For disease case detection, a number of studies have shown that coded data such as International Classification of Disease (ICD) codes were not sufficient or accurate enough [16–18]. Therefore, many studies involve data extraction from clinical text and NLP has been used for phenotype extraction in various studies. Penz et al. [19] found that ICD-9 and Current Procedural Terminology (CPT) codes only identified less than 11% of the cases in a study of detecting adverse events related to central venous catheters, while NLP methods achieved a sensitivity of 0.72 and a specificity of 0.80. Li et al. [20] compared the results of ICD-9 encoded diagnoses and NLP-processed discharge summaries for clinical trial eligibility queries. They concluded that NLP-processed notes provide more valuable data sources for clinical trial pre-screening as they provide past medical histories as well as more specific details about disease that are unavailable in ICD-9 codes. Savova et al. [21] used cTAKES to discover peripheral arterial disease (PAD) cases from radiology notes, and classified cases into four categories: positive, negative, probable, and unknown. There are a number of studies focused on cancer case detection. For example, Friedlin et al. [22] found either ICD-9 codes and NLP methods identified pancreatic cancer patient well from a cohort of pancreatic cyst patients. However, the ICD-9 code method had lower specificity and positive predictive value. Wilson et al. [23] evaluated the ability of an automated system to extract mesothelioma patients’ personal and family history of cancer from positive surgical pathological report and when patients are admitted to hospital after mesothelioma diagnosis. They tested two information extraction methods: Dynamic-window and ConText [15] for building cancer frames, which showed that both the methods performed much better than their human benchmark. Relevant to CRC patients, we have previously compared NLP methods to colonoscopy events to find completed colonoscopies, finding that billing records identified 67% of the events compared to NLP with 92% [24].
Despite the success of applying informatics approaches to case detection from EHRs, most prior research has focused on a limited number of document types (e.g., chest radiographs for pneumonia). Few studies have explored methods to extract, integrate, and utilize heterogeneous clinical data (including structured and narrative data from various types of notes) in entire EHR, for case detection purposes. In this study, we investigated informatics approaches for detecting CRC patients from entire EHR, in order to support an ongoing epidemiological study at Vanderbilt. Colorectal cancer (CRC) is the fourth most common incident cancer and the second most leading cause of cancer death in the United States [25], despite significant efforts to increase screening and improve treatment. About 1 in 18 individuals will develop colorectal cancer over their lifetime and 40% will die within 5 years of diagnosis, mainly due to diagnosis at a late stage [26, 27]. The goal of the study is to collect all CRC cases, including those with history of treated CRC from Vanderbilt University Hospital’s (VUH) EHR. We developed a 2-step approach for CRC case detection, which consists of 1) extraction of CRC concepts from various types of clinical documents, and 2) prediction of CRC cases by integrating evidences from both structured and narrative sources.
METHODS
Overview of the Case Detection Method
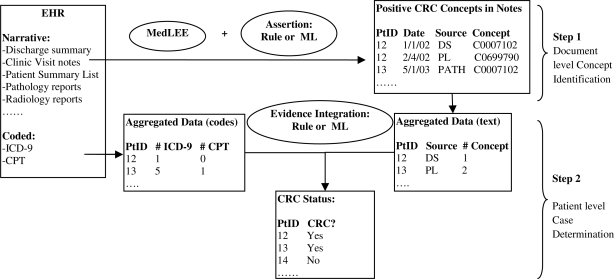
The case detection method proposed in this study consists of two components: 1) a module to identify positive CRC concepts from various types of narrative clinical documents (document-level concept identification); and 2) a module to determine CRC cases using aggregated information from both narrative data and coded ICD-9 and CPT data (patient-level case determination). Figure 1 shows an overview of our approach. The document-level concept identification component also consists of two steps. The first step is to identify all CRC concepts in clinical documents using the MedLEE NLP system; while the second step determined if a detected CRC concept is positive or negative. For the patient-level case determination module, we implemented and compared two different methods to combine evidence from heterogeneous clinical data: a heuristic rule-based approach and a ML-based approach.
Figure 1.
Overview of the 2-step case detection method from EHR.
Data Set
In this study, we used clinical data from the Synthetic Derivative (SD) database, which is a de-identified copy of the EHR at VUH. Different types of notes include DS – discharge summaries, CC – clinical communications, FORM – clinical forms, RAD – radiology notes, PATH – pathology notes, PL – patient summary lists, and OTHER – Other clinical notes (History & Physicals, clinic notes, progress notes). We limited the scope of this study to a 10-year period (1999–2008), in which there were 1,262,671 patients in total. Given the overall low incidence of CRC (about 5% individuals will develop CRC over their lifetime), we first selected an minimum data set that we expected to contain all patients with CRC. Domain experts defined a broad filter using ICD-9 codes, CPT codes, CRC and related drug keywords, to capture all possible CRC cases and form a data set of interest for this study. Table 1 shows the criteria of the filter. If a patient matched any one of the criteria in Table 1, he/she was included into the pool of possible CRC cases, which had 17,125 patients.
Table 1.
Criteria for selecting possible CRC patients from EHR.
| Inclusion Criteria | # of Patients |
|---|---|
| CRC related ICD-9 codes, including “153, 153.0, 153.1, 153.2, 153.3, 153.4, 153.5, 153.6, 153.7, 153.8, 153.9, 154.0, 154.1, 45.92, 45.93, 45.94, 48.5, 48.62” | 5,797 |
| CRC related CPT codes, including “44160, 44147, 44140, 44145, 44146, 44143, 44144, 44141, 44156, 44158, 44157, 44155, 44151, 44150, 38564, 38562, 38770, 4180F” | 2,090 |
| CRC keyword search: any sentence containing both a cancer term in “adenocarcinoma OR cancer OR carcinoma OR neoplasm OR tumor OR tumour” AND a body location term in “appendix OR bowel OR caecum OR cecum OR colon OR colorectal OR intestine OR rectosigmoid OR rectal OR rectum OR sigmoid OR splenic flexture”. This search is limited to four types of notes: discharge summaries, pathology notes, radiology notes, and patient summary list for outpatients. | 13,241 |
| CRC related drug keyword search, including “irinotecan, cpt11, camptosar, eloxatin, oxaliplatin” | 2,475 |
| Any of Above | 17,125 |
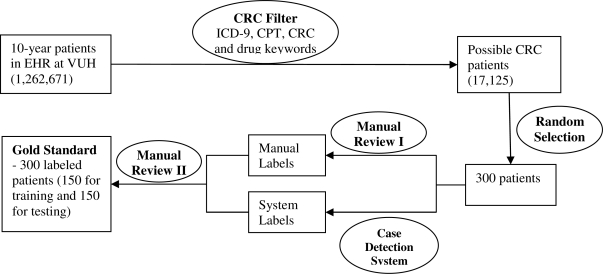
From the pool of possible CRC cases, we randomly selected 300 patients and collected all of their records in EHR. An oncologist (ZF) manually reviewed records of all 300 patients and decided if a patient had colorectal cancer (including those with history of CRC). After we developed our automated case detection system, we applied the system to the same 300 patients. Any discrepancy between the manually reviewed result and system generated result was submitted to another domain expert (NP) for a second review. The final decision was made by NP, with discussion with other physicians in the team. This labeled data set with 300 patients served as the gold standard for this study. We further divided the entire data into a training set of 150 patients, and a test set of 150 patients. The methods described below were developed and optimized using the training set, and evaluated on the independent test set. Figure 2 shows the workflow about the creation of the data sets for this study.
Figure 2.
The workflow of creation of the gold standard data set containing 300 patients with labels indicating their CRC status (Yes or No).
Document-level Concept Identification
Based on UMLS, domain experts in our team defined a list of concepts for CRC, including134 UMLS CUIs, manually selecting concepts from related CRC concepts using the KnowledgeMap web application [28]. The task is then to identify those CRC concepts that are about patients from clinical text, excluding those that are negated, hypothetical, or representing family history. We used the well-established NLP system MedLEE for concept extraction in this study, because it has the capability to detect contextual information associated with a concept. The MedLEE system has shown good coverage on concept extraction in many studies [29]; therefore our focus in this study was to improve the assertion determination – to decide if a CRC concept is positive or negative. A CRC concept is classified as positive if it is not negated, associated with other people, hypothetical, or possible. As we included all types of clinical notes in this study, the assertion determination was more challenging than focusing on only one type of clinical notes.
The first method for assertion determination was a heuristic rule-based approach. MedLEE detects some contextual information associated with a concept, including “certainty”, “status”, and “family” modifiers associated with concepts. These attributes can be used to identity negation and the experiencer of a clinical concept. Our manual review showed that MedLEE had good performance on recognizing contextual information overall, but sometimes it missed certain section headers or other context modifiers. For example, sometimes it did not recognize certain section headers indicating family history sections, or it missed the non-patient experiencer information (e.g. “M: Colon Ca” – it means that mother had colon cancer). Therefore, we developed an additional rule-based program to search context around CRC concepts and identify contextual information that were missed or incorrectly extracted by MedLEE. Some rules are specific to certain types of notes. For example, a section header in the Problem List often starts with four dashes, e.g., “----Social History” and MedLEE did not recognize that kind of format of section headers. Therefore we developed rules to remove dashes from section headers in Problem List.
The second method used SVM (Support Vector Machines) to determine assertion. We randomly selected 500 unique sentences containing CRC concepts, from the training set of 150 patients. We manually reviewed them and labeled each CRC concept as “positive” or “negative”. Based on this annotated data set, we developed a classifier using SVMs implemented via the LibSVM package [30]. The features for the classifier include modifier information from MedLEE’s output, words and bigram from contextual window around the CRC concept, as well as the distance and direction (left vs. right) of those words. The parameters of the SVM classifier were optimized using a 3-fold cross validation (CV) method on the 500 annotated sentences from the training set, before it was applied to the test set.
To assess the performance of the rule-based and ML-based assertion determination methods for CRC concepts, we developed an independent data set of 300 randomly-selected sentences. These were chosen from all CRC-concept-containing sentences in the test set, and then manually labeled. Performance of both methods on this independent data set was evaluated and reported as well.
Patient-level Case Determination
Once positive CRC concepts were identified from all types of clinical notes, aggregated information about counts of CRC concepts in each type of clinical documents (e.g., DS – discharge summaries) could be obtained (shown in Figure 1). Meanwhile, we also collected counts of ICD-9 and CPT codes from EHR. The goal of this step was to assess how such aggregated information could be used to identify CRC cases, as well as how to combine information from both unstructured and structured data to optimize the case detection task. We tested two types of methods here: rule-based and ML-based approaches.
For rule-based methods, we manually review examples in the training set and developed rules to determine if a patient was a CRC case. Concepts identified from the best algorithm in the document-level concept extraction task were used. Simple rules could be based on counts from a single source, e.g., if ICD-9 count >= 1, then it is a case. More complicated rules could take multiple sources into consideration, e.g., if CRC count from text >= 1 and patient has at least one ICD-9 or CPT code, then it is a case. However, we found that it was very difficult to define useful rules by manual review of the aggregated counts CRC concepts from different sources. Therefore we investigated ML-based approaches, which would automatically find useful patterns to determine if a patient is a CRC case or not. The input data for ML algorithms contain 12 columns, including counts of ICD-9 codes, CPT codes, CRC concepts from all types of notes, and CRC concepts from 9 individual types of clinical notes. In addition to raw counts, we also defined normalized counts, which were the ratio between the raw counts of CRC concepts from a single source and the counts of any concepts in the source documents. For example, if MedLEE identified 200 clinical concepts from entire discharge summaries of one patient and 10 of them were CRC concepts, the normalized CRC count would be 10/200=0.05 (raw count was 10). The intension was to normalize the CRC information by the length of available clinical records of a patient. Four different ML algorithms were tested, including Random Forest (RF) [31], Ripper [32], Support Vector Machine (SVM), and Logistic Regression (LR) [33]. RF is implemented via the R package [34, 35], Ripper is implemented via the WEKA package [36], SVM is implemented via the LibSVM package [30], and LR is implemented via the LibLinear package [33]. Parameters of each algorithm were optimized using a 3-fold cross validation method on the training set. Finally we trained ML-based models on entire training set and applied them to the test set. We reported both results from the training set and the test set below.
Evaluation
As both the document-level CRC concept identification and patient-level case determination were binary classification tasks, we reported standard classification metrics including precision, recall (sensitivity), specificity, accuracy, and F-measures, by comparing the system output with its corresponding manually annotated gold standard.
RESULTS
Characteristics of Data Sets
Based on the manual annotation, there were 121 CRC cases in the 300 patients who were randomly selected from the pool of 17,125 possible CRC patients. The detailed characteristics of data sets used in this study are shown in Table 2.
Table 2.
Distribution of cases and non-cases in testing and training data sets.
| Data Set | # of Patients | # Cases | # Non-cases | # Clinical notes | # Sentences with CRC concepts | # ICD-9 codes | # CPT codes |
|---|---|---|---|---|---|---|---|
| Training Set | 150 | 63 | 87 | 35727 | 1879 | 49 | 19 |
| Test Set | 150 | 58 | 92 | 34093 | 1099 | 39 | 10 |
| All | 300 | 121 | 179 | 69820 | 2978 | 88 | 29 |
Document-level CRC Concept Identification
The document-level CRC concept identification system was developed using 500 annotated sentences in the training set, and evaluated using 300 annotated sentences in the test set. These sentences were randomly selected from all types of notes. The rule-based method achieved high performance on both training and test data sets (F-measure of 0.996), indicating such rules were generalizable. Table 3 shows the results of detecting positive CRC concepts for both rule-based and ML-based methods using both the training and the test sets. The results of ML-based method on the training set were the averages from 3-fold cross validation.
Table 3.
Results of document level CRC concept identification using both rule-based and ML-based methods.
| Method | Data Set | Precision | Recall (Sensitivity) | F-measure | Specificity | Accuracy |
|---|---|---|---|---|---|---|
| Rule-based | Training (500) | 0.985 | 0.942 | 0.963 | 0.868 | 0.924 |
| Testing (300) | 0.996 | 0.996 | 0.996 | 0.969 | 0.996 | |
| ML-based | Training (500) | 0.951 | 0.993 | 0.971 | 0.479 | 0.945 |
| Testing (300) | 0.927 | 0.995 | 0.960 | 0.344 | 0.927 |
Patient-Level CRC Case Determination
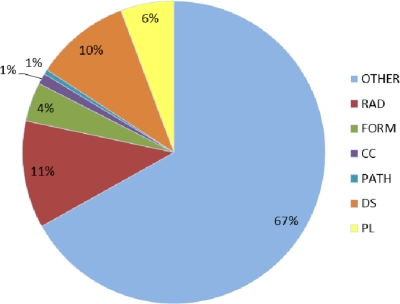
Results of patient-level case detection using the ML-based algorithms on the training set are shown in Table 4. The Ripper and RF methods achieved higher performance than SVM and LR, when normalized counts were used. Therefore, we only applied those two ML methods to the independent test data set. Results of patient-level case detection using the test set are shown in Table 5. CPT codes alone were not useful at all. When only ICD-9 codes were used (Count >=1), results were poor as well (Precision 0.77, Recall 0.52). When a single source of clinical documents was used (e.g., DS), it usually had good precision (e.g., 0.95 for DS); but its recall was poor (e.g., 0.36 for DS). However, using counts of CRC concepts from all types of clinical text achieved very good performance. The simplest rule (“if CRC count in text >=1, then it is a case”) achieved 0.84 precision and 0.97 recall. However, more complicated rules considering coded data (e.g., “if # CRC concept > 2 OR (# CRC concept >=1 AND (# ICD-9 or #CPT) >=1), then case”) did not improve much of the performance (precision 0.88, recall 0.91). However, the ML-based methods, both RF and Ripper algorithms using normalized counts achieved the better results. The Ripper algorithm achieved the best F-measure of 0.93 (0.90 precision and 0.97 recall). Also distribution of CRC concepts among different types of notes is shown in Figure 3.
Table 4.
Results of patient-level case determination using different ML algorithms (values are averages from a 3-fold CV on the training set)
| ML Algorithm | Count Representation | Pre | Rec | F-measure | Spec | ACC |
|---|---|---|---|---|---|---|
| RF | Raw | 0.90 | 0.90 | 0.90 | 0.93 | 0.92 |
| Normalized | 0.97 | 0.92 | 0.94 | 0.98 | 0.95 | |
| Ripper | Raw | 0.91 | 0.97 | 0.94 | 0.93 | 0.93 |
| Normalized | 0.95 | 0.95 | 0.95 | 0.97 | 0.96 | |
| SVM | Raw | 0.92 | 0.89 | 0.90 | 0.94 | 0.92 |
| Normalized | 0.86 | 0.70 | 0.77 | 0.92 | 0.79 | |
| LR | Raw | 0.95 | 0.79 | 0.86 | 0.97 | 0.89 |
| Normalized | 0.93 | 0.94 | 0.93 | 0.94 | 0.94 |
Table 5.
Results of patient-level case determination on the test set of 150 patients, when different data sources and methods (rule-based vs. ML-based) were used. Abbreviations of data sources: PATH – pathology notes, RAD – radiology notes, PL – patient summary list, DS – discharge summary, CC – clinical communication, and ALL – all types of notes including FORM – clinical forms and OTHER – Other clinical notes (History & Physicals, clinic notes, progress notes).
| Data Source | Method | Pre | Rec | F-measure | Spec | ACC | |
|---|---|---|---|---|---|---|---|
| Text | PATH | Rule: if # CRC concept >= 1, then case | 0.88 | 0.26 | 0.40 | 0.98 | 0.70 |
| RAD | Rule: if # CRC concept >= 1, then case | 0.88 | 0.36 | 0.51 | 0.97 | 0.73 | |
| PL | Rule: if # CRC concept >= 1, then case | 0.92 | 0.59 | 0.72 | 0.97 | 0.82 | |
| DS | Rule: if # CRC concept >= 1, then case | 0.95 | 0.36 | 0.53 | 0.99 | 0.74 | |
| CC | Rule: if # CRC concept >= 1, then case | 1.0 | 0.05 | 0.10 | 1.0 | 0.63 | |
| ALL | Rule: if # CRC concept >= 1, then case | 0.84 | 0.97 | 0.90 | 0.88 | 0.91 | |
| ALL | Rule: if # CRC concept >=2, then case | 0.84 | 0.92 | 0.88 | 0.89 | 0.90 | |
| ALL | Rule: if # CRC concept >=3, then case | 0.93 | 0.86 | 0.89 | 0.96 | 0.92 | |
| Code | ICD-9 | Rule: if # CRC ICD-9 > 0, then case | 0.77 | 0.52 | 0.62 | 0.90 | 0.75 |
| CPT | Rule: if # CRC CPT > 0, then case | 0.10 | 0.02 | 0.03 | 0.90 | 0.56 | |
| Mixed | Rule: if # CRC concept > 2 OR (# CRC concept >=1 AND (# ICD-9 or #CPT) >=1), then case | 0.88 | 0.91 | 0.90 | 0.92 | 0.92 | |
| ML: RF with normalized counts | 0.90 | 0.95 | 0.92 | 0.93 | 0.94 | ||
| ML: Ripper with normalize counts | 0.90 | 0.97 | 0.93 | 0.93 | 0.95 | ||
Figure 3.
Distribution of CRC concepts among different types of notes, including DS – discharge summaries, CC – clinical communications, FORM – clinical forms, RAD – radiology notes, PATH – pathology notes, PL – patient summary lists, and OTHER – Other clinical notes (History & Physicals, clinic notes, progress notes).
DISCUSSION
Identification of cases for a disease cohort from EHR is an important task for clinical and biologic research. Use of informatics approaches to identify cases using available EHR data may enable a broad new class of data using real-world patients. In this study, we scanned an entire EHR database for colorectal cancer cases, including both various types of free-text clinical notes and coded data such as ICD-9 and CPT codes. To handle the massive data extracted from EHR, we developed a 2-step framework for case detection, including a document-level concept identification module and a patient-level case determination module. Our evaluation using a collection of 300 possible CRC patients showed that such methods were superior, when compared with methods using a single source (e.g., pathology notes).). Overall, these methods achieved strong results with a case detection F-measure of 0.93.
Our results (see Table 4) showed that clinical narratives more reliably identified CRC cases than coded data such as ICD-9 and CPT. When CRC concepts were accurately identified from clinical text, simple rules that rely on the counts of CRC concepts could detect CRC cases with good performance. The generalizability of this finding needs further validation with other diseases. Using all types of notes in the EHR improved recall of CRC case detection and did not decrease precision significantly, when compared to the results from using pathology notes alone (0.88 for PATH and 0.84 for ALL, see Table 5). Figure 3 shows the distribution of CRC concepts among different types of clinical notes. Since CRC is largely an outpatient disease with the exception of surgical intervention, this distribution is logical. Indeed, it is notable that relevant CRC concepts were even found in nontraditional types of clinical documentation such as provider-staff interactions and clinic-patient messages through the MyHealthAtVanderbilt.com patient portal (collectively referred to as “clinical communications - CC” in Figure 3 below).
We investigated both rule-based and machine learning based methods for concept assertion case determination. For assertion determination of CRC concepts, the rule-based method achieved higher performance on the independent test set, indicating such customized rules derived from the corpus were generalizable within the given corpus. The well-established MedLEE system was another reason for the high performance of CRC concept identification. For case detection, ML-based methods outperformed rule-based methods, though both methods performed very well. Our experience was that it was very difficult to define effective rules by manual review of aggregated data based on the training set. However, ML algorithm could identify inexplicit patterns, thus improving the case detection. An interesting finding was that the normalization of the raw counts helped with the RF, Ripper, and LR algorithm, but not SVM. This finding indicates that methods used to normalize features are important to ML-based case detection, and that it could be ML algorithm-specific. We plan to look into this issue in the future.
We looked into failures of the rule-based system to accurately identify CRC cases in the test set. Two CRC cases were missed because MedLEE system did not identify any CRC concepts from the notes of those patients. In this study, we applied MedLEE to VUH clinical text without any changes and it showed very good performance on capturing CRC concepts overall (56 out of 58 cases in the test set had at least one CRC concept extracted by MedLEE from their notes). We reviewed clinical notes of those two cases and noticed that there were some scenarios where text was complicated and difficult for NLP systems. For example, MedLEE missed the CRC concept from this sentence “COLON, SPLENIC FLEXURE, PARTIAL COLECTOMY (***PATH-NUMBER[3], 8 SLIDES;**DATE[Jan 07 01]): MODERATELY DIFFERENTIATED ADENOCARCINOMA”. In this example, the word “COLON” was far away from the term “ADENOCARCINOMA”; therefore MedLEE did not link “COLON” and “ADENOCARCINOMA.” Concept linkage is difficult for any NLP system, and could be addressed with customization for these specific note types. False positives in case detection were often caused by false positives in concept identification phrase, highlighting the importance of accurate negation detection. Various reasons contributed to false positives in concept identification. For example, a misspelling of the word “screening” as “srceeing” resulted in a false positive case. In another case, a physician incorrectly entered “colon cancer” in a note for a lung cancer patient. These problems are likely very challenging for a computer system to resolve.
In this study, we evaluated the framework on colorectal cancer case detection only. More diseases have to be investigated in order to make generalizable conclusions on how to combine different data sources for case detection. Instead of using either rule-based or ML-based methods, we are planning to combine both methods to further improve the performance of case detection methods. Currently, we only used aggregated counts from different sources to determine cases. Future work would consider the temporal sequence of extracted concepts and develop more sophisticated methods for case determination.
CONCLUSION
In this study, we scanned the entire EHR to collect and integrate evidence for accurate identification of CRC patients. NLP techniques identify concepts from EHR entries are more reliable than using coded data like ICD-9 code and CPT code. The two-step case identification framework not only accurately extracts relevant concepts from clinical documents, but also provides a general approach to combine information from heterogeneous data sources in EHR for case detection.
Acknowledgments
This study was supported in part by NCI grant R01CA141307. The datasets used were obtained from Vanderbilt University Medical Center’s Synthetic Derivative, which is supported by institutional funding and by the Vanderbilt CTSA grant 1UL1RR024975-01 from NCRR/NIH. We also thank Dr. Carol Friedman at Columbia University for providing MedLEE, which is supported by NLM grants LM008635 and LM010016.
References
- 1.eMERGE Network. https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page.
- 2.Sager N, et al. The analysis and processing of clinical narrative. MedInfo. 1986 [Google Scholar]
- 3.Sager N, Friedman C, Lyman M. Medical language processing: computer management of narrative data. Reading, MA: Addison-Wesley; 1987. [Google Scholar]
- 4.Hripcsak G, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122(9):681–8. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]
- 5.Friedman C, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hripcsak G, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224(1):157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 7.Haug PJ, et al. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care. 1995:284–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Fiszman M, et al. Automatic identification of pneumonia related concepts on chest x-ray reports. Proc AMIA Symp. 1999:67–71. [PMC free article] [PubMed] [Google Scholar]
- 9.Haug PJ, et al. A natural language parsing system for encoding admitting diagnoses. Proc AMIA Annu Fall Symp. 1997:814–8. [PMC free article] [PubMed] [Google Scholar]
- 10.Savova GK, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng QT, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC, et al. Development and evaluation of a clinical note section header terminology. AMIA Annu Symp Proc; 2008. pp. 156–60. [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman WW, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 15.Harkema H, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42(5):839–51. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birman-Deych E, et al. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Medical care. 2005;43(5):480–5. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- 17.Kern EF, et al. Failure of ICD-9-CM codes to identify patients with comorbid chronic kidney disease in diabetes. Health services research. 2006;41(2):564–80. doi: 10.1111/j.1475-6773.2005.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmiedeskamp M, et al. Use of International Classification of Diseases, Ninth Revision, Clinical Modification codes and medication use data to identify nosocomial Clostridium difficile infection. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America. 2009;30(11):1070–6. doi: 10.1086/606164. [DOI] [PubMed] [Google Scholar]
- 19.Penz JF, Wilcox AB, Hurdle JF. Automated identification of adverse events related to central venous catheters. J Biomed Inform. 2007;40(2):174–82. doi: 10.1016/j.jbi.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Li L, et al. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc; 2008. pp. 404–8. [PMC free article] [PubMed] [Google Scholar]
- 21.Savova GK, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA ... Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2010. pp. 722–6. [PMC free article] [PubMed] [Google Scholar]
- 22.Friedlin J, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. AMIA ... Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2010. pp. 237–41. [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson RA, et al. Automated ancillary cancer history classification for mesothelioma patients from free-text clinical reports. J Pathol Inform. 2010;1:24. doi: 10.4103/2153-3539.71065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denny JC, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc. 2010;17(4):383–8. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ries LAG, EM, Kosary CL, Hankey BF, Miller BA. SEER Cancer Statistics Review. 2004 [Google Scholar]
- 26.Jemal A, TA, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 27.Hardy RG, MS, Jankowski JA. ABC of colorectal cancer. Molecular basis for risk factors. 2000;321:886–889. doi: 10.1136/bmj.321.7265.886. BMJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denny J, et al. “Where do we teach what?” Finding broad concepts in the medical school curriculum. J Gen Intern Med. 2005;20(10):4. doi: 10.1111/j.1525-1497.2005.0203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meystre SM, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 30.Chang C, Lin C. {LIBSVM}: a library for support vector machines. 2001.
- 31.Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 32.Cohen W. Learning Trees and Rules with Set-valued Features. 1996. pp. 709–716.
- 33.Fan R-E, et al. LIBLINEAR: A Library for Large Linear Classification. J. Mach. Learn. Res. 2008;9:1871–1874. [Google Scholar]
- 34.Kleiber A.Z.a.F.L.a.K.H.a.C. An {R} Package for Testing for Structural Change in Linear Regression Models. An {R} Package for Testing for Structural. 2002;7(2) [Google Scholar]
- 35.Hornik K. The {R} {FAQ} 2011.
- 36.Witten I, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2005.