Abstract
Clinical information is often coded using different terminologies, and therefore is not interoperable. Our goal is to develop a general natural language processing (NLP) system, called Medical Text Extraction, Reasoning and Mapping System (MTERMS), which encodes clinical text using different terminologies and simultaneously establishes dynamic mappings between them. MTERMS applies a modular, pipeline approach flowing from a preprocessor, semantic tagger, terminology mapper, context analyzer, and parser to structure inputted clinical notes. Evaluators manually reviewed 30 free-text and 10 structured outpatient clinical notes compared to MTERMS output. MTERMS achieved an overall F-measure of 90.6 and 94.0 for free-text and structured notes respectively for medication and temporal information. The local medication terminology had 83.0% coverage compared to RxNorm’s 98.0% coverage for free-text notes. 61.6% of mappings between the terminologies are exact match. Capture of duration was significantly improved (91.7% vs. 52.5%) from systems in the third i2b2 challenge.
Introduction
In the past three decades, natural language processing (NLP) has been a fertile area of research in biomedical informatics. Many NLP methods and systems have been developed for automatically extracting and structuring clinical information (e.g., medical problems and medications) from clinical text, which dramatically increases the amount and quality of information available to clinicians, patients and researchers.1–3
There is clear value to using NLP output as a data source for tasks such as medication reconciliation. Medication reconciliation is a process for creating the most complete and accurate medication list and comparing the list to all of the medications in patient records. Using NLP to pull information from textual records and then present that view alongside other data sources, such as structured medication list in Electronic Health Records (EHR) and prescription fill data in Pharmacy Information Systems, will make these tasks more efficient. A stumbling block has always been that the information from these sources is usually coded using different medical terminologies and therefore not interoperable, making information integration a great challenge. For example, the medication list may be coded using an institutional terminology, pharmacy data may be coded by a commercial terminology, and most existing NLP systems encode clinical text using standard terminologies (e.g., the Unified Medical Language System (UMLS)). At present, the real-time translations between these diverse terminologies, especially local terminologies to standard terminologies, using automated methods (such as NLP) have not been well established. We therefore developed a general NLP system, named Medical Text Extraction, Reasoning and Mapping System (MTERMS), which extracts clinical information from clinical text and encodes the extracted information using both local and standard terminologies. It also allows mapping between terminologies when appropriate.
Background
There are many NLP tools for processing biomedical textual data. Details can be found in review articles1–3 and the following examples. Linguistic String Project in 1960s–80s was one of the first comprehensive NLP systems for general English and was adapted to medical text.4 In 1990s, MedLEE5–7 and SPRUS/SymText/MPLUS8–10 were developed to process clinical reports and MetaMap11 was designed primarily for processing biomedical scholarly articles. Open-source clinical NLP systems such as HITEx12 and cTAKES13 were reported in 2006 and 2010, respectively. A few NLP tools handle specific issues, such as NegEx/ConText14, 15 for identifying negatives and contextual information, TimeText16, 17 for dealing with temporal information, and SecTag18 for identifying section headers. There are also commercial products19 as well as NLP tools for biomedical text mining.3
Most recently, multiple research efforts focused on medication information extraction, such as MERKI,20 MedEx21 and others.19, 22–24 In 2009, the Third i2b2 Workshop on NLP Challenges for Clinical Records, also referred as the medication challenge,25 focused on the extraction of medication information from discharge summaries. The top 10 systems that participated in this challenge processed discharge summaries provided by Partners Healthcare and achieved: precision (range: 0.78–0.90), recall (range: 0.66–0.82), and F-measure (range: 0.76–0.86).25 The top 10 systems applied rule-based, supervised machine learning, or hybrid approaches, with comparable results.25 The two remaining limitations of most systems were in recognizing durations (best F-measure is 0.525) and reasons.25
Many NLP applications in biomedical informatics automatically encode clinical text to concepts within a standard terminology. UMLS is widely used since it includes various controlled vocabularies and provides mappings among them.7, 10, 11, 13 Some studies map terms in clinical text to a specific terminology, such as MeSH terms,26 SNOMED_CT,27, 28 ICD-9-CM,29 or RxNorm.21 These studies applied approaches based on string matching, statistical processing, and NLP techniques (e.g., term composition, noun phrase identification, syntactic parsing, etc). In addition, many previous NLP studies reported evaluation of their systems’ performance using inpatient reports, such as radiology reports and discharge summaries. Outpatient clinical notes often contain unique formats and characteristics.
Taboada identified three techniques for mapping terminologies: name based30, structure based31, and linguistic resource based32 techniques.33 Barrows et al mapped diagnostic terms from a legacy ambulatory care system to a separate controlled vocabulary using lexical and morphologic text matching techniques.30 Kannry et al mapped pharmacy terms between Yale’s local terminology based on AHFS and Columbia’s Medical Entities Dictionary (MED) based on UMLS, ICD-9-CM, and local terms by looking at the terms, relationships between the terms, and attributes that modify terms using lexical matching.32
Our approach for designing and developing MTERMS represents a unique contribution to the field in that MTERMS encodes the terms in clinical text using different terminologies, and at the same time establishes dynamic mappings between these terminologies when appropriate. These mappings may not have existed elsewhere before. We believe this will improve data interoperability and integration and make the NLP output available to other EHR applications. In addition, we extended and integrated the Temporal Constraint Structure (TCS) tagger,34 which is part of TimeText,16, 17 into MTERMS to capture diverse temporal information, including date/time, duration, relative time, etc. We also applied a “sandwich” parsing method that packs in a deep parser between a Pattern Recognizer and a shallow parser to improve system’s efficiency. In this paper, we introduce MTERMS system design and demonstrate the term mapping method using the medication domain as an example. We report the system’s performance on processing medication information from outpatient clinical notes (e.g., office visit notes and specialist consultation reports) from an ambulatory EHR.
Methods and System Design
System Design
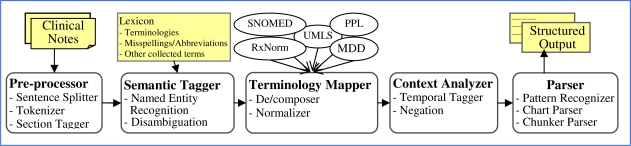
MTERMS is a modular system using a pipeline approach in which clinical free-text documents are entered into a preprocessor, to the semantic tagger, terminology mapper, context analyzer, and parser (see Figure 1). The output of MTERMS is a structured document in XML format. The preprocessor is used for cleaning, reformatting and tokenizing the text into individual sections, sentences, and word units. The semantic tagger uses lexicons to identify words or phrases to determine what categorical bucket they should be placed in (e.g. medication name and route). The terminology mapper translates concepts between different terminologies. The context analyzer looks for temporal context and other contextual information to further determine the meaning of a phrase in context with the rest of the text. The parser identifies the structure of phrases and sentences.
Figure 1.
MTERMS System Components (MDD: Partners Master Drug Dictionary; PPL: Partners Problem List)
Pre-Processor
The sentence splitter uses a set of rules mainly based on punctuations and carriage return (CR). MTERMS ignores a set of abbreviations (e.g., Dr., p.o., a.m., Jan., Mon.) as well as bullets and numbering symbols (e.g., A.) that have punctuation but don’t actually indicate the end of a sentence. The Sentence Splitter connects back if the next sentence starts with a lower case and the current sentence’s last character is not “., ?, or !”. The Section Tagger uses the Partners Notes Concept Dictionary for structured notes to extract the Section Headers of length less than 30 characters as the lexicon. The list was manually reviewed by a physician and a nurse to exclude ambiguous terms and to add additional common sense headers.
Semantic Tagger
MTERMS lexicon includes a subset of terms from standard terminologies (e.g., UMLS, RxNorm, and SNOMED CT), local terminologies (e.g. Partners Master Drug Dictionary (MDD), Partners Problem List (PPL)), HL7 value sets, regular expression rules, and manually collected terms from chart review, or literature review. The length of the string was used to sort the terminologies in decreasing order so that the most specific term available would match.
Medication Names and Drug Classes
Medication names were independently dual-coded using a local terminology source Partners Master Drug Dictionary (MDD) and a standard terminology (RxNorm).
RxNorm is created and maintained by the National Library of Medicine (NLM).35 It provides normalized names for clinical drugs, composed of ingredients, strengths and forms. These terms are then linked to many of the drug vocabularies commonly used in pharmacy management and drug interaction software, including First Databank (FDB) (NDDF), Micromedex (MMX), and Multum (MMSL). A subset of standard terminologies from the RxNorm (January 3, 2011 release) nomenclature was used to encode medication names. The following elements are used from RxNorm: drug name (STR), concept identifier (RXCUI), source terminology (SAB), concept identifier in source terminology (CODE), term type in source (TTY), and semantic type defined in UMLS Semantic Network (STY). Specifically, we use the following SAB-TTY combinations and terminology prioritization:
RXNORM: IN (ingredients), BN (brand name), PIN (precise ingredients), MIN (multiple ingredients), SCD (semantic clinical drug), SCDC (semantic clinical drug components), SCDF (semantic clinical drug form), SBD (semantic branded drug), SBDC (semantic branded drug component), SBDF (semantic branded drug form), GPCK (generic packs), BPCK (branded packs), SY (synonym)
SNOMED CT: FN (fully specified name), PT (preferred term), SY (synonym), PTGB (preferred term Great Britain), SYGB (synonym Great Britain)
MMX: CD (clinical drug);
VANDF: IN, PT, CD
NDDF: IN
MMSL: IN, BN, CD, BD (branded drug)
In order to limit our medication terminologies to medication concepts, terms with the STY’s (e.g., body part, organ, cell component, etc) were excluded from our lexicon on the advice of a pharmacist. Also, terms with an STY of food, fungus, or plant were included only if they were in the medication section of a clinical note. SNOMED CT fully specified name suffixes were extracted to further clarify matched SNOMED CT terms.
Partners Master Drug Dictionary (MDD) is used by ambulatory and inpatient EHR systems by providers at the time of ordering. MDD contains generic medication names, synonyms, and misspellings, which are used as pointers for providers but not for encoding. MDD also includes First DataBank (FDB) ingredient codes (HIC_SEQNO) and links are created to leverage FDB GCN-SEQNOs and Enhanced Therapeutic Classification (ETC) system.
While RxNorm normalized names for clinical drugs and information related to ingredients, strengths, and dose forms, MDD essentially de-normalizes names for the purpose of facilitating medication order entry. MDD names identify differences in medication concepts based on elements such as indication, specific route of administration, or protocol. To this effect, MDD serves as an interface terminology and is critical to a good interface design and Computerized Physician Order Entry (CPOE) adoption, whereas RxNorm serves as a reference terminology and is critical to data transmission and interoperability.
In addition, a “medication section only” exclusion list and total exclusion list were created by expert review to reduce ambiguity in the list and to remove concepts corresponding to commonly used English words or laboratory tests. A list of common misspellings from a drug information website36 and manual review was used to identify typing errors and were mapped by the corrected drug name to RxNorm.
Drug Signatures
A comprehensive dictionary along with regular expression rules were compiled for drug signature elements as defined in previous studies:21, 25 route, drug form, frequency, dispense amount, dose, strength, dose preparation, refill, intake time, necessity, duration, and drug status.
Route uses terms from HL7 value set of RouteOfAdministration37 and manual review. Drug form consists of terms from RxNorm, NDDF, and manual review. Frequency, dispense amount, dose, and strength use terms from manual review combined with regular expression rules that capture both a combination of numerical value and unit. Dose preparation, refill, intake time, and necessity use terms from manual review. Duration, date/time and other temporal information is captured using the TimeText system’s set of regular expression rules and lexicon.34 Drug status is captured using terms from a literature review22, 23 and manual review. Ambiguous drug signature terms (e.g., IS abbreviation for intravesicle injection) were identified and excluded by manual review.
Medical Problems and Other Medical Concepts
MTERMS also processes problems and other medical concepts (e.g. body locations). Partners local terminology for problems, called Partners Problem List (PPL), has been manually mapped to SNOMED CT and/or ICD-9-CM by the Partners Knowledge Management Team. We used this mapping as our lexicon and also extended it by including an additional subset of SNOMED CT in the UMLS (October 15, 2010 release).
Context Analyzer
The NegEx,14 TimeText,34 and ConText15 algorithms were implemented in their standard format using their lexicons to tag negated terms, temporal information, and other contextual information.
Terminology Mapper
MTERMS terminology mapper uses multiple levels of linguistic analyses and NLP techniques, including simple analysis (e.g., exact string match), morphological analysis (e.g., handling punctuation and other morphological variations), lexical analysis (e.g. handling abbreviations and acronyms), syntactic analysis (e.g. phrase segmentation and recombination), and semantic analysis (e.g. identifying meaning and assigning the terms to an appropriate semantic group). The algorithm begins by identifying elements tagged by the Semantic Tagger of the same semantic type (e.g., DrugName) that are at the same or overlapping word position in the text. Next, if the drug name is tagged by both terminologies, the algorithm compares the names to see if an exact string match exists. If not an exact string match, then additional analyses are conducted on the MDD term, and RxNorm is used as the reference to match against. Rules are applied to normalize difficult strings. If the drug name is only tagged by MDD, similar normalization rules are applied to find an appropriate match in RxNorm. The rules were developed through an iterative review process, and contain the following categories:
Morphological: Handle specific symbols. For example, MDD uses the symbol “/” or “+” to connect ingredients in a multiple ingredient product while RxNorm uses “ / ”.
Lexical: Replace abbreviations and acronyms with fully specified names. For example, APAP in MDD is converted to acetaminophen and search for an RxNorm term that matches.
Syntactic: Re-sequence term components. For example, the MDD multiple ingredients are not necessarily ordered alphabetically and therefore need to be normalized using RxNorm’s alphabetical order naming convention before mapping.
Semantic: Normalization. For example, normalize strength in MDD using RxNorm name conventions (e.g., % to mg/ml).
If no exact semantic match from above steps is found, then we attempt to find a close partial match by converting or removing drug signature elements from MDD names, for example, route information (e.g., p.o.) from MDD medication names, as RxNorm uses Dose Forms that may not always be equivalent semantically.
If nothing is found in the above steps, but the Semantic Tagger tagged an RxNorm term, then we indicate that this is an incomplete partial match.
The estimated computational complexity of the terminology mapper is O(nm), where n and m represent the number of terms in RxNorm and MDD respectively.
Parser
A parser is essentially a recognizer where a grammar verifies whether the structure of a particular sentence fits the grammar rules of the language. We adopted a semantic-based approach similar to MedLEE6, 7 and MedEx,21 with consideration of sequencing a Pattern Recognizer, a deep Chart Parser38 and a shallow Chunker in order to achieve an optimal system efficiency without affecting the semantic parsing performance, which we called a “sandwich” approach. Through manual chart review, we identified a set of common patterns (e.g., medication names and drug signatures) and put them into a pattern table. The Pattern Recognizer was implemented to determine if the sentence semantic pattern was known based on their presence in the pattern table. The deep Chart Parser was implemented using a grammar consisting of a set of syntactic and semantic rules. The Chart Parser applies a dynamic programming method and is computationally expensive due to its exponential nature. Therefore, a shallow Chunker parser was built upon regular expression rules (e.g., group drug signatures to the medication name based on distance and position in the sentence) and is used if the Chart Parser times out or fails.
Implementation
Microsoft SQL server is used as a backend for storing the Lexicon. The NLP output is in XML format. Microsoft .NET’s XML APIs are used for the XML parsing and C# language is used to build the NLP program.
Evaluation Methods
The mapping between SNOMED CT and PPL is already manually established, but the mapping between RxNorm and MDD does not currently exist, thus this paper focuses on the evaluation of the latter case only.
Corpus
This paper focuses on free-text outpatient clinical notes created mainly by patients’ primary care physicians and medical specialists, such as cardiologists. Patients with chronic diseases usually have rich medication information in their EHRs and their medication lists are challenging to maintain. Five common chronic diseases were included in this study: diabetes, hypertension, congestive heart failure, chronic obstructive pulmonary diseases, and coronary artery disease. We retrieved 2 years of data (2009 and 2010) that meet above criteria from Partners Ambulatory EHR system, called the Longitudinal Medical Records (LMR), through the Partners Research Patient Data Registry. A test set was set aside by a non-study staff member consisting of 40 randomly selected clinical notes that were stratified by cohort to ensure that all diseases were represented within the sample. Free-text notes and structured notes were evaluated separately as the structure and content are different, thus the system performance may also be different. Structured clinical notes refer to instances where the medication list was copied directly from the Structured Medication List (SML) in a machine specified format that differs from natural language. LMR allows physicians to copy medications from the structured medication list and paste them into the notes for further editing. 10 of the 40 notes contain the “copy-paste” medications and another 30 notes only contain free-text medication entries. We assessed the system’s performance on processing these two types of the notes separately. All developers were blinded to the test set and only had access to the training set.
Verification of System-extracted Medication and Temporal Information
A physician (NK) and a clinical Informatician (LZ) served as judges to manually review clinical notes and MTERMS annotations for medication names and drug signatures from MTERMS output for clinical notes in the test corpus, while a Doctor of Pharmacy candidate (DD) judged the corresponding temporal information related to medications. Raw agreement was used to compare the inter-rater reliability of reviewers for 10 randomly selected clinical notes from the test set. The two reviewers reached an agreement for the first 10 notes and then each assessed another 15 notes separately. The commonly used statistical metrics of Precision, Recall, and F-measure39 were calculated for each type of data.
Evaluation of Term Mapping
The Doctor of Pharmacy candidate (DD) manually reviewed the terminology mapping under the supervision of a pharmacist (LM). The performance of the term mapping is annotated using the following codes: Exact Match, Partial Match and Missing, as defined below, which is presented along with an example of simplified MTERMS xml output.
Exact Match: a concept in the source terminology matched to a concept in the target terminology that has exact same meaning. For example, “Botox” in MDD and “Botox” in RxNorm were an exact semantic match.
1. <DrugName sectionID="2" sentenceID="8" wordID="5” RxCUI="203279" RxNormSTR="Botox" RxTTY="BN" RxSAB="RXNORM"CODE="203279" RxSTY="Hazardous or Poisonous Substance" MDDSTR="Botox" MedType="SYNONYM" />
Partial match: a match between two terminologies is true but does not represent a full semantic match. Partial match can be further broken down into three classes; Broader, Narrower and Incomplete.
▪ Broader Partial Match: a term in the source terminology (MDD) is more specific than the best-matched term identified in the target terminology (RxNorm).
-
2. <DrugName sectionID="13" sentenceID="161" wordID="43" MDDSTR="NIZORAL CREAM" MedType="MISSPELLING" />
<DrugName sectionID="13" sentenceID="161" wordID="43" RxCUI="202692" RxNormSTR="Nizoral" RxTTY="BN"RxSAB="RXNORM" CODE="202692" RxSTY="Organic Chemical" />
▪ Narrower Partial Match: a term in the source terminology (MDD) is less specific than the best-matched term identified in the target terminology (RxNorm).
-
3. <DrugName sectionID="5" sentenceID="43" wordID="2" MDDSTR="FUROSEMIDE" MedType="GENERIC" />
<DrugName sectionID="5" sentenceID="43" wordID="2" RxCUI="315971" RxNormSTR="Furosemide 40 MG" RxTTY="SCDC" RxSAB="RXNORM" CODE="315971" RxSTY="Clinical Drug" />
▪ Incomplete Partial Match: this represents a match in which the missing piece is critical to the correct identification of the matching concept and so is not complete. For example, a multi-ingredient products in MDD mapped to a separate ingredient concepts in RxNorm.
-
4. <DrugName sectionID="25" sentenceID="468" wordID="3" MDDSTR="ROBITUSSIN WITH CODEINE" MedType="MISSPELLING" />
<DrugName sectionID="25" sentenceID="468" wordID="3" RxCUI="219702" RxNormSTR="Robitussin" RxTTY="BN" RxSAB="RXNORM" CODE="219702" RxSTY="Pharmacologic Substance" />
<DrugName sectionID="25" sentenceID="468" wordID="5" RxCUI="2670" RxNormSTR="Codeine" RxTTY="IN" RxSAB="RXNORM" CODE="2670" RxSTY="Organic Chemical" />
Missing: A term is classified as missing when there is no target identified by the tool but a match is available.
5. <DrugName sectionID="8" sentenceID="93" wordID="2" MDDSTR="MVI" MedType="SYNONYM" />
6. <DrugName sectionID="10" sentenceID="128" wordID="71" RxCUI="3567" RxNormSTR="diuretic" RxTTY="FN"RxSAB="SNOMEDCT" CODE="372695000" RxSTY="Pharmacologic Substance" FullySpecifiedNameSuffix="(substance)" />
Results
Overall, there were 1108 free-text note terms from 30 charts and 1035 structured note terms from 10 charts for a combination of findings types (medication names, drug signatures and temporal) analyzed with F-measures of 90.6 and 94.0 respectively. The raw agreement between the two evaluators was 86.3% for medication names and drug signatures on 10 charts. Table 1 further breaks down the total number of instances, precision, recall, and F-measure for the various findings types for the free-text notes and structured notes separately.
Table 1.
MTERMS System Performance on Processing Medication Related Information
| Free-Text Notes (n=30) | Structured Notes (n=10) | |||||||
|---|---|---|---|---|---|---|---|---|
| Findings Type | Total # | Precision (%) | Recall (%) | F-Measure (%) | Total # | Precision (%) | Recall (%) | F-Measure (%) |
| Drug Name | 455 | 92.5 | 91.6 | 92.1 | 271 | 93.8 | 93.5 | 93.7 |
| Dose | 177 | 92.1 | 91.6 | 91.8 | 171 | 89.3 | 93.8 | 91.5 |
| Frequency | 174 | 91.4 | 91.4 | 91.4 | 139 | 97.1 | 97.1 | 97.1 |
| Route | 50 | 88.0 | 100 | 93.6 | 141 | 97.2 | 100 | 98.6 |
| Strength | 20 | 29.4 | 55.6 | 38.5 | 116 | 82.8 | 98.0 | 89.7 |
| Necessity | 27 | 100 | 100 | 100 | 16 | 100 | 87.5 | 93.3 |
| Drug Form | 13 | 77.8 | 63.6 | 70.0 | 11 | 100 | 90.9 | 95.2 |
| Dispense Amount | 1 | 0 | - | - | 97 | 97.8 | 93.8 | 95.8 |
| Status | 79 | 70.5 | 67.2 | 68.8 | 26 | 87.5 | 91.3 | 89.4 |
| Duration | 20 | 100 | 100 | 100 | 52 | 76.9 | 100 | 87.0 |
| Date & Time | 34 | 97.1 | 100 | 98.5 | 6 | 100 | 100 | 100 |
| Relative Time | 15 | 93.3 | 100 | 96.6 | 2 | 100 | 100 | 100 |
| Temporal (Other) | 43 | 100 | 100 | 100 | 19 | 100 | 100 | 100 |
| Total | 1108 | 90.3 | 90.9 | 90.6 | 1067 | 92.4 | 95.6 | 94.0 |
Table 2 shows the coverage of RxNorm source terminologies ordered by their inclusion within MTERMS search sequence. 98.0% and 83.0% of terms in free-text notes are covered by RxNorm and MDD respectively, whereas 93.4% and 94.3% of terms in structured notes are covered by RxNorm and MDD respectively.
Table 2.
Coverage of RxNorm Source Terminologies Ordered By MTERMS Search Sequence and Coverage of local Master Drug Dictionary (MDD)
| RxNorm Source (SAB) | (%) in Free-Text Notes | (%) in Structured Notes |
|---|---|---|
| RxNorm | 88.1 | 93.1 |
| SNOMED CT | 9.4 | 5.9 |
| MMX (Micromedex) | 0 | 0 |
| VANDF (Veterans Administration) | 1.1 | 0 |
| NDDF (First DataBank) | 0.2 | 0 |
| MMSL (Multum) | 1.1 | 0.9 |
| Misspelling (Drugs.com) | 0 | 0 |
| Overall RxNorm Coverage | 98.0 | 93.4 |
| Overall MDD Coverage | 83.0 | 94.3 |
Table 3 shows the frequency of RxNorm type (TTY), indicating that most terms are ingredients (IN), or brand names (BN). RxNorm Types (TTY) that were not detected were not included in the table. Table 3 also shows the frequency of medication name types (Generic, Synonym, Misspelling) within Partners Master Drug Dictionary (MDD), indicating that generic terms are the most prevalent within both free-text notes and structured notes, which is closely followed by synonyms.
Table 3.
System Detected RxNorm by Type (TTY) and local Master Drug Dictionary (MDD) by Type
| (%) in Free-Text Notes | (%) in Structured Notes | |
|---|---|---|
| RxNorm Type (TTY) | ||
| IN (ingredients) | 43.6 | 43.4 |
| BN (brand name) | 34.8 | 26.8 |
| SCDC (semantic clinical drug component) | 11.1 | 18.5 |
| FN (fully specified name) | 6.4 | 1.9 |
| SY (synonym) | 2.9 | 1.7 |
| PIN (precise ingredient) | 0.8 | 7.8 |
| PT (preferred term) | 0.4 | 0 |
| MDD Type | ||
| Generic | 59.3 | 57.5 |
| Synonym | 31.7 | 35.2 |
| Misspelling | 8.9 | 7.3 |
Table 4 presents an analysis of the terminology mapping between MDD and RxNorm, which shows that 63.0% and 58.7% of mappings in free-text notes and structured notes respectively are exact matches and 14.7% and 27.9% are partial matches respectively. Overall, 61.6% of mappings are exact matches.
Table 4.
Mapping Medication Terms between local Master Drug Dictionary (MDD) and RxNorm
| Type | (%) in Free-Text Notes | (%) in Structured Notes |
|---|---|---|
| Exact | 63.0 | 58.7 |
| Broader (Partial)* | 2.3 | 6.3 |
| Narrower (Partial)^ | 12.1 | 21.2 |
| Incomplete (Partial) | 0.2 | 0.5 |
| Extraneous (Incorrect Match) | 0.2 | 0 |
| Missing | 22.1 | 13.5 |
resulting MDD terms have more specific meaning than RxNorm terms.
resulting MDD terms have a less specific meaning than RxNorm terms
Discussion
In this paper, we present a NLP system, called MTERMS, which conducts different levels of linguistic analysis on clinical notes, and can be used to create structured clinical documents and to map terminologies. MTERMS sequences shallow parsers and a deep parser to achieve optimal system efficiency, which is critical for the future potential integration with real-time EHR applications.
MTERMS achieved 90.3% precision and 90.9% recall for free-text clinical notes, and 92.4% precision and 95.6% recall for structured clinical notes on processing medication names, drug signatures, and temporal information. These results seem consistent with previous studies25 but may not be directly comparable as this study focused on outpatient clinical notes, whereas previous studies focused on discharge summaries or applied more stringent annotation guidelines.25 Medication information in structured notes is formatted automatically by the EHR system, for example, “Lipitor (ATORVASTATIN) 20 MG (20MG TABLET Take 1) PO QD x 60 days #60 Tablet(s).” NLP tools can be trained to capture these specific structures. However, the format may vary in different systems or different versions of a system and can be modified by physicians when copied to a free-text field. Although structured notes, on average, contained more medications than free-text notes, one would argue that this information might not be that useful due to readability issues, and redundancy with the structured medication list.
MTERMS uses a selective lexicon based on expert review of clinical charts and terminologies. MTERMS missed some medication abbreviations (e.g., INH) and vitamins (e.g., B12 which is Vitamin B12), which may have been on the exclusion list due to ambiguity concerns. MTERMS also incorrectly captured some non-drug terms such as “gel” which is from FDB NDDF as an ingredient and also misinterpreted “thymus” in “the patient has an enlarged thymus as “Thymus Extracts” which is from VANDF. MTERMS is integrated with the Temporal Constraint Structure (TCS) tagger and achieved high precision and recall on capturing temporal information. The TCS structure allows MTERMS to conduct temporal reasoning of clinical events. It is more difficult to capture strength and drug form information in free-text notes than structured notes as these terms in free-texts have more variations. One challenging areas for strength is the presence of ambiguous fractions with missing units. For example, percocet 2.5mg/325mg may be represented as Percocet 2.5/325 in the free-text notes. Another example is that it is unclear if vancomycin 3/10 is a valid strength or a date. The challenges with drug form are in disambiguating equipment (e.g., pill organizer), brand name of drugs (e.g., Timolol eye drops), and where it represents a class of products instead of a form in context of the sentence (e.g., eye drops, nasal spray). Status is still a challenging area and needs an expanded lexicon and further investigation. We adopted HL7’s value set to standardize the Route information. Standard structured output for frequency also needs further investigation.
MTERMS also encodes problems using Partners local problem terminology, ICD-9, and SNOMED. However, this feature needs further lexical refinements. MTERMS applies extensible general methods and a modular infrastructure, so it is extensible to process other types of clinical information such as procedures and laboratory results.
Approximately 60% of mappings between MDD and RxNorm were exact match for both types of notes, however 22.1% and 13.5% were missing for free-text notes and structured notes respectively (Table 4). A common reason for missing is abbreviations (e.g., MVI for multivitamin). Another reason for a missing match might be an obsolete name that is not maintained in RxNorm but is still in MDD (such as “pancreatic enzymes”). It is not surprising that MDD covered a greater percentage of terms in the structured notes than the free-text notes (Table 2) because MDD is used within the Computerized Physician Order Entry (CPOE) system.
Mapping medication terms from a local terminology source to a normalized standard requires a variety of simultaneously applied strategies in order to capture terms specified at various levels of complexity. The simplest and most effective strategy was a basic string match with no manipulation, indicating that terms were identical. In normalization, simpler sequenced strategies often yielded a greater volume of additional matched terms than more complex and targeted strategies. There is a difference in granularity across terminologies in terms of additional drug signature elements included within a drug name, which makes mapping of these terms difficult. Previous studies in mapping medical terminologies made the assumption that “the terminologies to be mapped are correctly designed, so the problems to map them come from the different design decision making in both terminologies.”33 This is not completely true for our homegrown evolved MDD. A potential extension of MTERMS terminology mapper would be to apply N-grams or statistical or machine learning methods. In addition, dynamic, real-time mappings provide up-to-date maps between continually changing terminologies. However, an alternative to the dynamic mapping would be to use a static knowledge base approach updated on a regular basis to keep the mapping up-to-date. The tradeoff between dynamic and static mapping is the optimization of speed, storage, and maintenance.
The feasibility for real-time clinical use of NLP in assembling the medication reconciliation list is strong. However, a real-life application will require change management. For example, a terminology management process is needed to review how updates to terminologies will affect the mappings and to track retired concepts. A common occurrence in electronic order entry systems is free text medication entries. These represent something of a black box to the systems that process them. NLP could be used to extract coded medications from these entries and allow duplication alerts or drug interaction system to catch potential medication errors.
Our study has several limitations. A limiting factor to external validation of this research is that the local Medication Drug Dictionary (MDD), the manually reviewed lexicon, and Partners’ notes corpus used for testing are not publicly accessible thus researchers outside of the organization cannot verify our results. The scope of our evaluation is limited to patients with chronic diseases, as their medication use may be different from patients in the general population. Another limitation to the evaluation is that one of the evaluators (LZ) was involved with the design and development of the system.
Conclusion
We present the Medical Text Extraction, Reasoning and Mapping System (MTERMS), and evaluated its performance using free-text clinical notes and structured clinical notes. Our main finding was that the combination of automated NLP methodologies for processing clinical notes with a terminology mapper and a temporal reasoning system can be used to extract, encode and reason about clinically relevant information. The gap in knowledge addressed is an automated approach to mapping terminologies that can be used to extract clinical concepts from notes to increase the interoperability and utility of clinical information.
Acknowledgments
This project was supported by grant number 1R03HS018288-01 from the Agency for Healthcare Research and Quality (AHRQ), U.S. Department of Health and Human Services, and Partners-Siemens Research Council.
References
- [1].Friedman C, Johnson SB. Natural language and text processing in biomedicine. In: Shortliffe EH, Cimino JJ, editors. Biomedical Informatics: Computer Applications in Health Care and Biomedicine. Springer; NY: 2006. [Google Scholar]
- [2].Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med. 1999 Aug;74(8):890–5. doi: 10.1097/00001888-199908000-00012. [DOI] [PubMed] [Google Scholar]
- [3].Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Yearbook of medical informatics. 2008. Extracting information from textual documents in the electronic health record: a review of recent research; pp. 128–44. [PubMed] [Google Scholar]
- [4].Sager, et al. Addison-Wesley; 1987. Medical language processing: Computer Management of Narrative Data. [Google Scholar]
- [5].Friedman C. Towards a comprehensive medical language processing system: methods and issues. Proc AMIA Annu Fall Symp; 1997. pp. 595–9. [PMC free article] [PubMed] [Google Scholar]
- [6].Friedman C. A broad-coverage natural language processing system. Proceedings / AMIA Annual Symposium; 2000. pp. 270–4. [PMC free article] [PubMed] [Google Scholar]
- [7].Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004 Sep-Oct;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000 Nov-Dec;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haug P, Koehler S, Lau LM, Wang P, Rocha R, Huff S. A natural language understanding system combining syntactic and semantic techniques. Proceedings / the Annual Symposium on Computer Application [sic] in Medical Care; 1994. pp. 247–51. [PMC free article] [PubMed] [Google Scholar]
- [10].Christensen LM, Haug PJ, Fiszman M. MPLUS: A Probabilistic Medical Language Understanding System. Proceedings of the Workshop on Natural Language Processing in the Biomedical Domain; Philadelphia. July 2002; pp. 29–36. [Google Scholar]
- [11].Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings / AMIA Annual Symposium; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- [12].Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, comorbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC medical informatics and decision making. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001 Oct;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- [15].Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of biomedical informatics. 2009 Oct;42(5):839–51. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhou L, Friedman C, Parsons S, Hripcsak G. System architecture for temporal information extraction, representation and reasoning in clinical narrative reports. AMIA Annual Symposium proceedings / AMIA Symposium; 2005. pp. 869–73. [PMC free article] [PubMed] [Google Scholar]
- [17].Zhou L, Parsons S, Hripcsak G. The evaluation of a temporal reasoning system in processing clinical discharge summaries. J Am Med Inform Assoc. 2008 Jan-Feb;15(1):99–106. doi: 10.1197/jamia.M2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Denny JC, Spickard A, 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. 2009 Nov-Dec;16(6):806–15. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jagannathan V, Mullett CJ, Arbogast JG, Halbritter KA, Yellapragada D, Regulapati S, Bandaru P. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. International journal of medical informatics. 2009 Apr;78(4):284–91. doi: 10.1016/j.ijmedinf.2008.08.006. [DOI] [PubMed] [Google Scholar]
- [20].Gold S, Elhadad N, Zhu X, Cimino J, Hripcsak G. Extracting structured medication event information from discharge summaries. AMIA Annual Symposium proceedings / AMIA Symposium; 2008. pp. 237–41. [PMC free article] [PubMed] [Google Scholar]
- [21].Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Jan-Feb;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Breydo E, Chu JT, Turchin A. Identification of inactive medications in narrative medical text. AMIA Annual Symposium proceedings / AMIA Symposium; 2008. pp. 66–70. [PMC free article] [PubMed] [Google Scholar]
- [23].Turchin A, Wheeler HI, Labreche M, Chu JT, Pendergrass ML, Einbinder JS. Identification of documented medication non-adherence in physician notes. AMIA Annual Symposium proceedings / AMIA Symposium; 2008. pp. 732–6. [PMC free article] [PubMed] [Google Scholar]
- [24].Levin MA, Krol M, Doshi AM, Reich DL. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annual Symposium proceedings / AMIA Symposium; 2007. pp. 438–42. [PMC free article] [PubMed] [Google Scholar]
- [25].Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cooper GF, Miller RA. An experiment comparing lexical and statistical methods for extracting MeSH terms from clinical free text. J Am Med Inform Assoc. 1998 Jan-Feb;5(1):62–75. doi: 10.1136/jamia.1998.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lussier YA, Shagina L, Friedman C. Automating SNOMED coding using medical language understanding: a feasibility study. Proceedings / AMIA Annual Symposium; 2001. pp. 418–22. [PMC free article] [PubMed] [Google Scholar]
- [28].Elkin PL, Tuttle M, Keck K, Campbell K, Atkin G, Chute CG. The role of compositionality in standardized problem list generation. Studies in health technology and informatics. 1998;52(Pt 1):660–4. [PubMed] [Google Scholar]
- [29].Pestian JP, Brew C, Matykiewicz P, Hovermale D, Johnson N, Cohen KB, Duch W. A shared task involving multi-label classification of clinical free text. Proceedings of ACL BioNLP; 2007. [Google Scholar]
- [30].Barrows RC, Jr, Cimino JJ, Clayton PD. Mapping clinically useful terminology to a controlled medical vocabulary. Proceedings / the Annual Symposium on Computer Application [sic] in Medical Care; 1994. pp. 211–5. [PMC free article] [PubMed] [Google Scholar]
- [31].Rocha RA, Rocha BH, Huff SM. Automated translation between medical vocabularies using a frame-based interlingua. Proceedings / the Annual Symposium on Computer Application [sic] in Medical Care; 1993. pp. 690–4. [PMC free article] [PubMed] [Google Scholar]
- [32].Kannry JL, Wright L, Shifman M, Silverstein S, Miller PL. Portability issues for a structured clinical vocabulary: mapping from Yale to the Columbia medical entities dictionary. J Am Med Inform Assoc. 1996 Jan-Feb;3(1):66–78. doi: 10.1136/jamia.1996.96342650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Taboada M, Lalin R, Martinez D. An automated approach to mapping external terminologies to the UMLS. IEEE transactions on bio-medical engineering. 2009 Jun;56(6):1598–605. doi: 10.1109/TBME.2009.2015651. [DOI] [PubMed] [Google Scholar]
- [34].Zhou L, Melton GB, Parsons S, Hripcsak G. A temporal constraint structure for extracting temporal information from clinical narrative. Journal of biomedical informatics. 2006 Aug;39(4):424–39. doi: 10.1016/j.jbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- [35].RxNorm: http://www.nlm.nih.gov/research/umls/rxnorm/.
- [36].www.drugs.com (accessed on 03/15/2011).
- [37].HL7 v3 Value Set - RouteOfAdministration. http://wwwhl7org/v3ballot/html/welcome/environment/indexhtml.
- [38].Russell S, Norvig P. Artificial Intelligence - A Modern Approach. Second Edition ed. 2003. [Google Scholar]
- [39].Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. New York: ACM Press, Addison-Wesley; 1999. [Google Scholar]