Abstract
Longitudinal treatment histories may offer valuable information about clinical practice patterns to the clinical researcher as part of data exploration, cohort identification, or discovery of potentially beneficial or harmful practices in the health care community. We present a novel approach to temporal clustering of patient treatment information based on the semantic similarity of longitudinal histories. Using combined breast cancer registry data from two neighboring health care institutions, we constructed a database of longitudinal treatment histories that included surgical procedures, radiation therapy, chemotherapy, and hormone replacement therapy. We then did pair-wise similarity comparisons of treatment histories, and used the similarity measures to cluster patients with machine learning methods. An evaluation of our results found that patients clustered on stage of breast cancer and type of treatment provided. We propose that this approach can be applied towards identification of similar cohorts, and for discovery of novel or anomalous clinical practice patterns.
Introduction
Background
A common use case among clinical and health services researchers is exploration of clinical data for temporal patterns of care. This kind of data mining allows for hypothesis generation, and enables the analysis of clinical practice patterns as well as comparative outcomes research. Recently, we presented LATCH, a computational algorithm that finds patients from clinical databases with similar drug treatment histories1. LATCH uses a hierarchy based similarity measure for individual drugs in a treatment regimen, which is then extended to a similarity measure between a treatment history and a query pattern. In this paper, we move beyond the query and search function of LATCH, and explore its application to cohort identification and discovery of clinically relevant practice patterns.
Cohort identification from electronic medical record (EMR) data and clinical databases is a common task of researchers. A standard approach is for the researcher to find patients based on a set of demographic or clinical features (such as age, gender, or co-morbidities) and then further constrain the dataset based on similar treatment histories. For example, in the breast cancer domain, a researcher may be interested in patients who received adjuvant chemotherapy after surgical removal of the tumor. The high level description of a surgical procedure followed by chemotherapy can be enumerated in multiple ways, given the availability of a number of surgical, as well as chemotherapeutic approaches. The different possible regimens would be the basis for a comparative outcomes analysis. In this scenario, all the patients in the cohort have a similar treatment history, differing primarily by the alternate regimen and measured outcome. As opposed to this standard method of cohort identification, the researcher could use computational clustering methods to quickly identify patients with similar longitudinal treatment histories from large databases.
In terms of discovery, in many clinical studies the investigator has an existing expectation of the treatment histories in the study cohort. Indeed, it is because of the expectation of similar histories that the researcher is able to make causal inferences between drug exposures and measured clinical outcomes2. Clustering of patients by treatment histories, however, would incorporate all of the treatment histories in the cohort. This could result in the discovery of previously unrecognized practice patterns that may be associated with relevant outcomes, such as adverse drug events, cost-effective use of resources, biomarkers, or improved mortality.
Related Work
Related work is in the areas of patient similarity, sequence analysis, and anomaly detection. Clustering methods based on a set of defined clinical features have been used to identify similar patients in the critical care setting, as well as for diabetes and schizophrenia3,4. A more robust approach uses the SNOMED-CT ontology to measure inter-patient semantic distances, allowing the researcher to find clusters of patients who are within a threshold distance5. These approaches, however, are agnostic to the temporal ordering of the clinical events, and may not cluster patients with similar treatment histories.
Sequence analysis includes quantitative and qualitative methods. Quantitative methods, such as with time series, include fast search techniques for similar temporal patterns and clustering of time series abstractions6,7,8. These methods are not directly applicable to medical treatment histories, which are often represented qualitatively (i.e. a patient had a modified radical mastectomy, which is a surgical procedure for breast cancer). Qualitative sequence analysis appears in the biological domain as sequence alignment algorithms for genetic and protein code9,10,11. Sequence alignment in biology, however, uses a scoring table like PAM or BLOSUM12. With clinical treatment histories, generating a similar scoring table would be much more difficult as there are a multitude of possible treatment combinations. Recently, a hybrid quantitative/qualitative approach uses temporal abstraction to dynamically determine short temporal patterns, and then clusters gene expression profiles according to the overall temporal pattern13. Gene-gene similarity, however, is not considered in the clusters.
Anomaly detection is relevant to a number of domains, including intrusion of computer systems, fraud detection, and the identification of novel patterns in industry and health. Detection methods using symbolic representations include the use of semantic technologies and rules based methods14. Limitations to these methods are the need for a reference model so that anomalies can be identified. Statistical outlier methods rely on a distributional analysis of quantitative data, and do not require a reference model for comparison. However, these methods have limited applicability to medical treatments, which cannot be measured quantitatively at an individual level.
Research Goals and Hypothesis
The goal of this work is to develop a method to analyze temporal patterns of care that 1) uses knowledge of hierarchical relationships among medical treatment events, 2) allows a quantitative semantic similarity measurement of treatment histories, and 3) conducts clustering of treatment histories agnostic to expected patterns of care. As a quantitative method of sequence analysis, LATCH allows a semantic comparison of longitudinal treatment history, and may have benefit when attempting to cluster patients based on clinical practice patterns. In this paper, we use the LATCH algorithm to generate a patient-to-patient similarity matrix, and then cluster the patients according to the treatment history similarity. We hypothesize that LATCH can be used to achieve meaningful clusters of patients based on their longitudinal treatment history, and that relevant clinical practice patterns can be identified from these clusters.
Methods
The Stanford-PAMF OncoShare Database
Started in 2009, the OncoShare database is a collaborative project between the Stanford School of Medicine (SOM) and the Palo Alto Medical Foundation (PAMF). The goals of OncoShare are to enable joint analyses of the breast cancer care provided in academic and community health systems. As a national benchmark for breast cancer care, OncoShare collects all available electronic data that is related to evidence-based practice guidelines. To support this effort, OncoShare also collaborates with investigators from the Cancer Prevention Institute of California (CPIC). CPIC operates the greater Bay Area Cancer Registry with the National Cancer Institute SEER program and the California Cancer Registry. For this paper, we used de-identified cancer registry data collected by CPIC from both SOM and PAMF, and shared with the OncoShare database. The cancer registry contains structured information on the tumor, tumor stage, pathology, patient age, procedures performed, as well as the date of the procedure. Table 1 shows a sample of the descriptive procedure terms that we used to construct longitudinal treatment histories for breast cancer. These terms, along with basic demographic and disease specific information are uniquely linked with a patient and a tumor identification number.
Table 1.
Sample of the descriptive procedure terms used in the OncoShare database
| Surgery | Chemotherapy | Radiation Therapy | Hormone Therapy | |
|---|---|---|---|---|
| Local tumor destruction | Lumpectomy | Not administered due to death | Not Administered | Not administered due to death |
| Partial mastectomy | Segmental mastectomy | Not administered due to refusal | Beam Radiation | Not administered due to refusal |
| Partial mastectomy with nipple resection | Total mastectomy | Single Agent Chemo | Radioactive Implants | Not recommended or administered |
| Multi-Agent Chemo | Strontium-89 | Hormone therapy provided | ||
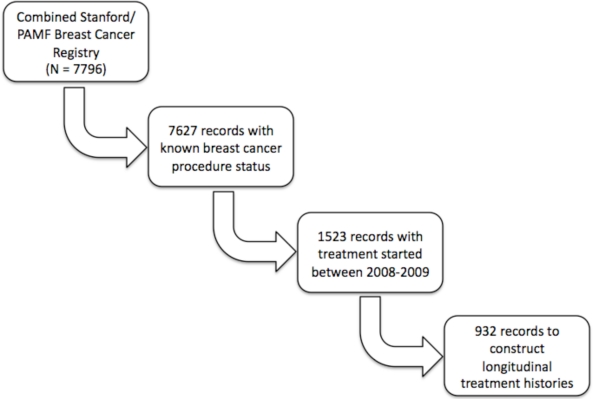
Figure 1 shows a schematic of how patients were selected for our study. We excluded from our analysis those patients for whom the status of any procedure was unknown. Although we have treatment information in the OncoShare database from 2001 – 2010, we specifically focused on patients with early stage breast cancer (stage I and II) for whom registry data was available in 2008 and 2009. We chose this time period because we intend to evaluate our methodology against the 2008 published guidelines for breast cancer care from the National Comprehensive Cancer Network (NCCN)15. Also, we focused on early stage breast cancer patients assuming that these patients would have the most variability in clinical practice patterns. With our defined cohort, we constructed a longitudinal treatment history for every unique patient, tumor-id pair. We did this because in the database some of the patients had more than one breast tumor, and we wanted to evaluate the treatment histories specific to each one.
Figure 1.
Selection of patient-tumor records from the OncoShare database
Thus, each unique patient, tumor-id pair received a code for a breast surgery, chemotherapy, radiation therapy, and hormone therapy procedure. Start dates for each procedure was recorded, and used to temporally sort each procedure history. When a start date for procedures was not available, we assumed that the treatment followed a standard pattern of surgery-radiation therapy-chemotherapy-hormone therapy. With this assumption, we sorted any procedure dates that we could, but otherwise kept the ordering of the other procedures per this template.
The Local Alignment Tool for Clinical Histories (LATCH)
The LATCH algorithm has been described previously. We provide in this paper a brief summary description. The parameters of LATCH are 1) the query, which consists of a temporally ordered sequence of treatment procedures, 2) a database of longitudinal treatment histories, and 3) a user-determined threshold score, S, such that 0 ≤ S ≤ 1, where a score of 1 signifies that the user wants a perfect match between the query regimens and the database sequences.
For each procedure node in the query, LATCH will determine a minimum procedure score (MPS) such that the overall average score for the treatment history meets the threshold score S. It then steps through every treatment regimen in every treatment history in the database. Any treatment procedure that satisfies the MPS is added to a set, along with the corresponding patient ID and the index position of that procedure.
The algorithm then uses the set of regimen nodes that has been determined on the first pass, and for each regimen in that set, does a pair-wise comparison of the subsequent regimen to the second regimen of the query. As before, the MPS is determined for each subsequent regimen node, and only those nodes that meet or exceed it are kept in the set. This continues sequentially until the end of the query is reached. The regimens left in the set will have an average score A such that A ≥ S. The pseudo-code for LATCH is as follows:
LATCH (query, database, threshold score)
For first node in query
Determine the MPS
For each patient in database
Find all matches that meet MPS
Add matches with start index to a Set
For each subsequent node in query
For each match in the Set
Determine the MPS
If the node at start index+1 ≥ MPS
Keep node in Set
Else discard the node
Return all remaining nodes and start indexes in Set
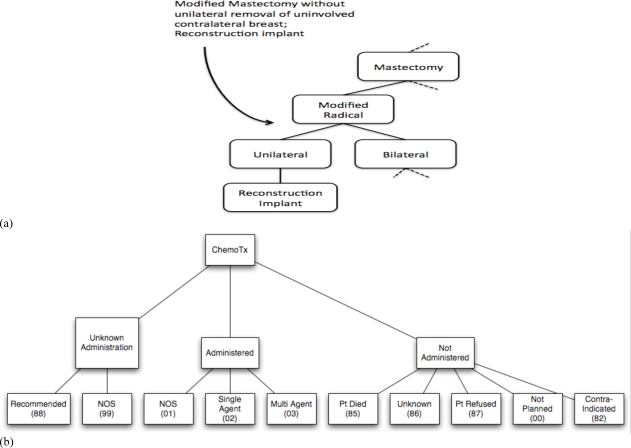
Scoring of pair-wise comparisons of procedure nodes used an ontology based semantic similarity measure to generate a similarity matrix that could be used as a look up table. The procedures consist of a defined set of surgical, radiation, chemo and hormone therapeutic options. The codes contain an implicit hierarchy, and we took used this to construct an ontology hierarchy for each of the procedure classes. For example, the surgical procedure code Modified Mastectomy without Removal of Contralateral Breast implies that Modified Mastectomy is a subclass under the Mastectomy. Figure 2 shows the ontology hierarchy developed for surgical procedures (2a), and chemotherapy procedures (2b). For each procedure class, we used the ontology based semantic similarity measure by Al-Mubaid16 to construct a similarity matrix, which we then used as a look up score table in the LATCH algorithm. A recent study had suggested good performance of Al-Mubaid’s cluster based similarity measure with human domain experts in the medical field17.
Figure 2.
Graphical representations of ontology hierarchies generated by the author WL from the cancer registry coded terms. In (a) a single term for a surgical mastectomy is interpreted by WL into a part of the ontology hierarchy. (b) shows the complete ontology hierarchy for chemotherapy codes that was used in the study.
Extending LATCH to generate an inter-patient similarity matrix
Because of the possibility of multiple matches in a single treatment history, we need to extend LATCH so that a single similarity score could be determined for each pair-wise comparison of patients. Our approach is to first look at pair-wise comparisons of patient treatment histories in which the index history, h, has fewer regimens than the comparison history, C. In this case, we set the LATCH threshold score to 0.0, ensuring that all possible LATCH scores are generated between h and C. We then determine the inter-patient LATCH score, PL such that
The weighting factor, ω, is the ratio of the number of procedure codes between the two histories, such that ω ≤ 1. In this study, every procedure modality had a code, so practically, ω = 1. We make the assumption that the similarity score PL is symmetric, and therefore we would arrive at the same score when comparing C as the index history and h as the comparison history. With the revised LATCH score, we then took the patients in our cohort, and generated a square similarity matrix in which every patient’s treatment history was compared to every other patient’s treatment history.
Clustering and analysis of the cluster assignments
We applied a hierarchical clustering method to cluster the patients based on the inter-patient similarity matrix. Hierarchical clustering provides a simple and intuitive way to group the patients based on their pair wise similarity and build a tree to represent the hierarchy of patient clusters in a bottom-up approach. In this work, we apply the average-linkage hierarchical clustering algorithm to cluster the patients [16]. The average-linkage algorithm starts with each patient in an individual cluster as the leaves of the hierarchal tree. It then iteratively merges the two closest clusters together till it ends with one cluster containing all the patients as the root of the hierarchical tree. In this method, the similarity between two clusters is considered as the average similarity between the two clusters’ elements. To estimate a reasonable number of clusters, we utilized the gap statistic for a range of clusters and then plotted of the gap score18. The gap statistic is determined by comparing the expected value of the within cluster sums of squares to a bootstrap generated reference curve. The difference between the log of these values is the gap score, which can then be plotted against a range of cluster numbers. The cluster number that results in the highest gap score is the estimated optimal number.
Results
Of 1523 patient records that were initially treated between 2008 and 2009 we analyzed the longitudinal treatment histories of 932 with early stage (stage I or II) breast cancer. Table 2 shows the baseline characteristics of the records from the initial cohort. The patients in this study were relatively young at the time of diagnosis, which likely reflects the fact that SOM is a referral hospital for the greater San Francisco bay area, and as a result will tend to have younger patients at the time of tumor diagnosis. Most of the patients (82%) had curable or early stage disease, between Stage 0 and Stage IIB. Most of the patients in our cohort were alive at the time the cancer registry data was updated in 2009. The cohort with early stage breast cancer is younger than those with late stage (stage III and IV) breast cancer (57 to 54.8 years, p-value = 0.046), and also had a substantially better survival rate by the end of the data collection period in 2010 (97.8% to 87.6%, p-value < 0.001).
Table 2.
Demographic features of the cohort
| Mean Age at time of tumor diagnosis (s.d.) | 55 years (13.2) |
| Vital Status (% Alive) | 1468 (96.4%) |
| Stage at Diagnosis | |
| Stage 0 | 317 (20.8%) |
| Stage I, IIA, IIB | 932 (61.2%) |
| Stage IIIA, IIIB, IIIC | 145 (9.5%) |
| Stage IV | 57 (3.7%) |
| Unknown Stage | 72 (4.7%) |
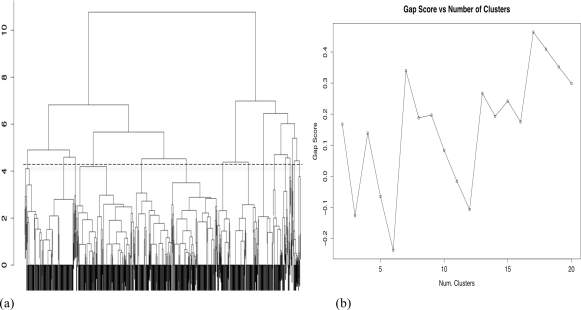
Figure 3a shows the results of the hierarchical clustering of the patient records based on the inter-patient LATCH score. The plot of the gap statistic (Figure 3b) suggests the optimal number of clusters to be seventeen, and the cutoff level in the dendrogram is shown with the dotted line. Table 3 shows the treatment practice patterns that were uncovered by clustering and the frequency of the most common guideline based pattern found in each cluster. For summarization purposes, we only show the 11 clusters (labeled A through K) that contained 20 or more records. Of the 932 patient records, 854 (91.6%) fell into a cluster dominated (prevalence rate between 88.7% – 100%) by a NCCN guideline based practice pattern for early stage breast cancer. Only the last cluster, K, had a practice pattern that could not be explained by reference to the NCCN guidelines. For these records (N = 51), we conducted a chart review with a breast cancer specialist, and discovered that all of these patients were seen at the Stanford SOM as geographically distant referrals. The review found that the patients were given hormone therapy as an outpatient prescription with the intention that it would be taken after radiation therapy was provided in the patient’s home location. The breast cancer specialist confirmed that this was acceptable practice for consultative care of patients from distant regions.
Figure 3.
(a) The dendrogram and (b) gap statistic from the hierarchical clustering of the patient-tumor records. The peak gap statistic was at k=17, and the dotted line in the dendrogram shows the level of the cut.
Table 3.
Descriptive pattern analysis of clusters.
| Cluster (Number) | NCCN Breast Cancer Guideline Pattern | Pct (%) |
|---|---|---|
| A (155) | Surgery Only | 100 |
| B (62) | Surgery → Radiation | 88.7 |
| C (124) | Surgery → Chemotherapy | 91.1 |
| D (128) | Surgery → Hormone Therapy | 91.4 |
| E (106) | Surgery → Radiation → Hormone Therapy | 99.1 |
| F (60) | Surgery → Chemotherapy → Radiation | 100 |
| G (70) | Surgery → Chemotherapy → Hormone Therapy | 100 |
| H (77) | Surgery → Chemotherapy → Radiation → Hormone Therapy | 98.7 |
| I (31) | Surgery → Chemotherapy → Hormone Therapy → Radiation | 96.8 |
| J (22) | Neo-Adjuvant Therapy | 100 |
| K (51) | Surgery → Hormone Therapy → Radiation | 100 |
Conclusion
In this study, we took the LATCH algorithm and extended it so that an inter-patient similarity score could be measured. LATCH was originally intended to search clinical databases for patients who are semantically similar to a query pattern, such as one from a clinical practice guideline. We hypothesized that the similarity matrix we created within a patient cohort would result in meaningful clusters being generated, and that interesting practice patterns could be discovered. Our results found that the patients separated based on the provision of specific patterns of surgery, radiation, chemo and hormone therapy. Nearly all of these patterns matched treatment recommendations found in the NCCN clinical practice guidelines for breast cancer care, and they suggest that both SOM and PAMF follow evidence-based practices in the care of early stage breast cancer. The finding of the anomalous practice pattern (K) supports our goal of discovering patterns of care that are agnostic to a particular clinical practice guideline.
Limitations to our study are that the LATCH algorithm is not yet equipped to deal with gaps or reversals in the temporal ordering of events. In addition, we did not incorporate a domain specific clinical outcome in our analysis, such as mortality or breast cancer remission rates. We are currently working on extensions to LATCH that will permit gaps and reversals, and we intend to apply our methods to a much larger group of breast cancer patients over a longer cohort period. Future work also includes the development of methods to explore temporal patterns of care within specific clusters as part of anomaly detection. The original implementation of LATCH was designed as a query tool over clinical databases – the LATCH score may be useful in conducting a distributional analysis of temporal patterns within a cluster.
In conclusion, we have developed a novel approach to temporal clustering that incorporates the semantic similarity of longitudinal treatment histories, and we have established its value for pattern discovery in the breast cancer domain. Using combined data from the breast cancer registries of two institutions, we have shown that the majority of clusters we discovered are meaningful when compared to recommended care in clinical practice guidelines. Finally, we have identified an anomalous practice pattern that supports our hypothesis that our method is agnostic to reference models of care, such as in clinical practice guidelines. We believe that our methods apply to other clinical domains, and are pursuing further efforts in that direction.
Acknowledgments
WL is supported as a medical informatics fellow in the Veterans Administration Health Care System of Palo Alto, Palo Alto, California. Work by the authors WB and AD is funded by grant R01LM09607-01. The OncoShare Database is supported by a generous gift from the Richard and Susan Levy Gift Fund. Views expressed are those of the authors and not necessarily those of the Department of Veterans Affairs.
References
- 1.Lee WN, Das AK. Local alignment tool for clinical history: temporal semantic search of clinical databases. AMIA Annu Symp Proc; 2010. pp. 437–441. [PMC free article] [PubMed] [Google Scholar]
- 2.Rothman K, Greenland S. Modern Epidemiology. Philadelphia, PA: Lippincott-Raven Publishers; 1998. [Google Scholar]
- 3.Cohen MJ, Grossman AD, Morabito D, et al. Identification of complex metabolic states in critically injured patients using bioinformatic cluster analysis. Crit Care. 2010;14:R10. doi: 10.1186/cc8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Copeland LA, Zeber JE, Wang CP, et al. Patterns of primary care and mortality among patients with schizophrenia or diabetes: a cluster analysis approach to the retrospective study of healthcare utilization. BMC Health Serv Res. 2009;9:127. doi: 10.1186/1472-6963-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melton GB, Parsons S, Morrison FP, et al. Inter-patient distance metrics using SNOMED CT defining relationships. J Biomed Inform. 2006;39:697–705. doi: 10.1016/j.jbi.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal R, Faloutsos C, Swami A. Efficient similarity search in sequence databases. Proc. of the 4th Conference on Foundations of Data Organization and Algorithms; 1993. pp. 69–84. [Google Scholar]
- 7.Keogh E, Pazzani M. An enhanced representation of time series which allows fast and accurate classification, clustering and relevance feedback. Proceedings of the 4th International Conference of Knowledge Discovery and Data Mining; 1998. pp. 239–241. [Google Scholar]
- 8.Keogh E, Smyth P. A probabilistic approach to fast pattern matching in time series databases. Proc. of the 3rd International Conference of Knowledge Discovery and Data Mining; 1997. pp. 24–20. [Google Scholar]
- 9.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 10.Smith TF, Waterman MS. Identification of Common Molecular Subsequences. J of Mol Biol. 1991;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 11.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Ewens WJ, Grant GR. Statistical methods in bioinformatics: An introduction. New York: Springer-Verlag; 2001. [Google Scholar]
- 13.Sacchi L, Bellazzi R, Larizza C, et al. TA-clustering: cluster analysis of gene expression profiles through temporal abstractions. Int J Med Inform. 2005 Aug;74:505–17. doi: 10.1016/j.ijmedinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Agyemang M, Barker K, Alhajj R. A comprehensive survey of numeric and symbolic outlier mining techniques. Intelligent Data Analysis. 2006;10:521–538. [Google Scholar]
- 15.http://www.nccn.org/professionals/physician_gls/f_guidelines.asp
- 16.Al-Mubaid H, Nguyen HA. A cluster-based approach for semantic similarity in the biomedical domain. IEEE Int Conf GR Proc; 2006. pp. 2713–2717. [DOI] [PubMed] [Google Scholar]
- 17.Lee WN, Shah N, Sundlass K, Musen M. Comparison of ontology-based semantic-similarity measures. AMIA Annu Symp Proc; 2008. pp. 384–388. [PMC free article] [PubMed] [Google Scholar]
- 18.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J. R. Statist. Soc. B. 2001;63:411–423. [Google Scholar]