Abstract
A source of semantically coded Adverse Drug Event (ADE) data can be useful for identifying common phenotypes related to ADEs. We proposed a comprehensive framework for building a standardized ADE knowledge base (called ADEpedia) through combining ontology-based approach with semantic web technology. The framework comprises four primary modules: 1) an XML2RDF transformation module; 2) a data normalization module based on NCBO Open Biomedical Annotator; 3) a RDF store based persistence module; and 4) a front-end module based on a Semantic Wiki for the review and curation. A prototype is successfully implemented to demonstrate the capability of the system to integrate multiple drug data and ontology resources and open web services for the ADE data standardization. A preliminary evaluation is performed to demonstrate the usefulness of the system, including the performance of the NCBO annotator. In conclusion, the semantic web technology provides a highly scalable framework for ADE data source integration and standard query service.
1. Introduction
There is an emerging interest in building algorithms on identifying clinical phenotypes relevant to symptoms and findings associated with adverse drug events (ADEs) from the electronic medical records (EMRs) [1]. Conventionally, the first step is to review the existing drug information resources which may help identify common phenotypes related to ADEs. However, performing this review can be time-consuming as the drug information resources are typically medication-oriented. For instance, the National Library of Medicine (NLM) DailyMed web site provides high quality information about marketed drugs derived from FDA Structured Product Labels (SPLs) [2]. The search functionality of the web site only supports a single drug name or NDC code input so a user can not query against multiple medications simultaneously. Furthermore, the adverse events are described in free text (i.e. non machine-readable) under a number of section headings (e.g. the Adverse Reaction Section).
To deal with these challenges, on the one hand, biomedical researchers have turned to ontologies and terminologies to structure and annotate their data with ontology concepts for better search and retrieval. For example, Duke et al [3] developed a system called ADESSA, in which the ADEs were extracted from the SPL labels and mapped to the MedDRA terms and concepts, then utilized the UMLS to generate mappings between extracted the MedDRA terms and the SNOMED CT concepts. On the other hand, Semantic Web technology can be useful to provide a scalable framework for facilitating semantic data integration of heterogeneous resources and enabling semantic sharing through the standard query services. Linked data technique, in particular, is such a method that aims to shape a general solution for information dissemination and integration [4]. Linked Data Source of Clinical Trials [5] and Linked Open Drug Data [6] are typical instances adopting the technology.
In this paper, we propose a comprehensive framework for building a standardized ADE knowledge base (called ADEpedia) through combining ontology-based approach with semantic web technology. The framework comprises four primary modules: 1) an XML2RDF transformation module; 2) a data normalization module; 3) a RDF store based persistence module; and 4) a front-end module for the review and curation. A prototype is implemented to demonstrate the capability of the system to integrate multiple ontology resources and open web services for the ADE data standardization. A preliminary evaluation is performed to demonstrate the usefulness of the system.
2. Background
2.1. Structured Product Labels
The Structured Product Labeling (SPL) is a document markup standard approved by the Health Level Seven (HL7) and adopted by the FDA as a mechanism for exchanging product information [7]. Since 2006, pharmaceutical manufacturers produce SPL labels for their products and FDA has released an increasing number of these labels through NLM’s DailyMed [8]. Usually, the contents of a SPL XML document are organized into section headings. In the SPL terminology, there are 76 section headings which are specified and coded in LOINC codes [9]. In this study, for the purpose of extracting ADEs, we focused on three sections, comprising the “ADVERSE REACTIONS SECTION” (LOINC code: 34084-4), the “WARNINGS AND PRECAUTIONS SECTION” (LOINC code: 43685-7) and the “WARNINGS SECTION” (LOINC code: 34071-1).
2.2. RxNorm and RxNav
RxNorm, a nomenclature for clinical drugs, is developed by the US National Library of Medicine [10]. It contains the names of prescription and many nonprescription formulations approved for human use (primarily in the USA). An RxNorm clinical drug name reflects the active ingredients, strengths, and dose form comprising that drug. When any of these elements vary, a new RxNorm drug name is created as a separate concept identified by a concept unique identifier (RxCUI). In addition, RxNorm contains mappings from its concepts to one or more concepts in external drug terminologies (or databases) including First DataBank, Micromedex, Medi-Span, Multum, and NDF-RT, and FDA SPL setId as well.
The RxNav is a browser for RxNorm. The RxNav uses a web service to access the RxNorm data. The API provides various functionality such as 1) searching for a name in the RxNorm dataset to get the RxCUI; 2) finding relationships between drug entities [11–12].
2.3. NCBO BioPortal Open Biomedical Annotator (OBA)
The National Center for Biomedical Ontology Annotator is an onotology-based web service for annotating the textual biomedical data with biomedical ontology concepts [13–14]. The biomedical community can use the Annotator service to tag datasets automatically with concepts from more than 200 ontologies coming from the two most important biomedical ontology & terminology repositories: the Unified Medical Language System (UMKS) Metathesaurus and NCBO BioPortal. Such annotations contribute to create a biomedical semantic web that facilitates translational scientific discoveries by integrating annotated data. In this study, two clinical terminologies SNOMED CT (the most comprehensive clinically oriented medical terminology system) and MedDRA are configured to annotate the ADE data.
2.4. Semantic Web Technologies
The World Wide Web Consortium (W3C) is the main international standards organization for the World Wide Web. Its goal is to develop interoperable technologies and tools as well as specifications and guidelines to lead the Web to its full potential. W3C recommendations have several maturity levels: Working Draft, Candidate Recommendation, Proposed Recommendation, and W3C Recommendation. The Resource Description Framework (RDF), a W3C recommendation, is a directed, labeled graph data format for representing information in the Web [15]. SPARQL is a query language for RDF graphs [13]. SPARQL queries are expressed as constraints on graphs, and return RDF graphs or sets as results. SPARQL 1.0 has been a W3C recommendation whereas SPARQL1.1 is a Working Draft [16–17]. Triplestore is a database for the storage and retrieval of RDF metadata, ideally through standard SPARQL query language. Some triplestores can store billions of triples. In addition, the XML Semantics Reuse methodology is a typical Semantic Web Technology, which focuses on moving meta-data from the XML domain to the Semantic Web [18].
Semantic Wiki provides the ability to capture (by humans), store and later identify (by machines) further meta-information or metadata about those articles and hyperlinks, as well as their relations [19] and has been demonstrated as an appropriate Semantic Web platform for knowledge engineering methods to work on the different levels of the continuum of formalism [20]. For instance, based upon semantic wiki technology, LexWiki has been developed as a collaborative authoring platform for large-scale biomedical terminologies [21].
3. Methods
3.1. System Architecture
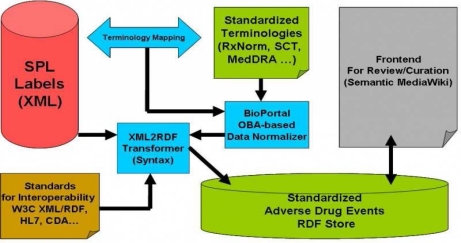
There are four primary modules in the system, comprising of 1) an XML2RDF transformation module; 2) a data normalization module; 3) a RDF store based persistence module; and 4) a front-end module for the review and curation. In the XML2RDF transformation module, the input will be any data resources rendered in the XML format, and the output will be in the RDF format through a transparent transformation service. In the data normalization module, the input will be any unstructured free-text data and the output will be in the XML format with the annotations and mappings from the standardized terminologies (e.g. SNOMED CT). In the persistence module, a RDF store will interact with the XML2RDF transformer and provides the RDF triple import functionality. Furthermore, a standard SPARQL endpoint will provide the standard SPARQL query service against the RDF store. In the review and curation module, a Semantic Wiki based environment, using the RDF store module as backend, on the one hand, will provide a user interface as the front-end to allow the users browse the standardized ADE data, query the customized data through a standard SPARQL query interface. On the other hand, a review and curation mechanism will support the collaborative review of the adverse event annotations generated by the automatic algorithm of data normalizer for the purpose of quality assurance. Fig. 1 shows the system architecture.
Fig. 1.
System Architecture
3.2. Prototype Implementation
In this prototype, we adopted the XML2RDF web service API implemented in the ReDeFer project as the core component of the XML2RDF transformation module [18]. For the data normalization module, we adopted the NCBO BioPortal OBA annotator which provides the annotation web service. For the persistence module, we adopted 4store which is a scalable open source RDF database developed at the Garlik [22]. We also used a third-party Java client wrapper API for the 4store httpd server which provides such functionality as add graph, append graph, delete graph, and query [23]. For the review and curation module, we implemented a Semantic MediaWiki [24] based environment as the front-end user interface. We created the scripts and realized the interactions between the XML2RDF transformer, the OBA normalizer, the Semantic MedicaWiki and the RDF store.
To build the ADE knowledge base, we utilized three different resources, comprising of 1) the drug information resource from all available Structured Product Labels; 2) the standard drug ontology RxNorm and its service API at RxNav; 3) the standard clinical terminologies SNOMED CT and MedDRA for the ADE annotations through the NCBO BioPortal OBA annotation web service. Specifically, we took the following steps for building a prototype of the ADE knowledge base.
Firstly, we downloaded and processed all available SPLs (n=17106, as of 11/18/2010) on the DailyMed. The contents of each label are rendered in a standard XML format, which is used to feed into the XML2RDF transformation module for the RDF transformation. Fig. 2 shows a SPL example in the transformed RDF format. Once a SPL document is transformed into the RDF format, it is loaded into the RDF store by an import script. We transformed and loaded all available SPLs into the RDF store.
Fig. 2.
A SPL example in the transformed RDF format
Secondly, we utilized the RxNav terminology service through an open web service provided by the NLM to retrieve the information about the standard drug ontology RxNorm. In this study, two services were invoked: 1) the service for getting the concept name of a given RxNorm Concept Unique Identifier (RxCUI); 2) the service for getting the structured product label set identifiers for a concept. As each terminology service is usually invoked by a RxCUI, we extracted a full list of RxCUIs (n=194179) from the data table MRCONSO.RRF of a RxNorm version of 10AA_100907F. The results of two services are all rendered in the XML format, which in turn are fed into the XML2RDF transformer. We transformed all those RxCUIs which have at least one mapping to the SPL label and loaded them into the RDF store.
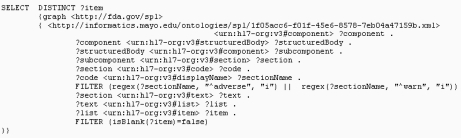
Thirdly, we used the NCBO BioPortal OBA annotation web service and annotated the adverse event data in unstructured free text which is extracted from the SPLs. As mentioned above, we focused on three sections: the “ADVERSE REACTIONS SECTION” (LOINC code: 34084-4), the “WARNINGS AND PRECAUTIONS SECTION” (LOINC code: 43685-7) and the “WARNINGS SECTION” (LOINC code: 34071-1). We identified four XML rendering patterns of the unstructured free text data from these sections and formed the SPARQL queries to extract the free text data from each SPL label (Fig. 3). In this way, the sections of a SPL label can have zero to many free text entries extracted and each entry will be fed into the OBA annotator for the ontology based annotation.
Fig. 3.
A SPARQL query example as one of four patterns to extract the free text ADE data.
Running the query illustrated in fig. 3 over the RDF store, for instance, it will result in five free text entries. Each of the entries will be fed into the OBA annotator. The annotator was configured for the annotations against SNOMED CT (ontologyId=44777) and MedDRA (ontologyId=42280) and the results were filtered by the Semantic Types T047 (Disease or Syndrome), T184 (Sing or Symptom), T033 (Finding). In addition, there is a parameter to set the output format as the XML so that the annotation results can also be fed into the XML transformer module.
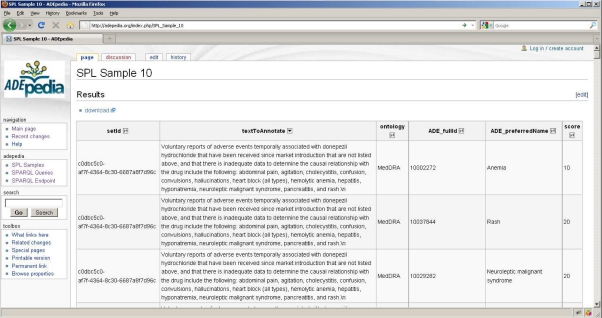
Finally, we implemented a Semantic MediaWiki based environment which provides a front-end to display the Adverse Drug Event data determined by a SPARQL query over the RDF store backend. Fig. 4 shows a screenshot of an ADEpedia front-end prototype implemented in a Semantic MediaWiki environment which uses a RDF store as the backend. The wiki page in the screenshot shows a sample SPL label with their annotations represented by the MedDRA or SNOMED CT concepts. This wiki page is actually generated by an underlying SPARQL query against its RDF store backend.
Fig. 4.
A screenshot of an ADEpedia front-end prototype implemented in a Semantic MediaWiki environment
3.3. System Evaluation
The system evaluation was performed in the following three aspects. The first aspect is to test whether the approach using multiple ontologies will result in better ADE coverage than the approach using a single ontology. We mapped the distinct SNOMED CT and MedDRA concept IDs in the knowledge base into the UMLS Concept Unique Identifiers (CUIs) and compared the CUIs across the two ontologies. The second aspect is to test the capability of the system for answering an ADE oriented question. For example, find all medications (represented by the RxNorm concepts) which have a common ADE represented by a SNOMED CT or MedDRA concept, e.g “pain”, “anemia”, “QT Prolongation”, etc. The third aspect is to evaluate the OBA annotation performance. A random set of 50 labels was selected and sent for the manual review by an experienced nosologist. Comparing the original free text ADE data of each SPL, the reviewer coded ADEs as either true positives (TP), false positives (FP) or false negatives (FN). The system recall, precision and F-measure were then calculated.
4. Results
We successfully built an ADE knowledge base (called ADEpedia) using the proposed framework. In the knowledge base, we created three graph models in the RDF store. The graph models are used as the containers of RDF triples generated from three different sources. For the first graph model, we successfully transformed all 17106 Structured Product Labels (as of 11/18/2010) from the XML format into the RDF, and loaded them into the RDF store that produced approximately 7.2 million triples. For the second graph model, we transformed and loaded 14206 out of 194179 RxNorm concept ids (i.e. RxCUI), concept names and their mappings to the SPLs (represented by 14206 distinct SPL setIds). For the third graph model, we first identified 8898 out of 17106 distinct SPL labels that contain the textual description in the target sections. And then we broke down the text description of each label into the paragraph entries and fed them into the OBA web service for the annotation purpose. In this annotation step, 79697 annotation XML files were generated by the OBA web service from the paragraph entries containing at least one SNOMED CT or MedDRA annotation. We then transformed all these XML files into the RDF format and loaded them into the RDF store. In total, we identified 4873 distinct SNOMED CT concepts and 3266 distinct MedDRA concepts from the knowledge base. Table 1 shows top 20 most common ADEs represented by the MedDRA concepts and top 20 common ADEs represented by the SNOMED CT concepts.
Table 1.
Top 20 common ADEs identified as the MedDRA concepts and top 20 common ADEs identified as the SNOMED CT concepts (ranked by the frequency)
For the system evaluation, firstly, we mapped the above distinct concept Ids from MedDRA and SNOMED CT to the UMLS Concept Unique Identifiers (CUIs). We identified 4364 CUIs for the SNOMED CT concepts, 2483 CUIs for the MedDRA concepts. There are 1825 CUIs overlapping across the two subsets, indicating the approach using multiple ontologies may have the better ADE coverage than the approach only using a single ontology.
Secondly, we successfully linked the ADEs coded by SNOMED CT and MedDRA with the drug ontology RxNorm. For example, using a SPAQL query, we identified 28 distinct RxNorm drugs linked with the ADE “Prolonged QT interval” represented by a SNOMED CT code “111975006” and the ADE “QT prolonged” represented by a MedDRA code “10037705” (Table 2).
Table 2.
A set of the example drugs linked with the ADE “Prolonged QT interval” represented by a SNOMED CT code “111975006” and the ADE “QT prolonged” represented by a MedDRA code “10037705”.
| Drug Name in RxNorm | RxCUI |
|---|---|
| Amiodarone hydrochloride | <http://rxnav.nlm.nih.gov/REST/rxcui/203114> |
| Amiodarone hydrochloride 50 MG/ML Injectable Solution | <http://rxnav.nlm.nih.gov/REST/rxcui/833532> |
| 3 ML Amiodarone hydrochloride 50 MG/ML Prefilled Syringe | <http://rxnav.nlm.nih.gov/REST/rxcui/834357> |
| 18 ML Amiodarone hydrochloride 50 MG/ML Prefilled Syringe | <http://rxnav.nlm.nih.gov/REST/rxcui/835910> |
| Citalopram 10 MG Oral Capsule | <http://rxnav.nlm.nih.gov/REST/rxcui/730440> |
| Citalopram 10 MG Oral Tablet | <http://rxnav.nlm.nih.gov/REST/rxcui/283672> |
| Citalopram 10 MG Oral Tablet [Celexa] | <http://rxnav.nlm.nih.gov/REST/rxcui/284591> |
| Citalopram 2 MG/ML Oral Solution [Celexa] | <http://rxnav.nlm.nih.gov/REST/rxcui/261342> |
| Citalopram 20 MG Oral Capsule | <http://rxnav.nlm.nih.gov/REST/rxcui/730441> |
| Citalopram 20 MG Oral Tablet | <http://rxnav.nlm.nih.gov/REST/rxcui/200371> |
| Citalopram 20 MG Oral Tablet [Celexa] | <http://rxnav.nlm.nih.gov/REST/rxcui/213344> |
| Citalopram 40 MG Oral Capsule | <http://rxnav.nlm.nih.gov/REST/rxcui/730442> |
| Citalopram 40 MG Oral Tablet | <http://rxnav.nlm.nih.gov/REST/rxcui/309314> |
| Citalopram 40 MG Oral Tablet [Celexa] | <http://rxnav.nlm.nih.gov/REST/rxcui/213345> |
| Citalopram hydrobromide | <http://rxnav.nlm.nih.gov/REST/rxcui/221078> |
| Memantine hydrochloride | <http://rxnav.nlm.nih.gov/REST/rxcui/236685> |
| {21 (Memantine hydrochloride 10 MG Oral Tablet [Namenda]) / 28 (Memantine hydrochloride 5 MG Oral Tablet [Namenda]) } Pack [Namenda 49 Titration Pack] | <http://rxnav.nlm.nih.gov/REST/rxcui/996634> |
| Memantine hydrochloride 10 MG Oral Tablet [Namenda] | <http://rxnav.nlm.nih.gov/REST/rxcui/996563> |
| Memantine hydrochloride 2 MG/ML Oral Solution [Namenda] | <http://rxnav.nlm.nih.gov/REST/rxcui/996742> |
| Memantine hydrochloride 5 MG Oral Tablet [Namenda] | <http://rxnav.nlm.nih.gov/REST/rxcui/996574> |
| Methadone Hydrochloride | <http://rxnav.nlm.nih.gov/REST/rxcui/218337> |
| Methadone Hydrochloride 0.333 MG/ML Oral Solution | <http://rxnav.nlm.nih.gov/REST/rxcui/991147> |
| Methadone Hydrochloride 0.333 MG/ML Oral Solution [Methadose] | <http://rxnav.nlm.nih.gov/REST/rxcui/991149> |
| Methadone Hydrochloride 0.333 MG/ML Oral Suspension | <http://rxnav.nlm.nih.gov/REST/rxcui/864978> |
| Methadone Hydrochloride 1 MG/ML Oral Solution | <http://rxnav.nlm.nih.gov/REST/rxcui/864761> |
| Methadone Hydrochloride 10 MG Oral Tablet | <http://rxnav.nlm.nih.gov/REST/rxcui/864706> |
| Methadone Hydrochloride 2 MG/ML Oral Solution | <http://rxnav.nlm.nih.gov/REST/rxcui/864769> |
| Methadone Hydrochloride 5 MG Oral Tablet | <http://rxnav.nlm.nih.gov/REST/rxcui/864718> |
Thirdly, we evaluated the NCBO annotator performance. The annotation evaluation set consisted of 50 labels which are mapped to 3997 ADEs. Of these, 3596 were true positives; 401 were false positives and 228 were not identified by the NCBO annotator. Based on these results, the precision was calculated as 90.0 %, the recall as 94.0 % and the F-measure as 92.0%.
5. Discussion
One of our goals is to figure out a source of semantically coded ADE data to support identifying common phenotypes related to the ADEs, however, no freely available comprehensive source of semantically coded ADE data exists. Duke et al. introduced an ADESSA system [3], in which the FDA SPLs were utilized as a main drug knowledge source to extract the ADEs. Motivated in part by their work, we utilized a different framework that combines ontology-based approach with semantic web technology.
The practical significance of our framework is many-fold. First of all, we were able to successfully identify a set of common ADEs standardized by the SNOMED CT or MedDRA ontologies (just like those illustrated in Table 1). These standardized ADEs are further linked with rich drug information provided by the original SPL labels. And more importantly, most of the drugs described in the SPL labels have been standardized by the RxNorm drug ontology. By having such an ADE knowledge base, we are able to build the semantic linkage between the standard drugs and the standardized ADEs. We consider the ADE knowledge base provides a useful and scalable source for facilitating the high throughput clinical phenotyping on common phenotypes relevant to the ADEs [1].
Second, we adopted the NCBO OBA open web service in the data normalization module. The open annotator not only provides a fast and accurate concept recognition tool, but also provides the access to the hundreds of ontologies from the NCBO BioPortal. We were able to annotate the ADE data using multiple ontologies, i.e. the MedDRA and the SNOMED CT. By mapping the identified concepts from the two ontologies to the UMLS CUIs, we noticed that there are only 1825 overlapping CUIs. This means that 2539 additional concepts were identified through using the SNOMED CT, in addition to using the MedDRA. We believe this approach provided a better ADE concept coverage than that of a single ontology source-based approach.
Furthermore, the annotator provides rich configuration parameters. For example, the parameter of Semantic Type is very helpful for the ADE extraction. In this study, we configured the parameter to just recognize the concepts with the semantic types T047 (Disease or Syndrome), T184 (Sing or Symptom), T033 (Finding). We consider this configuration improves the precision of ADE extraction. By a human based evaluation, the precision and the recall are calculated as 90.0% and 94.0% respectively. The preliminary results on the performance of the NCBO annotator are comparable with that of an in-house natural language processing (NLP) tool by Duke et al [3]. It seems clear to us that a future rigorous review and evaluation in the future would produce more reliable results. While we have demonstrated the merits for adopting the OBA open annotation service, our framework is highly extensible and does not exclude the possibility to utilize other sophisticated NLP annotation services [24].
Third, we introduced a Semantic Wiki based platform as the core of the review and curation module, which provides a mechanism for the community based ADE data quality management. As the wiki uses a RDF store as the backend, all ADE data in the RDF store are open to the reviewers. While we represent the ADE data in a customized way in the wiki, the reviewers will have full access to the data through a standard SPARQL query interface. More importantly, as all ADE annotations and the related provenance data are stored as the RDF triples, the reviewers can easily track the source of an ADE annotation for the purpose of quality checking.
Fourth, the XML Semantics Reuse methodology combines an XML Schema to the web ontology mapping, called XSD2OWL, with a transparent mapping from the XML to the RDF - XML2RDF [4, 18]. The ontologies generated by the former step are used during the XML to RDF step in order to generate semantic metadata that takes into account the XML Schema intended meaning. We consider this would provide the benefit for potential semantic reasoning extensions in future.
In addition, with the capacity of the RDF Store (e.g. the 4store implemented in our prototype), we were able to integrate multiple, large scale, heterogeneous ADE data and ontology resources easily in an agile manner. The underlying RDF model encoding of knowledge in the form of triples plays a key role on this as the RDF can be used as a schemaless data representation format. This ensures the flexibility, scalability of our system. For instance, the system can be extended to link with the Linked Data Source of Clinical Trials [5] to explore the eligibility criteria specific to the ADEs.
We encountered several challenges in the development of this system that should inform future work. First, while the NCBO OBA annotator provides a rich set of parameters for the configuration, we configured the service heuristically. For example, we set up the target ontologies as the SNOMED CT and the MedDRA, and configured three specific semantic types. For other parameters, we used default settings. To optimize the configuration for the ADE annotation, we consider that a systematic evaluation should be performed in the future. To link an annotation to a specific SPL label, we need an additional parameter to store meta-data like the SPL setId. Though we overcome this temporarily through using one of existing parameters, we consider it would be helpful for the OBA annotator to provide a mechanism for such meta-data parameters. In addition, for the annotations by the SNOMED CT concepts, we noticed that there is no way to filter out the concepts under the branches of “Duplicate concept” and “Ambiguous concepts” in current configuration.
Second, we used the RxNav service API to extract the mapping information between the RxNorm and the SPL labels. As mentioned in above section, the services have to be invoked by a RxCUI. However, there is no service available to get all RxCUIs which have the SPL mappings, so we had to seek the original RxNorm release to extract all RxCUIs. We expect that the RxNav could provide such a service in the future. An alternative solution is to use the latest LexEVS 6.0 CTS2 compatible terminology service which provides the value set definition and RDF export functionality [26].
Third, for the XML2RDF open web service, currently its API only accepts the GET method. To smooth the process of the RDF transformation of the annotated XML contents from the NCBO OBA web service, we consider a POST method will be very helpful if it can be available in the future. It is arguable that the capabilities of the SPARQL 1.0 query language are somewhat limited compared to typical SQL implementations. However, this can be offset by the convenience of the high degree of standardization between stores and the high degree of flexibility of data representation [4]. In addition, the latest W3C working draft on SPARQL 1.1 is providing many promising and powerful functionality enhancements including aggregate, update, service description, etc. [17].
Finally, we will make the ADEpedia knowledge base open online for public access in near future though we are aware that we will have to have an appropriate mechanism for dealing with the proprietary issue of the MedDRA.
In summary, we successfully integrated multiple heterogeneous drug data and ontology sources and developed a scalable, standardized ADE knowledge base (call ADEpedia) with a standard query service. Our next steps in the future will focus on a thorough examination of all ADE related sections in the SPL document, consolidating the review and curation mechanism of the front-end module, integrating other ADE resources and exploring the potential applications based upon the ADE knowledge base. The ADEpedia community wiki is accessible through the URL at http://adepedia.org, which will also be serving as an instance of the front-end module of the system.
Acknowledgments
The study is supported in part by the Mayo Clinic Center for Clinical and Translational Research (CTSA) grant – Common adverse event phenotyping related to pharmacogenomics (RR 24150). The authors are thankful to Dr. JD Duke, M.D. from the Regenstrief Institute, who provided helpful comments on the study.
References
- 1.THE ADE Phenotype Ontology project http://informatics.mayo.edu/adephont/index.php/Main_Page. last visited March 12, 2011.
- 2.The DailyMed URL http://dailymed.nlm.nih.gov/dailymed/about.cfm. last visited March 12, 2011.
- 3.Duke JD, Friedlin J. ADESSA: A real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Symposium Proceedings; 2010. pp. 177–181. [PMC free article] [PubMed] [Google Scholar]
- 4.Wood D. Linking Enterprise Data. Springer; 2010. ISBN-978-14419-7664-2. [Google Scholar]
- 5.The Linked Source of Clinical Trails http://linkedct.org/index.html. last visited March 12, 2011.
- 6.The Linked Open Drug Data http://esw.w3.org/HCLSIG/LODD. last visited March 12, 2011.
- 7.The FDA Structured Product Labeling URL http://www.fda.gov/ForIndustry/DataStandards/StructuredProductLabeling/default.htm. last visited March 12, 2011.
- 8.Schadow G. Assessing the impact of HL7/FDA structured product label (SPL) content for medication knowledge management. AMIA Symposium Proceedings; 2007. pp. 646–650. [PMC free article] [PubMed] [Google Scholar]
- 9.The SPL Section Headings http://www.fda.gov/ForIndustry/DataStandards/StructuredProductLabeling/ucm162057.htm. last visited March 12, 2011.
- 10.Pathak J, Chute CG. Analyzing categorical information in two publicly available drug terminologies: RxNorm and NDFRT. JAMIA. 2010;17:432–439. doi: 10.1136/jamia.2009.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pathak J, Peters L, Chute CG, Bodenreider O. Comparing and evaluating terminology services application programming interfaces: RxNav, UMLSKS and LexBIG. J Am Med Inform Assoc. 2010 Nov 1;17(6):714–9. doi: 10.1136/jamia.2009.001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The RxNAV http://mor.nlm.nih.gov/download/rxnav/index.html. last visited March 12, 2011.
- 13.Jonquet Clement, Shah Nigam H, Musen Mark A. The Open Biomedical Annotator. AMIA Summit on Translational Bioinformatics; March 2009; San Francisco, CA, USA. pp. 56–60. [PMC free article] [PubMed] [Google Scholar]
- 14.The NCBO Annotator service http://www.bioontology.org/annotator-service. last visited March 12, 2011.
- 15.The RDF http://www.w3.org/RDF/; last visited March 12, 2011.
- 16.The SPARQL http://www.w3.org/TR/rdf-sparql-query/; last visited March 12, 2011.
- 17.The SPRQL1.1 http://www.w3.org/2009/sparql/wiki/Main_Page. last visited March 12, 2011.
- 18.The XML2RDF http://rhizomik.net/html/redefer/xml2rdf/. last visited March 12, 2011.
- 19.Kamel Boulos MN. Semantic Wikis: A comprehensible introduction with examples from the health sciences. Journal of Emerging Technologies in Web Intelligence. 2009;(1):94–96. [Google Scholar]
- 20.Baumeister J, Reutelshoefer J, Puppe F. Engineering on the Knowledge Formalization Continuum. Proceedings of the Forth Semantic Wiki Workshop (SemWiki 2009), co-located with 6th European Semantic Web Conference (ESWC 2009); Hersonissos, Heraklion, Crete, Greece. June 1st, 2009. [Google Scholar]
- 21.Jiang G, Solbrig H, Chute CG. Mash-up of LexWiki and Web-Protégé for Distributed Authoring of Large-Scale Biomedical Terminologies. Proceedings of 18th Annula International Conference on Intellegent Systems for Molecular Biology (ISMB 2010) - Bio-Ontologies 2010: Semantic Applications in Life Sciences; Boston. July 9–10, 2010.pp. 132–135. [Google Scholar]
- 22.The 4store URL http://4store.org/; last visited March 12, 2011.
- 23.The 4store Java Client https://github.com/danhanley/4store-java-client; last visited March 12, 2011.
- 24.The Semantic MediaWiki http://semantic-mediawiki.org/wiki/Semantic_MediaWiki; last visited March 12, 2011.
- 25.The Open Health Natural Language Processing (OHNLP) Consortium URL . https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/OHNLP; last visited March 12, 2011. [Google Scholar]
- 26.The LexEVS 60 URL . https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/LexEVS_Version_6.0_Highlights; last visited March 12, 2011. [Google Scholar]