Abstract
The need for formal representations of eligibility criteria for clinical trials – and for phenotyping more generally – has been recognized for some time. Indeed, the availability of a formal computable representation that adequately reflects the types of data and logic evidenced in trial designs is a prerequisite for the automatic identification of study-eligible patients from Electronic Health Records. As part of the wider process of representation development, this paper reports on an analysis of fourteen Electronic Health Record oriented phenotyping algorithms (developed as part of the eMERGE project) in terms of their constituent data elements, types of logic used and temporal characteristics. We discovered that the majority of eMERGE algorithms analyzed include complex, nested boolean logic and negation, with several dependent on cardinality constraints and complex temporal logic. Insights gained from the study will be used to augment the CDISC Protocol Representation Model.
Introduction and Motivation
Identifying patients that match research criteria (that is, clinical phenotyping) is a major bottleneck in the successful and timely execution of clinical trials. The subject identification process currently used, requiring as it does the careful hand matching of patient to study, is time-consuming, inefficient and inconvenient for the busy clinician.1 Indeed, Patel et al. state that of the 80% of trials that suffer delays, 50% are delayed due to subject recruitment problems.2 Delays in clinical trials can have serious public health impact. For example, patients do not benefit from experimental treatments, or the treatment’s entry to market is delayed.
The current work has its roots in the eMERGE network3 (www.gwas.org) – a large scale, multi-site network of research organizations (Northwestern University, Vanderbilt University, Mayo Clinic, Marshfield Clinic, and Group Health Cooperative; University of Washington–Fred Hutchinson Cancer Research Center) dedicated to mining biobank resources (that is, collections of individual Electronic Health Records — EHRs — with their associated genomic data) for translational medicine. As part of this process, a series of phenotyping algorithms were developed. These phenotyping algorithms were designed to access the primarily semi-structured data fields in EHRs (for example, procedure codes, ICD-9 codes, laboratory results). Algorithms take the form of free text documents constructed of data elements (ICD-9, procedure codes and so on) combined with logical operators and are designed as “pseudocode” to identify study eligible patients from EHRs. However, these algorithms are normally stored as unstructured Microsoft Word and PDF documents, as opposed to a computable form that can be used in conjunction with EHR query methods. Additionally, phenotyping algorithms (as defined by the eMERGE network) often contain keywords (or indicative phrases linked to drug names or procedures) designed to facilitate Natural Language Processing (NLP) of the narrative (free text) sections of EHRs. Typically, administrative data (ICD-9 and CPT codes), laboratory data and medication data (RxNorm codes) form the core data elements of the algorithm, with NLP rules as an additional layer to disambiguate and refine the core data elements. It is important to note that these algorithms were developed at several different sites within the eMERGE network and without formal guidelines or specifications and therefore do not adhere to any formal protocol representation standards. Algorithm generation was very much a team effort, combining the knowledge and insights of clinicians, domain experts and informaticians. The algorithms were extensively tested over multiple iterations, and have proven to be robust over several sites.
The SHARP project (Strategic Health IT Advanced Research Projects: www.SHARPn.org) – again a consortium project with several different sites represented (including Northwestern University, Mayo Clinic and the University of Utah) plans to use the algorithms developed in the eMERGE network for the purposes of automatic phenotyping from EHRs with the goal of automating the phenotyping process. Currently, eMERGE algorithms are actually operationalized by informaticians, typically with informaticians developing queries (often in SQL) based on the eMERGE algorithms with the aid of a researcher or domain expert; informaticians are an intermediary between the algorithm and the EHR system. Under SHARP, instead of an informatician interpreting an algorithm document and executing queries against EHRs (that is, acting as an intermediary between algorithm and EHR), the algorithm (in a suitable representation) will be executed directly. SHARP takes human interpretation “out of the loop”. In order to facilitate the translation of eMERGE algorithms from their current unstructured state to a computable format — and indeed, to select or design such a representation — it is necessary to analyze the free text algorithms in terms of their data elements and logic.
The current research seeks to examine fourteen phenotyping algorithms generated by the eMERGE network in terms of their logical structure and data elements in order to:
Explore the types of algorithm available, and different strategies used in presenting data.
Identify common data elements, types of logic and formalisms used to represent eligibility criteria.
Assess the level of heterogeneity between algorithms in the use of data types and logic.
The overall goal and motivation for the study is to gain an improved understanding of EHR-oriented phenotyping algorithms which will in turn inform work on the development of a computable representation to support the processing of EHRs. Note that the eMERGE algorithms are publicly available at the eMERGE website (www.gwas.org).
The structure of this paper is as follows. First, we present some relevant background literature, then we describe the materials and methods used, before going on to set out the results of our analysis. We conclude the paper with a discussion of our results and a short conclusion.
Background
Recent years have seen a small number of papers on eligibility criteria and phenotyping, focused on both textual characteristics of eligibility criteria and on formal representations for eligibility requirements. Ross et al.4 analyzed one thousand clinical protocols from clinicaltrials.gov in terms of the complexity, semantic patterns, clinical content and data sources used in eligibility criteria and discovered that (among other findings) forty percent of eligibility criteria encode temporality in some way. In research following on from Ross, Tu et al.5 present an OWL-based annotation scheme for capturing the salient content of textual eligibility criteria.
In a detailed review paper, Weng et al.6 describes several eligibility criteria knowledge representations (including Aids27, Oasis8, Glif9, and Gello10) and classifies current eligibility criteria knowledge representations along five dimensions, including the expression language used (for example, Arden syntax, XML, OWL) and the type of eligibility criteria (for example, content topic, does the criterion require a boolean response?)
Until now, efforts at developing NLP techniques for the parsing of textual eligibility criteria in order to populate eligibility criteria oriented Knowledge Representations has been limited, although attempts have been made to develop annotation tools for converting raw text eligibility criteria to a computable form in the breast cancer domain.11 Additionally, there has been some experimental work on comparing the performance of an NLP system to an oncologist in the extraction of cancer diagnoses from eligibility criteria.12
The CDISC Consortium (Clinical Data Interchange Standards Consortium: http://www.cdisc.org) an international standards organization which aims to “to develop and support global, platform-independent data standards that enable information system interoperability to improve medical research and related areas of health-care”) has developed a Protocol Representation Model (PRM: http://www.cdisc.org/protocol) as a standard for the general representation of protocols. The PRM has an eligibility criteria module which is currently being implemented in XML for CDISC. While the PRM model is a general purpose eligibility criteria representation model unable to represent the data elements (for example, ICD-9 codes) and complex logic (for example, temporality) found in computable phenotyping algorithms, its status as a data standard and its adoption among clinical informatics software vendors make it a suitable choice for our basic model. The aim of the current paper is to describe the properties of eMERGE algorithms (in terms of their logical structure and data elements) in order to appropriately extend the CDISC PRM.
It is important to emphasize that none of the representations outlined above are designed to represent the kind of EHR oriented data elements (ICD-9 codes, CPT codes, and so on) found in eMERGE algorithms.
Materials and Methods
We used fourteen phenotyping algorithm documents generated by the eMERGE network in this work. The documents are in Microsoft Word format and from various eMERGE consortium institutions (see Table 1 for a list of phenotyping algorithms and their originating organizations). Phenotyping algorithms were translated to ASCII text format using the Unix utility antiword for automatic text processing.
Table 1:
Characteristics of phenotyping documents
| Name | Organizationa | WCb | Flowchartc | Tabulard | % Tabulare | # Sentencesf | SentLengg | |
|---|---|---|---|---|---|---|---|---|
| 1 | Alzheimers | GHCh | 1,317 | − | − | n/a | 40 | 32 |
| 2 | Dementia | GHC | 634 | − | − | n/a | 26 | 24 |
| 3 | Diabetic retinopathy | Marshfield | 324 | + | + | 19 | 18 | 18 |
| 4 | Height | Northwestern | 2,101 | − | + | 93 | n/a | n/a |
| 5 | Hypothyroidism | Vanderbilt | 1,351 | − | − | n/a | 28 | 40 |
| 6 | Serum lipid level | Northwestern | 1091 | − | + | 46 | 44 | 25 |
| 7 | Low HDLi cholesterol level | Marshfield | 2,579 | + | − | n/a | 126 | 21 |
| 8 | Peripheral arterial disease | Mayo | 1,353 | − | + | 8 | 49 | 28 |
| 9 | QRS duration | Vanderbilt | 26,695 | − | + | 78 | 43 | 620 |
| 10 | Red blood cell indices | Mayo | 2,857 | + | + | 62 | 91 | 31 |
| 11 | Resistant hypertension | Vanderbilt | 895 | − | + | 12 | 54 | 17 |
| 12 | Type 2 diabetes | Northwestern | 954 | + | + | 20 | 75 | 13 |
| 13 | White blood cell indices | GHC | 2,458 | − | + | 25 | 61 | 40 |
| 14 | Cataract | Marshfield | 3,152 | + | + | 81 | 108 | 29 |
Originating organization
Word count for document (indicates document length)
Does the document contain a flowchart?
Does the document contain tabular data?
Percentage of word tokens contained in tables (provides information on what proportion of the algorithm is in semi-structured tabular form)
Number of sentences (provides an estimate of syntactic complexity, identified using Perl CPAN sentence splitting module Lingua::EN::Sentence)
Mean sentence length in words (provides another indication of language complexity)
Group Health Cooperative in conjunction with the University of Washington and the Fred Hutchinson Cancer Research Center
High-density lipoprotein
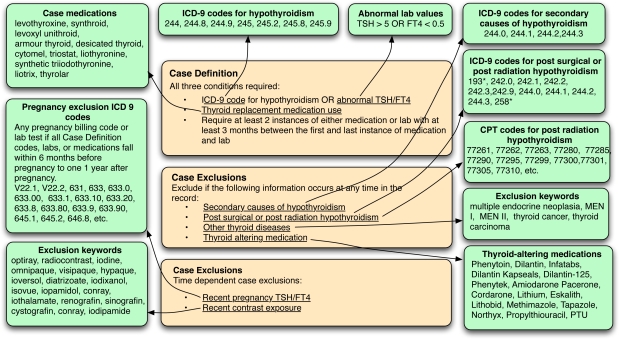
An example eMERGE algorithm for hypothyroidism (developed at Vanderbilt University) is presented in Figure 1 on the next page. The algorithm seeks to identify European-ancestry patients with autoimmune hypothyroidism (that is, Hashimoto’s hypothyroidism) and discard records where hypothyroidism is caused by:
surgery
radiological ablation
subclinical hypothyroidism
medication induced hypothyroidism (for example, lithium)
transient hypothyroidism (for example, during pregnancy)
Figure 1:
Hypothyroidism algorithm, developed by the eMERGE network
Note that for the hypothyroidism algorithm presented in Figure 1, criteria are divided into two categories, inclusions (case definitions) and exclusions (case exclusions). Case definitions are recognized first (for example, is one of a list of hypothyroidism ICD-9 codes present in the EHR?). Then, case exclusions are used to filter out records identified in the first stage (for example, is there a pregnancy ICD-9 code in the EHR? or does an “other thyroid disease” exclusion keyword occur?) Note that the algorithm as represented in Figure 1 is not complete and is provided for expositionary purposes only.
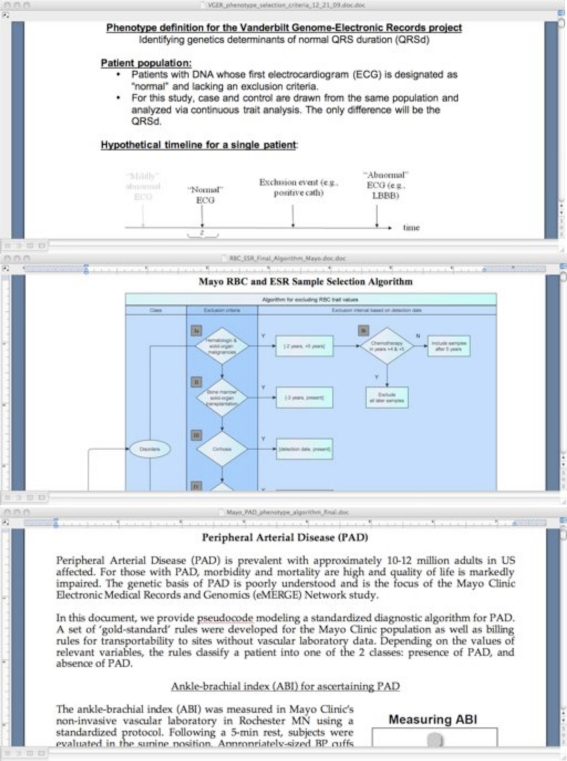
There is considerable heterogeneity of formatting among the fourteen algorithms, with some consisting primarily of narrative prose, and some encoding all logic and data elements in flowcharts or tabular form. Figure 2 on the next page shows fragments from three algorithms, each of which uses strikingly different methods of data representation. The three fragments shown are derived from the following algorithms:
QRS13 – largely constructed of semi-structured textual data (particularly bullet points)
Red blood cell indices14 – much of the data is encoded in a flowchart
Peripheral arterial disease15 – continuous text and diagram
Figure 2:
eMERGE algorithms for QRS duration13, red blood cell indices14 and peripheral arterial disease15
The eMERGE network did not stipulate formatting guidelines for algorithm authors, as for the purposes of eMERGE, algorithm standardization was an unnecessary overhead, with algorithms optimized for human rather than machine consumption. However, for the SHARP project, focused as it is on the automatic execution of phenotyping algorithms, standardized algorithm representation is a core concern.
We analyzed each of the fourteen phenotyping algorithms, both manually and using concordancing software16 and identified four broad areas of interest, partially based on Ross et al.4; phenotyping logic, temporality and document data. The first area logic focuses on the kinds and complexity of logic used.
Simple boolean – use of boolean operators to build phenotyping statements (for example, Lab test positive OR ICD-9 code)
Nested boolean – use of simple boolean statements to build arbitrarily complex statements (for example, [[[Lab test positive OR ICD-9 code A] OR [ICD-9 code B]] OR [Procedure code A]])
Cardinality – maximum or minimum number of data elements (for example, has been diagnosed >= 2 dates with T2DM in encounters or problems lists)
Negation – use of negation in complex boolean statements (for example, is currently prescribed only insulin or Symlin or insulin supplies AND does NOT have any T1DM diagnoses)
The second area of interest is temporality:
Temporal proximity – specifies a temporal period with respect to current time (for example, within the last three months, six months from now). Note that the notion of proximity encompasses the future as well as the past
Complex temporality – two or more criteria apply simultaneously, or at least two criteria have been applicable within a given time period (for example, has three simultaneous med classes mentioned on at least 2 occasions >= 1 month apart)
Family history – is the notion of family history used? (for example, AND IF the patient has family history data, there is no history of diabetes)
The third area is document data:
Metadata – includes the date the document was created, authorship, institution and contact information (for example, email addresses and telephone numbers)
Summary – does the document contain summary/introduction data?
Demographic data – includes age, sex, pregnancy status and ethnicity.
Vital signs – height, weight, BMI.
- Basic data types
- – Lab test results – including straightforward results of the form test > minimum and also criterion that appeal to a normal range or value
- – External codes – disease, drug, laboratory test and procedure codes (ICD-9, UMLS, CPT and RxNorm codes)
- – Indicative phrases – keywords used to facilitate pattern matching of narrative EHRs. Included in this category are concept classes designed to be used in conjunction with an NLP engine. (Note that this category also includes more complex NLP information. For example, keywords for laboratory tests must occur with an associated value)
Collections of datatypes – datatypes (like ICD-9 codes and indicative phrases are often stored in collections (sometimes stored in appendixes) that can be referred to using a collective name (for example, “the class of radiation exposure keywords”)
Meta-document knowledge – does any part of the algorithm require knowledge not specified in the document? (for example, no history of heart disease without specifying a procedure, list of codes or indicative phrases to evaluate it)
Complex calculation – are equations or other complex calculations present?
In addition to analysing the fourteen algorithms for the presence of the features listed above, we were also interested in the types of external codings used in the documents. We automatically extracted codes using regular expressions for ICD-9, CPT and UMLS. For RxNorm codes, we counted occurrences manually.
We also investigated how algorithms are organized. As the algorithms vary widely in structure, with some encoding algorithmic logic and data elements in flowcharts, some in tabular form and some in narrative text, we recorded the most important formatting characteristics of each algorithm (for example, Are flowcharts used? and What proportion of the document data is in tabular form?)
Finally, we surveyed the algorithms’ original writers on the algorithm development process (for example, how many iterations were required to produce the final algorithm?). We were particularly interested in the difficulty of adding new types of data to the algorithm, for instance, the difficulty of identifying NLP type information compared to the identification of ICD-9 codes.
Results
We analyzed fourteen phenotyping algorithms from five different sites. Table 1 shows the originating organization for each algorithm and several document level descriptive statistics. The algorithms vary considerably in length (that is, number of tokens) with the QRS algorithm’s prodigious length accounted for by its comprehensive listing disease and procedure code strings. Diabetic retinopathy is the shortest document at 324 words, yet this terseness is misleading when we consider that it “imports” (that is, reuses) the type 2 diabetes phenotyping algorithm from Northwestern University.
Most of the algorithms use tabular data, although this use is highly variable, with 93% of the tokens from the Height algorithm embedded in tables, but only 8% for the Peripheral arterial disease algorithm. Note also that four of the algorithms store their logic in flowchart diagrams, the semantics of which are extremely difficult to access computationally.
Table 2 shows the distribution of external vocabulary codes in the algorithms, where it can be seen that all algorithms rely on ICD-9 disease codes, and most use CPT procedure codes. Only two algorithms uses UMLS codes (due to site specific processing needs).
Table 2:
Distribution of codes across the fourteen eMERGE phenotyping algorithms
| Name | ICD-9a | CPTb | UMLSc | RxNormd | Total/WCe | Percentagef | |
|---|---|---|---|---|---|---|---|
| 1 | Alzheimers | 29 | 0 | 0 | 355 | 384/1,317 | 29 |
| 2 | Dementia | 30 | 0 | 0 | 20 | 50/634 | 7 |
| 3 | Diabetic retinopathy | 12 | 19 | 0 | 0 | 31/324 | 10 |
| 4 | Height | 156 | 0 | 0 | 11 | 167/2,101 | 8 |
| 5 | Hypothyroidism | 43 | 76 | 0 | 0 | 119/1351 | 9 |
| 6 | Serum lipid level | 11 | 0 | 0 | 0 | 11/1091 | 1 |
| 7 | Low HDLg cholesterol level | 41 | 10 | 0 | 0 | 51/2579 | 2 |
| 8 | Peripheral arterial disease | 90 | 112 | 0 | 0 | 202/1,353 | 15 |
| 9 | QRS duration | 50 | 157 | 595 | 0 | 802/26,695 | 3 |
| 10 | Red blood cell indices | 146 | 141 | 0 | 0 | 287/2,857 | 10 |
| 11 | Resistant hypertension | 35 | 0 | 0 | 0 | 35/895 | 4 |
| 12 | Type 2 diabetes | 25 | 0 | 0 | 0 | 25/954 | 3 |
| 13 | White blood cell indices | 18 | 131 | 0 | 0 | 149/2,458 | 6 |
| 14 | Cataract | 152 | 20 | 35 | 0 | 207/3,152 | 6 |
Number of ICD-9 (International Statistical Classification of Diseases, v9) codes present in the algorithm document
Number of CPT (Current Procedure Terminology) codes in document
Number of UMLS (Unified Medical Language System) codes in the document
Number of RxNorm (clinical drug) codes in the document
Total number of codes divided by the number of word tokens
Percentage of the document’s word tokens that are codes
High density lipoprotein
Table 3 on the next page shows the main result of this paper (note that “+” represents the presence of a feature and “−” its corresponding absence). It can be seen that most of the algorithms use some kind of nested boolean logic (along with negation), with cardinality important in six of the fourteen algorithms. The majority of the algorithms use temporal reasoning, and where temporal reasoning is present, it is always complex. Most of the algorithms have some notion of a “collection” of codes (allowing groups of codes to be referred to easily without enumerating every member of the collection) and three include complex equations. Indicative phrases to support NLP are included with the majority of algorithms.
Table 3:
Results of analysing eMERGE phenotyping algorithms
| Algorithm | Logic | Temporal | Document Data | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SBa | NBb | CRc | NGd | TPe | CTf | FHg | MTh | SMi | DMj | VSk | LRl | ECm | NLn | CLo | MKp | EQq | ||
| 1 | Alzheimers | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − |
| 2 | Dementia | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − |
| 3 | Diabetic retinopathy | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − |
| 4 | Height | + | − | − | − | − | − | − | + | + | + | − | + | + | − | + | − | − |
| 5 | Hypothyroidism | + | + | + | − | + | + | − | − | + | − | + | + | + | + | + | − | + |
| 6 | Serum lipid level | + | − | − | − | + | + | − | + | + | − | − | + | + | + | + | − | − |
| 7 | Low HDLrcholesterol level | + | + | + | + | + | + | − | + | + | − | + | + | + | + | − | − | − |
| 8 | Peripheral arterial disease | + | + | + | + | − | − | − | − | + | − | − | + | + | + | + | − | + |
| 9 | QRS duration | + | + | − | + | + | + | + | − | + | − | − | + | + | + | + | − | − |
| 10 | Red blood cell indices | + | + | − | + | + | + | − | − | + | − | − | + | + | + | + | − | − |
| 11 | Resistant hypertension | + | + | + | + | + | + | − | − | − | − | − | + | + | + | + | − | + |
| 12 | Type 2 diabetes | + | + | − | + | + | + | + | + | + | + | + | + | + | − | + | − | − |
| 13 | White blood cell indices | + | − | − | − | + | + | − | + | + | + | + | + | + | − | + | − | − |
| 14 | Cataract | + | + | + | + | + | + | − | + | + | + | − | − | + | + | + | − | − |
SB (Simple Boolean) — are simple boolean statements used (for example, OR, AND)
NB (Nested boolean) — are more complex, nested boolean statements used
CR (Cardinality) — is there evidence of cardinality (for example, at least three ICD-9 codes
NG (Negation) — is there evidence of negation in boolean statements?
TP (Temporal proximity) — specifies a temporal period with respect to a given time (for example, within the last three months, two weeks from now
CT (Complex temporal) — more complex temporal statements (for example, specifies the most recent event; specifies simultaneous events)
FH (Family History) — does the algorithm have a notion of family history?
MT (Metadata) — includes date of document creation, authorship, institution and contact information (only one of these is required)
SM (Summary) — does the document contain summary or introductory material?
DM (Demographic data) — includes age, sex, pregnancy status, ethnicity (only one of these is required)
VS (Vital signs) — defined for these purposes as height, weight or BMI (only one of these is required)
LR (Laboratory test results) — are laboratory test results used in the algorithm (with or without specified values or ranges)?
EC (External code) — are codes from external vocabularies used (for example, ICD-9, UMLS)
NL (NLP or indicative keywords) — are Natural Language Processing resources included? Note that these resources are typically keywords or phrases used for pattern matching.
CL (Collections) — are basic data types (ICD-9 codes, lab test results, indicative phrases) held in collections?
MK Meta-document knowledge — does the document rely on data outside the EHR?
EQ (Complex calculation) — are any complex calculations (for example, equations) included in the algorithm?
High-density lipoprotein
Note that the Alzheimer’s and Dementia algorithms (both very similar in content) are unusual when compared to the other eleven algorithms in that they simply list codes, the presence of which indicates the disease with no substantial overarching logic.
The eMERGE algorithm development process was (according to the results of our survey) a time consuming and difficult enterprise, which, for the more complex algorithms required more than six iterations. The respondents generally found adding laboratory test data relatively straightforward (as presumably there are only a limited number of tests relevant to the target phenotype). Adding codes was rated as slightly more difficult (perhaps because of the potentially large number of ICD-9 codes associated with a given condition). For example, the peripheral arterial disease algorithm uses ninety ICD-9 codes, requiring a lengthy manual selection process.
Survey responders identified the generation of NLP content as the most difficult aspects of algorithm construction (although not all algorithms contained NLP elements). We suspect that this difficulty arose from the requirement to extensively check existing EHRs for appropriate exclusion keywords and phrases.
Adding medication names was judged to be relatively straightforward, with the qualification that some of the algorithms required the inclusion of medications that while not currently used, are likely to occur in the older records. For some of the algorithms, the appropriate use of lab tests caused considerable debate (for example, in the case of Type 2 diabetes algorithm, it was difficult to determine in practice if a fasting glucose value was actually fasting or not).
The actual algorithm production process was lengthy, labor intensive and highly iterative, with usually the involvement of more than one algorithm author. The time needed to create an algorithm – in terms of person hours – was difficult to estimate by algorithm authors due to the iterative nature of the process.
Discussion
Our analysis of the fourteen eMERGE phenotyping algorithms, while demonstrating a significant degree of surface heterogeneity (in terms of document format) also suggests a certain underlying homogeneity of phenotyping logic and data elements. All algorithms use ICD-9 codes and – with the exception of Alzheimer’s and Dementia – use laboratory results and display relatively complex boolean and temporal logic.
This level of underlying logical homogeneity allows us some optimism with respect to the development of a target representation that can adequately reflect the complexity of phenotyping algorithms. However, the significant surface heterogeneity poses serious problems for any NLP approach to automatically converting raw text phenotyping algorithms to a computable form, in particular the near insuperable problem of using NLP to extract the semantic content of flowchart images.
A limitation of this study is that we are restricted to fourteen phenotyping algorithms. Clearly, claims based on only fourteen algorithms are necessarily qualified, but nevertheless, given the data available (which covers a range of diseases and populations) our study provides a firm foundation for building a computable representation appropriate for EHR-oriented phenotyping algorithms.
We suggest that a manual (or semi-automatic) approach is required to convert eMERGE algorithms into a computable representation. However, in the future, with the availability of a standardized representation language, it will be possible for clinicians, domain experts and informaticians to write new algorithms directly with the aid of special purpose authoring tools that can facilitate algorithm development. In this future scenario, algorithms developed using a standardized authoring tool could be run against EHRs at different sites without requiring retooling or modification.
Conclusions and Further Work
In conclusion, we have analyzed fourteen phenotyping algorithms (generated as part of the eMERGE project) in terms of their constituent data elements, types of logic used and temporal characteristics. We have discovered that while the surface forms of the document differ significantly, the underlying logic used is more homogeneous, with heavy reliance on nested boolean logic, complex temporality and ubiquitous ICD-9 codes.
We aim to use these results to develop a computable model for representing eMERGE style EHR-oriented phenotyping algorithms in the context of the SHARP automatic phenotyping project, and in order to conform to existing standards, we have plans to augment the CDISC-Protocol Representation Model to meet our representational needs.
Acknowledgments
This work was supported by the Strategic Health IT Advanced Research Projects (SHARP) Program (Research Area: 4 — Secondary Use of EHR Data). SHARP is a project of the Office of the National Coordinator for Health Information Technology (Award No: 90TR002/01). The eMERGE Network was initiated and funded by the National Human Genome Research Institute with additional funding from the National Institute of General Medical Sciences through the following grants: U01-HG-004610 (Group Health Cooperative); U01-HG-004608 (Marshfield Clinic); U01-HG-04599 (Mayo Clinic); U01HG004609 (Northwestern University); U01-HG-04603 (Vanderbilt University, also serving as the Administrative Coordinating Center).
Finally, we would like to express gratitude to all those algorithm writers who took the time to respond to our survey.
References
- 1.Raftery J, Kerr C, Hawker S, Powell J. Paying clinicians to join clinical trials: a review of guidelines and interview study of trialists. Trials. 2009;10:15. doi: 10.1186/1745-6215-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel C, Gomadam K, Khan S, Vivek G. TrialX: Using semantic technologies to match patients to relevant clinical trials based on their Personal Health Records. Web Semantics: Science, Services and Agents on The World Wide Web. 2010;8:342–347. [Google Scholar]
- 3.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010;2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 5.Tu SW, Peleg M, Carini S, Bobak M, Ross J, Rubin D, et al. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2010 Sep; doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010 Jun;43(3):451–67. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno-Machado L, Parra E, Henry SB, Tu SW, Musen MA. AIDS2: a decision-support tool for decreasing physicians’ uncertainty regarding patient eligibility for HIV treatment protocols. Proc Annu Symp Comput Appl Med Care. 1993:429–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond P, Sergot M. Computer support for protocol-based treatment of cancer. Journal of Logic Programming. 1996;26:93–111. [Google Scholar]
- 9.Boxwala AA, Peleg M, Tu S, Ogunyemi O, Zeng QT, Wang D, et al. GLIF3: a representation format for sharable computer-interpretable clinical practice guidelines. J Biomed Inform. 2004 Jun;37(3):147–61. doi: 10.1016/j.jbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Sordo M, Boxwala AA, Ogunyemi O, Greenes RA. Description and status update on GELLO: a proposed standardized object-oriented expression language for clinical decision support. Stud Health Technol Inform. 2004;107(1):164–8. [PubMed] [Google Scholar]
- 11.Harkema H, Kaur A, Lisovich A, El Saadawi G. Natural language processing for clinical trial protocol management. AMIA Clinical Research Informatics Proceedings. 2008 [Google Scholar]
- 12.Solti I, Gennari JH, Payne T, Payne T, Solti M, Tarczy-Hornoch P. Natural language processing of clinical trial announcements: exploratory-study of building an automated screening application. AMIA Annu Symp Proc; 2008. p. 1142. [PubMed] [Google Scholar]
- 13.Ramirez AH, Schildcrout JS, Blakemore DL, Masys DR, Pulley JM, Basford MA, et al. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm. 2011 Feb;8(2):271–7. doi: 10.1016/j.hrthm.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony L. AntConc: design and development of a freeware corpus analysis toolkit for the technical writing classroom. 2005 IEEE International Professional Communication Conference Proceedings; 2005. pp. 729–37. [Google Scholar]