Abstract
The CHA2DS2-VASc score is a 10-point scale which allows cardiologists to easily identify potential stroke risk for patients with non-valvular fibrillation. In this article, we present a system based on natural language processing (lexicon and linguistic modules), including negation and speculation handling, which extracts medical concepts from French clinical records and uses them as criteria to compute the CHA2DS2-VASc score. We evaluate this system by comparing its computed criteria with those obtained by human reading of the same clinical texts, and by assessing the impact of the observed differences on the resulting CHA2DS2-VASc scores. Given 21 patient records, 168 instances of criteria were computed, with an accuracy of 97.6%, and the accuracy of the 21 CHA2DS2-VASc scores was 85.7%. All differences in scores trigger the same alert, which means that system performance on this test set yields similar results to human reading of the texts.
Introduction
Unstructured clinical notes contain a wealth of information, some of which is absent from the structured part of the electronic patient record. This has motivated a long-running stream of work on natural language processing applied to clinical texts.[1,2,3] The bottom line of that work is the detection of information elements in clinical texts, encompassing the main types of concepts involved in clinical practice: diseases and other problems,[4] tests, treatments including medication,[5] and their component concepts such as anatomy.
A second level of target information aggregates this elementary information to compute clinical indicators for a patient, for instance their smoking status,[6] obesity status,[7] or the presence of a congestive heart failure (CHF).[8,9] Such information is useful for a variety of applications such as triggering alerts or recruiting patients for clinical trials.[8]
Project Akenaton[10] addresses the extraction of medical information from French free text patient reports in the domain of cardiology, focusing on patients who have a pacemaker. In this domain, a key information element is the CHA2DS2-VASc score,[11] a new recommendation of the European Society of Cardiology. It has been proposed to determine the stroke risk for patients with non-valvular fibrillation.[12] This score is computed from eight criteria. Each criterion counts for 1 or 2 points in the final score: (i) Congestive heart failure or left ventricular dysfunction, 1 pt, (ii) Hypertension, 1 pt, (iii) Age≥75, 2 pts, (iv) Diabetes mellitus, 1 pt, (v) Stroke, transient ischemic attack, or thromboembolism, 2 pts, (vi) Vascular disease (prior myocardial infarction, peripheral artery disease, aortic plaque), 1 pt, (vii) 65≤Age<75, 1 pt, (viii) Sex category, 1 pt if female gender. The final computed score varies from 0 to 9. In conclusion, the higher the score, the higher the risk of thromboembolism. This CHA2DS2-VASc score constitutes one of the elements taken into account when a clinician has to decide whether an anticoagulation therapy is required to prevent potential stroke. Some criteria such as age and sex can be extracted from the hospital information system, while others need a careful analysis of clinical documents. To the best of our knowledge, no previous work has addressed the automatic computation of this score based on clinical texts.
Furthermore, this task is harder than, e.g., computing a score such as the body mass index (BMI): whereas BMI only needs to find the two numeric measures of body weight and height, the CHA2DS2-VASc score needs to assess the presence or absence of concepts that have a more complex definition. For instance, the concept of peripheral artery disease (PAD) encompasses a range of diseases that are more specific than the generic description called PAD. Ontological knowledge is therefore useful to help link the specific disease names that can be found in patient reports to the generic detection of presence or absence of a peripheral arterial disease. We use a module[13] which computes the CHA2DS2-VASc formula based on concepts in an ontology,[14] which allows us to detect the presence of the relevant concepts at the right level of abstraction. The present paper focuses on the natural language processing part of the CHA2DS2-VASc computation pipeline. More detail on the ontology-based module can be found in Dameron et al.[13]
Since the score depends on the presence or absence of concepts for a given patient, a proper treatment of negation and other expressions of modality is important to avoid false detection of negated concepts. Negation and probability processing can be handled in English clinical texts by Chapman’s NegEx system.[15] A more complete detection of other modalities was addressed in the i2b2/VA 2010 challenge[4] task on the detection of assertions on medical problems, which additionally included categories conditional, hypothetical and not associated with the patient. However, NegEx works on English text, and there is no equivalent system available for French clinical texts. We therefore designed a version of NegEx extended to the i2b2 assertion categories and adapted to French.
In this paper, we present the system we designed and implemented to extract medical concepts from clinical records in cardiology, within the framework of thromboembolism risk. We then present its evaluation through a comparison of human reading vs. automatic extraction, focusing on the criteria that are used to compute the CHA2DS2-VASc score. We conclude with a discussion of current results and perspectives for further work.
Background
Previous work has addressed the classification of patients into predefined classes: such classes can be binary, e.g. CHF vs non-CHF,[8,9] or involve a choice among a few disjoint categories, e.g. five categories of smoking status (past smoker, current smoker, etc.).[6] In such cases, the problem can be modeled as a supervised classification task where the system must predict the correct class based on features which represent the patient cases. These features are extracted from the texts by natural language processing methods with a varying degree of sophistication, including some handling of negation.[9]
A characteristic of most work on the classification of patient information is the use of flat lists of terms, often obtained from existing controlled vocabularies (possibly through the UMLS Metathesaurus[16]), aggregated in such a way as to detect each relevant feature. This was the case for instance in the i2b2 medication extraction challenge,[5] where most systems used lists of drug names to detect medications and lists of findings and disease names to find the reason for a prescription. It would be possible to adopt the same approach to detect, e.g., peripheral arterial diseases. However, we opted for a more principled approach in which the natural language processing component deals with the recognition of specific concepts in the texts, and a separate ontological component decides whether a specific concept (e.g., “artérite des membres inférieurs” (lower limb arteritis)) is an instance of a generic concept (e.g., peripheral arterial disease) which instantiates a criterion or not. This avoids the overspecialization of concept detection in the texts, and allows the NLP component to output a more versatile representation where concepts can be used for a larger variety of tasks beyond the specific question of CHA2DS2-VASc score computation.
The supervised classification approach used by many recent systems also looses much relevance in the case of the computation of a formula. In the case of the CHA2DS2-VASc score, the number of possible values (10 values, from 0 to 9) would make it more difficult for supervised classification to take these values as discrete target classes. Besides, the mathematical relation of the score to the underlying features would be lost by a straightforward application of classical feature-based supervised learning methods. We found no compelling argument not to rely on the existing formula to compute the CHA2DS2-VASc score once the basic information elements (or features) are determined, that is, to apply a knowledge-based approach to that task. Our knowledge-based approach to the computation of the CHA2DS2-VASc score therefore departs from other works based on supervised classification of patient reports.
Negation processing has been extensively studied for English-language medical texts.[15,4,17] Recent work transferred NegEx to Swedish by transposing its trigger phrases from English to Swedish.[18,19] Although Swedish and English are both Germanic languages, a simple translation was not enough, because of differences in grammatical features (e.g., gender and number agreement in Swedish), constructs (e.g., do auxiliary in English negations), or word order. French is a Romance language, so transfer to French might raise other issues.
Methods
We need to determine the relevant information elements that will be used to compute the eight criteria on which the CHA2DS2-VASc score computation depends. We then need to design an information extraction system which will identify medical concepts in French clinical documents in this purpose. We consider as a relevant information element a piece of information obtained from a text, more specifically a clinical record, that could be of interest within the framework of thromboembolism risk assessment. The final objective of this framework is to automatically identify patients with risk of thromboembolism attack in case of atrial fibrillation.
With the help of a cardiologist, we defined a list of nine topics the system must focus on to deal with this framework. These are topics around which information elements needed to compute the eight CHA2DS2-VASc score criteria can be found, although there is not a one-to-one correspondence between these topics and the criteria: one topic may contribute to several criteria, and vice-versa. These topics are the following: (i) age of patient, (ii) atrial arrythmia episode, (iii) blood clot or thrombus formation, (iv) arterial embolism, (v) cardiovascular risk factors (tobacco addiction, diabetes, etc.), (vi) heart disease (aortic valve regurgitation, left ventricular ejection fraction (LVEF), left ventricular end diastolic diameter (LVEDD), mitral failure, etc.), (vii) atrial fibrillation duration and characteristics, (viii) rate-lowering drug/treatment and anticoagulation treatment, and (ix) pacemaker and defibrillator information. These topics are general categories which we have to expand to be able to extract the specific types of medical information that are relevant here. Example instances of information elements to extract are given in Table 1.
Table 1:
Relevant information elements for thromboembolism risk identification in French clinical documents.
| French examples | English translation |
|---|---|
| un tabagisme majeur, de l’ordre de 40 cigarettes/jour | a major tobacco addiction, about 40 cigarettes/day |
| poursuivi depuis l’âge de 12 ans... | continued since the age of 12... |
| un cholestérol totalà 2,9 g/l et des triglycéridesà 1,82 | a total cholesterol of 2.9g/l and triglycerides of 1.82g/l |
| g/l avec un LDLà 2,12 g/l. | with an LDL cholesterol of 2.12g/l. |
| la fraction d’éjection est retrouvéeà 44% avec un | the left ventricule ejection fraction is 44% with a left |
| diamètre télédiastolique du ventricule gaucheà 63 mm. | ventricular end diastolic diameter of 63mm. |
| il existe une insuffisance mitrale modérée 1,5/4 | there exists a moderate mitral valve regurgitation 1.5/4. |
Additionally, some drug prescriptions are also a precious clue of patient condition, such as hypertension or other coronary diseases. Extraction of drug prescriptions thus brings additional help to compute the CHA2DS2-VASc score. For instance, if hypertension is not explicitly mentioned but some antihypertensive medication is found in a patient report, the hypertension point can be added to the score.
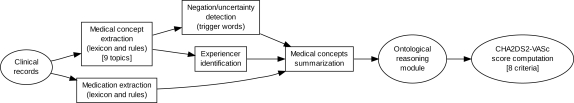
We created a system to extract these information elements using several NLP modules (see Figure 1). Each module of this system is based on two main characteristics: first, the use of a domain-restricted lexicon to identify important terms as well as trigger words, and second, extraction rules to refine concept identification.
Figure 1:
Global architecture of the system used to compute the CHA2DS2-VASc score. NLP modules are inside rectangle shape boxes.
The system first performs a basic sentence segmentation before applying the lexicon and extraction rules. This helps to process the documents at a linguistically sound level of granularity so that syntactic and semantic processes are applied to a controlled input. This system can be categorized as human-knowledge-based, in contrast to machine-learning-based systems, since it relies on human-defined lexicons and extraction rules.
Medical concept extraction
Lexicon
We created a global lexicon composed of 106,639 entries we gathered from three distinct lexicons. Not all entries focus on thromboembolism risk assessment (e.g., we gathered all existing drug names, not only those used in cardiology, assuming that ignoring information is easier than looking for missing information). The three lexicons are the following:
a drug name lexicon we gathered from both professional and general public sources (Vidal,* Doctissimo,† etc.);
a list of medical problems extracted from the Unified Medical Language System (UMLS) Metathesaurus;[16]
and a list of specific cardiological terms provided by a cardiologist.
Each entry in our lexicon contains a medical term, a general category the term belongs to (anatomy, disease, drug, family, laboratory results, procedure), the corresponding concept in a home-made ontology,[14] and the parent of the concept (as found in the ontology). Table 2 presents example medical concepts from each of these categories. The first step of the system uses this global lexicon to identify these medical terms.
Table 2:
Example entries of the global lexicon.
| Category | Examples | Number |
|---|---|---|
| Anatomy | oreillette gauche (left atrium), valve aortique (aortic valve), ventricule gauche (left ventricle), etc. | 5,792 |
| Disease | insuffisance de la valve mitrale (mitral valve regurgitation), lésion aortique (aortic lesion), myxome de l’oreillette gauche (left atrial myxoma), thrombose aortique (aortic thrombosis), etc. | 61,103 |
| Drug | atenolol (atenolol), avk (anti-vitamin k), coumadine (coumadin), héparine (heparin), etc. | 33,639 |
| Family | beau-père (father in law), jumeaux monozygotes (monozygotic twins), mère (mother), etc. | 147 |
| Laboratory results | débit cardiaque (cardiac output), ventilation pulmonaire (pulmonary ventilation), etc. | 602 |
| Procedure | pontage aortique (aortic bypass), valvulotomie mitrale (mitral valvulotomy), etc. | 5,357 |
Extraction rules
To refine medical concepts located in the documents, we defined specific extraction rules, which we implemented using regular expressions. We based these rules upon empirical observation of the clinical documents in our corpus. As we are focusing on thromboembolism risk, we defined a set of rules to deal with 25 cardiological cases.‡ These rules allow us to take into account variant expressions (full word, abbreviation, etc.) and/or different cases of precision (with adjectives or different formulations, etc.).
Negation and modality
As medical information is written in natural language in clinical documents, a basic identification of clinical concepts is not sufficient. Indeed, a system can detect a medical concept in a negated expression (the patient does not exhibit the mentioned problem); natural language can also express information with varying degrees of uncertainty; finally, within specific sections of the clinical documents, such as history of present illness or family antecedents, medical information can involve someone else than the patient (in the case of a hereditary disease). We created three modules to deal with these linguistic cases, as shown on Table 3.
Table 3:
Modules to handle negation and other modalities of the extracted information.
| Module | French examples | English translation |
|---|---|---|
| Negation | ne retrouve pas le moindre œdème des membres inférieurs | does not find the slightest lower limb edema |
| Uncertainty | afin de rechercher une éventuelle ischémie myocardique | in order to find a potential myocardial ischemia |
| Experiencer | un infarctus du myocarde chez son père | a myocardial infarction in her father |
In order to deal with negation, we used the NegEx algorithm.[15] To adapt this algorithm to French, we created a list of 318 negation triggers for French. This trigger list is based on a translation of the existing English triggers and empirical observation of the French cardiology corpus. We also used the 9 major categories of NegEx to categorize negation triggers. Table 4 displays examples of the triggers used in our adapted NegEx system to identify negation in French clinical documents.
Table 4:
French negation triggers used with the NegEx algorithm.
| Category | Trigger |
|---|---|
| Pre negation | absence de (lack of), jamais eu (never had), aucun (no), pas de signes de (no sign of), etc. |
| Pre possible negation | pour écarter (to rule out), etc. |
| Post negation | est écarté (is ruled out), ont été éliminés (have been eliminated), etc. |
| Post possible negation | peut être écarté (can be ruled out), sera écarté (will be ruled out), etc. |
| Conjunction | cependant (nevertheless), sauf (except), etc. |
| Pseudo negation | pas de changement significatif (no significant change), pas sûr de (not certain whether), ne cause pas (does not cause), etc. |
A second module allows us to determine the uncertainty of the expressed information within a sentence. We created a module based upon trigger words of two kinds:
Pre uncertainty trigger words:éventuel (eventual), hypothèse (hypothesis), possible (possible), probable (probable), risque de... (risk of...), dépistage du... (screening of...), prévention du... (prevention of...), recherche du... (search for...), etc.;
Post uncertainty trigger words: suspecté (suspected), comme hypothèse (as hypothesis), etc.
In case of uncertainty, we decided not to extract the medical concept, considering that a human interpretation of such a phrase is needed to decide whether this concept must be considered or not.
Finally, we also designed a module which tries to identify who is the experiencer of a medical problem within a window of 9 words before or after the studied medical problem. This module uses a list of 147 entries from our global lexicon to identify the subject of the disease. It detects whether the mentioned medical problem affects the patient or someone else from their family. If a problem affects someone else than the patient, we do not take this problem into account.
Medical prescription
Our medication extraction module is an adaptation to French of a system we designed for the 2009 i2b2 natural language processing challenge.[5] This challenge was dedicated to medication prescription extraction from clinical documents in English. It aimed at extracting drug names and all related information (dosage, mode of administration, frequency, duration, and reason for prescription). We took advantage of this challenge to develop a medication extraction system.[20] Using a rule-based system, we ranked 8th out of 22 participants with a 0.773 F-measure.
Our system relies on the use of lexicon and extraction rules based on trigger words (abbreviations and expressions for all related information classes). We built three lexicons: (i) a drug lexicon to detect drug names based upon drug names from the UMLS Metathesaurus and therapeutic classes, (ii) signs and symptoms lexicons to identify the reason why a given medication was prescribed, based upon the UMLS Metathesaurus using entries with the “Signs and Symptoms” semantic type and the “MetaMap NLP View” flagged terms, and (iii) lists of abbreviations and expressions to extract drug-related information, where each entry has been associated with the type of information it denotes.
We performed an extraction in several steps, as follows: (i) we split the document into sentences, (ii) we then applied the lexicon to identify drug names within each sentence as an exact match, (iii) we split the sentences into parts, where one part begins with a drug name, and (iv) we searched related information inside each part, considering that related information often follows a drug name, but we also extended the search to the sequence closely preceding the drug name.
We then adapted our system to the French language.[21] We kept the general architecture of the English system (sentence splitting, identification of drug names, and detection of associated information) using a lexicon and rules. We modified the lexicon (gathering drug names, pharmacological substances and abbreviations or spelling variants in French) and the rules (by adapting English rules and adding new rules designed by observation of the development corpus). We also kept the same classes of target information (medication, dosage, mode of administration, frequency, duration, and reason for prescription). We evaluated our French medication extraction system over a test corpus composed of 50 French patient records that we manually annotated as a reference (257 drug names to identify with their related information): it obtained a 0.867 F-measure.
Concept compilation
Having identified medical concepts and prescriptions within clinical documents, the system builds a global XML file that sums up all information extracted from the document; it also completes each kind of information with generic attributes that allow subsequent processing to easily access information items independently of the way information was given in the source clinical document.
For each medical concept, we add specific values denoting the type of concept and the corresponding concept in the Akenaton ontology. Table 5 lists examples of extracted concepts from a clinical document with the additional information we add for each medical concept.
Table 5:
Examples of concepts extracted from a clinical document and additional information for this concept.
| Concept in the document | Type | Concept in the ontology |
|---|---|---|
| Cardensiel | Bradycardisant | Not found |
| Mode AAI safe R | Pacemaker | AAI Mode |
| Pace maker double chambre | Pacemaker | Dual chamber pacemaker |
For each prescription, we also linked each drug name to the ATC (Anatomical Therapeutic Chemical) classification system, indicating both ATC code and ATC general class. Table 6 provides examples of medication prescriptions we extracted from a clinical document. The resulting XML file summarizes the results of the natural language processing modules of our text-based CHA2DS2-VASc computation pipeline.
Table 6:
Examples of prescriptions extracted from a clinical document and additional information for their concepts.
| Medication | ATC Code | ATC Class | Dosage | Frequency | Reason for prescription |
|---|---|---|---|---|---|
| Triatec (ramipril) | C09AA05 | Inhibiteurs de l’enzyme de conversion non associés (ACE inhibitors) | 5 mg | /jour (per day) | — |
| Zanidip (lercanidipine) | C08CA13 | Antagonistes calciques non associés (calcium channel blockers) | 10 mg | /jour (per day) | Hypertension (high blood pressure) |
Computation of CHA2DS2-VASc score
This computation is performed by an ontological reasoning module based on OWL and SWRL.[13] This score computation module completes our full CHA2DS2-VASc computation pipeline. In order to process the output of the NLP modules, the ontological reasoning module focuses on the concept in the ontology given for each extracted concept (see Table 5), and on the ATC code given for each drug name (see Table 6).
We give in Table 7 an example of medical information extracted by our system, and the computed CHA2DS2-VASc point obtained for each extracted concept.
Table 7:
Clinical information extraction processed by our NLP system.
| Clinical record | Antécédents médicaux : HTA. [...] Traitement à l’entrée : PREVISCAN (0-0-1/2). [...] A l’examen clinique de ce jour, il n’y a aucun signe d’insuffisance cardiaque. Medical antecedents: high blood pressure. [...] Treatment on admission: PREVISCAN (0-0-1/2). [...] On today’s physical examination, there is no sign of cardiac failure. |
|
| Medical concept | Extracted concept: | hta (high blood pressure) |
| Concept in the ontology: | hypertension (high blood pressure) | |
| CHA2DS2-VASc point: | 1 | |
| Medical concept | Extracted concept: | insuffisance cardiaque (cardiac failure) |
| Concept in the ontology: | heart-failure | |
| Assertion: | negated | |
| CHA2DS2-VASc point: | 0 | |
Evaluation corpus and setting
Our global corpus is composed of 62 files for patients that attended a cardiology hospital department. Each patient file includes clinical reports (neuro facial radiology and surgical reports), diagnoses (with the corresponding code), and a list of all medical procedures with the corresponding codes. We created a reference corpus composed of 21 patient files. First, the CHA2DS2-VASc score was automatically computed through our complete pipeline for each of the 62 patients. The computed scores ranged from 0 to 7. We then selected at least 2 patients for each value of this score for this reference corpus. This reference corpus was then read by a cardiologist who studied the source patient files and manually recorded all the CHA2DS2-VASc criteria necessary to compute the CHA2DS2-VASc score. These criteria were fed to the CHA2DS2-VASc computation module which produced the CHA2DS2-VASc scores for these patients. These scores constitute the reference we aim to reproduce.
We evaluated the natural language processing pipeline at two levels. First, by comparing its computed criteria with those obtained by human reading of the same clinical texts. Second, by assessing the impact of the observed differences on the resulting CHA2DS2-VASc scores.
Results
For each patient file of the reference corpus, we evaluated the performance of the natural language processing subset of the pipeline by comparing its results with human-based results.
Table 8 displays the differences between the human-based values and the automatically computed values for the criteria: 0 means identity while −1 or 2 is the difference between the two values. It lists, for the three patient files for which a difference was found, its identifier, the number of documents it contains, and the two information elements obtained from the structured part of the record: Age and Sex (although both are generally listed in the texts, their systematic presence in the structured record makes it less useful to use the values found in the texts). It also shows the two CHA2DS2-VASc scores obtained through human (Hum) and fully automatic processing (Auto).
Table 8:
CHA2DS2-VASc computed measures for both human-reading (Hum) and automatic method (Auto). CHF stands for “Congestive Heart Failure”, HTA for “Hypertension”, A2 for “Age>=75”, DIA for “Diabetes mellitus”, S2 for “Stroke”, PAD for “Peripheral Artery Disease”, A for “65<=Age<75, and Sc for “Sex category”.
| Patient | Score | Delta in CHA2DS2-VASc criteria | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Id | # doc | Age | Sex | Hum | Auto | CHF | HTA | A2 | DIA | S2 | PAD | A | Sc |
| 57 | 29 | 79 | M | 7 | 6 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 59 | 16 | 75 | F | 6 | 5 | 0 | 0 | 0 | 0 | 0 | −1 | 0 | 0 |
| 72 | 34 | 82 | F | 6 | 7 | −1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
Discussion
These results show that the CHA2DS2-VASc score computed using the automatically extracted medical concepts is very close to the scores computed through human reading of each document. At the level of criteria, 21 × 8 = 168 criteria had to be computed, among which 164 were exact. This corresponds to an accuracy of 164/168 = 0.976.
Over a total of 21 patients, 18 obtained the same CHA2DS2-VASc score through the fully-automatic and human-based methods, an accuracy of 0.857. Besides, all three different assessments of the score were distant of only one point. Moreover, all involved scores greater than two. All computed CHA2DS2-VASc scores vary from 0 to 7 in our corpus. Traditionally, cardiologists rely on three categories defined over the CHA2DS2-VASc score: a score greater than 2 (where anticoagulation therapy is permanent), a score of one (anticoagulation therapy is recommended), and a null score (anticoagulation therapy is not indicated). The three score errors computed a final score of 6 instead of 7 (patient #57), a score of 5 instead of 6 (patient #59), and a score of 7 instead of 6 (patient #72). Indeed, computational errors are always serious in medicine, but in these cases, the alert raised for these patients is similar; they must take a treatment and the pacemaker alert must be considered as serious. Therefore, system response for all 21 patients would be adequate in this test set: this means that system performance on this test set yields results identical to human reading of the texts.
Closer analysis shows that for patient #59, the automatic approach missed a medical concept which counts for one point (peripheral artery disease): “athérome calcifié non sténosant de la bifurcation” (non stenosing calcified atheroma in the bifurcation) that does not exist in our lexicon; this problem can be easily solved by adding this concept to the lexicon. For patient #72, the automatic approach leads to one excess point in the score. Looking in more detail, we find two errors: one error (linked to the criterion “congestive heart failure” that counts for one point in the score) concerns a medical concept that is not present in the lexicon: we have to add: “décompensation cardiaque globale” (global cardiac decompensation); the second error concerns a concept that has been extracted as being present in the history of the patient “une avc qui n’a pas été confirmée” (a stroke that has not been confirmed) whereas this concept (stroke) is not present; this counts for 2 points in the computed CHA2DS2-VASc score. In consequence, we have to improve the NegEx trigger list for French that we used to deal with this case of negation.
Looking at table columns, we can see that we missed one point twice in the criterion “Congestive Heart Failure” and one point in the criterion “Peripheral Artery Disease” due to medical concepts absent from our lexicon. We also added two points through the criterion “Stroke” because of a problem in our negation processing module.
We performed an evaluation of the negation detection module on the corpus of 21 patients which represents 424 clinical records to process. Over a total of 914 concepts, 59 are negated while 855 are not negated. Our system annotated 79 concepts as negated (among which 53 are correct) and 835 as not negated (among which 809 are correct). We obtained a global F-measure of 0.863.
Missing negations are due to non-existing trigger words in our lexicon (“non accompagnée de” not accompanied by) while false negations are due to an excessive factoring of the negation information from a concept to the following one: “sans gradient intraventriculaire, insuffisance mitrale toujours minime” (without any intraventricular gradient, mitral insufficiency still negligible); in this case, the negation “sans” (without) only focuses on the first concept “gradient intraventriculaire” (intraventricular gradient) while our adaptation also tagged the second concept “insuffisance mitrale” (mitral insufficiency) as negated without taking into account the comma as a clue separating two phrases in the sentence. Preventing the propagation of negation to several concepts is difficult because of the way some clinical records are written, especially when several successive concepts are listed without any punctuation mark to split these entries.
Conclusion
Within the framework of the thromboembolism risk, the CHA2DS2-VASc score allows cardiologists to identify easily potential stroke risk for patients with non-valvular fibrillation. We presented in this paper a system based on natural language processing which automatically extracts medical concepts and prescriptions from French clinical records, taking into account the expressed negation for each concept. These are then used as a way to identify relevant criteria that are part of the CHA2DS2-VASc score. When evaluating the NLP modules, the prescription extraction module obtained a global F-measure of 0.867 while the negation module obtained a global F-measure of 0.863.
We performed an overall evaluation on 21 patients files, based on a comparison of the computed CHA2DS2-VASc score, one score being computed based on criteria extracted by a cardiologist reading the documents, the other score being computed using the automatically extracted medical concepts. This evaluation showed similar results, with an accuracy of 0.976 at the level of the 186 individual criteria and of 0.857 at the level of the CHA2DS2-VASc scores. The observed differences are mainly due to two problems: first, a medical concept that is not present in a lexicon, and second, a lack of precision in negation handling. However, further consideration of practical implications of the obtained scores shows that the three differing scores remain in the same categories and that the same patients would raise alerts.
There is still room for improvement, especially for the adaptation of the NegEx algorithm and related linguistic resources to French language. This part of the work is crucial since a medical concept mentioned in the clinical document can be presented in a negative way (the problem is not present or the problem could occur under certain conditions), and could eventually change the patient results in a totally different way. We noticed that these assertion problems are a key asset to access the meaning of clinical records; in 2010, one task from the i2b2/VA challenge focused on the assertion annotation in order to detect if a medical concept was present, absent, possible, hypothetical, conditional, or associated with someone else.[4] While participating in this challenge,[22] we obtained high results using machine-learning approaches; further work is needed to better adapt the method we used in this challenge from English to French.
Acknowledgments
This work has been funded by the Akenaton project under grant number ANR-07-TecSan-001.
References
- 1.Sager Naomi, Lyman Margaret, Nhn Ng T, Tick Leo J. Medical language processing: Applications to patient data representation and automatic encoding. Methods Inf Med. 1995;34(1–2):140–6. [PubMed] [Google Scholar]
- 2.Friedman Carol, Alderson Philip O, Austin John HM, Cimino James J, Johnson Stephen B. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meystre Stefan M, Savova Guergana K, Kipper-Schuler KC, Hurdle JF. Yearb Med Inform. Shattauer, Stuttgart: 2008. Extracting information from textual documents in the electronic health record: a review of recent research; pp. 128–44. [PubMed] [Google Scholar]
- 4.Uzuner Özlem, South Brett R, Shen S, Duvall Scott L. i2b2/va challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2010 2011 Jun 16; doi: 10.1136/amiajnl-2011-000203. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzuner Özlem, Solti Imre, Cadag Eton. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzuner Özlem, Goldstein Ira, Luo Yuan, Kohane Isaac. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15(1):14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzuner Özlem. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc. 2009;16(4):561–70. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pakhomov Serguei V, Buntrock James, Chute Christopher G. Prospective recruitment of patients with congestive heart failure using an ad-hoc binary classifier. J Biomed Inform. 2005;38(2):145–53. doi: 10.1016/j.jbi.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Friedlin Jeff, McDonald Clement J. A Natural Language Processing System to Extract and Code Concepts Relating to Congestive Heart Failure from Chest Radiology Reports. AMIA Annu Symp Proc; 2006. pp. 269–73. [PMC free article] [PubMed] [Google Scholar]
- 10.Burgun Anita, Temal Lynda, Rosier Arnaud, Dameron Olivier, Mabo Philippe, Zweigenbaum Pierre, Beuscart Régis, Delerue David, Henry Christine. Integrating clinical data with information transmitted by implantable cardiac defibrillators to support medical decision in telecardiology: the application ontology of the Akenaton project. AMIA Annu Symp Proc; 2010. p. 992. (Poster). [Google Scholar]
- 11.European Heart Rhythm Association, European Association for Cardio-Thoracic Surgery. Camm A John, Kirchhof Paulus, Lip Gregory YH, Schotten Ulrich, Savelieva Irene, Ernst Sabine, Van Gelder Isabelle C, Al-Attar Nawwar, Hindricks Gerhard, Prendergast Bernard, Heidbuchel Hein, Alfieri Ottavio, Angelini Annalisa, Atar Dan, Colonna Paolo, De Caterina Raffaele, De Sutter Johan, Goette Andreas, Gorenek Bulent, Heldal Magnus, Hohloser Stefan H, Kolh Philippe, Le Heuzey Jean-Yves, Ponikowski Piotr, Rutten Frans H. Guidelines for the management of atrial fibrillation: the task force for the management of atrial fibrillation of the european society of cardiology (ESC) Eur Heart J. 2010 Oct;31(19):2369–429. doi: 10.1093/eurheartj/ehq278. [DOI] [PubMed] [Google Scholar]
- 12.Lip Gregory YH, Halperin Jonathan L. Improving stroke risk stratification in atrial fibrillation. Am J Med. 2010;123(6):484–8. doi: 10.1016/j.amjmed.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Dameron Olivier, Van Hille Pascal, Temal Lynda, Rosier Arnaud, Deléger Louise, Grouin Cyril, Zweigenbaum Pierre, Burgun Anita. Comparison of OWL and SWRL-based ontology modeling strategies for the determination of pacemaker alerts severity. AMIA Annu Symp Proc; 2011. [Google Scholar]
- 14.Temal Lynda, Rosier Arnaud, Dameron Olivier, Burgun Anita. Modeling cardiac rhythm and heart rate using BFO and DOLCE. International Conference on Biomedical Ontology; 2009. [Google Scholar]
- 15.Chapman Wendy W, Bridewell Will, Hanbury Paul, Cooper Gregory F, Buchanan Bruce G. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 16.Lindberg Donald A, Humphreys Betsy L, McRay Alexa T. The Unified Medical Language System. Methods Inf Med. 1993;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernhard Delphine, Ligozat Anne-Laure. Analyse automatique de la modalité et du niveau de certitude : application au domaine médical. Proceedings of TALN 2011; Montpellier. 2011. [Google Scholar]
- 18.Skeppstedt Maria. Negation Detection in Swedish Clinical Text. Proceedings of the NAACL HLT 2010 Second Louhi Workshop on Text and Data Mining of Health Documents; Los Angeles, California, USA. June 2010; Association for Computational Linguistics; pp. 15–21. [Google Scholar]
- 19.Dalianis Hercules, Skeppstedt Maria. Creating and evaluating a consensus for negated and speculative words in a Swedish clinical corpus. Proceedings of the Workshop on Negation and Speculation in Natural Language Processing; Uppsala, Sweden. July 2010; University of Antwerp; pp. 5–13. [Google Scholar]
- 20.Deléger Louise, Grouin Cyril, Zweigenbaum Pierre. Extracting medical information from narrative patient records: the case of medication-related information. J Am Med Inform Assoc. 2010;17(5):555–8. doi: 10.1136/jamia.2010.003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deléger Louise, Grouin Cyril, Zweigenbaum Pierre. Extracting Medication Information from French Clinical Texts. Stud Health Technol Inform. 2010;160:949–53. (Pt 2), [PubMed] [Google Scholar]
- 22.Minard Anne-Lyse, Ligozat Anne-Laure, Abacha Asma Ben, Bernhard Delphine, Cartoni Bruno, Delger Louise, Grau Brigitte, Rosset Sophie, Zweigenbaum Pierre, Grouin Cyril. Hybrid methods for improving information access in clinical documents: Concept, assertion, and relation identification. J Am Med Inform Assoc. 2011;18(5):588–593. doi: 10.1136/amiajnl-2011-000154. [DOI] [PMC free article] [PubMed] [Google Scholar]