Abstract
Evaluating the potential harm of a drug-drug interaction (DDI) requires knowledge of a patient’s relevant co-morbidities and risk factors. Current DDI alerts lack such patient-specific contextual data. In this paper, we present an efficient model for integrating pertinent patient data into DDI alerts. This framework is designed to be interoperable across multiple drug knowledge bases and clinical information systems. To evaluate the model, we generated a set of contextual DDI data using our local drug knowledge base then conducted an evaluation study of a prototype contextual alert design. The alert received favorable ratings from study subjects, who agreed it was an improvement over traditional alerts and was likely to support clinical management and save physician time. This framework may ultimately help reduce alert fatigue through the dynamic display of DDI alerts based on patient risk.
Introduction
Computerized physician order entry (CPOE) systems frequently employ clinical alerts as a means to influence physician prescribing behavior with the goal of improving patient safety.1–4 Such alerts can take a number of forms, from allergy warnings to overdose checking, but among the most widely implemented are drug-drug interaction (DDI) alerts. These warnings are designed to guide physicians in the appropriate monitoring, modification, or discontinuation of potentially interacting medications. Yet despite their widespread use, the potential value of DDI alerts has been limited by low rates of physician compliance.5,6 Doctors disregard the vast majority of drug interaction warnings, with 49%–96% of all such alerts overridden by the prescriber.6–9 One explanation for this high override rate is the issue of ‘alert fatigue’, in which physicians become less attentive to warnings in the setting of frequent, low-specificity alerts.2,10
Improving the specificity of DDI alerts may in part be achieved by identifying a subset of ‘clinically significant’ interactions and eliminating less critical alerts. Initiatives are currently underway to derive such a subset through expert consensus.11 Yet assessing the clinical significance of a DDI is difficult in the absence of patient context.12 An interaction of little relevance to one patient may be of great relevance to another. Ideally, a clinical decision support system would utilize patient context to dynamically alter the presentation of DDI alerts to highlight patients at higher and lower risk. Achieving this goal requires integration of pertinent, patient-specific data into interaction warnings. At present, the content of a DDI alert remains independent of patient characteristics, with the same information shown regardless of differences in patient age, co-morbidities, or medication history. We sought to address this gap, and in the following paper we present a model for creating customized, patient-specific drug interaction alerts through the dynamic integration of relevant clinical data.
Background
The integration of patient-specific data into clinical alerts is a well-established strategy in clinical decision support (CDS).2,13 Studies have shown that the delivery of relevant patient laboratory results at the time of medication order entry (e.g., displaying creatinine when ordering gentamicin) can improve compliance with drug dosing recommendations.3,14 Similarly, providing pertinent laboratory results on initiation of a new long-term medication (e.g., showing liver functions tests when ordering a statin) has been shown to improve compliance with drug monitoring.15 For such interventions to succeed, however, clinicians and information system developers must work together to define what data elements are considered relevant within the context of a given order. For example, a potassium value may be of great importance when ordering one medication (e.g., lisinopril) but have no relevance when ordering another (e.g., fluoxetine). Thus, for each medication or triggering order, a unique set of pertinent data elements must be defined.
Drug-drug interaction alerts operate under the same principle. In the setting of a potential interaction, what patient data is of interest to a physician will depend on the particular drugs involved and the associated adverse effects. Thus, to create DDI alerts containing meaningful patient-specific content, these relevant data elements must be defined for each interaction. Creating these definitions is not a simple undertaking however. We must consider the inconsistent format of existing drug interaction knowledge as well as sheer volume of interactions to be addressed.
Most CPOE systems rely on DDI data provided by a handful of large knowledgebase vendors. While the individual data elements that comprise a DDI alert vary slightly from one vendor to the next, the majority of alerts contain the following components: interacting drugs, severity of interaction, clinical effects, predisposing conditions, pharmacologic mechanism, recommended actions, and references.16 This information is generally provided in text-format rather than as coded “computable” data, and it lacks conceptual mappings to standardized terminologies such as SNOMED-CT. Due to this lack of standardization, a wide range of terms may be used to describe a given clinical effect. For example, a number of drug interactions may cause bleeding in a patient; but this effect is described variably as ”hemorrhage,” “bleeding risk,” “coagulation defect,” and so forth. The lack of consistent terminology makes it difficult to aggregate all interactions of a certain type. Such aggregation is of value because it allows the “batch” assignment of relevant data elements to entire classes of interactions rather mapping one interaction at a time. For example, we may wish to designate liver function tests as a “relevant lab” for all DDIs known to cause liver damage. But without a standard term for liver damage, we cannot aggregate this group of interactions in an automated fashion.
Anothe r significant challenge is the sheer number of interactions to be reviewed. Typical DDI compendia list well over 1000 drug interactions.16 For example, Drug Interaction Facts contains 1772 listings while the National Drug Data File contains 1846 DDI monographs.17,18 These numbers will continue to increase as new DDIs emerge in the literature. Given this large and growing body of information, attempting to define relevant data elements for every interaction is a daunting task. One approach is to focus solely on interactions labeled as ‘severe,’ yet prior research has shown a wide variability among drug information compendia in defining severity.16,19 Furthermore, interactions of even moderate severity may be clinically significant depending on patient context. Thus, we ideally want to assess the full breadth of known interactions but to do so in an efficient manner.
In the current paper, we address the aforementioned challenges by presenting a generalizable, sustainable model for integrating patient-specific data into drug-drug interaction alerts. This framework is known as the context-aware drug-drug interaction (CADDI) model and is designed to be interoperable across multiple drug knowledge bases and clinical information systems. We will describe the process for mapping drug interactions to relevant clinical data elements then present a web service that utilizes these data to generate patient-specific DDI alerts. Finally, we will report the results of an evaluation study of a prototype CADDI alert design.
Methods
Drug Knowledge Source
As the source of our drug interaction data, we used an in-house knowledge base developed and maintained by Regenstrief Institute for use in its CPOE system (‘Gopher’). This database is regularly reviewed and updated by a pharmacy team and has been an integral part of Gopher’s decision support operations for over 25 years. The database contains 601 distinct DDIs, including interactions between individual drugs and between drug classes.
Mapping Strategy
We sought to assign to each DDI a set of concepts specifying the patient data elements to be displayed when the alert is triggered. Rather than making these assignments on a per-DDI basis, we performed batch assignments according to clinical effect. So for example, all drug interactions causing ‘bleeding’ would be associated with relevant concepts such as hemoglobin, platelets, and PT/INR. To identify and standardize the effects of each interaction, we utilized the Medical Dictionary of Regulatory Activities (MedDRA), a hierarchical terminology used in drug safety. Because MedDRA contains some non-clinical concepts, we first reviewed the 76 MedDRA semantic types and eliminated 44 that were felt unlikely to be associated with adverse effects. Using the remaining 32 types, we selected 51599 MedDRA terms as our core set of clinical concepts. We then used natural language processing (regular expressions and stem matching) to search for these concepts in the ‘clinical effect’ section for each interaction in our knowledge base. We identified MedDRA terms in 489 of 601 interactions, with a total of 93 unique clinical effects represented. Table 1 shows the 12 most common effects, accounting for 199 (33%) of the interactions in our database.
Table 1.
Most common clinical effects extracted from drug interactions in the Gopher database.
| Clinical Effect | # of DDIs |
|---|---|
| Arrhythmia | 35 |
| Increased Anticoagulant Effect | 27 |
| Torsade de Pointes | 26 |
| Myopathy | 24 |
| Rhabdomyolysis | 15 |
| Digoxin Toxicity | 15 |
| Nephrotoxicity | 12 |
| Hypotension | 11 |
| Hypertension | 10 |
| Bleeding | 8 |
| Death | 6 |
| Cardiotoxicity | 5 |
| Hyperkalemia | 5 |
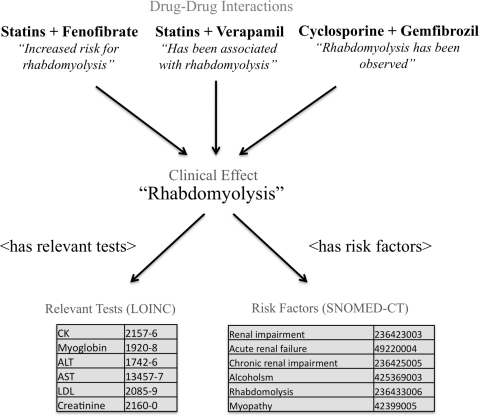
For 11 of these 12 clinical effects (‘death’ was excluded), we selected a set of relevant data elements appropriate for display at the time of physician alerting. Data elements were grouped into two categories: relevant tests and predisposing conditions. Tests were coded to the Logical Observation Identifiers Names and Codes (LOINC) terminology. Conditions were coded to Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT). Figure 1 shows an example of the interactions, concepts, and relationships for the clinical effect ‘rhabdomyolysis’.
Figure 1.
Sample relationships between drug interactions, clinical effects, and related data elements in the context-aware drug-drug interaction (CADDI) database
Prototype Service for Delivering CADDI Alerts
Once the interactions, clinical effects, and related data were mapped and stored in the database, we designed a prototype web service for generation of CADDI alerts. For our source of patient data, we utilized the Continuity of Care Document (CCD). The CCD is a patient summary specification based on the HL7 Clinical Document Architecture. CCDs play an important role in health information exchange and are increasingly offered as a format for export of patient data from electronic medical record systems. A fully semantically coded (‘Level 3’) CCD, such as that generated by our institution, provides patient medications, diagnoses, and laboratory results in structured form using standardized terminologies.
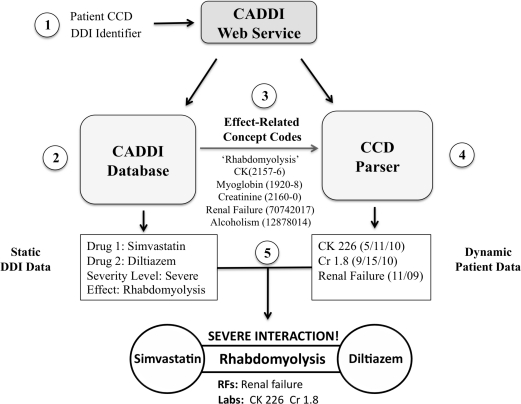
Figure 2 depicts the steps in the creation of a CADDI alert using the example of an interaction between simvastatin and diltiazem. First, the web service receives as its inputs a patient CCD and a drug interaction identifier (e.g., a Gopher DDI ID). Second, the service uses this identifier to query the CADDI database and retrieve static information about the DDI such as the interacting drug classes, severity level, and clinical effects. Third, based on the clinical effect (e.g., rhabdomyolysis), the LOINC and SNOMED-CT codes for all relevant laboratories and predisposing conditions are retrieved from the database and passed to a CCD parser. Fourth, the CCD parser uses these codes to search the patient’s record and retrieve any matching labs or diagnoses. Finally, the service combines the static DDI information from the CADDI database with the dynamic patient-specific data from the CCD to deliver an integrated contextual alert.
Figure 2.
Steps in the generation of a context-aware drug-drug interaction (CADDI) alert.
Prototype Alert Design
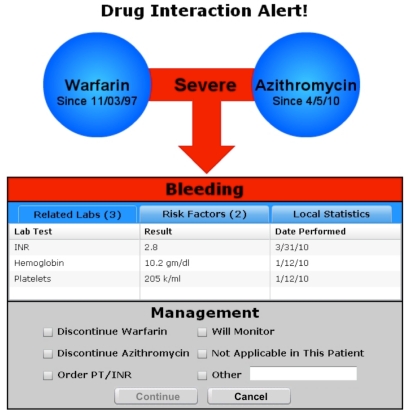
Having created a model for integrating patient specific data into DDI warnings, we developed and evaluated a prototype alert design. Few studies have explicitly explored the visual design of drug-drug interaction alerts, but several studies have looked at the design of clinical alerts in general. Feldstein et al found that physicians placed a high priority on alerts being brief, clear, and easy to navigate with minimal mouse clicks.20 A study by Krall and Sittig found that users were sensitive to multiple keystrokes, excess screen switching, and frequent movement from mouse to keyboard.21 Phansalkar et al performed a human factors review of alerting principles and made several recommendations including coding alert priority using color and position, perceptually grouping related information to assist in decision-making, and tailoring alerts to individual patients and users.22 After considering this literature, we began an iterative alert design process. Our goal was to create a succinct, textually sparse, and graphically strong alert. Our initial designs were refined through critiques by experts in human-computer interaction as well as review by clinical peers. An example of the final prototype design is shown in Figure 3.
Figure 3.
Prototype design of a context-aware drug-drug interaction alert.
Usability Evaluation
The goal of our evaluation study was to gather clinician perspectives on whether the addition of contextual data to drug interaction alerts would improve physician satisfaction, support clinical management, and save time. We also sought to measure usability factors specific to our prototype including its clarity, organization, and efficiency.
Participants consisted of 11 physicians and 1 senior pharmacist. Clinical experience ranged from 3 to 20 years, and all subjects except the pharmacist were currently engaged in clinical practice and were seeing patients at least one-half day a week. All subjects had prior experience with CPOE and electronic reminders. The usability evaluation consisted of task analysis using a think-aloud protocol. Subjects were informed that they were participating in an evaluation of a new clinical alert design and were thus aware of the focus of the study. After providing consent, the subjects were presented with an interactive mock CPOE interface and instructed to prescribe a set of medications for two different sample patients. During the course of ordering these medications, two contextual DDI alerts were triggered and displayed to the subject. The displayed alerts were embedded in the prototype environment and not generated dynamically via the CADDI web service. The triggering pairs of medications were 1) azithromycin and warfarin and 2) verapamil and simvastatin.
After the subjects provided their initial response to the alert, an observer asked them to perform a standardized set of tasks to evaluate the drug interaction. These tasks included identifying the interacting medications, the clinical effect and severity, and the relevant patient labs and risk factors. The subject was also asked to state their final clinical decision whether to accept or override the alert. For each task, a “correct” answer was established by the investigator (JDD) and validated with an independent physician. The observer questioned the participants regarding which data elements in the alert were most helpful, least helpful, and whether any information was missing. Upon completion of the two patient scenarios, the subjects were presented with a 17-question survey addressing the overall usability and utility of the CADDI alert prototype.
The types of data collected were 1) task success rates, 2) subjective user opinions as expressed during task performance, 3) responses to the post-test survey questions. We performed descriptive statistics on physician ratings of prototype usability and clinical utility. For the qualitative data, we aggregated subject responses and grouped them into design recommendations and new feature suggestions.
Results
Task Completion
The tasks and their completion rates are shown in Table 2. In the first scenario, subjects were able to find most information easily, but some had difficulty identifying the patient’s risk factors for the interaction. Successfully answering this question required clicking on the Risk Factors tab to reveal the data (see Figure 3). Some users did not see the tab or did not realize it was selectable. In the second scenario, users were more successful at identifying risk factors, but many missed a question regarding data in the alert that should reduce the level of clinical concern (‘attenuating information’). This question was an indirect reference to the duration of therapy—the patient had been taking both medications for many years and most physicians would be reassured by this fact. However, only 58% of subjects made this connection.
Table 2.
Success rate in retrieving information from context-aware drug-drug interaction alerts.
| Tasks | % Successful |
|---|---|
| Patient 1: Warfarin + Azithromycin (High Risk Scenario) | |
| 1. Identify the drugs involved | 100.00% |
| 2. Identify the clinical effect / severity | 91.70% |
| 3. Identify patient labs relevant to this DDI | 83.30% |
| 4. Identify patient risk factors for this DDI | 66.70% |
| 5. Identify any attenuating information provide by this alert | 91.70% |
| 6. Choose the appropriate management for this alert | 83.30% |
| Patient 2: Diltiazem + Simvastatin (Low Risk Scenario) | |
| 1. Identify the drugs involved | 100.00% |
| 2. Identify the clinical effect / severity | 91.70% |
| 3. Identify patient labs relevant to this DDI | 100.00% |
| 4. Identify patient risk factors for this DDI | 83.30% |
| 5. Identify any attenuating information provide by this alert | 58.30% |
| 6. Choose the appropriate management for this alert | 91.70% |
Post-Task Analysis of Usability and Clinical Utility
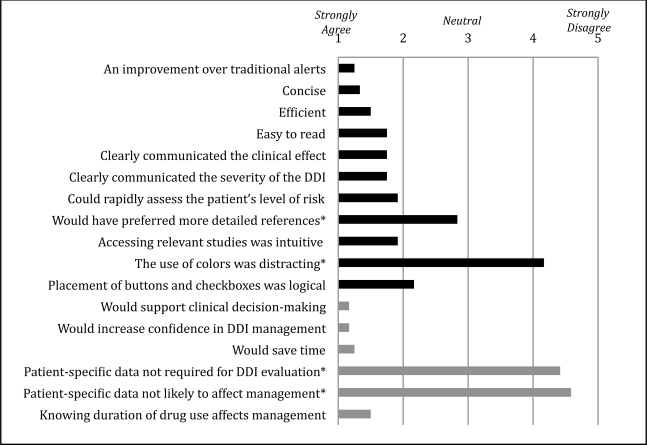
Figure 4 shows summary results of the post-task survey scored on a 5-point Likert scale with 1 being ‘Strongly Agree’ and 5 being ‘Strongly Disagree’. The survey found high levels of support for the integration of relevant patient data into DDI alerts. All subjects agreed or strongly agreed that CADDI alerts were an improvement over traditional alerts (mean score of 1.3). Looking specifically at the design and usability of the prototype, high marks were given to the alerts’ conciseness (1.3), efficiency (1.5), readability (1.8), and clear communication of data (1.8). Two areas of minor criticism were the layout of buttons and checkboxes and the lack of a more detailed reference section.
Figure 4.
Mean user responses regarding usability (black) and clinical utility (gray) of the context-aware drug-drug interaction alert prototype. * denotes a negatively worded question.
In exploring the utility of CADDI alerts, the survey focused on how the availability of patient-specific data would affect DDI evaluation and management. Here the physicians gave very high marks, agreeing strongly that CADDI alerts would support clinical decision making (1.2), increase confidence in management of drug interactions (1.2), and save time (1.3). As a consistency check, we also asked two negatively worded questions, dismissing the importance of patient-specific data in evaluating and managing drug interactions. Physician strongly disagreed with both these statements (4.4 and 4.6 respectively).
Analysis of User Comments
During the course of task completion, physicians were encouraged to make comments on the alert design. Overall response was highly positive (with statements such as “this is really cool” or “when will this be available?”), but the majority of comments were focused on design flaws and missing features. The most commonly cited design concerns were the unnecessary use of tabs (7 subjects), small size of the patient data box (3 subjects), and lack of salience of the clinical effect (3 subjects). The most common feature suggestions were to provide the normal ranges for labs (6 subjects), to offer alternative treatment options (4 subjects), to display whether a DDI is dose-dependent or idiosyncratic (4 subjects), and to provide more details about severity and clinical effect (3 subjects).
Discussion
In this paper, we demonstrated a successful model for integrating relevant, patient-specific data into drug-drug interaction alerts. We further showed that such warnings are of perceived value to physicians and are viewed as likely to save time and improve DDI management. Our prototype alert design, notable for its compact graphical form, was also viewed favorably, receiving high ratings for efficiency and clarity and was overall felt to be an improvement over traditional DDI alerts. In this section, we explore the specific strengths and limitations of the CADDI model as well as discuss its potential application to the challenge of reducing alert fatigue.
Replicability, Interoperability, and Maintainability
Our goal was to provide a way to integrate relevant patient data into DDI alerts in a manner that is replicable across multiple drug knowledge bases, interoperable with multiple electronic medical record systems, and maintainable as new drugs and interactions emerge. The CADDI model achieves these three objectives.
The replicability of the model stems from its use of natural language processing and a standardized adverse event terminology (MedDRA) to convert text-based descriptions into coded clinical effects. This process can be applied to any DDI knowledge base (vendor or homegrown) that provides a section on clinical effects. Thus, while in our particular project we linked local DDI identifiers to relevant patient data, the process would require only minimal additional time to link vendor DDI identifiers as well. Once a knowledge vendor’s DDIs have been mapped to their coded clinical effects, contextual alerts can be generated without further effort. The necessary mappings between the clinical effects and relevant laboratory and diagnosis codes have already been created and are independent of the DDI knowledge source.
The interoperability of the CADDI model stems from its use of standardized terminologies such as LOINC and SNOMED-CT to define the relevant data elements for each clinical effect. Thus, any system able to provide patient data using these standard terminologies should be capable of generating contextual DDI alerts. CCD-based decision support strategies, such as that employed by the Clinical Decision Support Consortium23, would be a natural fit for our model, but CADDI can also be integrated into traditional CPOE systems. Additionally, while we chose to use LOINC and SNOMED-CT to represent relevant data elements, the model is inherently extensible such that local laboratory or diagnosis concepts could be used if a particular vendor or institution wished to perform these mappings.
In terms of its long-term maintainability, the CADDI model benefits from the batch aggregation and mapping of relevant data elements according to clinical effect rather than by individual drug-drug interaction. As discussed, with thousands of known drug interactions it is impractical to create a system requiring ongoing review of every new and changed DDI as it emerges. However, all of these DDIs are associated with a comparatively small number of clinical effects (93 in our knowledge base). The CADDI model employs manual expertise only for the mapping of relevant data elements to this small number of clinical effects. And once these effects are mapped, changes are likely to be rare (primarily based on new LOINC or SNOMED-CT codes rather than changes to fundamental clinical relationships). Thus, ongoing manual maintenance of the CADDI database should be minimal even as new interactions emerge. When a DDI appears, the only required step is to extract the clinical effects by natural language processing. All data elements relevant to this DDI are then automatically mapped based on the existing relationships for these clinical effects.
Lessons from the Prototype Design
The evaluation study confirmed our expectation that physicians would see clinical value in integrating patient data into DDI alerts. Physicians rated CADDI as likely to save time and improve DDI management. Both findings are likely due to the reduced need to explore the patient’s chart to determine potential risk. Another factor that may have lead to the perception of time savings was the alert’s minimalist style. Textual content was sparse, reducing both scanning and reading time. Yet some providers rejected this approach and wanted more detailed information regarding clinical effects and interaction severity. This finding suggests that maintaining succinct ‘headline’ information while providing details on demand would be a more successful design strategy. This approach is consistent with that recommended by Phansalkar et al in their review of human factors principles in clinical alerting.22
Graphics were used in our prototype to augment the impact and clarity of the alert content. Based on the survey results, these additions were not a distraction and did not lessen readability. A few subjects commented that the visual aesthetics made the alert more prominent. However, the long-term benefit of incorporating graphical elements is unclear. Over time providers may become habituated to such designs, and further research is necessary to determine whether graphical images capture physician attention on a consistent basis.
Limitations
Several limitations of the CADDI model should be noted. First, the model is reliant on capturing coded clinical effects from drug interaction knowledge bases using natural language processing. However, some interactions do not list specific clinical effects or their effects may not be accurately captured by our NLP approach. We are currently conducting performance analysis to refine our data capture methodology. Another limitation of the model is the challenge of comprehensively identifying LOINC and SNOMED-CT codes for clinical concepts. This work requires both clinical expertise and an understanding of terminology structures and hierarchies. For this project we performed data mappings for only 11 of 93 clinical effects, so additional mapping would be necessary to create a comprehensive database.
Our prototype study had several limitations as well. It was relatively small in size with only 12 subjects. However, the data collected from this group showed remarkable consistency of themes and critiques, suggesting that we did indeed capture user perception accurately. Another study limitation is the lack of a direct comparator in the form of traditional DDI alerts. We relied on user’s existing perceptions of DDI warnings, which may have been highly variable, to characterize whether CADDI alerts were an improvement. Finally, our study was conducted in a laboratory environment rather than a clinical setting. While this may have sacrificed some clinical realism, the slower pace of the laboratory allowed for more detailed analysis of the prototype design and yielded robust commentary from study subjects.
Future Work
The CADDI model is a first step towards our goal of reducing alert fatigue through the dynamic display of clinical alerts. Currently, CADDI adds relevant patient data to an alert but does not affect whether or not the alert is displayed. The next stage of our work is to define thresholds for highlighting or suppressing warnings based on patient risk factors. For example, in a patient with a low calcium level, interactions causing arrhythmia may be more prominently displayed. Conversely, in a patient with a low potassium level, interactions causing hyperkalemia may be diminished or hidden altogether. Further research is necessary to define and validate these thresholds as well as determine the proper algorithms for dynamic suppression. Additionally, we are currently preparing a trial of CADDI alerts in the clinical environment to evaluate impact on physician adherence and alert fatigue. Ultimately, we hope the CADDI model will reduce the overall volume of clinical alerts while ensuring that the most important safety warnings for individual patients are preserved.
Acknowledgments
This work was performed at the Regenstrief Institute, Indianapolis, IN, and was supported in part by grant 5T 15 LM007117 from the National Library of Medicine. We would like to thank Dr. David Shepherd for his assistance with clinical validation.
References
- 1.Teich JM, Merchia PR, Schmiz JL, et al. Effects of Computerized Physician Order Entry on Prescribing Practices. Arch Intern Med. 2000;160(18):2741–2747. doi: 10.1001/archinte.160.18.2741. [DOI] [PubMed] [Google Scholar]
- 2.Kuperman GJ, Bobb A, Payne TH, et al. Medication-related clinical decision support in computerized provider order entry systems: a review. J Am Med Inform Assoc. 2007;14(1):29–40. doi: 10.1197/jamia.M2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garg AX, Adhikari NKJ, McDonald H, et al. Effects of Computerized Clinical Decision Support Systems on Practitioner Performance and Patient Outcomes: A Systematic Review. JAMA. 2005;293(10):1223–1238. doi: 10.1001/jama.293.10.1223. [DOI] [PubMed] [Google Scholar]
- 4.Kaushal R, Shojania KG, Bates DW. Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review. Arch. Intern. Med. 2003;163(12):1409–1416. doi: 10.1001/archinte.163.12.1409. [DOI] [PubMed] [Google Scholar]
- 5.Schedlbauer A, Prasad V, Mulvaney C, et al. What Evidence Supports the Use of Computerized Alerts and Prompts to Improve Clinicians’ Prescribing Behavior? J Am Med Inform Assoc. 2009;16(4):531–538. doi: 10.1197/jamia.M2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Sijs H, Aarts J, Vulto A, Berg M. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc. 2006;13(2):138–147. doi: 10.1197/jamia.M1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weingart SN, Toth M, Sands DZ, et al. Physicians’ decisions to override computerized drug alerts in primary care. Arch. Intern. Med. 2003;163(21):2625–2631. doi: 10.1001/archinte.163.21.2625. [DOI] [PubMed] [Google Scholar]
- 8.Nightingale PG, Adu D, Richards NT, Peters M. Implementation of rules based computerised bedside prescribing and administration: intervention study. BMJ. 2000;320(7237):750–753. doi: 10.1136/bmj.320.7237.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne TH, Nichol WP, Hoey P, Savarino J. Characteristics and override rates of order checks in a practitioner order entry system. Proc AMIA Symp. 2002:602–606. [PMC free article] [PubMed] [Google Scholar]
- 10.Ash JS, Berg M, Coiera E. Some unintended consequences of information technology in health care: the nature of patient care information system-related errors. J Am Med Inform Assoc. 2004;11(2):104–112. doi: 10.1197/jamia.M1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anon Advancing Clinical Decision Support |RAND. Available at: http://www.rand.org/health/projects/clinical-decision-support.html.
- 12.Glassman PA, Simon B, Belperio P, Lanto A. Improving recognition of drug interactions: benefits and barriers to using automated drug alerts. Med Care. 2002;40(12):1161–1171. doi: 10.1097/00005650-200212000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Teich JM, Osheroff JA, Pifer EA, Sittig DF, Jenders RA. Clinical Decision Support in Electronic Prescribing: Recommendations and an Action Plan. J Am Med Inform Assoc. 2005;12(4):365–376. doi: 10.1197/jamia.M1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raschke RA, Gollihare B, Wunderlich TA, et al. A computer alert system to prevent injury from adverse drug events: development and evaluation in a community teaching hospital. JAMA. 1998;280(15):1317–1320. doi: 10.1001/jama.280.15.1317. [DOI] [PubMed] [Google Scholar]
- 15.Raebel MA, Lyons EE, Chester EA, et al. Improving laboratory monitoring at initiation of drug therapy in ambulatory care: a randomized trial. Arch. Intern. Med. 2005;165(20):2395–2401. doi: 10.1001/archinte.165.20.2395. [DOI] [PubMed] [Google Scholar]
- 16.Vitry AI. Comparative assessment of four drug interaction compendia. Br J Clin Pharmacol. 2007;63(6):709–714. doi: 10.1111/j.1365-2125.2006.02809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comparisons F. Drug Facts and Comparisons 2009. Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 18.Anon . NDDF Database. San Francisco, CA: First Databank; 2010. [Google Scholar]
- 19.Fulda TR. Disagreement Among Drug Compendia on Inclusion and Ratings of Drug-Drug Interactions. Current Therapeutic Research. 2000;61:540–548. [1] [Google Scholar]
- 20.Feldstein A, Simon SR, Schneider J, et al. How to design computerized alerts to safe prescribing practices. Jt Comm J Qual Saf. 2004;30(11):602–613. doi: 10.1016/s1549-3741(04)30071-7. [DOI] [PubMed] [Google Scholar]
- 21.Krall MA, Sittig DF. Clinician’s assessments of outpatient electronic medical record alert and reminder usability and usefulness requirements. Proc AMIA Symp. 2002:400–404. [PMC free article] [PubMed] [Google Scholar]
- 22.Phansalkar S, Edworthy J, Hellier E, et al. A review of human factors principles for the design and implementation of medication safety alerts in clinical information systems. Journal of the American Medical Informatics Association. 2010;17(5):493–501. doi: 10.1136/jamia.2010.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middleton B. The clinical decision support consortium. Stud Health Technol Inform. 2009;150:26–30. [PubMed] [Google Scholar]