Abstract
Developing effective methods to enable the practice of personalized medicine is a national priority for translational science. By leveraging modern genotyping technology and health information technologies, prescribing therapies based on genotype becomes an achievable goal. Within this manuscript, we describe the development, implementation, and piloting of a surveillance tool to assure the quality of clinical decision making in the context of new pharmacogenetic information. The surveillance tool allows a quality assurance (QA) team to review significant genotyping results and deliver focused educational interventions to providers. We report on the first eight patients undergoing genotyping to support antiplatelet therapy selection after drug-eluting stent placement. The collected pilot data supports an informatics approach to QA process management, as our tool delivered actionable patient information. It also enabled providers to tailor antiplatelet therapy to individual patients’ genotypes. Our expectation is to continue collecting surveillance reports to perform an in-depth analysis of our tool.
Introduction
Genomic medicine has continued to rapidly grow, and is accompanied by burgeoning expectations of individualized patient therapy in the form of “personalized medicine.”1–8 The effective practice of personalized medicine requires deployment of robust translational science tools, which leverage modern genotyping technology and a sophisticated information architecture to deliver drug metabolism genotypes to the prescribing provider1, 3, 5–7, 9, 10. To provide patient-specific clinical decision support (CDS), electronic medical records (EMR) and order entry systems must accurately distill complex pharmacogenetic information into an actionable form of information. However, several practical barriers complicate the successful delivery of decision support to the right provider at the right time. First, the methods to measure drug metabolism genotypes frequently require one week or more to return a result, a period in which patients may transition to another site of care or undergo a hand-off to a second provider team. Secondly, providers are frequently unfamiliar with drug-genotype associations4, 6–8, 11 and may not respond reliably to direct CDS. As one potential solution to these scenarios, we developed a surveillance tool to assure the quality of drug decision-making in the context of new pharmacogenetic information. The tool enables a surveillance team to evaluate providers’ response to CDS, and enables researchers to retrospectively analyze the quality of clinical decision-making. In this paper, we describe our tool development process, the rationale for our design decisions, and results from a pilot of our system. We conducted the pilot on patients routinely genotyped prior to undergoing cardiac catheterization, with the expectation that patients would require post-procedure antiplatelet therapy and potentially require genotype-directed drug selection or dosing12–14. We conclude with a discussion of the challenges of implementing personalized medicine in routine practice and our expectations for the future role of information systems.
Background
With the increasing adoption of electronic health records and growing sophistication in various other health information technologies (HIT)9, 15–18, a deluge of information now awaits clinicians in current healthcare settings5, 15, 17–19. The data being reported by various pharmacy, laboratory, medical imaging, and other HIT systems is growing in both volume and complexity5, 9, 17–19. In an attempt to mitigate the cognitive burden generated by the rapid expansion of clinical data19, informaticians have developed diverse approaches and frameworks to better organize information 5, 7, 9, 10, 17, 18, 20–22. The emergence of clinically relevant genetic testing has only added to the exponential growth in clinically relevant data2, 5, 19, 20 mandating continued informatics advancements.
A national priority for translational science is the development of effective methods to empower care providers to practice personalized medicine3–5, 7, 8, 10, 20, 21, 23. Meeting the expectations of personalized medicine requires a feasible method for providing patient-specific pharmacogenetic information to clinicians in an actionable manner3, 4, 7, 20. Achieving this objective requires going beyond CDS applications that remain static in their functionality5. Personalized medicine requires a CDS backbone that is responsive to the user5, 6 and one that can dynamically adjust with changes in any of its associated HIT systems5. According to Kawamoto et al., “new genomic interventions, like any new medical intervention, will remain substantially underutilized for many years unless a robust infrastructure is established for supporting their appropriate use.”5 Specifically, informatics architecture for personalized medicine tools must provide adequate mechanisms for QA oversight, ongoing post-surveillance processes for all patients receiving personalized medicine “interventions,”5 and the ability to remain synchronized with pharmacogenomic research findings4–6, 21.
Moreover, as with other medical interventions,15–18, 24 QA informatics for personalized medicine must ensure that care providers receive clinical decision-making information in an efficient and timely manner. This is particularly challenging considering the clinician’s HIT landscape often lacks this functionality for even standard diagnostic testing18. Harnessing the capabilities of a clinical environment’s existing HIT architecture and then augmenting it to support the voluminous data generated for personalized medicine is a non-trivial task3–5, 20.
Our pilot study focuses on clinical decision-making surrounding antiplatelet therapy selection for patients receiving a drug-eluting stent (DES). Interventionalists typically prescribe clopidogrel to patients following the DES placement procedure12–14, 25–30. Notably, clopidogrel’s pharmacologic properties can be highly dependent on a patient’s genotype. Specifically, CYP2C19 allele variation affects clopidogrel metabolism13, 14, 29, 30. Patients lacking the *2, *3, or *17 allele for CYP2C19 are designated “extensive metabolizers.” Those with one *2 or *3 receive the “intermediate metabolizer” designation, while patients with two *2 or *3 alleles are categorized as “poor metabolizers.” Patients carrying a single *17 allele are assigned as “ultra-metabolizers” and a combination of *17 with either *2 or *3 reflects an “unknown” status14.
Early recognition of a patient’s CYP2C19 allele status can guide physicians toward selecting a more appropriate dosage or alternative antiplatelet medication31. Studies demonstrate an increased risk of major adverse cardiovascular consequences, such as in-stent thrombosis, for both “poor metabolizers” and “intermediate metabolizers” receiving sub-optimal dosages of clopidogrel13. Since March 2010, an FDA warning accompanies clopidogrel packaging, encouraging clinicians to account for CYP2C19 genotypes during antiplatelet therapy decision making13, 29. Typical recommendations include either switching the patient to a comparable medication, such as prasugrel13, 31, or increasing the prescribed clopidogrel dosage13.
Methods
Setting
We conducted our pilot at Vanderbilt University Medical Center (VUMC), a tertiary care, academic facility, with 50,000 annual admissions and ∼800 beds. Care providers at VUMC utilize a locally developed and maintained EMR and computerized provider order entry (CPOE) system. These systems have been evolving over the past 15 years and feature a highly integrative CDS architecture32–34.
System Overview
In designing our QA tool, we gave considerable thought to appropriate integration with the existing HIT infrastructure. We aimed to provide an application that was not intrusive or counter-productive to clinical workflows, but still offered sufficient oversight to all clinical decision making surrounding genotype directed therapy. We also wished to facilitate ongoing institution-wide genomic medicine research efforts, and the design emphasized scalability and interoperability.
Design Considerations
Currently, our institution has begun routine pharmacogenetic testing of patients preparing to undergo coronary angiography. Physicians generally prescribe post-procedure antiplatelet therapy immediately following the intervention, with expectation to continue the medication for 12 months. With test result turnaround averaging one week, a new workflow process was required to follow-up on patients who have transitioned to another setting or care team. A QA team was convened, consisting of a pharmacist and a nurse with extensive experience in goal directed therapy as leaders of the institution’s “Coumadin Clinic.” The QA process complemented traditional computerized reporting mechanisms in the EMR. However, EMR based reporting lacked the functionality to simultaneously inform the responsible care provider of relevant results and offer an efficient means for therapy modification on a discharged patient, who may receive longitudinal care within another health system. Consequently, meeting the ideal window for therapeutic intervention becomes complex. This is of particular concern for personalized medicine interventions, as the optimum benefit of genotype-driven prescribing depends on meeting specific time constraints4, 7, 12–14, 21.
Over time, pharmacogenetic assays will generate large volumes of results, requiring additional considerations in QA tool design. Manually parsing and interpreting the formidable amount of results is problematic and associated with considerable cognitive burden 19. Healthcare personnel tasked with QA need reliable, comprehensible, aggregate results information. Existing CDS services may provide a large part of this functionality, but additional QA-specific HIT components are still required to intercept errors for high-severity drug-genome interactions and facilitate educational efforts.
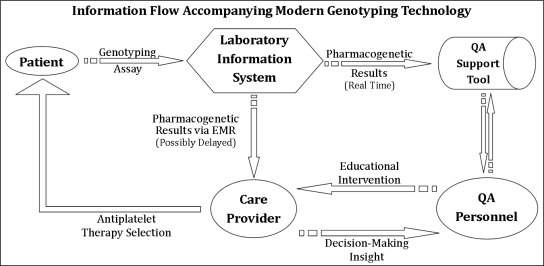
Essentially, the services delivering pharmacogenetic results and interpretations must also provide feedback mechanisms for QA personnel, to “close the loop” (Figure 1) of information flow17, 18, 35. For instance, the tools must provide a means for capturing care provider responses to QA recommendations. Understanding clinical decision-making with respect to provided pharmacogenetic information is central to optimizing personalized medicine interventions4, 7. With this feature set, care provider disagreement with recommended interventions or confusion surrounding intervention guidelines is readily identifiable. A more effective approach is then possible when modifying the underlying CDS system or other HIT processes.
Figure 1.
The laboratory information system broadcasts genotype results to our support tool in real-time, alerting QA personnel to potential antiplatelet therapy modification opportunities. The QA personnel can then perform an educational intervention, while observing decision-making insight.
QA Tool Development
With the above design considerations in mind, we elected to deploy a multi-user web-based application for our QA tools. We utilized Python and JavaScript in developing our tool’s core functionalities. The genotype data, patient demographics, and QA reports were stored in a secure MySQL database. We obtained institutional IRB approval for the repository, as it maintains identifiable patient information. To deploy our QA tool, we created an interface accessible directly through the EMR system’s ‘dashboard’ links. We limited access to QA personnel authorized by the institution to follow-up pharmacogenetic testing.
Through our web-based implementation, our tool integrates with other data services maintained by the Vanderbilt Informatics Center. This enables efficient pipelining of information provided by existing web services to our QA tool. For example, patient demographic information and relevant laboratory results are acquired via these web services and auto-populated into QA reporting as needed. Users fulfilling QA tasks do not need to hunt for specific findings, as the tool is either automatically capturing this information or displaying it for user verification. In uncommon instances where users need additional results, the user interface offers simple navigation links back to the EMR. Moreover, pharmacogenetic assay results are transmitted the instant they are available from the laboratory information system. In effect, the tool’s interface presents pharmacogenetic results as they become available in real time.
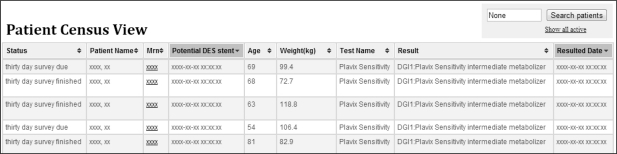
The tool’s interface offers two main views: a patient census view (Figure 2) and a detailed individual patient view (Figure 3). The summary census view orients the user to patients with newly resulted pharmacogenetic assays, as well as to other patients needing pharmacogenetic QA follow-up. Patients are sortable by follow-up status, name, MRN, and pharmacogenetic results, among other categories. The drilled down individual patient view offers QA personnel a user-interactive reporting interface.
Figure 2.
Our QA tool’s main user interface, featuring its patient census view.
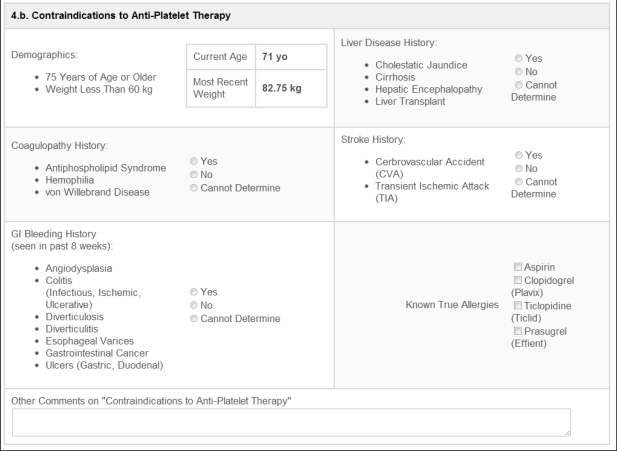
Figure 3.
Example of one section of the detailed patient view’s user interface. This demonstrates the guided prompts provided for contraindication identification.
The detailed patient view interface facilitates pharmacogenetic results’ follow-ups by offering a streamlined and standardized approach to QA processes. In this view, the tool provides QA personnel with a guided approach to determining optimal antiplatelet therapy for the patient of interest. For example, one view offers users the ability to record contraindications to specific antiplatelet therapies (Figure 3). Currently, users determine contraindication assessments manually, after reviewing records from the EMR. We expect future tool versions will make these determinations in an automated manner. On completion of all of the sections in the detailed view, and taking into context a patient’s metabolizer status, the QA personnel can more easily derive an optimal set of therapy recommendations. They then convey these informed therapy recommendations to patient providers for an ultimate decision in treatment modification. Specifically, care providers receive recommendations to maintain the standard clopidogrel dosage of 75 mg daily, to increase clopidogrel to 150 mg daily, and/or to switch the patient to prasugrel therapy. Collected report information during this process bolsters ongoing personalized medicine outcomes research.
Results
During a two-week pilot implementation period, we collected QA surveillance reports for eight unique patients. These patients had all undergone cardiac catheterization and been genotyped, as per protocol. Pharmacogenetic assays indicated that four of these patients are poor clopidogrel metabolizers, due to the presence of CYP2C19 *2/*2. The remaining four are intermediate clopidogrel metabolizers, possessing CYP2C19 *1/*2. Three of the eight patients had a drug-eluting stent placed during the catheterization procedure, or prior to undergoing pharmacogenetic testing. The five patients without a drug-eluting stent underwent coronary angiography alone, with interventionalists making the intra-procedure decision not to place any drug-eluting stents.
Providers for the three patients with drug-eluting stents received therapy modification recommendations based on the patient’s medical history. Specifically, QA personnel assessed presence of anti-platelet therapy contraindications. These three patients did not possess a history of coagulopathy, or recent (two months prior to catheterization) GI bleeding. Two patients also had no significant liver disease history, with the third patient’s liver disease history being unknown. For positive, negative, and unknown stroke or transient ischemic history, one patient met criteria for each category. Finally, none of the patients possessed allergies to the anti-platelet medications of interest. For the one patient with a stroke/TIA history, QA personnel recommended either maintaining the standard clopidogrel dosage of 75 mg daily, or increasing the dosage to 150 mg daily. Since the other two patients did not meet contraindication criteria, their providers received all therapy modification recommendations.
Discussion
We designed, implemented and conducted a pilot evaluation of a surveillance tool that facilitates the QA and research processes associated with personalized medicine interventions. Our tool’s feature set assisted QA personnel in reliably deriving patient genotype-specific antiplatelet therapy recommendations for care providers. It also helped capture the relatively complex clinical decision-making processes that accompany the reporting of pharmacogenetic results.
The surveillance approach is particularly applicable to the highest severity tier of drug-genome interactions, such as those associated with warfarin initiation and the efficacy of clopidogrel. Heightened concerns of patient privacy also frequently accompany the collection and dissemination of genetic information. To this end, we limited disclosure via the tool to FDA-approved genetic testing which impacts a drug currently prescribed for the patient.
Designing and implementing a CDS to automate the process of adjusting therapy based on genotype is one alternative to the system we propose. We have opted against implementing clinical decision support alone, as the ability for a CDS to deliver an informed recommendation considering all clinical factors such as contraindications is uncertain. Reasons for avoiding antiplatelet therapy could be present in an unstructured manner within the EMR, which most modern decision engines would overlook. At this stage of implementation, we considered the delivery of optimal therapeutic recommendations required direct contact with the care provider. Relying on the electronic health record to prompt therapeutic action by simply displaying pharmacogenetic results is unlikely to lead to optimal prescribing decisions. The surveillance approach facilitates the understanding of the nascent practice of genomic medicine in a HIT environment as well as introducing the pharmacogenetics to providers.
We intend to continue collecting QA surveillance reports through our tool for a more replete analysis of their impact on therapeutic decision-making. Expanding coverage to other drugs with pharmacogenomic implications, such as HMG-CoA reductase inhibitors, is another objective. Simultaneously, we expect to continue refining our tool to accommodate users’ needs and to reflect ongoing discoveries in genomic medicine. Our eventual goal is to achieve maximal process automation without loss of surveillance accuracy. To this end, we hope to optimize structured data usage in report completion and in instances of relevant CDS.
Conclusion
We created and deployed an informatics tool to support QA processes that are relevant to pharmacogenetic resulting. Our tool enables near real-time transmission of relevant genotype data to prescribing physicians. This implementation facilitates accurate and efficient clinical decision making necessary for applying pharmacogenomics in personalized medicine. The collected pilot data supports an informatics approach to managing QA processes associated with pharmacogenetic testing. Through our tool’s interface, we offer actionable, relevant patient information for QA personnel. Tool users can then communicate informed antiplatelet therapy recommendations to care providers. In turn, these clinicians can more readily identify patients that would benefit from pharmacogenetically cognizant antiplatelet therapy choices. Future work will also include designing specialized CDS architecture that will further automate surveillance processes.
Acknowledgments
This research was supported by NLM/NIH grant 5T15LM007450-09. We thank Ioana Danciu for her programming assistance. We thank Karla Davis and Tommy Meador for piloting the tool and for providing us with insightful feedback. We also thank the pharmacogenetic program coordinator, Jill Pulley, and other members of the institutional pharmacogenetic implementation team.
References
- 1.Bellazzi R, Larizza C, Gabetta M, et al. Translational bioinformatics: Challenges and opportunities for case-based reasoning and decision support. In: Bichindaritz I, Montani S, editors. Case-based reasoning research and development. Springer Berlin/Heidelberg; 2010. pp. 1–11. [Google Scholar]
- 2.Carlson RJ. The disruptive nature of personalized medicine technologies: Implications for the health care system. Public Health Genomics. 2009;12(3):180–4. doi: 10.1159/000189631. [DOI] [PubMed] [Google Scholar]
- 3.Hamburg MA, Collins FS. The path to personalized medicine. New England Journal of Medicine. 2010;363(4):301–4. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins MM, Ibarreta D, Gaisser S, et al. Putting pharmacogenetics into practice. Nat Biotech. 2006;24(4):403–10. doi: 10.1038/nbt0406-403. [10.1038/nbt0406-403]. [DOI] [PubMed] [Google Scholar]
- 5.Kawamoto K, Lobach D, Willard H, Ginsburg G. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Medical Informatics and Decision Making. 2009;9(1):17. doi: 10.1186/1472-6947-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maojo V, Kulikowski CA. Bioinformatics and medical informatics: Collaborations on the road to genomic medicine? Journal of the American Medical Informatics Association. 2003;10(6):515–22. doi: 10.1197/jamia.M1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Sanchez F, Iakovidis I, Nørager S, et al. Synergy between medical informatics and bioinformatics: Facilitating genomic medicine for future health care. Journal of Biomedical Informatics. 2004;37(1):30–42. doi: 10.1016/j.jbi.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Woodcock J. The prospects for “personalized medicine” in drug development and drug therapy. Clin Pharmacol Ther. 2007;81(2):164–9. doi: 10.1038/sj.clpt.6100063. [DOI] [PubMed] [Google Scholar]
- 9.Doebbeling B, Chou A, Tierney W. Priorities and strategies for the implementation of integrated informatics and communications technology to improve evidence-based practice. Journal of General Internal Medicine. 2006;21(0):S50–S7. doi: 10.1111/j.1525-1497.2006.00363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar I. Biomedical informatics and translational medicine. Journal of Translational Medicine. 2010;8(1):22. doi: 10.1186/1479-5876-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner MG, Embi PJ. Toward reuse of clinical data for research and quality improvement: The end of the beginning? Annals of Internal Medicine. 2009 2009 Sep 1;151(5):359–60. doi: 10.7326/0003-4819-151-5-200909010-00141. [DOI] [PubMed] [Google Scholar]
- 12.Eisenstein EL, Anstrom KJ, Kong DF, et al. Clopidogrel use and long-term clinical outcomes after drug-eluting stent implantation. JAMA: The Journal of the American Medical Association. 2007 2007 Jan 10;297(2):159–68. doi: 10.1001/jama.297.2.joc60179. [DOI] [PubMed] [Google Scholar]
- 13.Mega JL, Simon T, Collet J-P, et al. Reduced-function cyp2c19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for pci. JAMA: The Journal of the American Medical Association. 2010 2010 Oct 27;304(16):1821–30. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paré G, Mehta SR, Yusuf S, et al. Effects of cyp2c19 genotype on outcomes of clopidogrel treatment. New England Journal of Medicine. 2010;363(18):1704–14. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 15.Embi PJ, Kaufman SE, Payne PRO. Biomedical informatics and outcomes research: Enabling knowledge-driven health care. Circulation. 2009 2009 Dec 8;120(23):2393–9. doi: 10.1161/CIRCULATIONAHA.108.795526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickner JMF, Douglas H, Harris Daniel M, Poon Eric G, Elder Nancy C, Mold James W. Issues and initiatives in the testing process in primary care physician offices. Joint Commission Journal on Quality and Patient Safety. 2005;31:81–9. doi: 10.1016/s1553-7250(05)31012-9. [DOI] [PubMed] [Google Scholar]
- 17.Matheny ME, Gandhi TK, Orav EJ, et al. Impact of an automated test results management system on patients’ satisfaction about test result communication. Arch Intern Med. 2007 2007 Nov 12;167(20):2233–9. doi: 10.1001/archinte.167.20.2233. [DOI] [PubMed] [Google Scholar]
- 18.Poon EG, Wang SJ, Gandhi TK, Bates DW, Kuperman GJ. Design and implementation of a comprehensive outpatient results manager. J of Biomedical Informatics. 2003;36(1/2):80–91. doi: 10.1016/s1532-0464(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 19.Stead WWMD, Searle JRP, Fessler HEMD, Smith JWMDP, Shortliffe EHMDP. Biomedical informatics: Changing what physicians need to know and how they learn. Academic Medicine. 2011 doi: 10.1097/ACM.0b013e3181f41e8c. [DOI] [PubMed] [Google Scholar]
- 20.Gurwitz D, Lunshof JE, Altman RB. A call for the creation of personalized medicine databases. Nat Rev Drug Discov. 2006;5(1):23–6. doi: 10.1038/nrd1931. [10.1038/nrd1931]. [DOI] [PubMed] [Google Scholar]
- 21.Khoury MJ, Rich EC, Randhawa G, Teutsch SM, Niederhuber J. Comparative effectiveness research and genomic medicine: An evolving partnership for 21st century medicine. Genetics in Medicine. 2009;11(10):707–11. doi: 10.1097/GIM.0b013e3181b99b90. 10.1097/GIM.0b013e3181b99b90. [DOI] [PubMed] [Google Scholar]
- 22.Safran C, Bloomrosen M, Hammond W, et al. Toward a national framework for the secondary use of health data: An american medical informatics association white paper. Journal of the American Medical Informatics Association. 2007 2007 Jan 1;14(1):1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood L, Friend SH. Predictive, personalized, preventive, participatory (p4) cancer medicine. Nat Rev Clin Oncol. 2011;8(3):184–7. doi: 10.1038/nrclinonc.2010.227. [10.1038/nrclinonc.2010.227]. [DOI] [PubMed] [Google Scholar]
- 24.Singh H, Naik A, Rao R, Petersen L. Reducing diagnostic errors through effective communication: Harnessing the power of information technology. Journal of General Internal Medicine. 2008;23(4):489–94. doi: 10.1007/s11606-007-0393-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurbel PA, Bliden KP, Zaman KA, et al. Clopidogrel loading with eptifibatide to arrest the reactivity of platelets: Results of the clopidogrel loading with eptifibatide to arrest the reactivity of platelets (clear platelets) study. Circulation. 2005 2005 Mar 8;111(9):1153–9. doi: 10.1161/01.CIR.0000157138.02645.11. [DOI] [PubMed] [Google Scholar]
- 26.Hochholzer W, Trenk D, Bestehorn H-P, et al. Impact of the degree of peri-interventional platelet inhibition after loading with clopidogrel on early clinical outcome of elective coronary stent placement. J Am Coll Cardiol. 2006 2006 Nov 7;48(9):1742–50. doi: 10.1016/j.jacc.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 27.Saw J, Steinhubl SR, Berger PB, et al. Lack of adverse clopidogrel-atorvastatin clinical interaction from secondary analysis of a randomized, placebo-controlled clopidogrel trial. Circulation. 2003 2003 Aug 26;108(8):921–4. doi: 10.1161/01.CIR.0000088780.57432.43. [DOI] [PubMed] [Google Scholar]
- 28.Fox KAA, Mehta SR, Peters R, et al. Benefits and risks of the combination of clopidogrel and aspirin in patients undergoing surgical revascularization for non-st-elevation acute coronary syndrome: The clopidogrel in unstable angina to prevent recurrent ischemic events (cure) trial. Circulation. 2004 2004 Sep 7;110(10):1202–8. doi: 10.1161/01.CIR.0000140675.85342.1B. [DOI] [PubMed] [Google Scholar]
- 29.Sibbing D, Stegherr J, Latz W, et al. Cytochrome p450 2c19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. European Heart Journal. 2009 2009 Apr 1;30(8):916–22. doi: 10.1093/eurheartj/ehp041. [DOI] [PubMed] [Google Scholar]
- 30.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. New England Journal of Medicine. 2009;360(4):363–75. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 31.Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of cyp2c19 and cyp2c9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. Journal of thrombosis and haemostasis : JTH. 2007;5(12):2429–36. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 32.Miller RA, Waitman LR, Chen S, Rosenbloom ST. The anatomy of decision support during inpatient care provider order entry (cpoe): Empirical observations from a decade of cpoe experience at vanderbilt. Journal of Biomedical Informatics. 2005;38(6):469–85. doi: 10.1016/j.jbi.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giuse DA. Supporting communication in an integrated patient record system. Proc AMIA Annu Fall Symp. 2003:1065. [PMC free article] [PubMed] [Google Scholar]
- 34.Geissbuhler AM, Randolph A. A new approach to the implementation of direct care-provider order entry. 1996:689–93. [PMC free article] [PubMed] [Google Scholar]
- 35.Rigby M, Forsström J, Roberts R, Wyatt J. Verifying quality and safety in health informatics services. BMJ. 2001 2001 Sep 8;323(7312):552–6. doi: 10.1136/bmj.323.7312.552. [DOI] [PMC free article] [PubMed] [Google Scholar]