Abstract
The increasing availability of personal genome data has led to escalating needs by consumers to understand the implications of their gene sequences. At present, poorly integrated genetic knowledge has not met these needs. This proof-of-concept study proposes a similarity-based approach to assess the disease risk predisposition for personal genomes. We hypothesize that the semantic similarity between a personal genome and a disease can indicate the disease risks in the person. We developed a knowledge network that integrates existing knowledge of genes, diseases, and symptoms from six sources using the Semantic Web standard, Resource Description Framework (RDF). We then used latent relationships between genes and diseases derived from our knowledge network to measure the semantic similarity between a personal genome and a genetic disease. For demonstration, we showed the feasibility of assessing the disease risks in one personal genome and discussed related methodology issues.
Introduction
The rapid growth of genomic knowledge can enrich the annotation of personal genome sequences as well as drive the trend of applying whole genome sequencing in the clinical setting. However, the existence of knowledge does not necessarily guarantee its effective use. The current lack of knowledge resources and tools for understanding personal genomes remains a severe challenge in the translation from genomic research to genomic medicine. In addition, the existing genomic knowledge bases do not communicate with one another as they were originally developed for specific genomic research and not for integration into clinical use. Consequently, it remains a laborious task for researchers to search for and integrate scattered genomics knowledge manually for clinical decision-making
Recently, Ashley et al. analyzed a human genome sequence in the clinical context9, which is regarded as the first attempt to comprehensively identify the clinical significance of a whole genome sequence to a patient. In order to study the genetic risks of various diseases, drug responses, and pathogenicity of novel variants, they devoted substantial time and effort to querying and summarizing information from scattered knowledge bases. There is neither an established tool for annotating personal genome sequences nor an integrated knowledge network for genomic medicine despite the large number of databases available for genomic research. In addition, there are no resources for linking together disease, biological pathways, genes, Gene Ontology (GO)10 terms, and symptoms, which are necessary in order to establish the foundations of genomic medicine. The lack of annotation tools for personal genome sequences is partly due to this absence of an integrated knowledge resource.
Due to rapidly declining costs, whole genome sequencing technology has become more accessible and affordable to large health care facilities and the general public1–4. Some hospitals have started sequencing personal genomes for research use and are exploring methods to analyze and convey the clinical implications of personal genome information to clinicians and health consumers5–8. In early 2010, scientists at the Johns Hopkins Kimmel Cancer Center (JHKCC) began using data from whole genome sequencing of cancer patients to develop individualized treatments7. Currently, JHKCC provides only a limited range of genomic analysis that identifies translocations in solid tumors, and thus named the approach the Personalized Analysis of Rearranged Ends (PARE). PARE has been offered as a regular clinical genetic test at a cost of $5,000 per patient, which is relatively expensive compared to traditional diagnostic measurements. However, JHKCC believes that PARE will become more cost-effective as sequencing costs continue to decline and the amount of information provided to patients increases as we gain better understandings of the implications of these genetic sequences. Since conventional diagnostic tools have limited applicability for detecting the molecular signatures of cancer cells, we believe that the trend of whole genome sequencing for clinical applications will be increasingly common for predicting disease risks and individualized drug responses.
An integrated knowledge resource is needed to reveal disease relationships. Recently, researchers measured the genetic similarity between diseases using the accumulated disease-gene knowledge in the Online Mendelian Inheritance in Man database (OMIM).Goh et al. created a disease network consisting of diseases as nodes11. Two nodes were connected if they shared disease-related genes based on information obtained from OMIM. However, joining disease nodes only by gene overlap might miss some true associations because different genes can participate in the same function. Even if two diseases share no genes, they may have genetic relationships when their genes participate in the same biological pathway. For example, we can find 16 genes for prostate cancer (OMIM ID: #176807) and 6 genes for ovarian cancer (OMIM ID: #604370) in the OMIM knowledge base without a common gene. However, we identified 42 GO intersected terms between the two cancers when we changed the level of comparison from gene to GO terms. Table 1 shows the GO term intersection between prostate cancer and ovarian cancer. At the gene level, some people might think that the inherited bases for prostate and ovarian cancer do not overlap. However, there might be overlaps between inherited bases of the two cancers when comparing them from functional dimensions using GO rather than at individual gene level. In the absence of this latent information, it would be difficult to navigate the complex relationships between genes and diseases.
Table 1.
Intersection of Gene & GO between Prostate Cancer and Ovarian Cancer in OMIM.
| Prostate Cancer (OMIM ID: #176807) | Ovarian Cancer (OMIM ID: #604370) | |
|---|---|---|
| OMIM Gene | HPCX, TUSC3, MSR1, CHEK2, ELAC2, MXI1, HNF1B, BRCA2, MAD1L1, CD82, HIP1, PCAP, KLF6, PTEN, AR, ZFHX3 | PIK3CA, PARK2, MSH6, OPCML, AKT1, BRCA1 |
| Intersected Gene | - | |
| Intersected GO | endoplasmic reticulum, protein binding, integral to plasma membrane, nucleotide binding, protein serine/threonine kinase activity, transferase activity, ATP binding, metal ion binding, response to DNA damage stimulus, protein amino acid phosphorylation, cell cycle, DNA damage response signal transduction resulting in induction of apoptosis, nucleus, DNA binding, transcription activator activity, protein homodimerization activity, positive regulation of gene-specific transcription from RNA polymerase II promoter, negative regulation of apoptosis, positive regulation of transcription DNA-dependent, DNA damage response signal transduction by p53 class mediator resulting in transcription of p21 class mediator, double-strand break repair, response to estrogen stimulus, double-strand break repair via homologous recombination, protein complex, cytoplasm, nucleoplasm, spindle, cytosol, plasma membrane, apoptosis, regulation of apoptosis, Golgi apparatus, zinc ion binding, double-stranded DNA binding, intracellular, enzyme binding, magnesium ion binding, PDZ domain binding, negative regulation of protein amino acid phosphorylation, regulation of neuron projection development, central nervous system development, signal transduction | |
Abbreviations: GO, Gene Ontology; OMIM, Online Mendelian Inheritance in Man
To address these problems, we created a knowledge network that integrates knowledge about diseases, genes, and symptoms, and developed a tool to use this knowledge network to estimate potential disease risks associated with a personal genome. We hypothesize that the semantic similarity between the textual representation for a personal genome and a disease can indicate the disease risks in the person. In the rest of this paper, we present the design and results of this method for using rich genetic knowledge about diseases and genomics to interpret personal genomes.
2. Methods and Materials
2.1. Development of a ‘genome-disease’ knowledge network KNODGE
Our knowledge network is called the KNOwledge-extension for Disease-Gene Associations (KNODGE). KNODGE integrates six heterogeneous biomedical knowledge bases (OMIM12, GO10, Human Phenotype Ontology13, GO Annotation14, Entrez Gene15, and KEGG pathway database16). OMIM was our first choice for a knowledge source because it is the most widely accepted gene disease knowledge base and it shares common semantic entities with other knowledge sources. For example, OMIM has genes associated with various diseases, but the GO and KEGG pathway databases contain functional structure and regulatory information for disease and symptoms that overlap with disease components in OMIM. We used the semantic Web standard Resource Description Framework (RDF) to represent each knowledge source. All semantic relationships were represented as RDF triples in the “subject-predicate-object” format and merged to form an integrated knowledge network. This knowledge network harbors 545,008 triples. Example triples are ‘gene:FGFR-relation:hasGO-GO:skeletal system development’ or ‘gene:FGFR3-relation:hasDisease-disease:Bladder cancer.’ After converting all knowledge into such triples, we merged all the triples into one file so that we could query and obtain a novel association such as “bladder cancer has an association with gene function, skeletal stem development via anchor, FGFR3.” To facilitate easy interrogation of the knowledge base, we also designed a Web-based semantic query interface using the SPARQL query engine. We also considered other related knowledge sources for microRNA and DNA copy number variance, which are genomic components that have known strong associations with diseases. However, their associations with diseases were established only recently and have not been widely tested, and thus we did not include them in this first step of our proof-of-concept study. In addition, one advantage of the RDF format is that new knowledge can be easily inserted at anytime in the future; therefore, we can still incorporate these knowledge sources when they are more mature.
2.2. A similarity analysis algorithm GEAR
Having an integrated network of gene, disease, and GO, we developed a tool to predict associations between disease and abnormal signatures of personal genomes on the functional dimension (i.e. GO space). We developed the following 4-step procedure. First, we identified deleterious variants from personal genome sequences. Second, we mapped them to GO terms through genes that harbor the abnormalities. Third, we mapped disease-related genes to GO terms to make them comparable to GO terms from the abnormal signatures of personal genomes; and fourth, we compared the two lists of GO terms to evaluate their similarity.
We limited the input data type to a list of SNPs because the primary result of whole genome sequencing was presented as a list of SNPs1–4. From the input, SNPs were selected when the substitution of a nucleotide results in a structural change to a protein with deleterious effects on its function. The PolyPhen-2 algorithm17 was used for SNP selection. PolyPhen-2 calculated the naive Bayesian posterior probability that a given mutation was damaging. We downloaded an annotation file, HumVar, consisting of 110,939 SNPs and their status to list all SNPs that may have abnormal effects. Each of the selected SNPs was scored as 0, +1, or +2 according to the number of dysfunctional alleles in each SNP genotype.
For the SNPs in a gene, the scores were summarized to determine the gene disruption score (GDS). Genes with a GDS greater than ‘1’ were regarded as ‘abnormal genes’ and used for further analysis. Afterward, all GO terms annotated as abnormal genes were extracted. We included not only explicit but also implicit GO annotations for each gene, incorporating GO hierarchical information. Similarly, we extracted all of the GO terms of major diseases based on the integration of Entrez Gene (GO-gene annotations) and OMIM (gene-disease associations). This provided us with various lists of GO terms: one from abnormalities found in the personal genome data and others from disease-associated genes. We incorporated a GS2 algorithm18 to measure semantic similarities among the lists of GO terms. GS2 was originally developed to measure gene set similarity using GO semantics, and therefore we considered GS2 well suited to our scheme. Briefly, the measure quantified the similarity of the GO annotations by averaging the contribution of GO terms and their ancestral terms with respect to the GO vocabulary graph. The input of GS2 is two lists of GO terms, and it measured the similarities between the two lists by incorporating the hierarchical structure of GO. Briefly, for each GO term in the first list, it calculates how similar its ancestor set is in comparison to the ancestors of the terms in the second list. It normalizes and averages the values and returns a score between 0 and 1. The estimated relationships were visualized as a network at the end of the process. We took the negative logarithm of the similarity score in order to generate a better presentation of the network diagram. To facilitate user interaction with the knowledge base for understanding their personal genomes, we developed a Web-based system, Genetic Abnormalities Report (GEAR), to allow users to access the aforementioned analysis module.
3. Results
3.1. Example uses of KNODGE
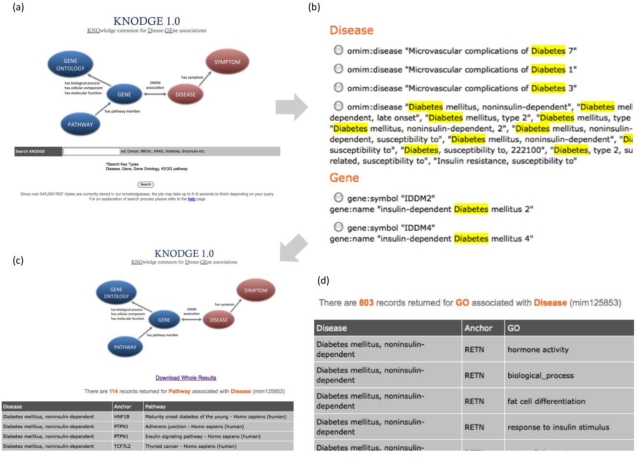
KNODGE is freely available at http://impact.dbmi.columbia.edu/~juw7003/KNODGE/index.html. The home page (Figure 1a) briefly describes the structure of the knowledge base and contains a search box to begin navigation. Users can query either disease or gene terms to obtain matching genes, diseases, gene pathways, and symptoms. Example terms are provided next to the search box. For example, users can type ‘BRCA1’ to search for other diseases and symptoms associated with the gene and those known to be associated with breast cancer. In addition, users can type ‘Diabetes’ to query related diseases, genes, pathways, or symptoms of this disease.
Figure 1.
Sample KNODGE screenshots:
(a) the schematic structure of the knowledge base and the semantic query interface;
(b) the query results for one search keyword ’Diabetes’, grouped by entity types, including disease, gene, gene terms, and gene pathways;
(c) the filtered query result for the entity ‘type 2 diabetes mellitus’; all the results can be downloaded in a structured format;
(d) a subset of GO terms that describe the genes associated with ‘type 2 diabetes mellitus’.
If a user enters the keyword ’Diabetes,’ KNODGE returns the records that match the keyword in their names, descriptions, or synonyms (Figure 1b). The matched term is highlighted so that users can understand why those records were retrieved. A user can further specify the particular term to query all semantic relationships related to the term. Assume that a user selects ‘Type 2 Diabetes Mellitus,’ one of the instances listed with the disease semantic type, and wants to obtain all related biomedical entities linked to this disease. Each query usually requires 4 to 5 seconds to complete. Figure 1c shows the screenshot of this query result. All entities related to this disease are returned, including 114 biological pathways, 803 GO terms, 28 genes, and 4 symptoms. The query results can be downloaded in a structured format. Some entities are connected via ‘anchor.’ For example, a GO term and disease are semantically related via a gene as an anchor. According to Figure 1d, the gene ‘RETN’ is associated with the ‘type 2 diabetes mellitus,’ and it serves a particular role in the biological pathway ‘hormone activity.’
3.2. Example uses of GEAR
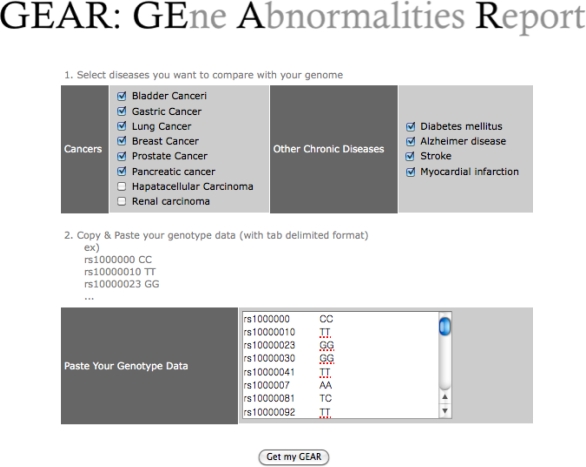
GEAR is a web-based system for evaluating the use of KNODGE for assessing personal genomes in the clinical setting. It is available at http://impact.dbmi.columbia.edu/~juw7003/GEAR/index.html. Personal genome information can be summarized as a list of sequence variants after comparisons with the reference sequence. Among the various types of sequence variants (e.g., copy number variations (CNVs), SNPs, insertions, and deletions), the current version of our module can only accept SNPs. Users may choose the diseases of interest (Figure 2) such as those for which they have concerns or those related to a family history of a certain disease, and they can compare the similarity between disease-related genes as specified in OMIM and their personal genetic abnormalities. The other input element would be a list of SNPs and their genotypes.
Figure 2.
The Web interface for GEAR.
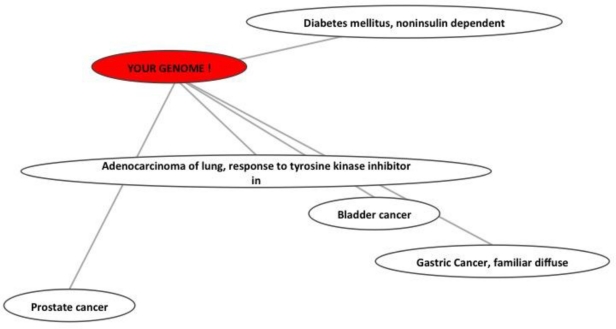
The SNP identifier should be a dbSNP identifier so that GEAR can determine which genes are potentially affected by the sequence change. Figure 3 shows the output of GEAR. Each node represents diseases of interest or the individual owner of the input genotype data. Edges indicate the genetic similarities among nodes. The genetic structure similarity between the two corresponding nodes increases as the edge becomes shorter. The topological structure reveals the relative distance between the ‘personal genome’ and ‘diseases’ but does not reveal the distance between diseases. For instance, it does not indicate that bladder cancer is more similar to gastric cancer than to prostate cancer, but that the abnormal signature of the test genome is more similar to diabetes and lung cancer than to prostate cancer or gastric cancer.
Figure 3.
GEAR result: Relative genetic distance between personal genome and diseases
4. Discussion
In this proof-of-concept study, we developed KNODGE by integrating various sources of genomic knowledge and demonstrated that this integration enables a preliminary clinical assessment of personal genomes. First, we integrated six knowledge resources related to genes and diseases. Then, we developed GEAR, which measures the genetic similarity between abnormal signatures of personal genomes and diseases. In comparison, we used the integrated knowledge, using GO instead of only specific genetic sequences, to assess the functional relationships among the objects. Briefly, GEAR selects potentially deleterious SNPs and summarizes the number of deleterious substitutions at the gene level. Next, we measured the genetic similarity between personal genomes and diseases using GO terms annotated to the significant genes that can be directly extracted from our knowledge network. Theoretically, this method has advantages over simply comparing the list of significant genes. If we search the relationships using only a single gene, we could miss additional relationships considering the fact the multiple genes are often involved in the same biological processes. Overall, our integrated genomic knowledge enables measurements of genetic similarity.
One other important characteristic of our design is that we provide only ‘similarity of genetic structure’ between a personal genome and diseases. Our scheme is fundamentally different from that of current direct-to-consumer companies. For example, GEAR does not provide any deterministic information such as ‘90% risk of breast cancer,’ which can often be a false positive that raises unnecessary concerns for the general public. Instead, our reports measure relative distances between personal genomes and diseases to inform people of their general risks. Cancer, diabetes, and heart disease can develop in anyone because their occurrence is largely influenced by environmental factors. Without these environmental factors, we can never measure ‘true disease risk.’ Therefore, we strongly believe that reporting imprecise disease risk based only on genetic information is not appropriate. Instead, people could use GEAR to determine which diseases have similar structures to their own genomes. In other words, GEAR can be used for prioritizing diseases that people should be alerted to. For instance, if one’s genome appears to be closer to colon cancer than gastric cancer in GEAR output, he or she might be suggested towatch for any symptoms related to colon cancer for early prevention of this disease. .
Additionally, our integrated knowledge base can be used to increase the sensitivity for identifying genetic relationships between diseases. Using this knowledge network, researchers could easily explore the genetic basis of diseases and generate hypotheses of disease similarity. This possibility is particularly important because molecular similarity of diseases could be used for finding new applications of existing drugs. For instance, Suthram et al. measured disease similarity based on gene expression profiles and identified a strong correlation between urothelial carcinoma and acute myeloid leukemia.19 They also found that the FDA approved drug Flouroucil, is used to treat both of the diseases. However, Suthram et al. used gene expression data only for measuring disease similarity. Our integrated knowledge base could enrich the disease similarity network by incorporating additional layers of knowledge. Another advantage of our knowledge network is that it is easy to use and extend. Any newly added knowledge to any knowledge source can be converted to the RDF format and appended to the integrated RDF knowledge network. This knowledge base can also be easily integrated with other knowledge bases using the Semantic Web standard such as RDF.
Our study has several limitations, which will be addressed in our future work. This knowledge network provides us with a new infrastructure to generate hypotheses, but we have not evaluated the validity of the novel associations recommended by our integrated knowledge network. Involvement of clinical and biological researchers in a pilot study can help us test some meaningful hypotheses generated by this novel knowledge network. In addition, we have not evaluated the performance of our similarity metric, which was primarily due to the lack of a gold standard to compare against. In the current version of GEAR, the list of diseases used for interrogating personal genome is too short to be useful in real clinical setting. We first listed over 50 diseases but it made GEAR user interface cluttered and confusing. Therefore, we only selected a handful diseases of higher prevalence rates in United States for this pilot version. The interface should be re-designed to deal with a larger number of diseases. The other limitation is that data cannot be entered to GEAR if there is no Reference SNP (rs) identifier. The input module should be revised to accept variants without rs number because personal genome sequence will contain not only known common variants but also novel and rare variants. There are also far more personal variants beyond SNPs (e.g., CNVs), which are currently our sole form of input. Additionally, other functional SNPs can affect splicing, transcriptional, and post-translational regulation. We only used SNPs that affect protein coding. Thus, we will need to extend our analysis coverage to other functional SNPs. Accordingly, our tool and algorithms need further constant upgrading as knowledge of the genome expands. The other limitation of our study is the coverage of our knowledge network. The current version of KNODGE only includes a limited number of entities (disease, gene, pathway, GO, and symptoms). Future versions of KNODGE will include other entities and semantic relationships such as ‘microRNA-gene target information’ or ‘protein-protein interaction’ that may be important to genomic medicine. Furthermore, the ‘gene-disease’ relationships will become more complex by adding ‘genomic variation’ (e.g., SNPs and CNVs) that establishes relationships in OMIM. Future work will include evaluating the plausibility and validity of these novel associations in a constrained medical domain using subject matter experts.
5. Conclusion
In this study, we contribute an integrated knowledge network that provides integrated access to knowledge of relations between genes, diseases, and symptoms. The creation of this new knowledge resource has the potential to enable biomedical researchers to query one single knowledge source to generate hypotheses about diseases, symptoms, and genes, as well as to generate hypotheses about their personal genome’s risk factors. We also implemented a similarity measurement algorithm to explore the interpretations of personal genomes. We conclude that this knowledge network is rich and extensible and has the potential to serve as an integrated knowledge resource for personal genome interpretation.
Acknowledgments
This study was supported by grants R01LM009886 and R01LM010815 from national library of medicine, CTSA awards UL1 RR024156, and AHRQ grant R01 HS019853. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
References
- 1.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–5. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Lee S, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Park H, Kim JY, Gokcumen O, Mills RE, Zaranek AW, Thakuria J, Wu X, Kim RW, Huntley JJ, Luo S, Schroth GP, Wu TD, Kim H, Yang KS, Park WY, Kim H, Church GM, Lee C, Kingsmore SF, Seo JS. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460(7258):1011–5. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 7.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med. 2011;364(4):340–50. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 9.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), A knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–7. doi: 10.1093/nar/gki033. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am J Hum Genet. 2008;83(5):610–5. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–7. doi: 10.1093/nar/gkq1237. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–49. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruths T, Ruths D, Nakhleh L. GS2: an efficiently computable measure of GO-based similarity of gene sets. Bioinformatics. 2009;25(9):1178–84. doi: 10.1093/bioinformatics/btp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suthram S, Dudley JT, Chiang AP, Chen R, Hastie TJ, Butte AJ. Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput Biol. 2010;6(2):e1000662. doi: 10.1371/journal.pcbi.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]