Abstract
Increasing use of electronic health records requires comprehensive patient-centered views of clinical data. We describe a prototype knowledge base and SMART app that facilitates organization of patient medications by clinical problems, comprising a preliminary step in building such patient-centered views. The knowledge base includes 7,164,444 distinct problem-medication links, generated from RxNorm, SNOMED CT, and NDF-RT within the UMLS Metathesaurus. In an evaluation of the knowledge base applied to 5000 de-identified patient records, 22.4% of medications linked to an entry in the patient’s active problem list, compared to 32.6% of medications manually linked by providers; 46.5% of total links were unique to the knowledge base, not added by providers. Expert review of a random patient subset estimated a sensitivity of 37.1% and specificity of 98.9%. The SMART API successfully utilized the knowledge base to generate problem-medication links for test patients. Future work is necessary to improve knowledge base sensitivity and efficiency.
Introduction
In early electronic health record (EHR) and health information technology (HIT) adopter organizations, patients may have over 10 years of clinical data covering all aspects of their care. These readily accessible, legible archives of clinical notes, laboratory test results, radiographic images, and provider correspondence can inform clinical care (1), but they also pose a challenge for time-pressured clinicians working in busy settings (2). As the number of clinicians using EHRs and the number of health information exchanges (HIEs) capable of exchanging patient-level data increases, the quantity of data those clinicians will need to review for safe and effective care increases exponentially (3). In addition, clinicians must integrate these data with their existing knowledge and fast-changing external scientific literature and guidelines, while still keeping in view the internal policies and procedures of their institutions (4). HIT is likely to be critical to management of these data and information needs, and solutions will require using explicit, unified, accurate, and comprehensive patient-centered models that reflect the true work domain ontology (5). A preliminary step in creating such models is automated linking of a patient’s medications with clinical indications, or problems. In this paper, we describe the generation of a clinical knowledge base and development of a prototype SMART app to facilitate the organization of a patient’s medications according to their clinical problems, which we believe is a better cognitive match for most tasks involving clinician review and integration than an alphabetical or chronological view of medications.
Background
The Unified Medical Language System Metathesaurus
The Unified Medical Language System (UMLS) Metathesaurus (6), developed over the last 25 years by the United States National Library of Medicine (NLM), is a large, integrated biomedical database of 158 source vocabularies containing more than 2.3 million concepts, 8.5 million unique concept names, and over 12 million relationships among concepts. The UMLS Metathesaurus is built from more than 100 source vocabularies, including RxNorm, SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms), and the Veterans Health Administration National Drug File-Reference Terminology (NDF-RT).
RxNorm is a standardized nomenclature that provides normalized names for drugs and drug delivery devices, aimed to facilitate interoperability among HIT systems by linking to drug vocabularies within the UMLS Metathesaurus (7). Also facilitating interoperability is SNOMED CT, a comprehensive clinical terminology maintained by the International Health Terminology Standards Development Organisation (IHTSDO) (8). NDF-RT is a reference terminology for medications, providing a formal content model to describe medications and definitional relationships (e.g., “may_treat”) (9). Many concepts, including medications and clinical problems, have numerous synonyms associated with them, creating a dense graph of inter-relationships.
The SMART Architecture
The SMART (Substitutable Medical Apps, Reusable Technologies) platform was developed by researchers and developers from Harvard Medical School as part of Strategic Health IT Advanced Research Projects (SHARP) program funded by the Office of the National Coordinator for Health IT. The platform’s goal is to facilitate “iPhone-like” substitutability for medical apps and provides an application programming interface (API) for data capture, storage, and retrieval (10). EHRs and other HIT systems may be “SMART-enabled,” allowing apps employing the SMART API to interact with those systems.
Significance
Substantially large patient records from various sources generated during years of EHR use combined with the need for busy clinicians to efficiently review such records calls for automated methods to summarize patient data (1–4). Researchers have previously described various methods for developing similar systems, such as automatic linking and identification of patient medications and clinical problems (11–14). While these methods are beneficial and serve as the basis for our knowledge base, use and evaluation of the systems with actual patient data are limited to local installations. A readily available methodology for summarizing patient data generalizable to any HIT system is necessary.
Methods
Study Setting
We developed and evaluated the knowledge base and SMART app at a large, multi-specialty, ambulatory academic practice that provides medical care for adults, adolescents, and children throughout the Houston community. Providers utilize Allscripts Enterprise Electronic Health Record (v11.1.7; Chicago, IL) to maintain patient notes and problem lists, order and view results of laboratory tests, and prescribe medications. Providers can manually link medications to a diagnosis within the patient’s clinical problem list, though this feature is seldom utilized. We randomly selected 5000 de-identified patients with an outpatient encounter between July 1, 2010 and December 31, 2010 and at least one active, coded clinical problem and medication. Because the knowledge base depends on RxNorm CUIs and SNOMED CT codes, we included only medications and problems that could be mapped to the respective vocabulary. This study was approved by The University of Texas Health Science Center at Houston’s Committee for the Protection of Human Subjects (HSC-SHIS-10-0238).
Design Considerations
The primary design and performance goals for this project aimed to:
Extract and evaluate a prototype clinical knowledge base for linking medications with clinical problems.
Utilize the knowledge base to automatically associate a patient’s medications with the clinical problem(s) for which they were prescribed.
Generate the list of patient problems and linked medications in less than 1 second.
Employ the SMART architecture to develop a patient summary view generalizable to various EHRs.
Knowledge Base Generation
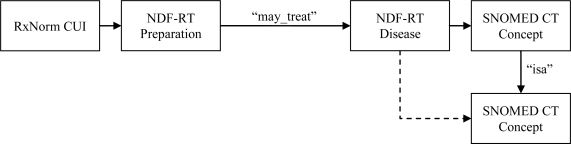
To facilitate organization of patient medications by clinical problems, we generated a knowledge base from concepts within the UMLS Metathesaurus. Figure 1 depicts the process by which we generated the knowledge. We first extracted medication indication information from NDF-RT using the “may_treat” relationship and mapped terms to SNOMED CT and RxNorm. To account for varying specificity in recording of problems and medications within the EHR, we allowed linking of medications and problems to child concepts within NDF-RT. For example, a “may_treat” relationship exists between “Insulin” and “Diabetes mellitus type 1”, but not between “Insulin” and “Insulin-dependent diabetes mellitus type IA”, a child concept of “Diabetes mellitus type 1.” We inferred relationships using the “isa” relationship between SNOMED CT problems, such that children of problems in a problem-medication pair also linked to the medication. We traversed child concepts for each problem until no new relationships were inferred, which occurred after twelve iterations of problem inferences. Our resulting knowledge base contained RxCUI and SNOMED CT Code pairs, which may be mapped to medications and problems within any ONC-approved EHR (15).
Figure 1.
Generation of knowledge base for linking patient medications with clinical problems using RxNorm, NDF-RT, and SNOMED CT. Dashed line indicates inferred relationship.
SMART App Development
To create a summary app, we employed the SMART Javascript Library. We developed and tested the app using the SMART Reference EMR, which contains records for 50 test patients. The records include allergy, allergy exception, demographics, prescription fulfillment, lab result, lab result panel, medication, note, and problem types that are mapped to coded vocabularies. Specifically, medication and clinical problems are mapped to RxNorm and SNOMED CT respectively. Using the SMART API, we retrieved medication and clinical problem data, displaying problem-medication links when possible.
Knowledge Base and SMART App Evaluation
To evaluate the coverage of the knowledge base, we first counted the number of unique medications within RxNorm for which we were able to identify one or more clinical problems within SNOMED CT using NDF-RT relationships. We also calculated the total number of problem-medication links identified within each level of the SNOMED CT hierarchies.
To simulate use of the newly developed clinical knowledge base within an EHR, we evaluated the problem-medication linking system against the 5000 de-identified Allscripts patients using a locally developed display tool and the SMART test patients using the SMART app within the SMART Reference EMR. For each, we calculated the percentage of medications linking to a clinical problem within the patient’s record. We measured the execution speed of the linking system on problem-medication links generated for a subset of 1000 randomly selected patients with at least three clinical problems and five medications, calculating the mean time to process each medication on the patient’s list.
To assess the accuracy of the problem-medication linking knowledge base, we first calculated the number of automatically linked problems and medications that matched links manually entered by ordering providers within Allscripts. We then randomly selected a subset of 25 Allscripts patients with at least three clinical problems and five medications documented in the EHR for expert review. For each problem-medication pair, a board-certified internist (AL) determined whether the problem was an appropriate indication for the medication. Based on these results, we calculated the estimated sensitivity and specificity of the problem-medication linking system.
Results
Allscripts Problem and Medication Mapping
For all patients experiencing an encounter during the study period, problems mapped to SNOMED CT concepts accounted for 4,978 of 16,713 distinct problems (29.79%) and 159,519 of 472,970 total problem instances (33.72%). Medications mapped to RxNorm concepts accounted for 11,974 of 17,410 unique medications (68.78%) and 360,900 of 455,609 total medication instances (79.21%). Providers manually linked one or more problems to 51,099 (14.16%) medication instances.
Knowledge Base Coverage and Speed
The knowledge base contained 7,164,444 problem-medication linked pairs. These pairs are comprised of 8,038 distinct RxNorm terms linked to 59,647 distinct SNOMED CT problems, encompassing 7.4% of medication terms within RxNorm and 20.4% of problems within SNOMED CT. Table 1 indicates the number of links included in the knowledge base by depth of inference through traversal of the “isa” SNOMED CT relationships.
Table 1.
Distinct links included in the problem-medication knowledge base. Relationships extracted directly from NDF-RT have a depth equal to zero.
| SNOMED CT Problem “isa” Depth | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| Distinct Links | 44,421 | 591,014 | 1,481,766 | 1,988,527 | 1,677,023 | 935,235 | 350,401 | 94,406 | 31,030 | 23,602 | 13,753 | 2,924 | 342 |
Using the new knowledge base, the system generated problem-medication summaries for the subset of 1000 randomly selected patients with at least three clinical problems and five medications at a mean rate of 1.84 seconds per patient (0.19 seconds per medication).
Knowledge Base Accuracy
The 5000 randomly selected patients for evaluation included 22,887 total problem instances and 23,424 total medication instances. Providers manually linked 7,635 (32.59%) medications to patient problems. The knowledge base linked 5,251 (22.42%) medications to patient problems. Of these, 2,148 problem-medication pairings occurred for both manual and knowledge base linking, indicating that the knowledge base linked 4,763 (46.47% of total links) additional medications not previously linked by providers.
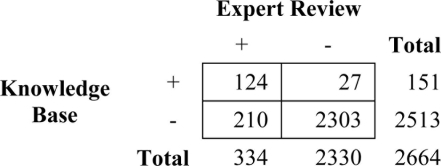
During expert review of the 151 problem-medication matches for the subset of 25 patients, we found that 124 were correct, indicating a positive predictive value for the knowledge base of 82.1%. Some causes of false positive matches included wrong dose (e.g. linking Aspirin 81 MG Tablet to Prepatellar Bursitis) and wrong route of administration (e.g. linking Prednisone 20 MG Tablet to Hypertrophic Scar). For those medications not linked, 210 (8.4%) should have matched to a problem on the patient’s list. These results indicate a sensitivity of 37.1% and specificity of 98.9% (Table 2).
Table 2.
Results of expert problem-medication link review.
SMART App
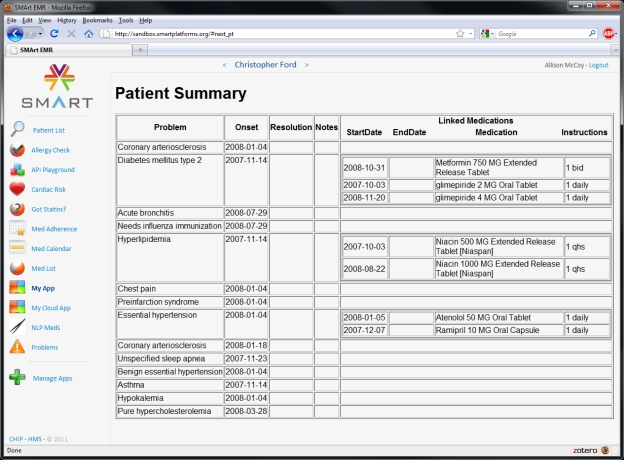
Figure 2 depicts a screenshot of the problem-medication linking app within the SMART Reference EMR. The app successfully retrieved patient problem and medication data using the SMART API and linked medications to relevant problems using the new knowledge base.
Figure 2.
Screenshot of SMART app for test patient in SMART Reference EMR.
Discussion
Summary of Findings
We aimed to first generate a knowledge base for linking medications to problems and then design a prototype SMART app to organize and display a patient’s active medications by associated clinical problem. Using the UMLS Metathesaurus, we successfully created a knowledge base that linked medications to patient’s problems with high specificity. The low sensitivity of the knowledge base may be due to more general or precise use of medications and problems by providers in patient records than the knowledge base covers, though this may be improved with a more expansive knowledge base. Likewise, the lower than expected efficiency may be improved by optimization to both the knowledge base and summary display code. The SMART app effectively linked medications and problems for test patients in the SMART reference EMR.
Others have described similar methods for automatically linking medications and problems. For example, Burton, et al. also mapped medications and diseases using RxNorm, NDF-RT, and SNOMED CT with reasonable sensitivity and specificity (11). Our attempt expands these methods by including related problems with the “isa” relationship within SNOMED CT to map problems with varying specificity in the EHR data. In addition, our employment of the SMART API to construct a patient summary using the knowledge base provides a generalizable product that may be implemented and evaluated by any entity with a SMART-enabled HIT system.
Limitations
This research has a number of limitations. First, it is necessary that any system utilizing the knowledge base have medications and clinical problems that are linked to RxNorm and SNOMED CT. Such mappings may be difficult, as internal vocabularies may contain terms that convey other semantic types, and no equivalent term exists in the corresponding vocabularies (16). Limitations still exist with internal medications and clinical problems completely mapped to existing vocabularies, as these have been found to be incomplete (17). Mapping between medications and clinical problems may also be limited by incompleteness or inaccuracies in a patient’s medication and problem lists. Providers may enter incorrect problems for patients or fail to enter some problems altogether.
Future Work
As our methods are limited by inaccurate or incomplete patient problem lists, an initial direction for our future work includes suggestion of problems to be added to the patient’s list. One approach might involve a previously described method for automatically associating patient medications and problems (12). Other approaches may use inferences based on frequent prior manual mappings or related problems within the new knowledge base with high utilization, which may be obtained from the SNOMED CT CORE problem list (18). Another future aim includes improvement of the existing knowledge base and summary display code. Curation of the knowledge base, such as addition of relationships known to exist but not occurring in NDF-RT or removal of frequently false positive relationships may improve sensitivity and specificity. Additionally, grouping of problems within the knowledge base may allow better summarization of the data. Finally, we aim to add vocabularies and relationships to the knowledge base to include linking of laboratory results and other relevant data to the summary.
Conclusion
The newly generated knowledge base performed with a high specificity to organize patient’s medications by their clinical problems, and the SMART app effectively served as a generalizable patient summary display. Further research is necessary to improve the sensitivity and efficiency of the knowledge base.
Acknowledgments
This project was supported in part by Grant No. 10510592 for Patient-Centered Cognitive Support under the Strategic Health IT Advanced Research Projects Program (SHARP) from the Office of the National Coordinator for Health Information Technology, the Houston VA HSR&D Center of Excellence (HFP90-020), and NCRR Grant 3UL1RR024148. The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs or any of the other funding agencies.
References
- 1.Singh H, Naik AD, Rao R, Petersen LA. Reducing diagnostic errors through effective communication: harnessing the power of information technology. J Gen Intern Med. 2008 Apr;23(4):489–494. doi: 10.1007/s11606-007-0393-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh H, Thomas EJ, Mani S, Sittig D, Arora H, Espadas D, et al. Timely follow-up of abnormal diagnostic imaging test results in an outpatient setting: are electronic medical records achieving their potential? Arch Intern Med. 2009 Sep 28;169(17):1578–1586. doi: 10.1001/archinternmed.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sittig DF, Singh H. Legal, ethical, and financial dilemmas in electronic health record adoption and use. Pediatrics. 2011 Apr;127(4) doi: 10.1542/peds.2010-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feblowitz JC, Wright A, Singh H, Samal L, Sittig DF. Summarization of Clinical Information: A Conceptual Model and Case Study. J Biomed Inform. 2011 doi: 10.1016/j.jbi.2011.03.008. (In Press). [DOI] [PubMed] [Google Scholar]
- 5.Sittig DF, Teich JM, Osheroff JA, Singh H. Improving clinical quality indicators through electronic health records: it takes more than just a reminder. Pediatrics. 2009 Jul;124(1):375–377. doi: 10.1542/peds.2009-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UMLS® Reference Manual. Bethesda (MD): National Library of Medicine; 2009. [Google Scholar]
- 7.Liu S, Ma Wei, Moore R, Ganesan V, Nelson S. RxNorm: prescription for electronic drug information exchange. IT Professional. 2005;7(5):17–23. [Google Scholar]
- 8.SNOMED Clinical Terms® (SNOMED CT®) [Internet] 2009. Jul 20, [cited 2011 Mar 8]; Available from: http://www.nlm.nih.gov.ezproxyhost.library.tmc.edu/research/umls/Snomed/snomed_main.html.
- 9.Carter JS, Brown SH, Erlbaum MS, Gregg W, Elkin PL, Speroff T, et al. Initializing the VA medication reference terminology using UMLS metathesaurus co-occurrences. Proc AMIA Symp. 2002:116–120. [PMC free article] [PubMed] [Google Scholar]
- 10.Mandl KD, Kohane IS. No small change for the health information economy. N Engl J Med. 2009 Mar 26;360(13):1278–1281. doi: 10.1056/NEJMp0900411. [DOI] [PubMed] [Google Scholar]
- 11.Burton MM, Simonaitis L, Schadow G. Medication and indication linkage: A practical therapy for the problem list?. AMIA Annu Symp Proc; 2008. pp. 86–90. [PMC free article] [PubMed] [Google Scholar]
- 12.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform. 2010 Dec;43(6):891–901. doi: 10.1016/j.jbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Brown SH, Miller RA, Camp HN, Guise DA, Walker HK. Empirical derivation of an electronic clinically useful problem statement system. Ann Intern Med. 1999 Jul 20;131(2):117–126. doi: 10.7326/0003-4819-131-2-199907200-00008. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Q, Cimino JJ, Zou KH. Providing concept-oriented views for clinical data using a knowledge-based system: an evaluation. J Am Med Inform Assoc. 2002 Jun;9(3):294–305. doi: 10.1197/jamia.M1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Establishment of the Permanent Certification Program for Health Information Technology [Internet] Office of the National Coordinator for Health Information Technology, Department of Health and Human Services; Available from: http://origin.www.gpo.gov/fdsys/pkg/FR-2011-01-07/pdf/2010-33174.pdf. [PubMed]
- 16.Van Vleck TT, Wilcox A, Stetson PD, Johnson SB, Elhadad N. Content and structure of clinical problem lists: a corpus analysis. AMIA Annu Symp Proc; 2008. pp. 753–757. [PMC free article] [PubMed] [Google Scholar]
- 17.Pathak J, Chute CG. Analyzing categorical information in two publicly available drug terminologies: RxNorm and NDF-RT. J Am Med Inform Assoc. 2010 Aug;17(4):432–439. doi: 10.1136/jamia.2009.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fung KW, McDonald C, Srinivasan S. The UMLS-CORE project: a study of the problem list terminologies used in large healthcare institutions. J Am Med Inform Assoc. 2010 Nov 1;17(6):675–680. doi: 10.1136/jamia.2010.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]