Abstract
We present an extension to literature-based discovery that goes beyond making discoveries to a principled way of navigating through selected aspects of some biomedical domain. The method is a type of “discovery browsing” that guides the user through the research literature on a specified phenomenon. Poorly understood relationships may be explored through novel points of view, and potentially interesting relationships need not be known ahead of time. In a process of “cooperative reciprocity” the user iteratively focuses system output, thus controlling the large number of relationships often generated in literature-based discovery systems. The underlying technology exploits SemRep semantic predications represented as a graph of interconnected nodes (predication arguments) and edges (predicates). The system suggests paths in this graph, which represent chains of relationships. The methodology is illustrated with depressive disorder and focuses on the interaction of inflammation, circadian phenomena, and the neurotransmitter norepinephrine. Insight provided may contribute to enhanced understanding of the pathophysiology, treatment, and prevention of this disorder.
Introduction
Sophisticated methods are needed to supplement traditional information retrieval tools for effectively exploiting the large amount of online textual resources currently available. An active area of research in biomedicine in this regard is literature-based discovery (LBD), the primary goal of which is to help researchers make new discoveries by generating novel hypotheses. As pioneered by Swanson,1 the basic underlying principle of the LBD paradigm is that relations A − B and B − C may be known, yet relation A − C has gone unnoticed. Earlier LBD systems2, 3, 4 used concept cooccurrence as their primary mechanism for representing relations. Since only some cooccurrences underlie “interesting” relations, this has drawbacks, which have been addressed first by Hristovski et al.5 and later by Cohen et al.6 with the use of semantic relations. The use of discovery patterns5 is a further refinement for focusing on useful relations. One such pattern5 is Maybe_treats, which says (in part) that a therapeutic agent C maybe treats disease A if the level of an important measurement B is typically increased in patients with disease A and if C is able to reduce the level of B. Additional discovery patterns have been investigated.7, 8
We present a novel LBD methodology incorporating semantic predications and graph-based methods in order to guide researchers through the relevant literature on a user-specified biomedical phenomenon. The motivation is to extend LDB methodology beyond making discoveries to a principled way of navigating through selected aspects of some research area.9 An additional goal is to go beyond document retrieval in response to a query by revealing crucial relationships in the domain, which may evolve as the user exploits the method. Related work in network analysis of microarray data is becoming widely used in systems-based research for drug discovery.10
LBD provides the ability to uncover previously implicit or unnoticed relationships in the research literature, but has been primarily used when component relationships of the final discovery are already known. In the method we propose, it is not necessary to know ahead of time which relationships may be useful for guiding the research process. Only the general content area need be specified. The method might be thought of as “discovery browsing,” using graph theoretic paths to generalize Hristovski’s discovery patterns, not with the purpose of necessarily making a discovery, but of explicating poorly understood relationships, by providing novel points of view on some research problem. The method involves an interaction between user decisions and system results. Such “cooperative reciprocity” focuses system output iteratively based on stipulations that bring relevant relations into clearer focus by narrowing choices, thus controlling the explosion of potential relationships often generated in LBD.
The underlying technology depends on semantic predications extracted from MEDLINE citations using SemRep11 and represented as a large graph of interconnected nodes (predication arguments) and edges (predicates). The graph-theoretic constructs degree centrality and path analysis are used to suggest paths in this graph, which represent chains of relationships that may guide the research process. The methodology is illustrated with selected aspects of depressive disorder.
Background
Semantic predications
The predications for this study were provided by the SemRep system,11 which relies on biomedical domain knowledge in the Unified Medical Language System (UMLS). Access to the UMLS Metathesaurus is provided by MetaMap,12 while a set of semantic relationships (predications) is extracted based on the UMLS Semantic Network. For example, SemRep extracts the relationship: “Leptin STIMULATES Serotonin” from text “CNS serotonin activated by leptin modulates sympathetic outflow to the skeleton.” Although coverage of text processed by SemRep is a limiting factor in this work, the expressiveness of semantic predications positively contributes to the discovery process. Several evaluations of SemRep (e.g. Ahlers et al.13) indicate that precision is near 75%. SemRep has been used to extract nearly 25 million semantic predication instances from some 7 million MEDLINE citations (titles and abstracts dating from 1999 through 2010). These are stored in a MySQL database, which was exploited for this work.
Literature-based discovery
Swanson’s1 paradigm is based on concepts A and C coming from two different, nonoverlapping, domains. The goal is to find an intermediate concept B, which occurs with both A and C, and validates a new, earlier unknown, A − C relationship. Such a discovery is called an open discovery. Another type of discovery, a closed discovery, assumes that a relationship A − C is known. Then, a common concept B and relations A − B and B − C are to be found in order to explicate the relationship A − C.
The aim of our work is to expand the B element of the A − B − C paradigm. Our methodology considers B not as a single concept, but as a subchain of intermediate concepts, where A − B − C has the form in (1), where n ∈ [1, ∞).
| (1) |
Investigation of possible chains of relationships may result in a discovery (either open or closed). Swanson1 introduced the possibility of having a chain of Bs between A and C, but this has not been extensively exploited. We use semantic predications to represent these relationships, and we decided to use graphs as a medium for representing the predications. Referring to graph theory terminology, we implement discovery chains as paths in a graph.
Graph theory
A graph is a representation of connections (edges) between objects (nodes). Graphs, also known as networks, are extensively studied in social network analysis and the Semantic Web. Graph theory is a set of functions and measures pertaining to graph properties. One such measure used in this paper is degree centrality, which measures the connectedness of nodes in a graph. A node with more connections (relationships) to other nodes has higher degree centrality. Freeman14 describes degree centrality as an indicator of the communication activity in a social network. The importance of high connectivity is also seen in gene interaction networks (p53, for example).15 In our case degree centrality may be considered as an indicator of the principal substances in the domain for which the graph was constructed. The formula for degree centrality of node v in a graph with n nodes is:14
| (2) |
In graph theory, a path is a sequence of edges connecting any two nodes in the graph. Paths may be of any length. The shortest is of length 1:
| (3) |
The longest is of length N − 1, where N is the number of nodes in the graph:
| (4) |
In Semantic Web research on ranking paths of semantic associations Anyanwu et al.16 exploit the notion of “predictability.” In their results longer paths more likely reveal rare and uncommon associations.
Dupont et al.17 discuss many walking approaches in a graph (edge passages), which may be also understood as extraction of paths from the graph. The definitions of maximal length of the edge passage (k-walk) and nodes of interest are based on this work. The nodes of interest are the start and end points of a walk in a graph. For them, length of the walk is the number of intermediate nodes visited during a walk between nodes of interest. We measure path length by the number of edges between the start and end nodes.
Methods
Overview
The procedure for exploiting paths in a graph to facilitate discovery browsing involves several steps: creating a graph of relevant predications, extracting and ranking paths, and finally, inspecting a small subgraph based on selected paths. At several steps in the process, system output is filtered based on user stipulation, representing the cooperative reciprocity involved in uncovering research insights in the domain. A crucial assumption of the system is that the user brings to bear domain knowledge as part of the process of navigating and focusing in the selected area of interest.
Creating the initial graph is an iterative process in which the user specifies a seed concept to extract predications from the SemRep predication database. (For this project, extracted predications were limited to those with one of the substance interaction predicates: STIMULATES, INHIBITS, INTERACTS_WITH, and COEXISTS_WITH.) Concepts in the graph are ranked by degree centrality, and a new seed concept is selected from those highest on the list, which is used to extract additional predications to be added to the growing graph. When a graph of sufficient size to produce “interesting” results has been generated, paths between stipulated concepts are extracted and ranked, also based on degree centrality. Finally, the user selects paths for further analysis. We will illustrate system processing with depressive disorder as the domain of interest.
Create graph
Serotonin was selected as the seed concept for investigating depression, since it is known to be a prominent neurotransmitter in this disorder. We extracted all predications from the database that had an argument containing the string “seroton” (ignoring case). In addition to “Serotonin” this included 183 concepts, such as “serotonin receptor,” “Serotonin Agonists,” “Serotonin Agents,” and “Serotonin 5-HT-3 Receptor.” (This was an implementation expedient. In the future, ontology resources will be exploited.) Retrieved predications were loaded into a graph consisting of 1561 nodes (concepts) and 7061 edges (predications) using the NetworkX18 software package written in the Python programming language. A path of length one in this graph represents all the predication instances in the database having the two nodes as either subject or object. For example, the path “Estradiol-stimulates-Serotonin” represents two instances of the corresponding predication extracted from MEDLINE citations (PMIDs: 16736471 and 19168037). Links between an edge in the graph and sentences in citations from which corresponding predications were extracted are maintained by the system.
As a first step in expanding this graph, we calculated degree centrality and ranked the results. Ignoring the concepts containing “serotonin,” melatonin was high on this list, and was chosen to expand the serotonin graph. This was a user choice which focused the subsequent graph on a particular aspect of depression. Other substances high on the degree centrality list, which could have been selected, were “Estrogens,” “Dopamine,” and “Corticosterone.” We then retrieved all predications from the SemRep predication database that had an argument containing the string “melato” (ignoring case). In addition to “Melatonin,” this included 126 concepts, such as “Melatonin Receptors” and “Receptor, Melatonin, MT2.” The resultant graph (containing both serotonin- and melatonin-related concepts) consisted of 2207 nodes (concepts) and 11,752 edges (predications).
The growing graph of predications for depression was expanded a third (and final) time. Degree centrality was again calculated and the results ranked. Before prominent concepts were selected from this list, a stop list (based on user domain knowledge) was applied to remove uninformative concepts (e.g. “Pharmaceutical Preparations”), classes of both drugs and body substances (e.g. “Antidepressive Agents,” “agonists”), and physiologically general terms (e.g. “Water,” “Oils”). Serotonin- and melatonin-related concepts were also ignored since predications with them were already in the graph. The top 140 concepts from this filtered degree centrality list were used in another query to the predication database and the retrieved predications were added to the graph. The resulting graph of predications for the depression study, consisting of 22,828 nodes (concepts) and 435,437 edges (predications), formed the basis for further processing.
Extract paths
The next step was to extract paths from the graph, which, as in constructing the graph, involved an interaction of system output and user stipulations. Serotonin and melatonin were selected as anchors, and all paths of length four between them were extracted from the graph using the depth-first algorithm.19 This value was selected as a compromise. Longer paths are likely to provide more revealing results for discovery;16 however, considerations of processing time with the current implementation imposed this limitation. The total number of paths extracted was 4,206,647, and all had the form:
| (5) |
Before proceeding, we removed all paths in which one of the five concepts was on the stop list of general and un-informative concepts noted above. (Such concepts “crept” into the graph by being arguments of predications with non-stopped concepts.) 3,840,958 remained, and a composite degree centrality score was computed for each. This was calculated as the arithmetic sum of the degree centrality values for all five nodes in the path:
| (6) |
The list of paths was ranked based on the composite degree centrality score.
As a further step in limiting the number of paths for analysis, we selected only those containing the concept “CLOCK,” based on domain knowledge that recent research implicates the clock genes in depression.20 The remaining paths (16,141) had one of the following patterns:
| (7) |
| (8) |
| (9) |
We further focused this list by eliminating concepts that we chose not to consider at this point. Paths with concepts such as “Glucose,” “Antibodies,” “Lipopolysaccharides,” “Kinases” etc. were removed (again, based on user domain knowledge), leaving 5,406 paths.
Analyze paths
From this list we chose the top twenty paths (sorted by composite degree centrality) for further analysis.
[Melatonin]-[Interleukin-1 beta]-[Glutamate]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Glutamate]-[Interleukin-1 beta]-[Serotonin]
[Melatonin]-[Insulin]-[Glutamate]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Glutamate]-[Insulin]-[Serotonin]
[Melatonin]-[Interleukin-6]-[Glutamate]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Glutamate]-[Interleukin-6]-[Serotonin]
[Melatonin]-[Interleukin-1 beta]-[Norepinephrine]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Norepinephrine]-[Interleukin-1 beta]-[Serotonin]
[Melatonin]-[Cholesterol]-[Glutamate]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Glutamate]-[Cholesterol]-[Serotonin]
[Melatonin]-[Insulin]-[Norepinephrine]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Norepinephrine]-[Insulin]-[Serotonin]
[Melatonin]-[Interleukin-1 beta]-[Interferon Type II]-[CLOCK]-[Serotonin]
[Melatonin]-[Interleukin-6]-[Norepinephrine]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Norepinephrine]-[Interleukin-6]-[Serotonin]
[Melatonin]-[Insulin]-[Interferon Type II]-[CLOCK]-[Serotonin]
[Melatonin]-[CLOCK]-[Dopamine]-[Interleukin-1 beta]-[Serotonin]
[Melatonin]-[Interleukin-1 beta]-[Dopamine]-[CLOCK]-[Serotonin]
[Melatonin]-[Interleukin-6]-[Interferon Type II]-[CLOCK]-[Serotonin]
[Melatonin]-[Insulin]-[Dopamine]-[CLOCK]-[Serotonin]
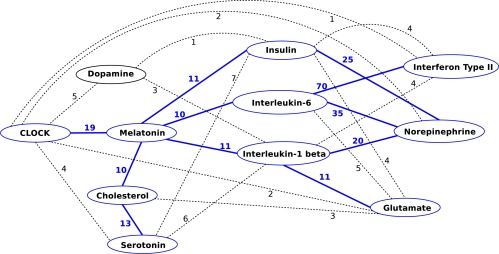
Eleven unique concepts are involved in these paths: “Interleukin-1 beta,” “Interleukin-6,” “Glutamate,” “Dopamine,” “Norepinephrine,” “Insulin,” “Cholesterol,” and “Interferon type II” (in addition to the stipulated “Serotonin,” “Melatonin,” and “CLOCK”). The graphical representation of these paths is shown in Figure 1. Numbers on the edges in Figure 1 show the number of predication instances (limited to substance interaction predicates) represented by that edge. Links to the corresponding citation sentences are maintained by the system and underpin the guidance provided to the user. It should be noted that there is considerable overlap among the predications represented in the paths. Edges with fewer than ten predications are dashed in this graph. We then filtered the subgraph further to include only those connections that occurred ten times or more, thus eliminating “Dopamine.” This filtering also highlights the more direct connection between CLOCK and melatonin, which was not the initial intent of this exploration, but nevertheless is an indication that this visualization highlights important known relationships.
Figure 1:
Graph representing the top twenty paths. Numbers on the edges show the number of predication instances.
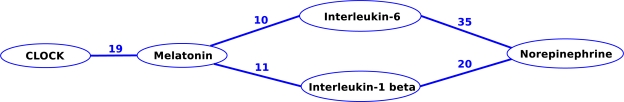
There are several “stories” implicit in Figure 1. We further decided to concentrate on just one, namely the interactions among melatonin, the clock genes, interleukin-1 beta (IL-1 beta), interleukin-6 (IL-6), and norepinephrine, as shown in Figure 2.
Figure 2:
User selected subgraph.
Results
Each of the 95 predications found in Figure 2 was analyzed for SemRep accuracy, and it was determined that 88% were correct. Each of the citations from which these predications had been extracted was then inspected. In general we found that even incorrect predications often reveal relevant, perhaps unknown, relationships that can in turn lead to further research. Below we give examples of predications drawn from citations that provide insight into the three aspects of depression covered by Figure 2: melatonin and CLOCK, melatonin and the two proinflammatory cytokines, the two cytokines and norepinephrine.
CLOCK stimulates Melatonin:
Although this predication is wrong, the citation from which it was extracted provides important information about the relationship. “This interaction, . . . may reflect the central role of melatonin, i.e. in synchronising peripheral clock cells that require unique phasing of output signals with the master clock in the brain.”21
Melatonin interacts with CLOCK / CLOCK interacts with Melatonin:
“. . . whereas an ‘internal coincidence model’ best explains the way melatonin affects the phasing of clock gene expression . . . ”22 “In mammals, the nocturnal rise in pineal melatonin is regulated by signals from the endogenous clock . . . ”23
Melatonin inhibits Interleukin-1 beta / Melatonin inhibits Interleukin-6 / Melatonin stimulates Interleukin-1 beta / Melatonin stimulates Interleukin-6:
“Further melatonin repressed the upregulated levels of expression of proinflammatory cytokines like, TNF-alpha, IL-1beta and IL-6 in RE.” (in experimental reflux esophagitis)24 “Treatment with melatonin significantly increased the levels of IL- 1beta, IL-6, . . . ” (in collagen-induced arthritis)25
Several predications are about the effect of norepinephrine on either interleukin-1 beta or interleukin-6. We did not pursue this relationship in this project.
Interleukin-6 stimulates Norepinephrine:
For the effect of IL-6 on norepinephrine, all of the predications were incorrect. Some of them are nonetheless useful in reporting on a reciprocal relationship between IL-6 and norepinephrine, even if a direct interaction is not noted. “In addition, plasma levels of IL-6 and IL-2 were increased in four stress groups, serum norepinephrine and dopamine were decreased dramatically in stress group and stress low-dose GTPs modulation group.”26
Interleukin-1 beta inhibits Norepinephrine:
“These results suggest that IL-1 beta could decrease NE levels”27 “. . . IL-1beta-induced suppression of the LH surge is most probably mediated through an increase in GABA levels in the MPA which causes a reduction in NE levels.”28
Interleukin-1 beta stimulates Norepinephrine:
“While acute treatment with IL-1beta increased NE concentrations in both the paraventricular nucleus and the median eminence (ME), chronic treatment increased NE concentrations only in the ME.”29 “These results indicate that IL-1beta increases NE levels both in the PVN and in the ME and this could be a possible mechanism by which it stimulates the HPA axis.”30 “We observed that IL-1beta increases the release of NPY, norepinephrine (NE), and epinephrine (EP) from human chromaffin cells.”31
Discussion
The methodology presented was illustrated with selected aspects of depressive disorder and focuses on the interaction of inflammation (particularly the proinflammatory cytokines interleukin-1 beta and interleukin-6), circadian phenomena (clock genes and melatonin), and the neurotransmitter norepinephrine. Below we give an overview of the extent of current research on these aspects of depression (PubMed queries issued on 03/10/2011) and suggest how our results may contribute to an understanding of the pathophysiology of this disorder.
There is considerable research investigating circadian phenomena and depression. The PubMed query “(circadian rhythms[mh] OR clock OR melatonin) AND depression[mh]” returns 331 citations. For example, Kennaway20 reports on the clock genes and behavioral disorders, including depression. Melatonin figures prominently in this review, but a mechanism is not proposed. Rosenwasser32 reviews research on the clock genes and psychiatric disorders more generally, but mechanisms and the connection with inflammation are not highlighted. Our results provide considerable detail on the mechanisms involved.
Far less research has investigated the interaction of circadian phenomena and inflammation with respect to depression. The PubMed query “cytokine[mh] AND (circadian rhythms[mh] OR clock OR melatonin) AND depression[mh]” only returns 18 citations. When limited to reviews, only 8 are retrieved with this query; several are concerned with specific disorders, such as rheumatologic disorders33 and cancer.34 Only one covers general considerations of the interaction of circadian phenomena and inflammation.35 Our results suggest details concerning the interaction of melatonin and the two proinflammatory cytokines interleukin-1 beta and interleukin-6.
The role of inflammation in depression has been extensively studied. The PubMed query “(inflammation OR cytokines) AND depression[mh]” returns 1500 citations (41 reviews). Raison et al.36 and Anisman,37 for example, provide particularly lucid overviews. Our results are not to be thought of as uncovering the insight that inflammation is intimately connected with depression (for which there is considerable evidence), but rather they provide additional information about the interaction of specific cytokines (IL-1 beta and IL-6) and norepinephrine.
Although the noradrenergic system is known to be involved in depression, the mechanistic details are still being investigated.38 Norepinephrine has not been targeted as intensely as serotonin in therapeutic approaches.39, 40 Only one norepinephrine reuptake inhibitor (duloxetine) is currently prescribed (in Europe, not in the U.S.).39 Recently, combined serotonin and norepinephrine reuptake inhibitors are being used.41, 42 Although it is known that the cytokines interact with norepinephrine,43 the particular mechanism of IL-1 beta and IL-6 has not been intensively studied (PubMed query “norepinephrine AND (interleukin-1 beta OR IL-1 beta OR interluekin-6 OR IL-6) AND depression” returns 35 citations). Our results point to considerable evidence of the interaction of IL-6 and IL-1 beta (in particular) in a variety of contexts beyond cerebral structures, and thus may suggest new avenues for research in explicating the details.
Finally, there is very little research investigating the comprehensive interaction of inflammatory processes, circadian phenomena, and noradrenergic aspects of depression. The specific PubMed query “(interleukin-1 beta OR IL-1 beta OR interluekin-6 OR IL-6) AND melatonin AND norepinephrine AND depression” returns no citations. The more general “cytokines AND (circadian rhythms[mh] OR clock OR melatonin) AND norepinephrine AND depression[mh]” returns three citations. One of these39 is a clinically oriented review and does not discuss any of the mechanisms addressed in this paper. The other two44, 45 cover research more generally, and report substance levels consistent with our results, but do not suggest mechanisms.
We approached high connectedness by filtering to include only those connections with 10 or more occurrences. One weakness of this strategy is that it focuses on the more often studied relations at the expense of others. An alternative approach would be to look at this from the opposite angle, and calculate degree centrality using relations with fewer occurrences each. This may highlight areas that are highly connected but have not been explored in a coordinated manner. A second aspect that can be explored is by taking into account the directionality of the relations – incoming and outgoing relations – and calculating directed paths.
Conclusion
We introduced a novel LBD methodology incorporating semantic predications and graph analysis that guides researchers through the research literature on a user-specified biomedical phenomenon. A type of “discovery browsing” exploits graph theoretic paths in order to elucidate poorly understood relationships by providing novel points of view on some research problem. The user is not required to specify which relationships may serve as a useful guide. A core aspect of the method is that the user brings to bear domain knowledge as part of the process of navigating in the selected area of interest. Such “cooperative reciprocity” focuses system output iteratively, thus controlling the explosion of potential relationships often generated in LBD.
In the method, relationships in MEDLINE citations are represented as a (large) graph of interconnected semantic predications. The system suggests paths in this graph, which represent interesting chains of relationships. The underlying technology depends on semantic predications provided by SemRep, and the graph theoretic constructs degree centrality and path analysis are particularly exploited.
We illustrated our methodology with depressive disorder, and there are three major components of our results: 1) inflammation and depression, 2) circadian phenomena and depression, 3) noradrenergic aspects of depression. Varying amounts of research have been devoted to each of these components, but little (if any) has considered all three together. Our results do not constitute a discovery in the sense of something previously not noticed by anyone. However, in several respects they contribute to various aspects of depression that are currently incompletely understood and have not been extensively studied. Insight into the interrelationships among all these components may materially contribute to unraveling the underlying pathophysiology of depression, thus underpinning more effective treatment (and prevention).
Acknowledgments
This research was supported in part by an appointment to the NLM Research Participation Program. This program is administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the National Library of Medicine. This research was also supported in part by the Intramural Research Program of the NIH, National Library of Medicine.
The first author also gratefully acknowledges funding from the Lundbeckfonden through the Center for Integrated Molecular Brain Imaging (Cimbi.org), Otto Mønsteds Fond, Kaj og Hermilla Ostenfelds Fond, and the Ingeniør Alexandre Haynman og hustru Nina Haynmans Fond.
References
- 1.Swanson DR. Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspectives in Biology and Medicine. 1986;30(1):7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 2.Swanson DR, Smalheiser NR. An interactive system for finding complementary literatures: a stimulus to scientific discovery. Artificial Intelligence. 1997;91(2):183–203. [Google Scholar]
- 3.Hristovski D, Stare J, Peterlin B, Dzeroski S. Studies in Health Technology and Informatics. IOS Press; 2001. Supporting discovery in medicine by association rule mining in Medline and UMLS; pp. 1344–1348. [PubMed] [Google Scholar]
- 4.Weeber M, Kors JA, Mons B. Online tools to support literature-based discovery in the life sciences. Briefings in Bioinformatics. 2005;6(3):277–286. doi: 10.1093/bib/6.3.277. [DOI] [PubMed] [Google Scholar]
- 5.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery. AMIA Annual Symposium Proceedings. 20062006:349–353. [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen T, Whitfield GK, Schvaneveldt RW, Mukund K, Rindflesch T. EpiphaNet: an interactive tool to support biomedical discoveries. Journal of Biomedical Discovery and Collaboration. 2010;5:22–44. [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlers CB, Hristovski D, Kilicoglu H, Rindflesch TC. Using the literature-based discovery paradigm to investigate drug mechanisms. AMIA Annual Symposium Proceedings. 2007:6–10. [PMC free article] [PubMed] [Google Scholar]
- 8.Hristovski D, Kastrin A, Peterlin B, Rindflesch T. Combining semantic relations and DNA microarray data for novel hypotheses generation. Linking Literature, Information, and Knowledge for Biology. 2010:53–61. [Google Scholar]
- 9.Smalheiser NR, Torvik VI, Bischoff-Grethe A, Burhans LB, Gabriel M, Homayouni R, Kashef A, Martone ME, Perkins GA, Price DL, et al. Collaborative development of the arrowsmith two node search interface designed for laboratory investigators. Journal of Biomedical Discovery and Collaboration. 2006;1(1):8. doi: 10.1186/1747-5333-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley JT, Schadt E, Sirota M, Butte AJ, Ashley E. Drug discovery in a multidimensional world: Systems, patterns, and networks. Journal of Cardiovascular Translational Research. 2010;3:438–47. doi: 10.1007/s12265-010-9214-6. [DOI] [PubMed] [Google Scholar]
- 11.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. Journal of Biomedical Informatics. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahlers CB, Fiszman M, Demner-Fushman D, Lang F, Rindflesch TC. Extracting semantic predications from MEDLINE citations for pharmacogenomics. Pacific Symposium on Biocomputing 2007. 2007:209–220. [PubMed] [Google Scholar]
- 14.Freeman LC. Centrality in social networks conceptual clarification. Social Networks. 1979;1(3):215–239. [Google Scholar]
- 15.Vogelstein B, Lane D, Levine AJ, et al. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 16.Anyanwu K, Maduko A, Sheth A. SemRank: ranking complex relationship search results on the semantic web. Proceedings of the 14th International Conference on World Wide Web; 2005. pp. 117–127. [Google Scholar]
- 17.Dupont P, Callut J, Dooms G, Monette JN, Deville Y. Relevant subgraph extraction from random walks in a graph. Research Report RR. 2006;38016:7. [Google Scholar]
- 18.Hagberg AA, Schult DA, Swart PJ. Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science Conference (SciPy2008); 2008. pp. 11–15. [Google Scholar]
- 19.Tarjan R. Depth-first search and linear graph algorithms. Conference Record 1971 Twelfth Annual Symposium on Switching and Automata Theory; 1971. pp. 114–121. [Google Scholar]
- 20.Kennaway DJ. Review: Clock genes at the heart of depression. Journal of Psychopharmacology. 2010;24(2 suppl):5–14. doi: 10.1177/1359786810372980. [DOI] [PubMed] [Google Scholar]
- 21.Stehle JH, von Gall C, Korf HW. Organisation of the circadian system in melatonin-proficient C3H and melatonin-deficient C57BL mice: a comparative investigation. Cell and Tissue Research. 2002;3091:173–182. doi: 10.1007/s00441-002-0583-2. [DOI] [PubMed] [Google Scholar]
- 22.Lincoln G, Messager S, Andersson H, Hazlerigg D. Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: evidence for an internal coincidence timer. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(21):13890–5. doi: 10.1073/pnas.212517599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang CS, Mulnier C, Pang SF, Yang JCS. Effects of halothane, pentobarbital and ketamine on serum melatonin levels in the early scotophase in New Zealand white rabbits. Neurosignals. 2001;10(5):310–316. doi: 10.1159/000046898. [DOI] [PubMed] [Google Scholar]
- 24.Lahiri S, Singh P, Singh S, Rasheed N, Palit G, Pant KK. Melatonin protects against experimental reflux esophagitis. Journal of Pineal Research. 2009;46(2):207–213. doi: 10.1111/j.1600-079X.2008.00650.x. [DOI] [PubMed] [Google Scholar]
- 25.Jiménez-Caliani AJ, Jiménez-Jorge S, Molinero P, Guerrero JM, Fernández-Santos JM, Martín-Lacave I, Osuna C. Dual effect of melatonin as proinflammatory and antioxidant in collagen-induced arthritis in rats. Journal of Pineal Research. 2005;38(2):93–99. doi: 10.1111/j.1600-079X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen WQ, Zhao XL, Hou Y, Li ST, Hong Y, Wang DL, Cheng YY. Protective effects of green tea polyphenols on cognitive impairments induced by psychological stress in rats. Behavioural Brain Research. 2009;202(1):71–76. doi: 10.1016/j.bbr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Sirivelu MP, Shin AC, Perez GI, MohanKumar PS, MohanKumar SMJ. Effect of l-dopa on interleukin-1-induced suppression of luteinizing hormone secretion in intact female rats. Human Reproduction. 2009;24(3):718–725. doi: 10.1093/humrep/den434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirivelu MP, Burnett R, Shin AC, Kim C, MohanKumar PS, MohanKumar SMJ. Interaction between GABA and norepinephrine in interleukin-1beta-induced suppression of the luteinizing hormone surge. Brain Research. 2009;1248:107–14. doi: 10.1016/j.brainres.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MohanKumar SMJ, Smith CL, MohanKumar PS. Central adaptation to chronic administration of interleukin-1beta (IL-1beta) in rats. Brain Research Bulletin. 2003;62(1):71–6. doi: 10.1016/j.brainresbull.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 30.MohanKumar SMJ, MohanKumar PS. Systemic Interleukin-1 [beta] stimulates the simultaneous release of norepinephrine in the paraventricular nucleus and the median eminence. Brain Research Bulletin. 2005;65(5):451–456. doi: 10.1016/j.brainresbull.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Rosmaninho-Salgado J, Araújo IM, Álvaro AR, Mendes AF, Ferreira L, Grouzmann E, Mota A, Duarte E, et al. Regulation of catecholamine release and tyrosine hydroxylase in human adrenal chromaffin cells by interleukin-1β: role of neuropeptide Y and nitric oxide. Journal of Neurochemistry. 2009;109(3):911–922. doi: 10.1111/j.1471-4159.2009.06023.x. [DOI] [PubMed] [Google Scholar]
- 32.Rosenwasser AM. Circadian clock genes: Non-circadian roles in sleep, addiction, and psychiatric disorders? Neuroscience & Biobehavioral Reviews. 2010;34(8):1249–1255. doi: 10.1016/j.neubiorev.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Abad VC, Sarinas PSA, Guilleminault C. Sleep and rheumatologic disorders. Sleep Medicine Reviews. 2008;12(3):211–228. doi: 10.1016/j.smrv.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Miller AH, Ancoli-Israel S, Bower JE, Capuron L, Irwin MR. Neuroendocrine-immune mechanisms of behavioral comorbidities in patients with cancer. Journal of Clinical Oncology. 2008;26(6):971. doi: 10.1200/JCO.2007.10.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasan V, Spence DW, Trakht I, Pandi-Perumal SR, Cardinali DP, Maestroni GJ. Immunomodulation by melatonin: its significance for seasonally occurring diseases. Neuroimmunomodulation. 2008;15(2):93–101. doi: 10.1159/000148191. [DOI] [PubMed] [Google Scholar]
- 36.Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends in Immunology. 2006;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anisman H. Cascading effects of stressors and inflammatory immune system activation: implications for major depressive disorder. Journal of Psychiatry & Neuroscience: JPN. 2009;34(1):4. [PMC free article] [PubMed] [Google Scholar]
- 38.Itoi K, Sugimoto N. The brainstem noradrenergic systems in stress, anxiety and depression. Journal of Neuroendocrinology. 2010;22(5):355–361. doi: 10.1111/j.1365-2826.2010.01988.x. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy SH, Rizvi SJ. Emerging drugs for major depressive disorder. Expert Opinion on Emerging Drugs. 2009;14(3):439–53. doi: 10.1517/14728210903107751. [DOI] [PubMed] [Google Scholar]
- 40.El Mansari M, Guiard BP, Chernoloz O, Ghanbari R, Katz N, Blier P. Relevance of norepinephrine–dopamine interactions in the treatment of major depressive disorder. CNS Neuroscience & Therapeutics. 2010;16(3):e1–e17. doi: 10.1111/j.1755-5949.2010.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Berardis D, Conti CM, Serroni N, Moschetta FS, Olivieri L, Carano A, Salerno RM, Cavuto M, Farina B, Alessandrini M, et al. The effect of newer serotonin-noradrenalin antidepressants on cytokine production: a review of the current literature. International Journal of Immunopathology and Pharmacology. 2010;23(2):417–22. doi: 10.1177/039463201002300204. [DOI] [PubMed] [Google Scholar]
- 42.Nichols AI, Tourian KA, Tse SY, Paul J. Desvenlafaxine for major depressive disorder: incremental clinical benefits from a second-generation serotonin-norepinephrine reuptake inhibitor. Expert Opinion on Drug Metabolism & Toxicology. 2010;6(12):1565–74. doi: 10.1517/17425255.2010.535810. [DOI] [PubMed] [Google Scholar]
- 43.Dunn AJ, Wang J, Ando T. Cytokines, Stress, and Depression. 1999. Effects of cytokines on cerebral neurotransmission; pp. 117–127. [DOI] [PubMed] [Google Scholar]
- 44.Irwin M, Clark C, Kennedy B, Christian GJ, Ziegler M. Nocturnal catecholamines and immune function in insomniacs, depressed patients, and control subjects. Brain, Behavior, and Immunity. 2003;17(5):365–372. doi: 10.1016/s0889-1591(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 45.Rief W, Mills PJ, Ancoli-Israel S, Ziegler MG, Pung MA, Dimsdale JE. Overnight changes of immune parameters and catecholamines are associated with mood and stress. Psychosomatic Medicine. 2010;72(8):755. doi: 10.1097/PSY.0b013e3181f367e2. [DOI] [PMC free article] [PubMed] [Google Scholar]