Abstract
It is widely believed that different clinical domains use their own sublanguage in clinical notes, complicating natural language processing, but this has never been demonstrated on a broad selection of note types. Starting from formal sublanguage theory, we constructed a feature space based on vocabulary and semantic types used in 17 different clinical domains by three author types (physicians, nurses, and social workers) in both the in- and outpatient settings. We supplied the resulting vectors to CLUTO, a robust clustering tool suitable for this high-dimensional space. Our results confirm that note types with a broad clinical scope, e.g, History & Physicals and Discharge Summaries, cluster together, while note types with a narrow clinical scope form surprisingly pure, disjoint sublanguages. A reasonable conclusion from this study is that any tool relying on term statistics or semantics trained on one clinical note type may not work well on any other.
Introduction
Clinical natural language processing (NLP) is enjoying a surge of interest among researchers and informatics practitioners.1,2 Researchers in this field might be tempted to use an existing NLP system developed in one clinical domain or in one clinical setting (i.e., in- or outpatient) in a new domain or setting without modification. However, previous research has suggested that clinical language is not homogeneous but consists of several narrow specialized domains that exhibit the characteristics of sublanguages.3,4 For example, Hyun et al. noted in their study of nursing narratives using the MedLEE system (trained on chest X-ray reports) that “For better NLP performance in the domain of nursing, additional nursing terms and abbreviations must be added to MedLEE’s lexicon.”5 Natural language processing across clinical domains is challenging precisely because of the differences in the language characteristics across those domains. This effect is compounded by a diversity of note-author roles, such as nurses, pharmacists, social workers, physicians, as well as by the clinical setting.
Clinical sublanguage
The term “sublanguage” was first introduced into linguistic theory by Zellig Harris.6,7 According to Harris, languages in specialized domains exhibit certain characteristics that set them apart from general language. Since Harris’ first publication on the subject, clinical NLP researchers very quickly decided that the language used by clinicians is a proper sublanguage. Numerous research projects have compared clinical language to the language of biomedical literature,8 to general English,9 and to newspaper language.10,11 Refining that notion, NLP researchers speculated that clinical language itself is subdivided into multiple sublanguages that are likely not uniform, varying in syntactic and semantic characteristics across notes of different types.3,12
An important implication of this phenomenon is that one could expect a significant decrease in the performance of an NLP system developed in one clinical domain as applied in another clinical domain. Some authors, such as Hyun and Bakken referenced above, study this effect between two note types, but there have been no systematic studies across a broad array of note types and note authors to formally assess the extent of the sublanguage phenomenon. Such a study would help address important clinical NLP strategies. Perhaps some subset of clinical note types cluster into families similar enough to be treated as one? Or perhaps the differences between the sublangauges can be extracted automatically to assist re-purposing a tool from one note type to another? In this paper we present a formal sublanguage analysis of 17 note types from three kinds of authors (plus MEDLINE) and we discuss how the results may be used to inform clinical NLP strategies.
Clinical language processing systems
Over the past decade or so work on processing clinical text has resulted in several well-known systems that are optimized for a subset of clinical notes. Mesytre et al. provides an excellent review paper on the topic,2 so here we will briefly describe three representative tools: MedLEE,3 cTAKES,13, and HITEx.14 Each of these systems was designed for a different purpose and they vary in their design approach, but all of them rely in part on a statistical analysis of a subset of clinical narratives.
The model used by MedLEE was designed by analyzing chest x-ray reports generated at Columbia Presbyterian Medical Center (CPMC). The clinical terms and their semantic types MedLEE uses were derived from the Medical Entities Dictionary developed at CPMC. A large portion of the knowledge base for the system was manually curated from statistical analysis of 8̃,000 reports. After the initial system was proven proficient, it was extended into new clinical domains, notably discharge summaries, which required additional manual effort.3
cTAKES was designed for the purpose of information extraction from clinical text. Its knowledge base was derived through machine learning using general English annotated text. The acquired statistical language model was then adapted to clinical language using a small manually annotated corpus of 273 clinical notes of various types. In addition, manually created rules were used to identify negation contexts of the extracted concepts.13
The original purpose of HITEx was specific to a research study on airway diseases such as asthma and chronic obstructive pulmonary disease, but it sees general purpose use as an NLP “cell” module in the i2b2 “hive” architecture. The typical goal of the system is to extract principle diagnoses, co-morbidities, and smoking status. The knowledge base for the system includes a set of manually designed regular expressions, as well as machine learning models trained on a corpus consisting of discharge summaries of the patients that had one or more related admission diagnosis defined by ICD9 codes.14
Like many of the excellent clinical NLP tools mentioned in the review by Meystre et al., these three systems are very good at processing the texts they were designed to handle. We are not criticizing these tools, but we are suggesting that they could be misapplied. If an NLP tool relies on the statistical properties of terms and semantics in a training language model, then to the extent that a new target language differs from that training language, the tool will perform less well on the target. This is one particularly important reason to study sublanguages systematically.
Document Clustering
After considering several modeling approaches, we decided to use document clustering to discover how closely related notes from diverse clinical domains and settings were. There are other possible approaches to discover similarities between documents, such as analysis of syntactic structure. However, such approaches rely on manually annotated corpus, whereas the approach that we chose does not. Document clustering is a commonly used unsupervised text mining technique that has been used for a range of natural language processing tasks such as information retrieval, question answering and others. The goal of document clustering is to find a set of “natural” patterns within a large amount of unlabeled data inside the documents and then to organize similar documents into groups using some measure of similarity.15 Because understanding something about the basics of clustering will aid in the interpretation of our results, we provide a brief overview of clustering below. This section will be especially useful to anyone who wants to replicate our approach with their own note corpora.
Cluster analysis typically consists of the following main stages:
Feature selection and extraction
Selection or design of a clustering algorithm
Cluster quality evaluation.16
Feature selection and extraction:
The most popular data set format for document clustering is a bag-of-words vector-space model. This method represents the entire set of documents as a T × D matrix, where T is the size of the vocabulary used in the document set; and D is the total number of documents in the data set.17 Each document is represented as a vector of length T, and since most terms do not appear in any given document, these vectors are sparse. Typically, each value in these vectors represents the importance of the particular term (t) in the particular document (d). In order to minimize the effect of the document size and extremes in the frequency of a specific term, the well known “term-frequency inverse-document-frequency (tf-idf)” measure is often used as the weight of each term in a document vector, calculated as follows:
where
ti.j - is the frequency of term i in document
∑tj - is the document length as a total count of terms in the document
D - is the total number of documents in the data set, and
di - is the number of documents in the data set that contain the term i.
Clustering algorithm:
The goal of cluster analysis is to place each document into one of K disjoint or overlapping clusters. Each cluster usually is defined by its centroid, which is the most representative vector in the cluster. Depending on the clustering algorithm used, the centroid can be either the average point for each dimension of the feature vector, or an actual point in the data set that is the closest to the average point.
Most clustering algorithms have three main components:
Similarity measure, used to measure vector relatedness;
Clustering method, the computational approach taken during the clustering process; and
Clustering criterion function, which is used for the optimization of the final clustering solution.18
Most commonly, one measures similarity between two vectors by calculating a Euclidean distance, a cosine distance, or a correlation coefficient. The general clustering method can be either partitional, agglomerative, density-based, or grid-based. Depending on the final specific solution desired, the clustering methods can be either hierarchical or non-hierarchical.19 The simplest and most widely used clustering method is k-means. Prior research concluded that a bisecting, K-means algorithm performs quite well despite its simplicity and lower computational complexity.20 This hierarchical algorithm iteratively splits the data set until the predefined number of clusters is reached. Selection of a clustering criterion function influences the final clustering solution by putting more emphasis on cohesion or on separation of the resulting clusters.
Cluster quality:
The measure of cluster quality can be classified as either internal or external. Internal measures of cluster quality aim to assess how closely the elements in each cluster are related to each other, evaluating “cohesion” and “separation” of the clustering results. Cohesion can be measured as the average similarity of the members of the cluster to each other or to the cluster centroid. Separation evaluates the average dissimilarity of the members of a particular cluster to all other elements in the data set. The external measures rely on knowing a true labeling of each of the documents. Clustering output can be measured externally in terms of purity and F-score. Purity is the proportion of each cluster that consists of the majority class. F-score evaluates precision and recall of each document type with respect to its cluster assignment. In evaluation of document clustering output, precision for each document type compares the largest number of documents that are assigned to a specific cluster to the total number of documents assigned to that cluster. Recall for each document type compares the fraction of the largest group assigned to the same cluster to the total number of documents of that type. The F-score is a harmonic mean of precision and recall.
An optimal clustering solution will have 100% purity, which means that each cluster contains elements that belong to a single class.18 Such purity can be achieved trivially when the number of clusters is equal to the number of elements in the data set. On the other hand, the perfect F-score will be achieved only if all documents of each type are grouped into a single cluster (100% recall) and no document types share a cluster label (100% precision). Using the note type as the true class labels, we exploit purity and F-score measures in our analysis below.
Methods
The complete list of all clinical narrative types in current use at our medical center (a large tertiary care teaching hospital) was analyzed by a clinical expert external to the study team to determine a study subset that was diverse yet representative across domains. Note types that consisted mostly of templated information, scanned hand-written documentation, or non-clinical documents were excluded. As a result, a set of 17 representative note types was selected for this study. These note types represented a cross-section of clinical narratives created by clinical personnel that varied by clinical role (physicians, nurses, social workers), specialty (Cardiology, Dermatology, Ob/Gyn, Oncology, etc.), and clinical setting (emergency, inpatient, outpatient).
The set of notes from these 17 types that were created between January 2007 and December 2008 comprised a corpus of 683,125 notes as extracted from the University Hospital Electronic Data Warehouse. Files that were less than 100 bytes in length were excluded because random sampling showed that they did not contain clinically relevant information. Out of the remaining 559,029 files, 48,685 were randomly selected (all 685 of the Emergency Department Reports and 3,000 each of the other note types). These were processed by the MetaMap binary (v.2009),21 running locally on a secure, HIPAA-compliant, high-performance compute cluster. In addition to the clinical narratives, a random set of 3,000 MEDLINE abstracts larger than100 bytes published between 2000–2008 was selected and also processed by MetaMap as a general biomedical comparator corpus.
A feature vector file was created where each note was represented by the standard term frequency-inverse document frequency(tf-idf) value for each term that MetaMap matched to at least one UMLS concept. To decrease the feature space, multi-word phrases were split into individual tokens and the base form of all tokens was obtained from SPECIALIST lexicon using the Norm tool.22 In addition to the lexical attributes, semantic types of those terms that were unambiguously mapped to a UMLS concept by MetaMap were also used as attributes. The derivation of what constitutes an unambiguously mapped term is more complex than simply choosing those terms with only one MetaMap semantic mapping. Those can be enriched with an algorithm that exploits the mapping scores provided by MetaMap. We describe that algorithm elsewhere.23 Using only those terms that MetaMap successfully mapped to at least one concept minimizes the size of the feature vector and focuses on only those tokens that are potentially relevant in the clinical setting, thus excluding misspellings, unrecognized locally specific abbreviations, and other language characteristics, which are artifacts of the local practices rather than being typical of the clinical domain.
This study addresses the following research questions:
- Will an unsupervised clustering of clinical notes result in clearly differentiated document clusters?
- Will an unsupervised clustering of clinical notes result in document clusters that correspond to the source note types?
To perform clustering we chose bisecting k-means clustering using a cosine similarity measure with the “internal criterion function,” which maximizes similarity between each document and the centroid of the cluster that it is assigned to. The clustering tool we chose was the CLUTO clustering toolkit.18 This software package offers a set of clustering algorithms that approach clustering as an optimization process aiming to minimize or maximize the selected clustering criterion function. It is written in C, and thus is quite fast. It also manages memory well. The Java-based Weka cluster toolset was unable to process the full feature space, and was too slow to be practical for even small subsets. The selected clustering algorithm requires the number for clusters to be specified a priori. In the current study each clustering experiment used the same number of clusters as the number of the analyzed note types.
Results
Our initial experiment using a subset of 685 documents of each type (i.e., the size of the smallest note type, Emergency Department Reports) as clustered into 18 clusters resulted in 74.8% average cluster purity. Analysis of the most descriptive and discriminating features (produced optionally by CLUTO) showed that several provider names in one type produced an unwarranted impact on clustering. After these names were identified, the feature vectors were recalculated and new clusters were analyzed. Review of the most important features showed that clinically irrelevant words, such as “phone” and “fax” were responsible for inflated cluster purity for Case Management Discharge Plan, thus skewing the clustering results.
The results of these two experiments led us to conclude that in order to acquire the most reasonable clusters, we had to exclude the lexical noise that resulted from the artifacts of the local practices and templates. So we manually designed a short stopword list that consisted of the most frequent person names and also the words “phone” and “fax”. This stopword list also included 5 semantic types that were identified as the most common for all note types in our previous work.23 These semantic type are: Findings, Temporal Concept, Qualitative Concept, Quantitative Concept, and Functional Concept.
After those stopwords were excluded, the new data set was analyzed and the average cluster purity of the resulting solution dropped to 73.3%. This confirmed that the artifact terms were artificially improving the clustering for some note types, for example terms that occurred frequently in section headers. To eliminate noisy terms more systematically, we calculated an additional set of stopwords that aimed to reduce the lexical artifacts for all the note types. Our new stopword list excluded any term in a specific note type if that term appeared in more than 95% of all documents of this note type. These terms were eliminated from the feature set for that particular note type but not for the other note types. Processing of the new data set resulted in an even lower average purity of 70.0%. Even though eliminating artifacts of the local practices resulted in lower cluster purity, we believe that by doing so we achieved clustering that more faithfully reflects the lexical patterns of the analyzed clinical domains rather than lexical noise due to local practice.
Purity is calculated in terms of the majority class for each cluster and reflects how well each cluster is represented by one of the document classes. Lower purity indicates that the cluster groups together notes of different classes, thus showing that those document classes have some documents that lexically are related among each other. For example, Table 1 shows that cluster 13 mostly has documents from three note types - Admission History and Physical, Case Management Discharge Plan, and Emergency Department Reports. On the other hand, cluster 6 is mostly represented by Rheumatology Clinic notes.
Table 1.
Clustering results of the data set consisting of 685 documents per document type. Values that represent less than 1% of the total note type count were excluded for clarity
Comparing the cluster assignment for Discharge Summaries and Admission History and Physical it is notable that out of 18 clusters 8 have similar counts of these note types. This is indicative of the large overlap in the lexical and semantic patterns appearing in the documents of these note types.
The next set of experiments evaluated the effect of larger sample size on clustering. Emergency Department Reports had only 685 notes available to us, so they were excluded from further processing. The feature vectors representing the remaining16 note types and MEDLINE abstracts with 3,000 documents in each set were clustered into 17 groups. As Table 2 illustrates, most document types were grouped each in its own cluster. Several note types are shown to be more general than others, such as, Admission History and Physical, Ambulatory Nursing Notes, Discharge Summary, Family Practice Clinic Notes and, not surprisingly, MEDLINE abstracts. Case Management Discharge Plan, Dermatology Clinic, and Plastic Surgery Notes exhibited a dichotomy in the lexical patterns. As the cluster hierarchy shows, despite such a split, each pair of the clusters are closely related, indicating similarity between the clusters. Increased sample size and removal of a more general document set (Emergency Department Reports) resulted in increase of the average purity to 76%.
Table 2.
Results of hierarchical cluster analysis of the set of 17 document types (n=3,000 notes per set). The values are the counts of all documents of the particular type that were grouped into each of the clusters. Cells with the value between 25% and 50% of the total count per note type are green, 50%–75% are yellow, and >75% are red. Values that represent less than 1% of the total note type count were excluded for clarity.
We recognized that the general note types, which are not specific to any clinical domain, span different topics and we excluded them for the next experiment. We processed the new data set consisting of the documents of those twelve note types, which are more focused on a specific clinical domain. The resulting twelve clusters had an impressive level of purity, 95.5%. Average F-score was also 95.5% (Table 3). This indicates that the overwhelming majority of the notes of each note type exhibit lexical patterns that are characteristic of that note type.
Table 3.
Results of clustering 12 note types with 3000 documents of each type. Values that represent less than 1% of the total note type count were excluded for clarity.
Analysis of a slightly lower F-score for Orthopedic (OCN) and Plastic Surgery Clinic Notes (PSC) and Operative Report (OPR) indicated a topic overlap for a portion of these notes as pointed out by the descriptive features for cluster 11 (Table 3), which are {fracture, orthopedics, motion, knee, splint, radiographic}.
Discussion
Applying document clustering to a large set of clinical narratives allowed us to expose the differences in the lexical and semantic patterns used within different clinical environments as well as among different author types. Our broad, systematic survey formally establishes what many clinical NLP researchers have suspected for a long time, namely that clinicians in different domains and in different settings use language in a highly idiosyncratic way.
It came as no real surprise that general-purpose notes such as Admission History & Physicals and Discharge Summaries bore a greater similarity to each other than they did to clinically narrow notes like those in a Dermatology Clinic. We were surprised, however, at how pure narrow domain notes proved to be. Take for example Dermatology Clinic Notes (DCN) and Plastic Surgery Clinic (PSC) notes (see Table 3). The DCN notes achieved a purity of 99% and had no overlap with PSC notes, although one might reasonably expect them to be somewhat similar because much of what they both deal with is skin. PSC notes did have a small amount of overlap with Burn Clinic Notes (which were 97% pure), but Burn Clinic Notes had the same amount of overlap with Orthopedic Clinic Notes. The three outpatient units that treat patients and share skin as a common clinical denominator (PSC, BCN, and DCN) were on average 97.3% pure – readily distinguishable from each other. This suggests to us that caution must be used when assuming, without any formal valuation, that natural language processing tools developed in any one of these units should be expected to work well with any of the others.
Document clustering using a bag-of-terms and bag-of-semantics approach has an advantage over a black-box technique such as support vector machines: it uncovers the importance of key features that are descriptive and discriminating. Descriptive features are the set of features which contribute the most to the average similarity of a document in a specific cluster. Discriminating features are those features that are more frequent in the particular cluster compared to other clusters. Given how high F-scores were for the clusters we studied, we can reasonably extrapolate at least some of the key sublanguage features in our notes. This could be useful, potentially, in guiding future work to re-purpose an NLP tool from note type to another. Analysis of the hierarchical tree produced by CLUTO(see Table 2 for an example) might help identify families of note types that are linguistically related and share vocabulary and term co-occurrence patterns. Table 2, for example, suggests that Family Practice Clinic Notes are more related to Neurology Clinic Notes than they are to Social Service Notes. One might expect that family practice and social services, as domains, might be related but the table suggest otherwise. Clearly one should be cautious when assuming relationships that are not well established empirically.
On a more detailed level, our clustering approach gives very specific insight about how sublanguages from different but related domains manifest. For example, analyzing descriptive features of cluster 4 with a high F-score (Table 3), we can conclude that terms describing that cluster are also descriptive of the Cardiology Clinic Notes because most of the documents of this type were categorized as that cluster. In this case the discriminating features are the terms, their co-occurrences within a document, and the percentage of the within-cluster similarity that these particular terms can explain. Examples from the Cardiology Clinic Notes are:
24.53% cardiology ventricular daily
20.84% cardiology coronary daily
11.95% cardiology tachycardia arrythmia
11.56% atrial fibrillation cardiology ventricular arrythmia
11.00% cardiology pacemaker arrythmia
10.67% atrial cardiology cardiomyopathy
We can make similar conclusion about other clusters and its corresponding majority document type.
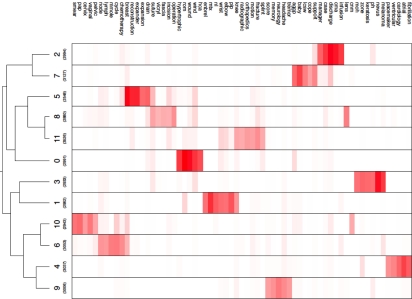
Figure 1 is a hierarchical cluster tree that allows us to visualize occurrences of descriptive and discriminating terms across all notes in the data set. In that diagram, the darker the color, the more discriminating are the terms that occur most often in the documents that belong to a particular cluster. Studying such clustering results should help to inform where to start lexicon building when constructing a natural language system being adapted into a new clinical domain. We limited the number of display terms to make a reasonably sized diagram here, but in principal one could drill down arbitrarily deep into these data when constructing a custom lexicon.
Figure 1.
Visualization of the hierarchical cluster tree demonstrating how clusters are related to each other by showing a color-intensity plot of the various values in the cluster centroid vectors. The height of each row is proportional to the log of the corresponding cluster’s size. The columns correspond to the union of the 5 descriptive and discriminating features of each cluster.
Conclusion
We hoped to address two questions in this study. The first, “Will an unsupervised clustering of clinical notes result in clearly differentiated document clusters?” can safely be answered “yes.” This is most clearly demonstrated in Figure 1. Each of the 12 narrow clinical notes are defined by readily distinguishable groups of lexical and semantic features (marked by red lines of different intensity). When the broader domain notes, like Discharge Summaries, are added to the mix, such as in Table 2, the clustering effect is less pronounced. However, even these clustering data are useful: they show which notes overlap as sublanguages. One could reasonably hypothesize that an NLP tool optimized for Discharge Summaries could be expected to work well with Admission History & Physical notes as well as with Family Practice Clinic Notes, an important thing to know if one is studying outpatient records.
The second question we addressed was, “Will an unsupervised clustering of clinical notes result in document clusters that correspond to the source note types?”, and again we can answer “yes.” Since we have the true labels for each note, we can verify that the clusters produced by CLUTO belong, with very high purity, to specific note types.
A number of research questions in clinical and clinical research informatics projects can be addressed through the use of natural language processing. Researchers might be tempted to use one of the large and growing set of existing clinical NLP systems to perform note analysis. However, one needs to be cognizant of the very real possibility of sub optimal performance across clinical domains and settings. At the same time, as this study shows, some note types are intrinsically more similar to some others, which suggests that less effort might be needed to transfer an existing system between these domains.
Acknowledgments
This research has been supported by the NLM under grants T15LM007124 (fellowship), 5R21LM009967-02, and 3R21LM009967-01S1(ARRA). An allocation of computer time from the Center for High Performance Computing at the University of Utah is gratefully acknowledged.
References
- 1.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association : JAMIA. 2010;17(5):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Yearbook of medical informatics. 2008. Extracting information from textual documents in the electronic health record: a review of recent research; pp. 128–44. [PubMed] [Google Scholar]
- 3.Friedman C. A broad-coverage natural language processing system. AMIA Annu Symp Proc; 2000. pp. 270–274. [PMC free article] [PubMed] [Google Scholar]
- 4.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. Proceedings / AMIA Annual Symposium AMIA Symposium; 2002. pp. 742–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Hyun S, Johnson SB, Bakken S. Exploring the Ability of Natural Language Processing to Extract Data From Nursing Narratives. Computers, informatics, nursing : CIN. 2009;27(4):215–23. doi: 10.1097/NCN.0b013e3181a91b58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris ZS. Mathematical structures of language. Interscience Publishers; 1968. [Google Scholar]
- 7.Harris ZS. A Theory of Language and Information: A Mathematical Approach. Clarendon Press; 1991. [Google Scholar]
- 8.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. Journal of biomedical informatics. 2002;35(4):222–235. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 9.Campbell DA, Johnson SB. Comparing syntactic complexity in medical and non-medical corpora. Proceedings / AMIA Annual Symposium AMIA Symposium; 2001. pp. 90–4. [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang G, Sato H, Endoh A, Ogasawara K, Sakurai T. Extraction of specific nursing terms using corpora comparison. AMIA Annu Symp Proc; 2005. p. 997. [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn U, Wermter J. High-performance tagging on medical texts. Proceedings of the 20th international conference on Computational Linguistics; 2004. [Google Scholar]
- 12.Bakken S, Hyun S, Friedman C, Johnson SB. A comparison of semantic categories of the ISO reference terminology models for nursing and the MedLEE natural language processing system. Medinfo. 2004;11(Pt 1):472–6. [PubMed] [Google Scholar]
- 13.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association : JAMIA. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC medical informatics and decision making. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T, Liu S, Chen Z. An Evaluation on Feature Selection for Text Clustering. Proceedings of the Twentieth International Conference on Machine Learning; 2003. pp. 488–495. [Google Scholar]
- 16.Xu R, Wunsch D. Survey of clustering algorithms. IEEE transactions on neural networks a publication of the IEEE Neural Networks Council. 2005;16(3):645–78. doi: 10.1109/TNN.2005.845141. [DOI] [PubMed] [Google Scholar]
- 17.Jayabharathy J, Kanmani S, Parveen AA. A Survey of Document Clustering Algorithms with Topic Discovery. Journal of Computing. 2011;3(2):21–27. [Google Scholar]
- 18.Zhao Y, Karypis G. Data clustering in life sciences. Molecular biotechnology. 2005;31(1):55–80. doi: 10.1385/MB:31:1:055. [DOI] [PubMed] [Google Scholar]
- 19.Zaane O, Foss A, Lee CH, Wang W. On Data Clustering Analysis: Scalability, Constraints, and Validation. In: Chen MS, Yu P, Liu B, editors. Advances in Knowledge Discovery and Data Mining. vol. 2336 of Lecture Notes in Computer Science. Springer Berlin; Heidelberg: 2002. pp. 28–39. [Google Scholar]
- 20.Steinbach M, Karypis G, Kumar V. A Comparison of Document Clustering Techniques. 6th ACM SIGKDD, World Text Mining Conference; 2000. [Google Scholar]
- 21.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association : JAMIA. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browne AC, Divita G, Lu C, McCreedy L, Nace D. Lexical Systems: A report to the Board of Scientific Counselors. 2003. [Google Scholar]
- 23.Patterson O, Igo S, Hurdle JF. Automatic acquisition of sublanguage semantic schema: towards the word sense disambiguation of clinical narratives. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2010; 2010. pp. 612–6. [PMC free article] [PubMed] [Google Scholar]