Abstract
We analyzed a longitudinal collection of query logs of a full-text search engine designed to facilitate information retrieval in electronic health records (EHR). The collection, 202,905 queries and 35,928 user sessions recorded over a course of 4 years, represents the information-seeking behavior of 533 medical professionals, including frontline practitioners, coding personnel, patient safety officers, and biomedical researchers for patient data stored in EHR systems. In this paper, we present descriptive statistics of the queries, a categorization of information needs manifested through the queries, as well as temporal patterns of the users’ information-seeking behavior. The results suggest that information needs in medical domain are substantially more sophisticated than those that general-purpose web search engines need to accommodate. Therefore, we envision there exists a significant challenge, along with significant opportunities, to provide intelligent query recommendations to facilitate information retrieval in EHR.
Introduction
The overload of information has created a significant challenge in accessing relevant information. It is not only a challenge for common web users to find relevant webpages, but also a challenge for medical practitioners to find electronic health records (EHR) of patients that match their information need. As by far the most effective tools of information retrieval, search engines have attracted much attentions in both industry and academia. The analysis of search engine query logs is especially useful for understanding search behaviors of users,1 categorizing users’ information need,2 enhancing document ranking,3 and generating useful query suggestions.4,5
Search behaviors of common Web users have been excessively studied through query log analysis of general purpose commercial search engines (e.g., Yahoo!,1 Altavista6). However, little effort has been made to understand search behaviors of medical specialists even though substantially different characteristics of the search behaviors can be anticipated. Indeed, although there has been some existing work in analyzing how clinical specialists search for medical literature,5,7,8 little has been done for analyzing how they use EHR search engines, i.e., retrieval systems that search for patient records stored in EHR systems. Part of the reason is the lack of specialized EHR search engines and longitudinal collections of query logs. Although there has been much effort on the design and construction of search engines of healthcare information which target at patients,9–17 there is little discussion about EHR search engines that target at medical specialists. To the best of our knowledge, this is the first large scale query log analysis of an EHR search engine. The closest work is Natarajan et al., 2010,18 which analyzes the query log of an EHR search engine of a much smaller scale (i.e., two thousand queries). Other related work includes the analysis of health-related queries submitted to commercial search engines19–22 and the analysis of queries submitted to PubMed and MedlinePlus.5,7,8 Neither of the two types of queries represent the information need of Medical practitioners in accessing electronic health records.
We study a large scale query log dataset collected from an EHR search engine running in University of Michigan Hospital for over 4 years. The analysis allows us to directly understand the information need of medical practitioners in accessing the electronic health records: the categorization, distributions, and temporal patterns. We aim at answering questions like “What are the most commonly searched concept?” and “When would the users adopt system suggested queries?”, and “Was the temporal pattern of search volume consistent with the clinic schedule?”
We start with a description of the environment of the EHR search engine.
The Medical Search Environment
A. The EMERSE Search Engine
We first describe a full-text EHR search engine that we previously developed called Electronic Medical Record Search Engine (EMERSE).23–25 The system is implemented in our institutional EHR environment to allow users to retrieve information from over 20 million unstructured or semi-structured narrative documents stored. Since its initial introduction in 2005, EMERSE has been routinely utilized by frontline clinicians, medical coding personnel, quality officers, and researchers, and has enabled chart abstraction tasks that would be otherwise difficult or even impossible.26–28
A user has to log in with his unique user ID to submit queries. Like a general purpose search engine (e.g., Google), EMERSE supports free text queries as well as boolean queries. Our observation of how the EMERSE users had interacted with the system in the past indicated several severe issues undermining its performance, such as poor quality of user submitted queries and a considerable amount of redundant effort in constructing the same or similar search queries repeatedly (by the same user or by different users).24 Motivated by the observation, EMERSE provides two mechanisms to assist users compose their queries:
Query Suggestion. The EMERSE system provides a primitive search term suggestion feature based on customized medical dictionaries and open-source phonetic matching method.29 This query recommendation function detects and suggests corrections and expansions of search queries submitted by the users. To date, EMERSE has recorded many variations of how a medical term could be spelled either in user submitted search queries or in the patient care documents that were searched. The customized dictionary is maintained by a manual process.25
Collaborative Search. Motivated by the concept of “wisdom of the crowd,” we implemented a social-information-foraging paradigm in EMERSE which allows users to share their expertise of query construction and search collaboratively.25 A user can save any effective queries he constructed as “search bundles,” and share the bundles to the public or to particular users he knows of. A user can adopt any bundles that are shared with them or made available to the public. Our previous study has shown that this collaborative search paradigm has provided the potential to improve the quality and efficiency of information retrieval in healthcare.25
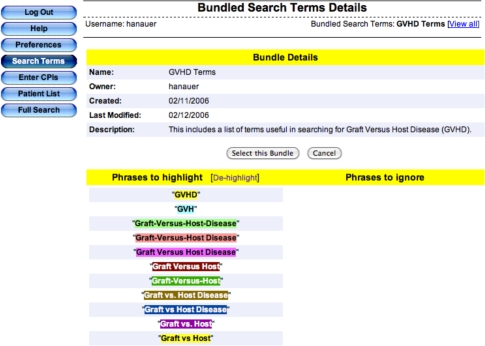
An example of a search bundle used in the context of EMERSE is shown in the following figures:
Additional screenshots illustrating the use of search bundles can be found in Zheng et al., 2011 (Appendix 1).25
B. Experiment Setup
We have logged every query submitted to the EMERSE search engine from 08/31/2006 to 12/21/2010. For each query, we recorded the query string, the time stamp when it is submitted, the user who submitted the query, and whether or not the query is a user created “bundle.” Note that the system did not log whether a query is manually typed in or selected from system generated suggestions. Instead, we infer this information based on several effective heuristics (the time lag from the previous query, comparison to the previous query, etc).
The 4-year query log data provides us a set of 202.9k queries submitted by 533 physicians in 21 departments of the University of Michigan Hospital. The basic statistics of the query log is presented in Table 1.
Table 1:
Basic Statistics of the EMERSE query log
| Number of Queries | 202,905 |
|---|---|
| # of Unique Query Terms | 16,728 |
| # of Users | 533 |
| proportion of user typed-in queries | 54.9% |
| proportion of system suggested queries | 25.9% |
| proportion of bundled queries | 19.3% |
| # of search bundles | 1,160 |
In the next section we present results of in-depth analysis of this query log collection.
Analysis and Results
In this section, we present the descriptive characteristics and patterns extracted from the query log data. We apply simple but effective heuristics to label each query as either “typed-in” by a user, “recommended” by the system, or an existing search “bundle.” Specifically, if a new query is logged within a very short time (e.g., a couple of seconds) of a previous query submitted by the same user, we label it as a system suggested query. We admit this heuristic, although well performing, underestimates the system suggested queries. Below we start with the analysis of queries.
A. Queries
Query Frequency
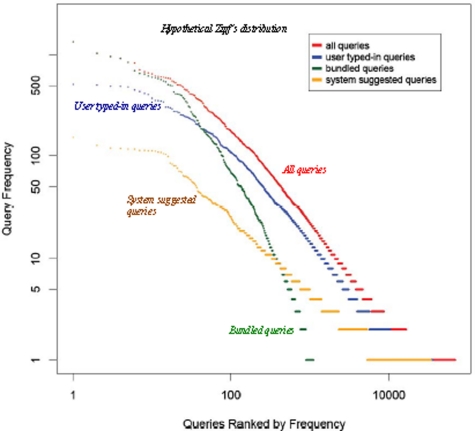
We first compute the frequency distribution of queries. In Figure 3, we plot the distribution of the frequency of all queries, “typed-in” queries, “bundles,” and system “recommended” queries with a log-log scale. Data points at the upper left corner (the head) represent queries that are most frequently used by the users; and data points at the lower right corner (the tail) represent queries that are only used for a few times.
Figure 3:
Query frequency distribution: long tail but no fat head.
It is commonly believed in Web search that the frequency of queries follow a power-law distribution (or Zipf’s distribution depending on how it is plotted).30 Some queries are excessively used, which contribute to the “fat head” of the distribution. Most of those queries are categorized as navigational queries (e.g., “facebook,” “pubmed”), where the information need of the user is to reach a particular site/document that he knows of.2 Non-navigational queries are categorized as either informational queries (to find information from one or more documents) and transactional queries (to perform web-mediate activity, like shopping).2 Many such queries are used infrequently, which contribute to the “long tail” of the power-law/Zipf’s distribution. In a log-log plot, a Zipf’s distribution presents a straight line (see the dash line in Figure 3).
From Figure 3, however, we see that the frequency distributions of medical search queries do not fit a power-law/Zipf’s distribution: a power-law/Zipf’s distribution will fit a straight line in such a plot with a log-log scale. Although the distributions do have a long tail, the head is not sufficiently “fat” and apparently doesn’t fit a straight line. This suggests that there aren’t many queries that are “excessively” used, like the common navigational queries in Web search. The distribution of search bundles has a higher skewness than the distribution of other types of queries, implying a rich-gets-richer phenomenon in the adoption of search bundles.
To further analyze this, we list the most frequent queries in Table 2. Interestingly, a large proportion of most frequently used queries are search bundles (17 out of 20). Overall, search bundles accounted for 19.3% of the total queries submitted to EMERSE. These results suggest that the collaborative search paradigm in EMERSE effectively influenced the search behaviors of users, by enhancing the sharing and reuse of effective queries.
Table 2:
20 Most Frequently Used Queries/Search Bundles.
| Rank | Query/Bundle Name | Query Type | Rank | Query/Bundle Name | Query Type |
|---|---|---|---|---|---|
| 1 | Cancer Staging Terms | Bundle | 11 | PEP risk factors | Bundle |
| 2 | Chemotherapy Terms | Bundle | 12 | cigarette/alcohol use | Bundle |
| 3 | Radiation Therapy Terms | Bundle | 13 | comorbidities | Bundle |
| 4 | Hormone Therapy Terms | Bundle | 14 | radiation | Typed-in |
| 5 | CTC Toxicities | Bundle | 15 | lupus | Bundle |
| 6 | GVHD Terms | Bundle | 16 | Deep Tissue Injuries | Bundle |
| 7 | Performance Status | Bundle | 17 | Life Threatening Infections | Bundle |
| 8 | Growth Factors | Bundle | 18 | CHF | Bundle |
| 9 | Other Toxicity Modifying Regimen | Bundle | 19 | relapse | Typed-in |
| 10 | Advanced lupus search | Bundle | 20 | anemia | Typed-in |
Note: a query is a set of words typed in the search box and submitted to the search engine. A Bundle is a query saved by a user, with a bundle name as a label (never used in search). A bundle usually contains a much longer list of terms than the bundle name as the actual query. We refer to Zheng et al. 201125 for details about search bundles.
It is readily apparent that none of the top queries are “navigational” queries or “transactional” queries. In fact, there are seldomly navigational queries in the whole collection of queries. Indeed, most searches of EHRs are trying to identify one or more patients that matches particular situations in order to further investigate their clinical information.
Query Length
We then measure the average length of queries of various types. From Table 3, we see that in average, a query submitted to the EHR search engine contains 5 terms, significantly larger than the average length of queries submitted to commercial web search engine (e.g., 2.356,31). Note that this long average length might be affected by the system generated recommendations and search bundles. In average, a user typed-in query contains 1.7 terms, which is shorter than the average length of Web queries but comparable to the average length of queries submitted to MedlinePlus.8 However, the average length of system recommended queries the users adopt are much longer (13.8), considerably longer than the query suggestions made by web search engines. This suggests that although the users tend to start a search task by typing in short queries, they are likely to end up with reformulating their information need with much longer queries. Indeed, the average length of search bundles, which are considered as effective queries by the creators of the bundles, is even significantly larger (58.9). This result indicates that the information need of EHR users is much more difficult to be represented comparing to the information need in Web search. There is a considerable gap between the “organic” queries the users start with and the well-formulated queries that effectively retrieve relevant documents.
Table 3:
Average Length of Query Types (by terms).
| All | Typed-In | System Recommended | Bundle Name | Bundle |
|---|---|---|---|---|
| 5.0 | 1.7 | 13.8 | 2.6 | 58.9 |
It is also interesting to note that although a search bundle tends to contain many query terms, the users create short names for the bundles so that they can be easily shared and adopted.
Acronyms Usage
We have also analyzed the usage of acronyms in the queries. We counted 2, 020 unique acronyms which appeared 55, 402 times in the query log. Among the 202.9k queries, 18.9% contain at least one acronyms. This coverage is significantly larger than existing findings in Web search.32
Dictionary coverage
To further understand the broadness and quality of the queries, we compute the coverage of the query terms by various specialized dictionaries. Note that terms that are not covered by the given dictionaries are commonly considered as spelling errors in existing literature.8,33 We choose three dictionaries: the term list (including synonyms) of the SNOMED-CT ontology (a clinical terminology), the term list of the UMLS (a comprehensive vocabulary of biomedical science), and a meta-dictionary that combines a common English dictionary, a medical dictionary, and all terms used in UMLS. Note that the SNOMED-CT dictionary is a subset of the UMLS dictionary, which is a subset of the meta-dictionary. To accommodate multiple forms of the same word, light stemming is done using Krovetz stemmer.34
Interestingly, even the comprehensive meta-dictionary can only cover 68% of the query terms. This coverage is much smaller than the dictionary coverage of Web search queries (e.g., 85%–90%)33 as well as the coverage of MedlinePlus queries (e.g., > 90%).8 This result, along with the result of acronym analysis, suggests that queries in EHR search are more specialized, noisier, and thus potentially more challenging to handle.
B. Session Analysis
In Web search, a session is defined as a sequence of consecutive queries submitted by the same user in a sufficiently small time period. A session is considered as a basic mission of the user to achieve a single information need.6,35 The analysis of sessions is very important in query log analysis, which helps people understand the dynamics of search behaviors and how users modify their queries to fill their information need.6
Following the common practice in Web search log analysis, we simply identify sessions using a 30-minute timeout.35 This means if a query is submitted at least 30 minutes later than the previous query, we consider it starts a new session. By doing this, we managed to identify 35,928 search sessions. In average, each session lasts for 14.80 minutes, which contains 5.64 queries. The length of a session in EHR search is considerably longer than a session in Web search, as reported in literature.6 A longer session length suggests that a search task in EHR is generally more difficult than a search task on the Web. Indeed, many Web queries are navigational queries, which are usually associated with shorter sessions. Interestingly, 27.7% sessions ended with a search bundle, comparing to the overall bundle usage of 19.3%. This suggests that users tend to seek bundles after a few unsuccessful attempts to formulate the queries by their own.
On average, a user spends 14.8 minutes on a search session, comparing to a slightly shorter duration off-work (13.1 minutes) and on weekends (12.9 minutes).
C. Search Categorization
To further understand the semantics of a user’s information need, we attempt to categorize search log queries according to the ontology structure of SNOMED clinical terms.36 SNOMED CT is a comprehensive clinical terminology that is comprised of concepts, terms, and relationships between these concepts.36 In this hierarchical ontology structure, there are 19 top-level concepts which we utilize as the semantic categories of queries. There is a unique path from each lower level concept to one of the top-level concepts in this hierarchical structure. Our task is to map each search query into one of these 19 top-level categories.
The categorization is done with the following procedure: 1. We represent each concept in SNOMED CT using the preferred terms and synonyms of this concept according to SNOMED CT. 2. For each of the 19 categories, we create a profile by aggregating the representation of the top-level concept and the representation of all its descendants in SNOMED CT. We remove the duplicated terms in such a profile. 3. For each query, we count the number of terms shared by the query and the profile of each of the 19 categories. The top-level concept that covers the most query terms will be assigned as the category of this query. Queries that don’t overlap with any of the 19 concepts are categorized as “Unknown.” The distribution of queries in each semantic category is shown in Table 5.
Table 5:
Category distribution of queries.
| Category | Coverage | Category | Coverage |
|---|---|---|---|
| Clinical finding | 28.0% | Physical object | 2.2% |
| Pharmaceutical / biologic product | 12.2% | Event | 1.7% |
| Procedure | 11.7% | Substance | 1.5% |
| UNKNOWN | 10.4% | Linkage concept | 1.0% |
| Situation with explicit context | 8.9% | Physical force | 0.6% |
| Body structure | 6.5% | Environment or geographical location | 0.6% |
| Observable entity | 4.3% | Social context | 0.5% |
| Qualifier value | 3.7% | Record artifact | 0.4% |
| Staging and scales | 3.2% | Organism | 0.3% |
| Specimen | 2.2% | Special concept | 0.03% |
Around 28% of queries are categorized as clinical findings, which includes subcategories such as diseases and drug actions. Pharmaceutical / biologic product accounts for 12.2% of the queries, followed by Procedure (11.7%) and Situation with explicit context (8.9%). There are also 10.4% queries that are not covered by any of the categories.
We also count the distribution of categories in different types of queries. According to Table 6, the distribution is similar in bundled queries and typed-in queries, except that Procedure queries are much more salient (18.4% of bundled queries comparing to 8.0% of queries directly typed in) and Pharmaceutical / biologic product queries are significantly underrepresented (3.5% of bundled queries comparing to 13.1% of queries directly typed in). Most system recommended queries adopted by the users are distributed in three categories, Clinical finding (49.2%), Pharmaceutical / biologic product (17.2%), and Procedure (14.6%). Note there is a substantially high rate of adopting system recommended queries in Clinical finding category, resulting in a 49.2% coverage of all system recommended queries. This indicates that the queries about clinical findings are more difficult than queries in other categories.
Table 6:
Top 5 categories of typed-in, bundled, and system recommended queries.
| Rank | Typed-In | Bundles | System |
|---|---|---|---|
| 1 | Clinical finding (20.7%) | Clinical finding (20.0%) | Clinical finding (49.2%) |
| 2 | UNKNOWN (16.5%) | Procedure (18.5%) | Pharm. / biologic product (17.2%) |
| 3 | Pharm. / biologic product (13.1%) | Body structure (11.4%) | Procedure (14.6%) |
| 4 | Situation with explicit context (11.1%) | Situation with explicit context (10.2%) | Body structure (4.4%) |
| 5 | Procedure (8.0%) | Observable entity (9.0%) | Situation with explicit context (3.0%) |
D. Temporal Patterns
Finally, we also analyze the temporal patterns of search queries submitted to EMERSE. Our goal is to answer questions like “Is the search volume lined up with the work hours?” and, “Will users search EHRs off work?” and, “Do users have different information need in clinic and off work?”
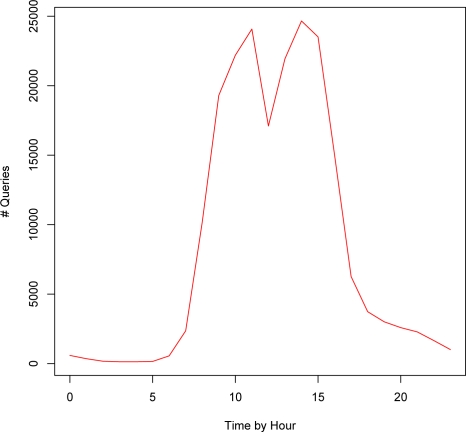
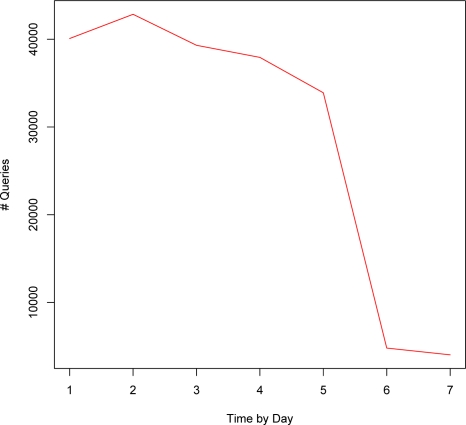
Figure 4 represents the temporal patterns of queries over hours-of-day. Clearly, the search volume correlates well with the common work hours. The search volume increases rapidly after the spike at 8am, drops during lunch hours, then climbs to the peak between 2pm and 3pm. Interestingly, there’s still a considerable volume after work which extends into midnight. Similar patterns can be observed in the distribution over days-of-week (Figure 5). Weekdays accumulate a significantly higher volume of queries than weekends, with Tuesdays the highest and Fridays the lowest. There is still a considerable proportion of queries coming in on Weekends.
Figure 4:
Search volume over hours-of-day (0–23 refers to midnight-11pm).
Figure 5:
Search volume over days-of-week (1–7 refers to Monday–Sunday).
To further understand whether there are different needs of information on and off work, we compared some statistics in and out of work hours. We found that searches in clinic have slightly longer sessions than searches off-work (14.6 minutes compared to 13.1 minutes), searches on weekdays have slightly longer sessions than searches on weekends (14.9 minutes compared to 12.9 minutes). It is also noticeable that the proportion of queries of clinical findings is larger on weekends (31.4%) and in off-work hours on weekdays (33.2%).
Discussion
What can we learn from the results of the query log analysis of this EHR search engine? What insights does it provide to the development of effective information retrieval systems for electronic health records? What particular aspects do we need to consider beyond the existing wisdom from Web search engines?
First, we see that EHR search is substantially different from Web search. There is a need of a new categorization schema of search behaviors. The well established categorization of Web search queries into “navigational,” “informational,” and “transactional” doesn’t apply to medical information retrieval. Deeper insights have to be obtained from the real users of the EHR search engine to categorize their information need. One possible categorization could discriminate high-precision information need and high-recall information need. We see that a clinical ontology can well categorize the semantics of queries into a few high-level concepts. To what extent this semantic categorization will be useful in the redesign of the search engine is an interesting research question.
Second, we find that EHR search queries are in general much more sophisticated than Web search queries. It is also more challenging for the users to well formulate their information need with appropriate queries. This implies an urgent need to design intelligent and effective query reformulation mechanisms and social search mechanisms to help users improve their queries automatically and collaboratively.
Moreover, we observe that the coverage of query terms by medical dictionaries and vocabularies is rather low. This not only implies the substantial difficulty to develop effective spelling check functionalities for EHR search, but also suggests that we need to seek beyond the use of medical ontologies to enhance medical information retrieval. One possible direction leads towards deeper corpus mining and natural language processing of electronic health records.
Finally, the effectiveness of collaborative search encourages us to further explore social-information-foraging techniques to bring together and utilize the collective expertise of the users.
Conclusion
We present the analysis of a longitudinal collection of query logs from a full-text search engine of electronic health records. The results suggest that users’ information needs and search behaviors are substantially different in EHR search from those in Web search. We present a primitive semantic categorization of EHR search queries into clinical concepts as well as an analysis of temporal search patterns. This analysis provides important insights on how to develop an intelligent information retrieval system for electronic health records.
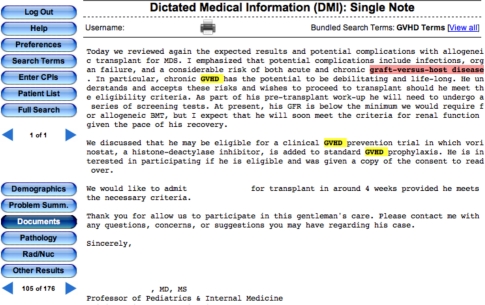
Figure 1:
Example of a selected bundle in EMERSE consisting of 11 permutations of how graft versus host disease (GVHD) might be worded in clinical documentation. Selecting this bundle will automatically search the documents for all of these variations.).
Figure 2:
Example of a clinical document that has two variations of GVHD highlighted in the text, making the concepts easy to find in the document. All protected health information has been removed from this example.).
Table 4:
Dictionary Coverage of Query Terms.
| Dictionary | Size of Vocabulary | Query Term Coverage |
|---|---|---|
| SNOMED-CT | 30,455 | 41.6% |
| UMLS | 797,941 | 63.6% |
| Meta-dictionary | 934,400 | 68.0% |
Acknowledgments
This project was supported in part by the National Library of Medicine under grant number HHSN276201000032C, the National Institute of Health under grant number UL1RR024986, and the National Science Foundation under grant number IIS-1054199.
References
- 1.Kumar R, Tomkins A. A characterization of online search behavior. IEEE Data Eng. Bull. 2009;32(2):3–11. [Google Scholar]
- 2.Broder A. A taxonomy of web search. ACM Sigir forum. 2002;36:3–10. ACM, [Google Scholar]
- 3.Joachims T. Optimizing search engines using clickthrough data. Proceedings of the eighth ACM SIGKDD international conference on Knowledge discovery and data mining; 2002. pp. 133–142. ACM, [Google Scholar]
- 4.Mei Q, Zhou D, Church K. Query suggestion using hitting time. Proceeding of the 17th ACM conference on Information and knowledge management; 2008. pp. 469–478. ACM, [Google Scholar]
- 5.Lu Z, Kim W, Wilbur WJ. Evaluation of query expansion using MeSH in PubMed. Information retrieval. 2009;12(1):69–80. doi: 10.1007/s10791-008-9074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverstein C, Marais H, Henzinger M, Moricz M. Analysis of a very large web search engine query log. ACM SIGIR Forum. 1999;33:6–12. ACM, [Google Scholar]
- 7.Herskovic JR, Tanaka LY, Hersh W, Bernstam EV. A day in the life of PubMed: analysis of a typical days query log. Journal of the American Medical Informatics Association. 2007;14(2):212. doi: 10.1197/jamia.M2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowell J, Zeng Q, Ngo L, Lacroix EVEM. A Frequency-based Technique to Improve the Spelling Suggestion Rank in Medical Queries. Journal of the American Medical Informatics Association. 2004;11(3):179–185. doi: 10.1197/jamia.M1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo G, Tang C, Yang H, Wei X. MedSearch: a specialized search engine for medical information retrieval. Proceeding of the 17th ACM conference on Information and knowledge management; 2008. pp. 143–152. ACM, [Google Scholar]
- 10.Liu RL, Huang YC. Medical query generation by term-category correlation. Information Processing & Management. 2011;47(1):68–79. [Google Scholar]
- 11.Luo G. Design and evaluation of the iMed intelligent medical search engine. Data Engineering, 2009 ICDE’09. IEEE 25th International Conference on; 2009. pp. 1379–1390. IEEE, [DOI] [PubMed] [Google Scholar]
- 12.Luo G, Tang C. Challenging issues in iterative intelligent medical search. Pattern Recognition, 2008 ICPR 2008 19th International Conference on; 2008. pp. 1–4. IEEE, [Google Scholar]
- 13.Zhang Y. Contextualizing consumer health information searching: an analysis of questions in a social Q&A community. Proceedings of the 1st ACM International Health Informatics Symposium; 2010. pp. 210–219. ACM, [Google Scholar]
- 14.Luo G. Lessons learned from building the iMed intelligent medical search engine. Engineering in Medicine and Biology Society, 2009 EMBC 2009 Annual International Conference of the IEEE; 2009. pp. 5138–5142. IEEE, [DOI] [PubMed] [Google Scholar]
- 15.Crain SP, Yang SH, Zha H, Jiao Y. Dialect Topic Modeling for Improved Consumer Medical Search. AMIA Annual Symposium Proceedings; 2010. p. 132. volume 2010, American Medical Informatics Association, [PMC free article] [PubMed] [Google Scholar]
- 16.Can AB, Baykal N. MedicoPort: A medical search engine for all. Computer methods and programs in biomedicine. 2007;86(1):73–86. doi: 10.1016/j.cmpb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Zeng QT, Crowell J, Plovnick RM, Kim E, Ngo L, Dibble E. Assisting consumer health information retrieval with query recommendations. Journal of the American Medical Informatics Association. 2006;13(1):80–90. doi: 10.1197/jamia.M1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain Samat, Natarajan Karthik, Daniel Stein, Elhadad Nomie. An analysis of clinical queries in an electronic health record search utility. International Journal of Medical Informatics (IJMI) 2010;79:515–522. doi: 10.1016/j.ijmedinf.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2008;457(7232):1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 20.Eysenbach G, Kohler C. Qualitative and quantitative analysis of search engine queries on the internet. American Medical Informatics Association; 2003. What is the prevalence of health-related searches on the World Wide Web? [PMC free article] [PubMed] [Google Scholar]
- 21.Eysenbach G, K’ohler C. How do consumers search for and appraise health information on the world wide web? Qualitative study using focus groups, usability tests, and in-depth interviews. Bmj. 2002;324(7337):573. doi: 10.1136/bmj.324.7337.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spink A, Yang Y, Jansen J, Nykanen P, Lorence DP, Ozmutlu S, Ozmutlu HC. A study of medical and health queries to web search engines. Health Information & Libraries Journal. 2004;21(1):44–51. doi: 10.1111/j.1471-1842.2004.00481.x. [DOI] [PubMed] [Google Scholar]
- 23.Hanauer DA. EMERSE: the electronic medical record search engine. AMIA Annual Symposium Proceedings,; American Medical Informatics Association; 2006. p. 941. volume 2006, [PMC free article] [PubMed] [Google Scholar]
- 24.Hanauer DA, Zheng K, Mei Q, Choi SW. Full-text search in electronic health records: Challenges and opportunities. Search Engines: Types, Optimization and Effectiveness. 2010;7:1–15. [Google Scholar]
- 25.Zheng K, Mei Q, Hanauer DA. Collaborative search in electronic health records. Journal of the American Medical Informatics Association. 2011;18(2) doi: 10.1136/amiajnl-2011-000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SW, Kitko CL, Braun T, Paczesny S, Yanik G, Mineishi S, Krijanovski O, Jones D, Whitfield J, Cooke K, et al. Change in plasma tumor necrosis factor receptor 1 levels in the first week after myeloablative allogeneic transplantation correlates with severity and incidence of GVHD and survival. Blood. 2008;112(4):1539. doi: 10.1182/blood-2008-02-138867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Englesbe MJ, Lynch RJ, Heidt DG, Thomas SE, Brooks M, Dubay DA, Pelletier SJ, Magee JC. Early Urologic Complications After Pediatric Renal Transplant: A Single-Center Experience. Transplantation. 2008;86(11):1560. doi: 10.1097/TP.0b013e31818b63da. [DOI] [PubMed] [Google Scholar]
- 28.DeBenedet AT, Raghunathan TE, Wing JJ, Wamsteker EJ, Dimagno MJ. Alcohol use and cigarette smoking as risk factors for post-endoscopic retrograde cholangiopancreatography pancreatitis. Clinical Gastroenterology and Hepatology. 2009;7(3):353–358. doi: 10.1016/j.cgh.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White T. Can’t beat jazzy: Introducing the java platform’s jazzy new spell checker api. 2010.
- 30.Xie Y, O’Hallaron D. Locality in search engine queries and its implications for caching. INFOCOM 2002 Twenty-First Annual Joint Conference of the IEEE Computer and Communications Societies. Proceedings. IEEE; 2002. pp. 1238–1247. volume 3, IEEE, [Google Scholar]
- 31.Jansen BJ, Spink A, Bateman J, Saracevic T. Real life information retrieval: a study of user queries on the Web. ACM SIGIR Forum. 1998. pp. 5–17. volume 32, ACM,
- 32.Jain A, Cucerzan S, Azzam S. Acronym-expansion recognition and ranking on the web. Information Reuse and Integration, 2007 IRI 2007. IEEE International Conference on; 2007. pp. 209–214. IEEE, [Google Scholar]
- 33.Cucerzan S, Brill E. Spelling correction as an iterative process that exploits the collective knowledge of web users. Proceedings of EMNLP; 2004. pp. 293–300. volume 4, [Google Scholar]
- 34.Krovetz R. Viewing morphology as an inference process. Proceedings of the 16th annual international ACM SIGIR conference on Research and development in information retrieval; 1993. pp. 191–202. ACM, [Google Scholar]
- 35.Jones R, Klinkner KL. Beyond the session timeout: automatic hierarchical segmentation of search topics in query logs. Proceeding of the 17th ACM conference on Information and knowledge management; 2008. pp. 699–708. ACM, [Google Scholar]
- 36.Stearns MQ, Price C, Spackman KA, Wang AY. SNOMED clinical terms: overview of the development process and project status. Proceedings of the AMIA Symposium; 2001. p. 662. American Medical Informatics Association, [PMC free article] [PubMed] [Google Scholar]