Abstract
Knowledge of medication indications is significant for automatic applications aimed at improving patient safety, such as computerized physician order entry and clinical decision support systems. The Electronic Health Record (EHR) contains pertinent information related to patient safety such as information related to appropriate prescribing. However, the reasons for medication prescriptions are usually not explicitly documented in the patient record. This paper describes a method that determines the reasons for medication uses based on information occurring in outpatient notes. The method utilizes drug-indication knowledge that we acquired, and natural language processing. Evaluation showed the method obtained a sensitivity of 62.8%, specificity of 93.9%, precision of 90% and F-measure of 73.9%. This pilot study demonstrated that linking external drug indication knowledge to the EHR for determining the reasons for medication use was promising, but also revealed some challenges. Future work will focus on increasing the accuracy and coverage of the indication knowledge and evaluating its performance using a much larger set of drugs frequently used in the outpatient population.
Introduction
Studies have reported on the frequency of inappropriate prescribing1, 2, and in order to improve patient safety, it is essential to know the reasons why medications are prescribed to patients. However, expert knowledge is required to determine the reasons a medication may be prescribed based on information contained in the Electronic Health Record (EHR) because physicians generally do not explicitly document the reasons medications are prescribed, and there are usually no direct connections between medications and the corresponding indications. Therefore, a knowledgebase of indications for medications would be valuable for automated clinical applications addressing patient safety, such as computerized physician order entry(CPOE), clinical decision support systems (CDSS)3, question answering system4, and post-marketing pharmacovigilence5,6. However, for the indication knowledge to be widely applicable it must be interoperable with information in the EHR, which contains pertinent information concerning medication prescriptions as well as a wide range of other types of patient data, including problems, diagnoses, procedures, and historical data. Typically, a clinical note is divided into several sections, such as History of Present Illness, Medications, Assessment and Plan, and Problems, and typically, there is no explicit linguistic relationship between the medication and indication. Therefore, the relationship between a medication and indication can only be inferred using expert knowledge. A manual chart review of a sample of 120 outpatient notes in our outpatient notes showed that in 76.7% of the notes physicians did not explicitly document the reasons medications were prescribed. Figure 1 shows a portion of a typical outpatient note where a physician included the drug sinemet in the Medications section and the reason for prescribing sinemet (i.e. Parkinsonism) in the Problem section.
Figure 1.
An example of an outpatient note (not from an actual patient)
Recently, as part of the I2B2 natural language processing (NLP) challenge7, methods were developed to automatically identify the reasons for medication mentions in discharge summaries using NLP and machine learning methods. The best NLP system obtained an F-measure of 0.4598 for that task, demonstrating that it was challenging and non trivial task. That task was only one of the tasks in the I2B2 challenge and not the focus, and therefore the different methods that were used for that task and the associated issues were not discussed much. In other related work, Chen and colleagues acquired knowledge of disease-drug associations from clinical documents using text mining and statistical approaches based on co-occurrence of the medication and disease in the same patient record9. The limitation of the statistical method based on co-occurrence is that the relation denoted by the drug-disease association may denote a treat relation, an adverse drug event (ADE), or may be due to an indirect relation, and statistical co-occurrence is not able to differentiate between the three different types of relations. For example, based on co-occurrence in the EHR an association between Warfarin and myocardial infarction would likely be detected, which corresponds to a treat relation, but another association between Warfarin and hemorrhaging would also likely be detected, which corresponds to an adverse event relation. Rindflesch and colleagues also calculated the co-occurrences of medications and diseases using 16 million patient notes at the Mayo clinic, and suggested a cutoff of 80 co-occurrences to determine that the drug condition relationship was probably a treat relation10. The constructed repository of drug indications based on these patient records was then used to validate predicates concerning drug treatments for diseases produced by SemRep as a result of extracting the information from PubMed abstracts using NLP.
Another challenge associated with finding reasons for medications in clinical notes is the lack of availability of a comprehensive source of executable indication knowledge. Multiple knowledge sources contain complementary drug knowledge of indications but they are either in the form of text, such as the FDA drug labels11 and therefore not in a computable form, or they are proprietary. Commercial databases such as Micromedex12 or privately owned databases belonging to pharmaceutical companies provide accurate drug knowledge but they are not accessible publicly, and they are not always up-to-date. Sharp et al. examined 23 public available drug knowledge sources for researchers to select sources for a given use case13. Among them, 6 sources provided drug indication information, but most of them cannot be directly used for automatic applications because they are in free-text. NDF-RT14 in the MRREL file of the UMLS was the only structured data source containing drug indication knowledge. However, Charter and colleagues showed that only 80% of the drug indication knowledge was correct15, but NDF-RT has been updated several times since Charter’s paper, and the accuracy may have improved. SemMED, another source providing drug indication knowledge, is generated from Medline citations using the NLP system SemREP to extract semantic predications from the medical literature16. However, none of the knowledge bases mentioned above reflect actual physician usage, and therefore they may not be adequate for obtaining reasons for medications based on information in the EHR notes. The Adverse Event Reporting System (AERS) is another source that has not been exploited yet to determine indications of medications. It is an important early warning system containing case reports of adverse drug reactions, where the reports come from health care professionals, consumers and manufacturers. In addition to reporting adverse events, other information, such as indications for use of the drugs, are also reported for each case. Indications named in AERS reflect actual usage by physicians, and therefore contain many variants of the indications and many off label uses. Most importantly, because the indications in AERS reflect actual uses of the medications, they are similar to conditions reported in the patient record. In AERS database, the indications and the suspected adverse events for a drug are coded by the corresponding preferred terms in the MedDRA terminology17, but the drug names are entered as free text, such as ESOMEPRAZOLE (NEXIUM) 40 MG, ESOMEPRAZOLE AT HOME and NEXIUM TB. RxNorm is a resource providing standardized nomenclature for clinical drugs. In our work, we use it to link drug brand names to the corresponding generic names8. However, the stand-alone RxNorm could only link standard drug brand names to corresponding generic names, while the drug data entry in AERS contained many variations of standard drug brand names. In order to link these variations to their standard brand names and then acquire generic names, we used the NLP system, MedLEE12 first, and then RxNorm. The Food and Drug Administration (FDA) provides the AERS data files from 2004 to 2009 on its website when we started our study18.
A number of approaches have emerged using the AERS data set that look for interesting associations through the adverse drug event (ADE) distribution19. Reporting ratio, proportional reporting ratio, reporting odds ratio and information component are four major association measurements based on two dimensional contingency tables. All four of the measures detect particular drugs that are more likely to cause particular adverse events than other drugs. In this study, we used the idea of interesting association measurement for ADEs, and chose reporting ratio as the preliminary method to measure the association between a drug and medical condition in the indication distribution and rank order of the association.
No single source provides complete up-to-date drug indication information. Wang et al. integrated three data sources (NDF-RT, SemMED and AERS), and results showed that the coverage of the drugs in the integrated knowledge base was higher than that of the individual ones. The coverage was 96% for the integrated knowledge base, 95% for AERS, 60% for SemMED and 72% for NDF-RT20. In addition, in order to get an increased accuracy of the indications in the knowledgebase, she selected drug indication knowledge based on a conditional probability with the cutoff of 0.2% from AERS and SemMED since both sources corresponded to case reports or to articles, and therefore contained much noise. However, Wang did not study how useful the indications in the knowledgebase were for determining indications for medications in the EHR, and therefore this study builds upon her work.
The aim of this study is to acquire a comprehensive structured and coded knowledge base of indications for medications, and to develop methods that determine the reasons for medication uses in the EHR using the knowledge base. We also assess the challenges associated with this task.
Data
We used three sources of data to acquire drug indication knowledge: (1) the textual FDA approved indications listed in Micromedex, (2) the structured drug indication knowledge in NDF-RT from the UMLS 2010AA, (3) the case reports concerning medication indication usage in the Adverse Event Reporting System (AERS) reported by health care providers during 2007 to 2009.
After IRB approval, the outpatient free text clinical notes during 2004 to 2009 were extracted from New York Presbyterian Hospital (NYPH) clinical data warehouse for this study, which includes 179,027 primary clinical notes. We used the notes of a population of patients in the Associates in Internal Medicine Practice (AIM), which is a general internal medicine clinic for adults. We used clinical notes associated with visits to primary care physicians only, and not to specialists because they often contained only special medical conditions for a few drugs.
Methods
Choosing drugs of interest
We obtained a list of generic drugs that were being prescribed to the outpatient population by processing the patient records using the NLP system MedLEE12 to extract and encode the drugs and to obtain their UMLS codes. RxNorm was then used to map the brand names to their generic names. For this preliminary work, we focused on 20 drugs, chosen by a medical expert (the fourth author, HC), representing a broad range of commonly used drugs. For the evaluation study, six of the 20 drugs were chosen based on the number and similarity of medical conditions for which the drug could be prescribed (indications). For example, Carbidopa is associated with very few indications and is mainly used to treat Parkinson disease, Metoprolol is associated with several related indications and is mainly used to treat hypertension, congestive heart failure and angina, and Diphenhydramine is associated with many different unrelated indications, such as allergic rhinitis, common cold, insomnia, motion sickness, Parkinson disease and pruritus of skin (see Table 1). We reasoned that the accuracy of the methodology described in this paper, to ascertain the presence or absence of indications for drug prescribing in the EHR notes, might be a function of the number and similarity of indications for a drug.
Table 1.
Categorization of drugs according to the indications
| Number of indications | Drugs |
|---|---|
| Very few indications | Carbidopa, Iron, Esomeprazole |
| Several indications | Metoprolol, Warfarin |
| Many different indications | Diphenhydramine |
Overview
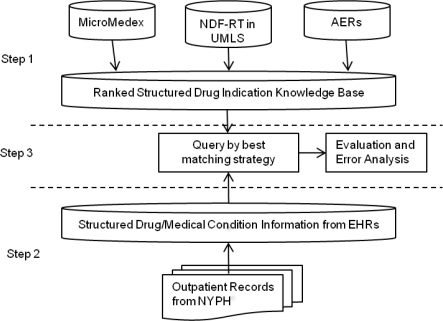
Figure 2 illustrates the overall process for finding reasons medications are prescribed, which consists of three steps: (1) creating the knowledgebase of indications from three sources: Micromedex, NDF-RT, and AERS, (2) using the MedLEE NLP system to process patient records in order to retrieve the relevant drug and medical conditions, (3) matching the drug-condition co-occurrences from the patient record with the knowledgebase drug-indication pairs, and choosing the highest ranked matches as the reason for the medication use.
Figure 2.
The overall process for determining reasons for medication usage
Generating the drug indication knowledgebase: we generated the knowledgebase for the selected drugs by acquiring the knowledge from each of the resources, obtaining the codes for the drugs and the corresponding indications so that the format consisted of paired drug and indication codes, and the source. We used Micromedex to acquire the FDA approved indications by searching for the 20 chosen generic names in Micromedex, manually copying each item under the list of FDA labeled indications section, and then using MetaMap21 to automatically map the terms to the corresponding UMLS codes. For the terms which MetaMap mapped to more than one concept, we chose the highest scoring concept which corresponded to a UMLS semantic type Disease and Syndrome or Pathologic Function. After that, we checked each code manually to make sure the code was correct, so that we could correct it if necessary. For the next source, we used MRREL in the UMLS to obtain drug indication knowledge corresponding to NDF-RT using the two relations: may_treat and may_prevent. These relations already consisted of UMLS codes for the drugs and indications. For the third source, the AERS database, we used the indications file, and chose only indications that were reported by health care professionals in order to reduce noise. The file consisted of free text drug names and MedDRA17 codes for the indications. We first mapped the drug names to UMLS codes using MedLEE, and then obtained the corresponding generic names using RxNorm. It was straightforward to map the indication terms to UMLS codes because they were in the form of MedDRA preferred terms, which are in the UMLS. Since the AERS database is composed of many case reports as described above, we combined the information consisting of the same drug-indication pairs from all the reports in AERS to obtain a reporting frequency for each pair. Because infrequent medication-indication pairs can be due to noise in AERS, we used the reporting ratio (RR)22, which is defined by Formula 1 below, with a cutoff of RR > 1 (according to the definition of RR, RR = 1 means no association) as a measurement of “interesting” drug indication pairs in order to filter out noisy pairs automatically. RR is the ratio of the observed drug indication co-occurrence to its expected co-occurrence under the assumption of independence, and the larger RRs are indicative of the more interesting drug indication combinations. We also compared the performance using this measurement to filter out noise with that of the conditional probability measurement used in previous work20, which measured the frequency of the indication for the specific drug over all the indications for that specific drug. Once we obtained indications from the three sources, we combined them and saved the sources. In addition, for AERS, we also saved the measurements of the RR corresponding to each entry. Then we ranked the indications for each drug so that the most established and frequent uses ranked higher than the other uses. More specifically, drug indication knowledge from Micromedex was always given the highest rank because that knowledge corresponded to FDA approved uses. Drug indication knowledge from NDF-RT was given the next best rank, and the indication knowledge from AERS was assigned different ranks based on the value of the RR measure. The RRs which had the same value were considered to be tied.
| (1) |
Processing the EHR: the clinical notes were processed by the NLP system MedLEE12, which extracted and mapped clinical terms to their UMLS codes along with modifiers, such as temporal and certainty modifiers. Furthermore, the extracted codes were filtered by the UMLS semantic types, such as Disease or Syndrome, Pathologic Function or Clinical Drug in order to obtain only the relevant drug entities and medical conditions. In addition, drug and condition mentions were filtered out if they were not asserted as being related to the patient or if they occurred in the past based on the modifier types and values generated by MedLEE. Brand names of drugs were mapped to generic names using RxNorm as described above. The processed data was stored in a relational database, which was then used as shown in Step 3 of Figure 2 to find the reasons for the medications mentioned in the EHR.
Finding the reasons: each drug was paired with all relevant medical conditions within a single clinical note to obtain a drug-condition dataset. We then obtained an intersection between the EHR based drug-condition set and the the knowledgebase dataset. The highest ranking drug-condition pair in the resultant intersection set was chosen as the most likely reason for prescribing the drug. If the drug indications were tied, the system chose all tied matches. The matching drug-indication pairs were considered positive matches. Unmatched cases were those notes which mentioned a drug in our study but had no matching indication according to the knowledgebase. In order to evaluate performance, we recorded the results of the matched and unmatched cases, and identified the corresponding notes, which were used later to evaluate performance.
Evaluation
We performed an evaluation of the method for a sample of 6 drugs out of the 20 drugs. At the sampling stage, we assumed all medications extracted by the NLP system in step 2 were prescribed to the patient. For each of the 6 drugs, we randomly sampled 10 patient records associated with a positive match between the medication and the indication (e.g. the system found a matching medical condition for the given drug), and 10 associated with a negative match according to our system (e.g. the system could not find a matching medical condition for the given drug), amounting to a total of 120 records. We established a criterion for true positive (TP), true negative (TN), false positive (FP) and false negative (FN) matches as shown in Table 2. A gold standard for these 120 records was obtained using a medical expert who reviewed the patient notes manually according to this criterion.
Table 2.
Definitions for TP, TN, FP, FN
| Definition | Description |
|---|---|
| True Positive | The medication extracted by the system is truly prescribed to the patient according to the patient note, and the matched medical condition by the system is the true indication for this taken medication in the note. |
| True Negative | The medication extracted by the system is truly prescribed to the patient according to the patient note, and the system could not match a medical condition and the tine indication for this medication is not documented in the note. |
| False Positive |
|
| False Negative | The medication extracted by the system is truly prescribed to the patient, but the system could not find a medical condition matched with this medication. The true indication is documented in the note. |
The medical expert was given two kinds of patient notes, one showed the medication and the matched medical condition (positive matches), and the other only showed the medication (negative notes). The expert reviewed each record associated with positive matches to determine if the matched pair was correct, and if it was incorrect, the expert provided an explanation for this decision. For records which only showed the medication the expert determined whether the case was correct, and if it was not, the expert provided an explanation. Sensitivity, specificity, precision as well as F-measure were computed, and an analysis of the problems was performed. In order to evaluate the accuracy of the drug indication knowledgebase itself, a reference standard was created by a medical expert (the second author, HS) for the 6 drugs. The expert reviewed the entire list of the indications collected from the three sources, and determined if the drug-indication pair was appropriate according to his clinical knowledge. The accuracy of the combined knowledgebase as well as the two different F-measures based on using the conditional probability with a cutoff of 0.2% and the RR with a cutoff of 1 was also computed.
Results
There were 2210 unique generic drugs mentions based on the outpatient set of records during 2004 to 2009. NDF-RT, AERS and the combined NDF-RT and AERS covered 43%, 67% and 69% respectively of the drugs used in the outpatient records. We did not report the coverage of Micromedex for the entire set of 2210 drugs because we could not acquire that information for all those drugs automatically. The 20 drugs in our sample were included in all the sources, and therefore the coverage for the 20 drugs was 100% for each source. The number of indications for the 20 drugs in the combined knowledgebase varied from Milrinone with 9 indications to Warfarin with 209 indications, where the average was 88. The total number of unique medication-indication pairs for the 20 drugs was 1760. Among them, 1541 drug-indication pairs were uniquely from AERS. The positive matching rate of a medication is the number of positive matches divided by the number of patient records mentioning the same medication. The average positive matching rates for the 20 chosen drugs were 42% when using Micromedex, 51% when using NDF-RT, 83.5% when using AERS and 84.5% when using the combined sources. The average accuracy of the combined knowledgebase for the 6 sampled drugs based on the gold standard was 63.1%. The indications from Micromedex using MetaMap and from NDF-RT were all correct for the 6 sampled drugs, and the errors all corresponded to indications from AERS. For the evaluation of performance of the overall process that determined the reasons for use of the 6 chosen drugs using the knowledgebase and patient record, the sensitivity was 62.8%, specificity was 93.3%, precision was 90% and F-measure was 73.9%. Furthermore, the average F-measures for retrieving drug indication knowledge from AERS were 61.5% when using RR with a cutoff of 1 compared to 38.3% for conditional probability with a cutoff of 0.2%.
Discussion
The aim of this pilot study was to see if we could automatically determine the reasons for drugs mentioned in the textual patient notes using external drug knowledge and NLP. Additionally, we wanted to identify and analyze the challenges. Overall, our results were promising, but also suggested areas of improvement. The F-measure for this study was 0.739, which was mainly due to the use of indications in AERS because it contained the largest amount of indications and corresponding variations, and was based on actual usage expressed by health professionals. A condition in the outpatient notes that matches an indication in the knowledgebase can be categorized into four groups: (1) an FDA approved indication, such as iron deficiency anemia which is treated by Iron; (2) a symptom closely related to an approved indication, such as tremor which is treated by Carbidopa. Tremor is a symptom of Parkinson disease which is an approved indication of Carbidopa; (3) a condition, such as essential hypertension, is a more specific term than the FDA approved indication hypertension for Metoprolol; (4) a condition is an off-label use, such as migraine headache which was treated by Metoprolol. The positive matching rate in our 6 chosen drug sample was substantially improved after using AERS because the number was 37.9% when using the drug label, 83.4% when using AERS, and 84%, when using the combined sources. AERS captured more indications than the two other sources and included many appropriate conditions that could be matched to the patient record. This is probably because the indications from AERS were reported by physicians who also generally write patient notes, and therefore the two groups are similar.
Although AERS captured many variants of the indications as well as off-label uses, it was not complete, which was a reason for some of the negative cases. Table 3 categorizes the most frequent errors according to their characteristics. Overall, 23.7% of the problems were because medical conditions documented as occurring in the past were filtered out in our matching process. Most of the medical conditions associated with this type of problem were chronic diseases, such as hypertension, or acute diseases where a recurrence could be prevented by a treatment, such as a stroke. However, we did not differentiate between the cases where conditions occurring in the past should be considered as an appropriate reason for prescribing a medication, and the cases where past conditions are not appropriate. One way to handle this would be to include information concerning temporality of indications in the knowledgebase, which would involve additional manual effort. Misspelling of the indications amounted to 15.8% of the problems, such as skin with excoration (excoriation), which is known to be a challenging NLP problem. Another 13.2% of the problems were due to drugs which were prescribed because a certain procedure was performed, and we missed these cases because we only looked at conditions when trying to find a matching indication. For example, Warfarin was prescribed for mitral valve replacement. Including the semantic type Therapeutic or Preventive Procedure in the information selection phrase of the EHR data process would be a simple way to solve this problem. The drug which was not currently taken by patient, such as s/p sinemet, was another type of problem that was not handled correctly. It could be addressed by looking at the temporality modifier of the medication. Another type of error, which would be a challenge to address, was due to implied reasons, which physicians were able to infer when reading the patient record. For example, Iron and irregular bleeding were mentioned in a patient record, and the expert inferred that irregular bleeding caused anemia, and therefore, irregular bleeding was a likely reason that Iron was being prescribed. Finally a number of incorrect indications were found in AERS, which generally involved infrequent indications. One type of problem was probably due to errors in reporting where the indications should have been entered as adverse events and not indications. For example, hypertension was listed several times as an indication for Esomeprazole, but it is actually an adverse event. Another problem was caused by indications which were too broad, which were seen both in the knowledgebase and patient record. An example is aneurysm, which was included as an indication for Warfarin in the knowledgebase from the source AERS. However, aneurysm is too broad an indication, and requires an associated anatomical location in order to determine whether the condition is appropriate for Warfarin. For example, Warfarin is given to prevent clots associated with atrial septal aneurysm which may lead to a stroke. At the same time, Warfarin should not be given to a patient with a brain aneurysm since Warfarin could cause a fatal hemorrhage in that situation. The third problem associated with the drug indication knowledgebase is that some infrequent but correct indications in AERS, such as cough as an indication for Diphenhydramine, were incorrectly eliminated by the RR measurement with the cutoff of 1.
Table 3.
Qualitative Analysis of Problems in Detecting Reasons for Medication Uses
| Types of Error | Examples | Cases | Percentage |
|---|---|---|---|
| *Past medical conditions were ignored | Warfarin: stroke-8/07 Iron: h/o Ferropenic anemia |
9 | 23.6% |
| Misspelling and typos in the patient record | Diphenhydramine: skin with excoration (excoriation) Metoprolol: elevated, blood pressure |
6 | 15.8% |
| *Procedures were ignored | Warfarin: mitral valve replacement Warfarin: aortic valve replacement |
5 | 13.2% |
| ***Indication were not in the knowledge base | Diphenhydramine: nasal congestion Metoprolol: migraine |
4 | 10.5% |
| **The drug was no longer prescribed | Carbidopa: s/p sinemet | 3 | 7.9% |
| Implied reasons | Iron: irregular bleeding Diphenhydramine: sleep) |
3 | 7.9% |
| ***Too broad indication | Warfarin: Aneurysm | 1 | 2.6% |
| Others | 7 | 18.4% |
The error could be fixed by changing the query to the output of NLP
The error could be fixed by improving the performance of NLP
The error could be fixed by improving the coverage and the accuracy of the drug indication knowledgebase
Some errors could be solved by fixing the query logic, improving the performance of NLP and the knowledge base. However, it would be a big challenge to identify implied reasons which do not match the appropriate indications, but which physicians could infer using their knowledge. This issue concerns adequate documentation, and the issue is whether other physicians should have to infer the reason for using a medication.
Limitation and Future Work
There were several limitations to our study. First, we did not evaluate the sensitivity of the NLP system itself when extracting medication mentions in the outpatient clinical notes, and therefore may have missed some notes where medications were actually mentioned but not detected, which would result in overestimating the sensitivity of our method. However, the focus of this study was exploring issues associated with finding reasons for a medication once it was specified. A second limitation concerned our evaluation, which involved a small sample consisting of 6 drugs corresponding to 535 drug-indication pairs and one medical expert. We will perform a larger evaluation with many more drugs in the future to see if the method is scalable. In addition, we will use more than one physician to determine the reason for a prescription when obtaining the gold standard. A third limitation was that we used AERS reports from 2007 to 2009 in our preliminary study set, but FDA provided AERS reports from 2004 to 2009, therefore some indications during the year 2004 to 2006 could be missed from the drug indication knowledge base. We will use the whole data set provided by the FDA in the future. Another limitation is that we used RR as an association measurement in order to eliminate certain indications from AERS; however, RR is subject to sampling variability and this is particularly the case with large, sparse database like AERS. Some of the literature describes different approaches such as the Gamma Poison shrinker (GPS) and multi-item Gamma Possion shrinker (MGPS) to deal with this problem, and we will investigate whether these methods could be adapted to the indication distribution.
Conclusions
Linking an external knowledgebase to clinical notes is a promising approach for determining likely reasons medications mentioned in the patient records are used. Acquiring the knowledge from available sources and standardizing and integrating the knowledge resulted in substantially improved performance than that which was obtained when using only one source. Using the indications in AERS enabled the capture of many reasons for the medication prescriptions in the patient record, and substantially increased sensitivity because AERS is based on actual use, and includes the many varied ways of expressing the uses as well as the many off label uses whereas the other sources do not. To the best of our knowledge this is the first time AERS has been explored to specify indications of medications. Future work is needed to improve coverage of the knowledgebase, to increase accuracy of use of indications in AERS, and to improve the use of NLP information.
Acknowledgments
The authors thank Lyudmila Shagina for assistance with MedLEE and the relational database. This research was supported in part by grants 1R01LM010016, 3R01LM010016-01S1, 3R01LM010016-02S1 and 5T15-LM007079-19(HS) from the National Library of Medicine.
Reference
- 1.Fick DM, Cooper JW, Wade WE, et al. Updating the Beers criteria for potentially inappropriate medication use in older adults: results of a US consensus panel of experts. Archives of Internal Medicine. 2003;163:2716–24. doi: 10.1001/archinte.163.22.2716. [DOI] [PubMed] [Google Scholar]
- 2.Curtis LH, Ostbye T, Sendersky V, et al. Inappropriate prescribing for elderly Americans in a large outpatient population. Archives of Internal Medicine. 2004;164:1621–25. doi: 10.1001/archinte.164.15.1621. [DOI] [PubMed] [Google Scholar]
- 3.Cowie MR. Improving the management of chronic heart failure. The Practitioner. 254:29–32. [PubMed] [Google Scholar]
- 4.Walton SM, Schumock GT, Lee KV, et al. Prioritizing future research on off-label prescribing: results of a quantitative evaluation. Pharmacotherapy. 2008;28:1443–1452. doi: 10.1592/phco.28.12.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman C. Discovering Novel Adverse Drug Events Using Natural Language Processing and Mining of the Electronic Health Record. Artificial Intelligence in Medicine. 2009:1–5. [Google Scholar]
- 6.Walker AM. Confounding by indication. Epidemiology (Cambridge, Mass.) 1996;7:335. [PubMed] [Google Scholar]
- 7.https://www.i2b2.org/NLP/.
- 8.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010;17:514–18. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ES, Hripcsak G, Xu H, et al. Automated acquisition of disease-drug knowledge from biomedical and clinical documents: An initial study. Journal of the American Medical Informatics Association. 2008;15:87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rindflesch TC, Pakhomov SV, Fiszman M, et al. “Medical facts to support inferencing in natural language processing,”. 2005:634–38. [PMC free article] [PubMed] [Google Scholar]
- 11.http://dailymed.nlm.nih.gov/dailymed.
- 12.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. Journal of the American Medical Informatics Association. 2004;11:392. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp M, Bodenreider O, Wacholder N. “A framework for characterizing drug information sources,”. 2008:662–66. [PMC free article] [PubMed] [Google Scholar]
- 14.Pathak J, Chute CG. Analyzing categorical information in two publicly available drug terminologies: RxNorm and NDF-RT. Journal of the American Medical Informatics Association. 2010;17:432–39. doi: 10.1136/jamia.2009.001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter JS, Brown SH, Erlbaum MS, et al. Initializing the VA medication reference terminology using UMLS metathesaurus co-occurrences. 2002:116–20. [PMC free article] [PubMed] [Google Scholar]
- 16.Fiszman M, Demner-Fushman D, Kilicoglu H, et al. Automatic summarization of MEDLINE citations for evidence-based medical treatment: A topic-oriented evaluation. Journal of Biomedical Informatics. 2009;42:801–813. doi: 10.1016/j.jbi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA) Drug Safety. 1999;20:109–117. doi: 10.2165/00002018-199920020-00002. [DOI] [PubMed] [Google Scholar]
- 18.http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm083765.htm.
- 19.Hauben M, Madigan D, Gerrits CM, et al. The role of data mining in pharmacovigilance. 2005:929–948. doi: 10.1517/14740338.4.5.929. [DOI] [PubMed] [Google Scholar]
- 20.Chase Xiaoyan Wang Herbert S, Li Jianhua, Hripesak George. Integrating Heterogeneous Knowledge Sources to Acquire Executable Drug-Related Knowledge. American Medical Informatics Association. 2010:852–56. [PMC free article] [PubMed] [Google Scholar]
- 21.Aronson AR. “Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program,”. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 22.DuMouchel W. Bayesian Data Mining in Large Frequency Tables, with an Application to the FDA Spontaneous Reporting System. The American Statistician. 1999;53:177–202. [Google Scholar]