Abstract
The analysis of disease using protein-protein interaction networks and network pharmacology has enabled better understanding of disease etiology and drug action. New insights into disease etiology and a better understanding of biological subsystems have opened up the possibility of finding new uses for existing drugs besides their original medical indication. We present an approach which makes use of the biological processes associated with diseases along with their known drugs and drug targets to predict Biological Process-Drug relationships. Network analysis is used to further refine these associations to eventually predict new Disease-Drug relationships. The approach is validated by the observation that, out of 2078 predicted disease-drug relationships, 401 (18.1%) have been used in a clinical trial.
Introduction
There have been fewer drug approvals in the past decade compared to the past, along with frequent drug recalls [1]. The high cost of new drug development is partly to blame for this. Drug repositioning, making alternative uses of drugs outside their original indication, has the potential to drastically offset drug development costs. Although drug repositioning has been pursued for a long time, many of them have been serendipitous discoveries [2] or on observable clinical phenotypes. Making clinical decisions based solely on observable phenotype can be risky and lead to sub-optimal treatment. On the positive side, new high-throughput techniques and better methods for data analysis have enabled a detailed understanding of disease etiology and its underlying cellular subsystems. Biological knowledge such as protein-protein interaction (PPI) networks and biomedical ontologies have accelerated the development of network-based approaches to understanding disease etiology [3, 4] and drug action (network pharmacology) [5]. This has increased the possibility of finding new disease-drug relationships for existing drugs (drug repositioning) [2]. If the underlying pathophysiology of the disease and knowledge of the mechanism of drug action is utilized to make decisions on drug repositioning, it can potentially lead to better treatment with lower side effects.
A disease is usually caused by congenital or acquired mutations, or by the action of external agents that disrupt gene regulation [6]. This in turn disrupts biological processes in which the genes participate. Disruption of the biological processes results in phenotypes that characterize each disease, with the phenotype depending on the influence of the affected biological processes on the larger biological network. Reproducibility implicating a set of genes across multiple microarray analyses [7] of a disease state is often low. Representations that summarize the contributions of groups of genes such as GO-Processes have helped determine signatures in meta-analysis of studies on breast cancer [8]. If the affected biological processes can be identified, then suitable drugs can be used to offset the anomalies (over/under regulation). In principle, the same drug or set of drugs could be used for other diseases that share the affected biological processes.
In related work on drug repositioning, Gloecker et al [9] used informed insights and high-throughput assays to test the drug closantel resulting in its being used for onchocerciasis. Chiang & Butte [10] connected all diseases that shared a drug and made inferences on new drug-disease pairs using guilt-by-association, and verified the associations against clinical trails. Qu et al [11] present an rdf-framework with controlled vocabulary using various knowledge sources on pathways, drugs and diseases. Dasika et al [12] identified all proteins affected by targeting a specific protein in the network and used a constrained downstream problem to find if a GO-Process is affected. Additionally, network-based methods have been employed to find combinations of drug to treat a disease, discovering new drug targets [13] and finding potential drug side effects. Network properties like degree, centrality, cutsets, articulation points can be used to quantify a gene’s influence on a network [14] and used to study the extent of a drug’s influence on the network [15]. Swanson et al [16] introduced methods for learning new relations between entities from bibliographic databases and applied them to learn new therapies for existing diseases.
Ontologies have been helpful in knowledge representation and inferring relationships between different entities. The GO-Process ontology [17] is used to represent the cellular and biological processes that genes participate in whereas UMLS vocabularies like SNOMED-CT, MeSH and RxNorm [18] have been used to integrate clinical and biological information.
In this paper, we present a novel approach to highlight new applications for known drugs that is based on existing annotated data resources and protein interaction network analysis. We find that several of the predictions are supported by ongoing and completed clinical trials.
Method
We first determine genes involved in a disease from various high quality sources of annotation and then represent the disease in terms of potentially affected biological processes (based on GO annotation of genes). These Disease-Process associations and existing Disease-Drug-Drug Target mappings are then used to compile a list of biological processes affected by each drug. The drug target’s role in each biological process is then estimated using protein-protein interaction networks by computing its centrality (vide infra for section on ‘Estimation of centrality’) among the gene products in the GO-Process subnetwork. A list of refined Process-Drug pairs is obtained after selecting for drug targets that exhibit high centrality in the respective processes. Using these pairs and Disease-Process pairs, new Disease-Drug pairs are predicted. The predictions are validated by searching for the presence of corresponding clinical trials listed at http://www.clinicaltrials.gov.
Data Sources:
Disease-Gene associations were derived from Swisprot disease text (3000 lines), disease-gene associations from GeneRIF, and heading and sub-headings from OMIM disease records. Disease Ontology (DO) [19] and UMLS vocabularies (SNOMED-CT, NCI, MeSH, ICD-9) were used as controlled vocabulary for disease names. GO-Process ontology (March 2010) was used for the biological processes associated with diseases. To increase the precision of the predictions, only experimentally derived annotations were used; the annotation of genes to GO-Processes inferred from electronic annotation (IEA evidence code) was not considered. Drugbank [20] was used to find Disease-Drug and Drug-Drug Target (genes) associations. The ‘Indication’ field of approved drugs from Drugbank was used as it consistently provides unambiguous text mentioning diseases being treated with each drug. Protein-Protein Interaction network was extracted from Pathway Commons that integrates information from 1400 pathways in humans. A total of 85,254 clinical trials having ‘Drug Intervention’ as an XML tag were downloaded from http://clinicaltrials.gov/. The <condition> and <intervention_name> fields were used for diseases and drugs respectively. The dataset included ongoing, completed or discontinued clinical trials.
Associating Genes to Diseases:
Gene-disease associations were pooled from multiple sources since there is variable coverage of disease terms in different ontologies. For example, though MeSH has broad coverage on a variety of subjects, it has several missing terms and lacks detail in the disease section. To overcome this, the Disease Ontology (DO ver. 3) vocabulary was augmented using UMLS (MeSH, SNOMED-CT, ICD9) as described in [21]. The DO consisting of 12082 terms was augmented with synonyms to a total of 33085 terms. To increase the amount of annotated data available, Disease-Gene associations from OMIM, Swissprot and GeneRIF were pooled together. Swissprot records were matched against UMLS using MetaMap. GeneRIF identifiers that were previously mapped to DO terms were mapped to the corresponding Entrez genes using available associations at NCBI (ftp://ftp.ncbi.nih.gov/gene/GeneRIF/ftp://ftp.ncbi.nih.gov/gene/GeneRIF/). OMIM records were mapped against UMLS AUI records from which Concept Identifiers (CUIs) were extracted. Protein identifiers from Swissprot, gene identifiers from OMIM, and GeneRIF identifiers were matched with corresponding Entrez gene identifiers. In the final annotation, each DO identifier was annotated with Entrez gene identifiers.
Mapping Text to Controlled Vocabulary:
MetaMap was used to map disease text to UMLS identifiers with 7 semantic types indicated in [21]. The disease text included text in the ‘Indication’ field from Drugbank and text from the <Condition> field in clinical trial records. A score of >=850 was used as a filter to minimize the occurrence of false positives while sacrificing recall. The diseases terms were mapped back to DO identifiers. The drug names in clinical trials and Drugbank were mapped to UMLS’s RxNorm vocabulary using MetaMap. Thus disease and drug names from Drugbank were converted into controlled vocabulary terms in DO and RxNorm.
Association of Drugs with GO-Processes:
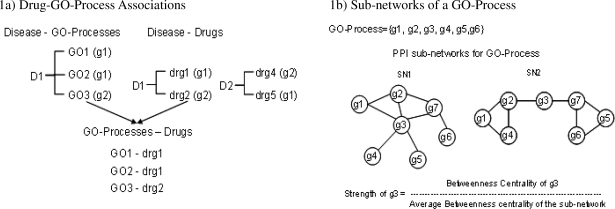
GO-Processes associated with each disease were identified by measuring the over-representation of GO-Processes in the corresponding gene set by using the hypergeometric test, and correcting for multiple testing using the Benjamini-Hochberg test. To minimize false-positives, a minimum membership of at least 3 genes was required for a GO-Process to be considered significant. To avoid abstract terms (low resolution information), only GO processes having a depth greater than 5 were chosen. GO-Processes with a p-value cut-off of 0.005 were extracted and only the most specific were associated with each disease. For example, in figure 1a, disease D1 is shown to be associated with Processes GO1, GO2 and GO3.
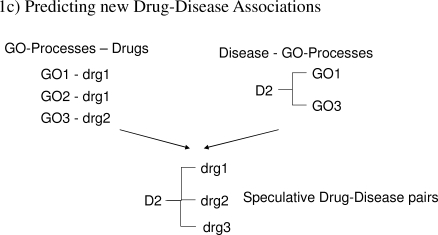
Figure 1.
Methods involved in predicting Drug-Disease associations. ‘GO’=GO-Process; ‘g’=Gene, ‘D’=Disease and ‘drg’=Drug. Figure 1a. Disease D1 is associated with GO1 and gene g1, the latter is also the drug target identified from Drugbank. Process GO1 is inferred to be associated with drg1. Figure 1b. Example of 2 subnetworks for a GO-Process with genes g1, g2, g3, g4, g5, g6 where g3 is the drug target. Figure 1c. Predicting new Drug-Disease associations from GO-Process – Drugs and Disease-GO-Process associations.
The disease-drug-drug target triplets from Drugbank and disease-GO Process pairs were used to associate drugs with GO-Processes. The drug was mapped to a GO-Process if it contained the drug target. To avoid false pairings, the association was made only if the drug targeted the same disease as the GO-Process was enriched in. For example, in figure 1a&b, using information from Disease D1, associations GO1-drg1, GO2-drg1 and GO3-drg2 were derived. However, GO3-drg4 or GO1-drg5 are not accepted since, even though the drug target is contained in the GO-process, the drugs are associated with a different disease.
Extraction of GO-Process subnetwork:
As the hypergeometric test assumes independence of entities (genes in the disease), many GO-Processes tend to get associated with a disease resulting in numerous drug-GO-Process associations. This increases the possibility of false associations. To overcome this, we use protein-protein interaction networks to find the influence of the drug target in the GO-Process. The sub-network relating to a GO-Process is extracted by computing shortest paths between every pair of genes in the PPI network. Intermediate genes (g7 in figure 1b) are also considered to be part of the sub-network. The assumption is made that each gene in a subnetwork has potential to regulate or disrupt it and a pair of genes is most likely to interact through the shortest path.
Estimation of Centrality:
If a sub-network is viewed as a biological module involved in a disease, the gene through which a lot of communication occurs can be considered as a potential drug target [22]. Betweenness centrality [14] gives the centrality measure of a vertex in a graph. The strength of a gene is defined as the ratio of its centrality over the average centrality of genes in the sub-network. For example, in figure 1b, centrality of gene ‘g3’ is high in both scenarios, even though the degree is lower in the 2nd case. The Drug-GO-Process associations were filtered by using a cut-off of strength >1.
Prediction of new Drug-Disease pairs:
Exploiting the Drug-GO-Process pairs and Disease-GO-Process pairs, drugs were associated with diseases. In figure 1c, given that disease D2 is associated with processes GO1and GO3, it is subsequently associated with drg1 and drg2.
GO-Process Information Content:
A common denominator of various diseases is the set of genes that participate in the immune response such as B/T-cell proliferation, chemotaxis and regulation of isotype switching. These processes, though not very specific to disease etiology, can be associated with many palliative drugs. To minimize the confounding effect of this, each GO-Process was normalized by its information content in the GO-Process graph and its information content in disease space.
where NF is the Normalizing Factor for the GO-Process, ICGO is the information content of the GO-Process P in the entire GO-Graph, and ICDIS is its information content in disease space. MaxICGO and MaxICDIS are maximum information contents in GO-Graph and Disease Space respectively. In effect, the effect of promiscuous association was mitigated.
Validation using Clinical Trials:
Drug-Disease pairs already present in Drugbank were removed from the list of predicted Drug-Disease pairs, leaving only purely predicted pairs. The remaining predictions were checked against the Disease-Drug pairs in clinical trials.
Results
Mapping Text using MetaMap:
Disease-related terms from the disease text in the ‘Indication’ field in Drugbank and <condition> entry of clinical trials were extracted using MetaMap. A random sample of 50 mappings from each was taken and the precision was found to be 94% in case of Drugbank and 98.7% in case of clinical trials. The high precision is attributable due to the high score cutoff (>=850) and other improvements suggested in [21]. The disease text from clinical trials consisted mostly of 1–2 words, hence the higher precision. The text related to drugs from Drugbank and clinical trials were mapped to RxNorm terms. The precision was 100% in case of Drugbank and 99% in clinical trails. Since the drug text mostly included only drug names, recall was 98% and 96% respectively. 1322 of 1398 approved drugs had a corresponding RxNorm identifier. Recall was not calculated for the disease text due to the vastness of disease vocabulary. From a previous study involving disease text from Swissprot [21], we estimate it to be approximately 72% in disease text. Recall was sacrificed for precision, as the focus is on short-listing Drug-Disease mappings to the more actionable predictions.
Disease, Drug, GO-Process Associations:
A total of 1698 drug-disease associations between 948 approved drugs and 581 diseases (DO terms) were extracted from Drugbank. A total of 40,731 clinical trials were determined to be associated with 1920 diseases and 792 drugs. 1188 diseases from DO were found to be enriched in 1480 GO-Processes - totaling 75,195 associations after filtering described in the Methods section. Using the drug-disease and disease-GO-Process associations, 1424 associations between 63 diseases, 147 GO-Process and 226 drugs were made. After the GO-Process sub-network analysis, where the betweenness centrality of the drug target was required to be higher than the average centrality of all genes, a list of 288 Drug-GO-Process associations between 45 GO-Processes and 95 drugs was created.
Predicted Drug-Disease Associations:
Using the final list of Drug-GO-Process associations, drugs were associated to diseases enriched in the corresponding GO-Processes (figure 1c). 2222 Drug-Disease associations were made between 169 diseases and 76 drugs. Out of these, 144 (6.4%) were found to be in Drugbank leaving 2078 new drug repositioning candidates.
Validation using Clinical Trials:
The 2078 Drug-Disease associations were checked against drug-disease associations in clinical trials. A total of 401 out of 2078 (19.3%) were found to be either a completed or ongoing clinical trial (involving 167 diseases and 67 drugs). The GO-Process information content was used to find the optimal cutoff as shown in Table 1.
Table 1.
Drug-Disease pairs found in clinical trails with corresponding GO-Process p-value cutoffs
| GO-Process Information Content | Number of Disease-Drug Associations | % found in clinical trials |
|---|---|---|
| 0.14 | 3000 | 15. 13% |
| 0.16 | 2512 | 16. 14% |
| 0.18 | 2365 | 17.09% |
| 0.2 | 2078 | 19.25% |
| 0.22 | 2010 | 19.25% |
| 0.28 | 1384 | 13.06% |
| 0.3 | 1173 | 15.15% |
| 0.35 | 507 | 12.86% |
It can be observed that at a p-value cutoff of 0.2, 19% of predicted drug-disease associations were found in clinical trials. To find if this cutoff is better than random association of drug and diseases a significance test was performed using randomly paired drugs and diseases (2078 pairs). These random pairs were checked against clinical trails and the number of matches recorded. A P-value was estimated as the number of matched clinical trials by random disease-drug pairs exceeding those found from the described method. This was done 1000 times and the resulting p-value was effectively 0. This shows that the drug-disease association prediction is not happenstance.
A representative sample of the Drug-Disease associations that had a clinical trial, together with the associated GO-Process, are presented in Table 2.
Table 2.
Drug-Disease pairs found in clinical trails with corresponding GO-Process p-value cutoffs
| Disease | Drug | GO-Process |
|---|---|---|
| Alzheimer’s Disease | Aripiprazole | Synaptic transmission, dopaminergic |
| Obesity | Metoprolol | G-protein signaling, coupled to cAMP nucleotide second messenger |
| Polycystic Ovary | Metformin | Regulation of fatty acid metabolic process |
| Coronary Arteriosclerosis | Telmisartan | Regulation of vasoconstriction |
| Cerebrovascular Accident | Alteplase | Negative regulation of blood coagulation |
Discussion
The drug industry has been adversely affected by fewer drug-approvals and increase in recall rates of drugs. There has been a recent push towards repositioning of existing drugs and targeting the underlying etiology of diseases rather than merely treating their effects. New techniques in data generation and analysis have enabled a better understanding of disease mechanisms. In this paper, we have presented a novel approach to drug repositioning by combining biological information of a disease and drug-disease relationships by using ontologies and network analysis. Ontologies are helpful not only in unambiguous data representation but also in finding relationships between entities and their importance [23]. They have helped to bridge the gap between biology and medicine (e.g, UMLS and GO). Protein-protein interaction networks that are based on physical interactions offer insight into biological modules. This has led to the development of network pharmacology through which methods have been developed to find new drug targets, disease modules and make advances in polypharmacology [24]. We have used the betweenness centrality measure to find the relevance of a gene in the GO-Process sub-network. This appears to help in improving the accuracy of predicted disease-drug associations. Without using this step, only 8% of predictions were found to correspond to a clinical trial (data not shown). In contrast, incorporation of the betweenness centrality resulted in a verification rate of 19.3%. Low information content of GO-Processes is helpful in filtering out abstract terms, rather than relying only on a rigid cutoff like being greater than 5 levels deep. To eliminate palliative drug associations, which have been extensively studied, the information content of diseases was combined with that of GO-Processes.
The gene target of aripiprazole (used to treat schizophrenia [19]) has been found to have high betweenness centrality in the GO-Process “Synaptic transmission, dopaminergic.” This GO-Process is also involved in bipolar disorder, dementia and Alzheimer’s disease, making it an attractive drug for repositioning. Telmisartan is an angiotensin II receptor antagonist (ARB) used in the management of hypertension. Regulation of vasoconstriction is a process affected in myocardial infarction, coronary arteriosclerosis and other cardiac disorders. Metformin improves glycemic control by improving insulin sensitivity and decreasing intestinal absorption of glucose and is used to improve glycemic control in type 2 diabetics [19]. Its target in regulation of fatty acid metabolic process was found to have high centrality and could potentially be used to treat PCOS. Some of the limitations of this approach are lower recall rates among disease text, which leads to lower disease-drug associations. The betweennness centrality measure eliminates false positives, but is a harsh threshold as it causes a decrease in recall.
Conclusion
In this paper, we have used the biological processes affected in a disease and existing disease-drug associations to predict new disease candidates for existing drugs. This is done using ontologies to match between biomedical text and network analysis of existing drug targets. Future work includes the use of additional network measures in conjunction with pathway data to check the importance of drug target genes in the network, thus increasing recall. Further investigation of similar biological processes involved in diseases of similar etiology (unpublished data) along with evidence of drug action can be used to quantify the use of existing drugs to treat a disease outside its original use.
Acknowledgments
We would like to thank Pablo Andrade for useful feedback on the clarity and organization of the manuscript.
References
- 1.Horrobin DF. Realism in drug discovery-could Cassandra be right? Nat Biotechnol. 2001;19(12):1099–100. doi: 10.1038/nbt1201-1099. [DOI] [PubMed] [Google Scholar]
- 2.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–83. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 3.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–72. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452(7186):429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ein-Dor L, et al. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21(2):171–8. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 8.Wirapati P, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gloeckner C, et al. Repositioning of an existing drug for the neglected tropical disease Onchocerciasis. Proc Natl Acad Sci U S A. 2010;107(8):3424–9. doi: 10.1073/pnas.0915125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang AP, Butte AJ. Systematic evaluation of drug-disease relationships to identify leads for novel drug uses. Clin Pharmacol Ther. 2009;86(5):507–10. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu XA, et al. Inferring novel disease indications for known drugs by semantically linking drug action and disease mechanism relationships. BMC Bioinformatics. 2009;10(Suppl 5):S4. doi: 10.1186/1471-2105-10-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasika MS, Burgard A, Maranas CD. A computational framework for the topological analysis and targeted disruption of signal transduction networks. Biophys J. 2006;91(1):382–98. doi: 10.1529/biophysj.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klamt S, et al. A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinformatics. 2006;7:56. doi: 10.1186/1471-2105-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barabasi AL, Bonabeau E. Scale-free networks. Sci Am. 2003;288(5):60–9. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 15.Yildirim MA, et al. Drug-target network. Nat Biotechnol. 2007;25(10):1119–26. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 16.Swanson DR. Somatomedin C and arginine: implicit connections between mutually isolated literatures. Perspect Biol Med. 1990;33(2):157–86. doi: 10.1353/pbm.1990.0031. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphreys BL, et al. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborne JD, et al. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10(Suppl 1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–6. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathur S, Dinakarpandian D. Automated Ontological Gene Annotation for Computing Disease Similarity. AMIA Summit on Translational Bioinformatics 2010. 2010. [PMC free article] [PubMed]
- 22.Estrada E. Protein bipartivity and essentiality in the yeast protein-protein interaction network. J Proteome Res. 2006;5(9):2177–84. doi: 10.1021/pr060106e. [DOI] [PubMed] [Google Scholar]
- 23.Mathur S, Dinakarpandian D. IEEE International Conference on Bioinformatics and Biomedicine. 2007. A New Metric to Measure Gene Product Similarity; pp. 333–338. [Google Scholar]
- 24.Boran AD, Iyengar R. Systems approaches to polypharmacology and drug discovery. Curr Opin Drug Discov Devel. 2010;13(3):297–309. [PMC free article] [PubMed] [Google Scholar]