Abstract
Electronic Health Records (EHRs) provide a real-world patient cohort for clinical and genomic research. Phenotype identification using informatics algorithms has been shown to replicate known genetic associations found in clinical trials and observational cohorts. However, development of accurate phenotype identification methods can be challenging, requiring significant time and effort. We applied Support Vector Machines (SVMs) to both naïve (i.e., non-curated) and expert-defined collections of EHR features to identify Rheumatoid Arthritis cases using billing codes, medication exposures, and natural language processing-derived concepts. SVMs trained on naïve and expert-defined data outperformed an existing deterministic algorithm; the best performing naïve system had precision of 0.94 and recall of 0.87, compared to precision of 0.75 and recall of 0.51 for the deterministic algorithm. We show that with an expert defined feature set as few as 50–100 training samples are required. This study demonstrates that SVMs operating on non-curated sets of attributes can accurately identify cases from an EHR.
Introduction
Electronic Health Records (EHRs) are valuable tools designed to assist care providers in treating patients; they also serve an increasingly important role in research. At Vanderbilt, a de-identified version of their EHR, called the Synthetic Derivative (SD)1, allows for privacy-preserving research. This has been used in conjunction with the Vanderbilt DNA biobank, BioVU, which accrues DNA samples from discarded blood samples. Together, these create a powerful tool for genomic science that requires no additional patient recruitment. SD-based research has also been successfully applied to clinical research.2
One primary limitation to EHR-based research is accurately finding cases and controls for certain phenotypes. Genomic studies in particular benefit from large sample sizes given typically small effects sizes of most individual genetic variants and requirement to adjust for the large numbers of hypotheses tested. Use of EHRs linked to DNA biobanks has provided a new resource for genomic and clinical investigation beyond that provided by clinical trials and observational cohorts. Rheumatoid arthritis (RA) was among the early diseases to be investigated through EHR-based genomic analysis.3–5 Current phenotype identification algorithms combine multimodal information including billing codes, natural language processing, laboratory data, and medication exposures to accurately identify cases and controls in selective populations. Such algorithms take significant time and expert knowledge to develop.
Current phenotype identification algorithms tend to be phenotype-specific, and require separate evaluation and multiple iterations of development by manual review with each new phenotype pursued. The first algorithms deployed for large-scale phenotype identification were designed using curated attributes deterministically combined with Boolean operators, and showed the practical effectiveness of automated phenotype identification.4 Phenotype identification algorithms that require no physician design in conjunction with training sets could allow for greater portability among systems and diseases.
In this study, we used a cohort of physician-identified RA patients to evaluate the performance of a support vector machine (SVM) to accurately identify cases.6 We also compared the use of different categories of information contained with the EHR. We show that an SVM can be trained on a set of attributes containing most ICD-9 codes and NLP-derived information and predict RA status with high sensitivity and recall without a need to significantly filter the attributes.
Methods
Patient Cohort:
The cohort used in this analysis was a gold standard reviewed set of 376 individuals from the SD. Based on the foundation of prior work5, we selected patients who had at least one 714.* ICD-9 billing code, where the asterisk represents a wildcard for the following digits, which includes “Rheumatoid arthritis and other inflammatory polyarthropathies,” but not including those patients with only codes in the 714.3* block, representing juvenile rheumatoid arthritis (JRA). These selections were made from a patient pool containing approximately 10,000 patients. A rheumatologist classified these individuals as definite RA, possible RA, or not RA based on test results and the treating physician’s observations in notes. To enable the use of two level classification methods in this analysis, we followed the methods in Liao, et al. and grouped possible RA patients with the not RA patients.5
Development of attributes for machine learning:
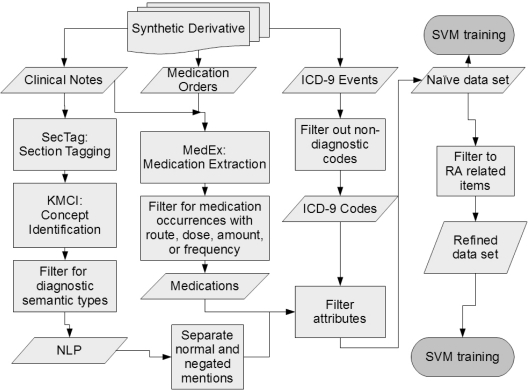
Two sets of attributes were prepared for each of the patients. The first data set contained no disease specific attribute limitations, referred to as the naïve data set, and the second data set contained only attributes clinically relevant to RA and related conditions, referred to as the refined data set. Both sets of attributes included the age and gender of the patient. The attributes each belonged to one of three subsets: ICD-9 codes, NLP results in the form of Unified Medical Language System (UMLS) Concept Unique Identifiers (CUIs), and medication names. All three subsets of attributes were gathered from the SD, and each attribute was represented as the natural log of one plus the total number of occurrences for that ICD-9 code, concept, or medication in the patient record. Each CUI was represented by two attributes; the first attribute corresponded to normal mentions of the concept, such as “patient has rheumatoid arthritis”, and the second attribute corresponded to negated mentions of the concept, such as “patient did not have RA”. The NLP was performed in two stages. First, the notes were processed by SecTag, which identifies the sections of clinical notes to which the text belongs.7 This allows occurrences of concepts identified later in the pipeline to be removed based on location; one example is the family history section of notes which may not apply directly to the individual. The notes were then processed by the KnowledgeMap Concept Identifier (KMCI)8 which processes clinical notes and returns CUIs and any qualifiers, such as negation status, which is implemented via a modified form of NegEx.9 Concepts were filtered based on their semantic type in the UMLS to include only concepts relating to patient presentation and diagnosis. CUIs were also removed from the attribute list if they appeared in the EHR of at least 50% of the approximately 10,000 patient records from which the cohort was selected. Total note counts for each patient were also included as an attribute in the NLP and Full models. Medication attributes were generated from medications found by MedEx, an NLP medication extraction tool, and filtered to those instances containing at least one of the following: dose, route, amount, or frequency, a heuristic for improving sensitivity.10 All three subsets were also filtered to attributes appearing in at least five patients in the cohort.
To create the refined data set, the naïve set was filtered to contain only attributes relevant to RA and related conditions. We selected all of the codified, NLP, and medication data specified in Liao, et al., while aggregating each category independently.5 The ICD-9 codes 714.*, representing RA and JRA, were retained, as well as the codes 696.0 and 710.0, representing Psoriatic Arthritis (PsA), and Systemic Lupus Erythematosus (SLE) respectively. The CUIs for those four terms and their neighbors in the UMLS tree were compiled into a list using a web-based tool to generate related terms based on relationships defined with the UMLS.11 The list was reviewed, and clinically relevant entries were retained as attributes in the refined data set. Finally, a list of medications commonly used in treating RA was included. These lists were constructed via consultation with rheumatologists through a prior project.3
Evaluation:
Two classes of algorithms were analyzed. The first was the deterministic algorithm found in Ritchie, et al, selected as it has been previously evaluated and shown to be able to replicate genetic associations known from previous genome wide association studies.3 The implemented algorithm selects patient records with at least one RA ICD-9, one RA text mention, one RA drug, and does not contain any ICD-9 codes or text matches for juvenile rheumatoid arthritis, inflammatory osteoarthritis, or reactive arthritis. The second class was comprised of two Support Vector Machines (SVMs). One SVM was trained using the naïve data set and the other was trained using the refined data set. All SVMs were trained using a Gaussian Radial Basis Function (RBF) kernel.
The main assessment was a comparison of the ability of the three algorithms to predict the disease status of individuals. Ten-fold cross validation was employed across the entire cohort to calculate the performance metrics. Individuals were stratified based on their disease status. The deterministic model required no training and was evaluated using the testing set of each fold of the cross validation. Both the SVM models were trained using another nested 10-fold cross validation to select the cost and gamma parameters, as applicable, for their kernels. The parameters were selected using a grid search across exponential sequences (e.g., 2−1, 20, 21). The gamma parameter search was across nine values centered on the value nearest 1/number of attributes, the default gamma in libSVM, and cost centered on one. An additional, localized search of the adjacent half units was performed in the region of the grid surrounding the best parameters from the first search (e.g., 20.5, 21, 21.5). If the best parameter selected in a majority of folds was on the border of the search space, the search space was expanded.
Additional measures were compared between the components of the SVM models. Three categories of attributes as the sole input were tested in addition to the full data sets: ICD-9 based, NLP based, and medication based. Both the SVM trained on the naïve data set and the SVM trained on the refined data set were tested using these attribute subsets. In total, eight SVM algorithms were trained and tested in the 10-fold cross validation: six subset SVM models in addition to the original two. To generate the predictive measures, a ranked list was created using the LibSVM probability option which fits the predictions to a logistic distribution.12 Comparisons were made using the area under the curve (AUC) of the Receiver Operating Characteristic (ROC) curve. In addition, the precision, recall, and F-measure were reported after adjusting the threshold to produce 95% specificity. To graphically compare the three algorithms, precision-recall curves were generated for the two SVMs, and a single point was plotted for the deterministic algorithm.
We compared the SVM models to the previously-published deterministic model for which we calculated the precision, recall, and F-measure. Precision was calculated as true positives divided by total algorithm predicted positives. Recall was calculated as true positives divided by total gold standard positives. The F-measure was calculated as the first harmonic mean of precision and recall.
Finally, the ability of both the SVMs trained on naïve and refined data sets to classify the cohort based on training set size was assessed using 10-fold cross validation. Twenty stratified samples of the training set were taken at intervals of 5% of the training set size within each fold of the cross validation. The mean and standard error of AUCs for each fold and subsample were recorded.
Analysis was performed using the R statistical package version 2.13.1.13 LibSVM was employed to train and test the SVMs using the package e1071 for R.12,14 ROC curves and performance measures were created using the package ROCR.15 Parallel processing was handled by the packages foreach, doMC, and multicore.16–18
Results
The demographics for the cohort are shown in Table 1. The original split of disease status was 185 definite RA individuals, 22 possible RA individuals, and 169 non-RA individuals. After merging to two categories, the patient population is nearly evenly split between cases (true RA patients) and controls (possible and not RA patients). The case group displays a much higher average number of RA ICD-9 codes. The case patients also have been followed for RA over a longer period of time on average.
Table 1:
Demographic details for the population (n=376).
| Cases | Controls | |
|---|---|---|
| n (%) | n (%) | |
| Total | 185 (49.2%) | 191 (50.8%) |
| Female | 141 (76.22%) | 148 (77.49%) |
| Ethnicity | n (%) | n (%) |
| Caucasian | 143 (77.3%) | 155 (81.15%) |
| African American | 14 (7.57%) | 26 (13.61%) |
| Hispanic | 1 (0.54%) | 1 (0.52%) |
| Asian | 2 (1.08%) | 1 (0.52%) |
| Other | 1 (0.54%) | 1 (0.52%) |
| Unknown | 24 (12.97%) | 7 (3.66%) |
| Attributes | Mean (SD) | Mean (SD) |
| Age (years) | 52.88 (13.06) | 56.2 (16.53) |
| Follow up (years) | 8.16 (4.17) | 1.74 (2.99) |
| Number of 714.* | 34.1 (31.12) | 5.42 (9.68) |
| ICD-9 Codes | ||
The results for the model comparisons are shown in Table 2. The bolded rows titled “Full” represent the results for each of the three main algorithms. The highest scoring algorithm based on AUC was the SVM trained on the refined data set including ICD-9s, NLP, and medications, though the difference in AUC between the best refined and the best naïve models was only 1%. In both data sets, the ordering of subsets on performance was ICD-9, NLP, and Medications. Interestingly, the best naïve data set trained model was based on ICD-9 codes only, which performed slightly better than model trained on all attributes, which had a much larger number of attributes (17110 vs. 795).
Table 2:
The number of attributes and cross-validated performance of an SVM trained on each data set. The performance of the deterministic model is also reported. Performance measures are mean ± standard error.
| Naïve | Precision | Recall | F measure | AUC | Attributes |
| Full | 93.3 ± 0.5 | 79.7 ± 5.2 | 85.1 ± 3.7 | 94.2 ± 1.3 | 17110 |
| ICD-9 | 94.1 ± 0.2 | 87.1 ± 2.8 | 90.3 ± 1.6 | 95.6 ± 1.0 | 795 |
| NLP | 92.2 ± 0.6 | 68.2 ± 5.6 | 77.4 ± 4.1 | 90.4 ± 2.1 | 15171 |
| Medication | 88.9 ± 1.8 | 51.0 ± 5.4 | 63.5 ± 5.5 | 84.6 ± 2.6 | 1148 |
| Refined | Precision | Recall | F measure | AUC | Attributes |
| Full | 93.7 ± 0.6 | 85.8 ± 5.7 | 88.6 ± 4.0 | 96.6 ± 1.1 | 59 |
| ICD-9 | 93.2 ± 0.5 | 78.1 ± 5.2 | 84.2 ± 3.5 | 95.5 ± 1.3 | 12 |
| NLP | 91.8 ± 1.0 | 68.8 ± 7.5 | 76.8 ± 5.7 | 89.5 ± 2.1 | 33 |
| Medication | 86.6 ± 1.6 | 40.5 ± 5.4 | 53.8 ± 5.2 | 83.3 ± 2.5 | 18 |
| Deterministic | Precision | Recall | F measure | AUC | Attributes |
| Full | 75.2 ± 2.5 | 51.6 ± 2.6 | 60.5 ± 2.6 | N/A | N/A |
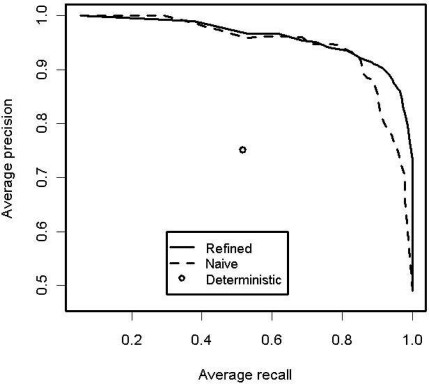
Figure 2 shows the averaged precision-recall curves for the SVMs trained using the refined and naïve data sets. The two SVM methods were very similar at low recall while the deterministic algorithm is well within the curves.
Figure 2:
Averaged precision-recall curves for the full SVM models. Also included is the precision-recall point for the deterministic algorithm.
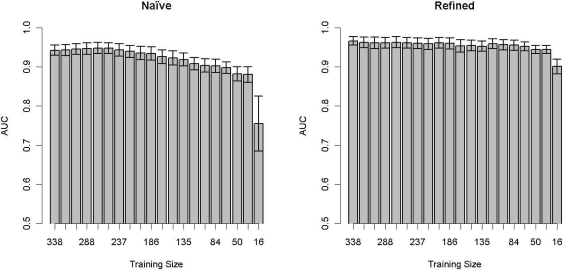
Figure 3 shows the relationship between the AUC measure and training set size. The SVM trained with the naïve data set displays a direct relationship between training set size and AUC, while the SVM trained with the refined data set maintains a more constant performance across training set sizes.
Figure 3:
The average AUC ± SE versus training set size for the naïve and refined data sets.
Discussion
This paper demonstrates that it is possible to create a high performance algorithm to detect cases of RA using machine learning techniques without significant manual selection of attributes. Indeed, the number of cases needed to train such a system appears very low, and the SVM trained on the naïve data set with only collections of all ICD-9 billing codes received by these patients performed well. This study has important implication is the design of future phenotype identification algorithms and suggests that future algorithm development may be possible merely with machine learning techniques applied to relatively small sets of manually tagged records (about 50–100 cases).
The SVM trained with a naïve data set performed very similarly to the SVM trained with a refined data set, although the recall and AUC of the refined SVM were better than with the naïve data set. Interestingly, the benefit of manual attribute selection manifested primarily in the ability to find more of the true cases (i.e., recall) from the population. ICD-9 codes were the best predictors amongst the variable sets. With respect to the naïve data set, training on ICD-9 codes alone outperformed training on the full set. The relationship of the number of ICD-9 codes in cases and controls foreshadows the strong performance of the ICD-9 based algorithms.
The SVMs trained using subsets of the naïve data set outperformed each of their respective refined subsets, but the SVM trained on the full refined set outperformed the SVM trained on the full naïve set. This suggests that information important to RA identification is missing from each of the refined subsets, but the sets are complementary. In the case of the naïve data set, the combination of information actually decreases the performance from the ICD-9 only subset. This may be related to the addition of irrelevant interactions among the large number of attributes.
NLP and medication methods showed more variability than the ICD-9 based models. One example source of error would be situations where patients may have RA in their notes or be prescribed a medication for a period of time before they see a rheumatologist and the diagnosis is contradicted only once. The total number of notes for each patient was included as an adjustment for this factor, but patients can have long-standing care before being diagnosed with true RA which could bias the effect of this adjustment. Weighting the findings in more recent notes or measuring the time since the last mention of RA may provide a way to decrease this aspect of the variability.
The recall and precision of medication-only based algorithms was lower than the other subsets. RA is a chronic condition, so patients who have a verified diagnosis will acquire many RA drug prescriptions and mentions over time. Predicting on these counts alone would leave out many individuals with more recently diagnosed cases of RA, however, partially explaining the low recall. Not all patients with RA are prescribed with RA specific medications, another contributor to low recall. The relatively higher precision is most likely due to the unlikely nature of a patient being on a RA medication for a long period of time without the condition. Some of the false positives are also explained by medications shared with other chronic autoimmune disorders.
Considering Figure 3, the SVM trained on the refined data was more robust with respect to a smaller training set size than the SVM trained on the naïve data set. The refined SVM performs well until the number of patients in the training set reaches the number of attributes, around 20. This suggests that an accurate classification model for RA could be generated with as few as 20 manually tagged patients with a refined set of attributes, providing a potential model for rapid phenotype identification algorithm development. The predictive power of the SVM trained on the naïve data set is based much more strongly on the number of patients trained. As with machine learning methods in general, increasing the number of attributes increases the required number of training samples for stable performance.
The deterministic algorithm had a lower performance in this study than its original publication. The performance decrease in precision and recall is related to the difference in the gold standards. This study used an independent evaluation of the patient record, while the previous study was based on what the patient’s physician said in their record.
This study was limited in several ways. First, only performance for one phenotype that is well-represented in the ICD9 codes was established. This study was also performed on the data from only one institution; reporting habits and writing styles can vary among physicians and institutions. The algorithm also preselected patients with at least one ICD9 code as a “minimum requirement” to be in the set, significantly increasing the prevalence of RA in the population to about 50%. Such criteria may also reduce recall somewhat, though this is not expected given the chronicity of RA and its associated morbidity. The gold standard was also generated based on the review of only one physician. Finally, methods that rely on count data are not as likely to be as efficacious for acute diseases as they are for chronic ones; the margin between patients with a long-standing chronic disease and a misdiagnosis is larger than that between a patient with a treatable infection and a misdiagnosis. More research is needed into multi-modal methods for both chronic and acute diseases.
Conclusion
This study demonstrates that application of an SVM to non-curated collections of attributes can classify patients with RA, although the SVM model based on a refined set of all attributes perform slightly better and can be trained with relatively fewer cases. Both performed significantly better than a previously-published deterministic algorithm. Future research deriving cases and controls for EHR data may be able to leverage machine learning techniques without variable selection to simplify the process of case selection. Further investigation with other disease phenotypes is needed.
Figure 1:
Flow chart showing data set creation.
Acknowledgments
We would like to thank Drs. Abel Kho, Robert Plenge, Katherine Liao, Chad Boomershine, and Will Thompson for their discussions on identification of RA patients in their medical record systems. This work was funded in part by 1 U01 GM092691-01 of the Pharmacogenomics Research Network from the National Institute of General Medical Sciences and the NLM Training grant: 3T15LM007450-08S1.
References
- 1.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramirez AH, Schildcrout JS, Blakemore DL, Masys DR, Pulley JM, Basford MA, et al. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm. 2011 Feb;8(2):271–277. doi: 10.1016/j.hrthm.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010 Apr 9;86(4):560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010 May 1;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010 Aug;62(8):1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995 Sep;20(3):273–297. [Google Scholar]
- 7.Denny JC, Spickard A, 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. 2009 Dec;16(6):806–815. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny JC, Smithers JD, Miller RA, Spickard A., 3rd “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc. 2003 Aug;10(4):351–362. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 10.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Feb;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny JC, Smithers JD, Armstrong B, Spickard A., 3rd “Where do we teach what?” Finding broad concepts in the medical school curriculum. J Gen Intern Med. 2005 Oct;20(10):943–946. doi: 10.1111/j.1525-1497.2005.0203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C-C, Lin CJ. LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3):27:1–27:27. [Google Scholar]
- 13.Team RDC. Vienna, Austria: 2011. R: A Language and Environment for Statistical Computing [Internet] Available from: http://www.R-project.org. [Google Scholar]
- 14.Dimitriadou E, Hornik K, Leisch F, Meyer D, Weingessel A. e1071: Misc Functions of the Department of Statistics (e1071), TU Wien [Internet] 2011. Available from: http://CRAN.R-project.org/package=e1071.
- 15.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing the performance of scoring classifiers. [Internet] 2009. Available from: http://CRAN.R-project.org/package=ROCR. [DOI] [PubMed]
- 16.Analytics R. foreach: Foreach looping construct for R [Internet] 2011. Available from: http://CRAN.R-project.org/package=foreach.
- 17.Analytics R. doMC: Foreach parallel adaptor for the multicore package [Internet] 2011. Available from: http://CRAN.R-project.org/package=doMC.
- 18.Urbanek S. multicore: Parallel processing of R code on machines with multiple cores or CPUs [Internet] 2011. Available from: http://CRAN.R-project.org/package=multicore.