Abstract
Patients presenting to Emergency Departments may be categorised into different symptom groups for the purpose of research and quality improvement. The grouping is challenging due to the variability in the way presenting complaints are recorded by clinical staff. This work proposes analysis of the presenting complaint free-text using the semantics encoded in the SNOMED CT ontology. This work demonstrates a validated prototype system that can classify unstructured free-text narratives into patient’s symptom group. A rule-based mechanism was developed using variety of keywords to identify the patient’s symptom group. The system was validated against the manual identification of the symptom groups by two expert clinical research nurses on 794 patient presentations from six participating hospitals. The comparison of system results with one clinical research nurse showed 99.3% sensitivity; 80.0% specificity and 0.9 F-score for identifying “chest pain” symptom group.
Introduction
The patient’s presenting complaint at the time of arrival in the Emergency Department(ED) is routinely recorded in an unstructured free-text format on a computerized hospital database [1]. Clinicians and researchers sometimes need to retrieve, group and analyse these presenting complaints for the purpose of quality improvement and research. However, variability in the way these data are recorded poses a significant challenge to any attempts to categorize them into symptom groups. Chest pain for example, can be recorded in various ways including chest tightness, chest discomfort, heartburn, pleuritic pain, and angina. These patient’s observations constitute a valuable source of key information for clinical research [2]. In the current ED set up, the identification of patient symptom groups is highly inefficient due to manual tasks. The process can also be very subjective due to lack of standard clinical terminologies used to describe patient observations.
This research aims to address these issues through the application of clinical terminology and semantic technologies. The authors report on the application of a semantic technology tool developed to analyse the patient observations. Specifically, this research demonstrates the application of Systematized Nomenclature of Medicine – Clinical Terminology (SNOMED CT) [3] combined with a rule-based mechanism for automatic identification of patient symptom groups from the free-text observations. This research demonstrates an approach to analyse patient presenting complaints using a commonly accepted set of clinical terminologies in an ED practice. The important computational challenge faced in this research is the automatic mapping of various terms describing a patient symptom to a standard clinical concept in SNOMED CT. This research reports an innovative use case of semantic technology application for Emergency Medicine.
Background
SNOMED CT is a systematically organised standard set of clinical terminology covering all the areas of clinical information such as clinical findings, procedures, diseases, body concepts, and micro-organisms. SNOMED CT consists of medical concepts arranged in a IS-A hierarchy. It is reported that SNOMED CT has 311,000 active concepts with unique meanings [4]. Due to the standard set of clinical terminology; SNOMED CT facilitates semantic interoperability between different healthcare providers, clinicians as well as clinical researchers. The important challenge in the application of SNOMED CT is appropriate mapping of existing clinical terms to SNOMED concepts. There has been mapping efforts between ICD10 codes to SNOMED CT. These efforts report that the process of mapping the ICD10 codes to SNOMED CT concepts can be subjective [5]. The author argues that the development of maps will not eliminate the need for expertise in code selection, because of the inherent differences between a terminology and a classification [6]. The implementation of SNOMED CT has progressed from research spaces to the actual information systems in hospitals [7]. These efforts have resulted into a need of effective mapping strategies [8]. These strategies suggest the need for better mechanisms to map patient data to standard clinical terminologies such as SNOMED CT. There also have been efforts to map the patient data to SNOMED CT concepts [9]. In these efforts, the terminology analysts analysed the patient data manually and identified the relevant SNOMED CT concepts in the patient data. This work relied on the judgment of terminology analysts to map patient data to SNOMED CT concepts. Some automated methods were suggested to map patient data to SNOMED CT concepts [10, 11]. There have been efforts for ED case detection using Natural Language Processing (NLP) algorithms [12]. TOPAZ demonstrated application of NLP algorithms for identifying 55 respiratory-related conditions from ED reports. The system was evaluated on a smaller evaluation set of 60 ED reports. It is not clear whether this system models the variations such as institution-specific acronyms of clinical terms.
There are many other clinical NLP systems developed for semantic analysis of clinical text. The systems such as clinical text analysis and knowledge extraction (cTAKES) [13], Health Information Text Extraction (HITEx) [14] and MetaMap [15] focus on mapping the clinical terms to a clinical concept using vocabularies in unified medical language system (UMLS®) [16]. cTAKES has the capability of identifying clinical named entities from unstructured clinical text. MetaMap is another example of a configurable system that can be used to map the clinical text to the UMLS vocabulary. The research shows that these existing systems have the capability to map the clinical text to a standard clinical concept. However, the main pragmatic challenge in real-life implementation of any clinical NLP system is to map the institution-specific clinical data to a standard clinical terminology. The existing systems need improvement in modeling institution-specific clinical text. Our research showed that the practice of recording clinical data in a hospital environment is institution-specific. The clinical data is recorded using local terms including specific acronyms and other key words. Due to the local practices of clinical data recording it is important to build a rule-base for converting local terms into terms that can be mapped to standard clinical concept. We demonstrate the application of a rule-based approach for the analysis of ED patient problem descriptions. Our research aims to contribute to the existing state-of-the-art and extend its application to a symptom classification problem in Emergency Medicine. Our research also presents validation of our proposed system by multiple clinicians.
Methods
An ethics approval was granted by the Human Research Ethics Committee at Queensland Health to use the non-identifying data. The following main steps were taken to conduct the research using our proposed system.
Symptom Groups: ED clinician identified chest pain, abdominal pain, dysponea and trauma as one of the critical ED problems. The task was to identify patient symptoms that can be related to these conditions and classify the patient presentations to one of these category.
Data Extraction: The non-identifying free-text descriptions of patient presenting complaints were extracted from the hospital Emergency Department Information Systems (EDIS). The data included patient presentations for 1 day at 6 participating hospital EDs. Total 794 ED presentations were extracted from the source system.
Data Analysis: The patient data were imported into a clinical terminology mapping tool called Snapper [17]. The tool was developed by the Australian e-Health Research Centre (AeHRCTM) at Commonwealth Scientific and Industrial Research Organisation (CSIRO). Snapper is designed to map the terms from the existing terminology to SNOMED CT. Snapper allowed importing list of free-text presenting complaint descriptions involving various clinical terms.
Rule-base development: A rule-base of various possible definitions of the target symptom group was developed. The rules were specified by the ED clinician using various synonyms and acronyms.
Concept Mapping: The rules were then implemented into the “Automap with script” feature of Snapper. This feature can process large set of free-text data involving thousands of free-text clinical records. This feature allowed developing scripts for mapping various synonyms and acronyms of a clinical term to a single SNOMED CT concept. This feature enabled identification of clinical terms in the free-text and mapping those terms to SNOMED CT concepts.
Manual Identification of symptom groups: Two expert clinical nurses were engaged to manually identify the patient symptom groups from each patient presenting complaint description in the data set of 794 ED presentations. In this paper, the two nurses will be referred as Nurse1 and Nurse2 respectively. The Nurse 1 and Nurse 2 outcomes were validated by an emergency department staff specialist. There was total three clinical staff members involved in manual identification of patient symptom groups.
The following table shows the examples of various synonym terms as well as acronyms used to develop the rule-based algorithm for identifying the keywords and thus detecting the four main symptoms groups.
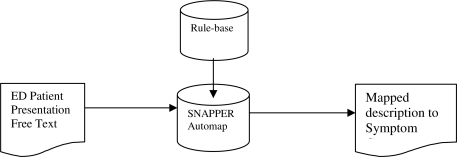
The rule-based algorithm was executed to map the free-text patient observation to the target symptom group. Figure 1 shows the process of mapping the patient narratives to the symptom groups.
Figure 1.
Process of mapping free-text descriptions to target symptom group
Our system is written in Java using the Eclipse platform. The Snapper tool is packaged as a collection of plug-ins that allows the easy addition of further functionality. The functioning of our tool consists of following main stages –
Importing Source Text: The tool allows importing a source terminology from a structured file such as tab or comma-separated value or an SQL data base.
Identifying SNOMED concepts: The tool allows identifying a concept from SNOMED CT through a search box as well as through mapping function. The search results are displayed in the list of possible matching concepts. The tool retrieves full information of the selected concept and displays the full information about the concept in the Ontology View.
Mapping the text to concepts: The tool displays a Mapping Table after the ED presenting problem free text is imported. The mapping table is an important component of the tool. The first column table displays the source term. In this research it is the presenting problem description. The third column shows the concept or expression that the term from the source text maps to. The second column allows the user to select the relationship between the source term and the mapping expression. The available relationships are specializes, generalizes, or is equivalent (the default) and should represent the semantic relationship between the term and the concept/expression. This then allows the mapping to be treated as a set of definitions of new concepts (one for each source term) and to generate an extension of the base ontology.
The Automap Feature: This functionality will perform an automatic search for the source term in the SNOMED CT terminology. Any mappings which are made are marked as “Automap” in the mapping table. This allows the user to proceed quickly through auto-mapped terms, check if the identified correspondence is correct, and mark the mapping as “Complete”. The Automap function in Snapper also provides various configuration options for the user to enhance its accuracy or usefulness.
Automap with Script Feature: This is the exclusive feature of our tool which is used to create an “If-then” type rule-base. This research used this feature to model institution-specific rules using local terms, synonyms and acronyms as mentioned in Table 1. This functionality provides the capability of mapping the source free text with SNOMED concepts based on institution-specific rules. This capability is provided by writing a script using Java-based scripting language Groovy. This allows for special treatment of repetitive phrases or special formatting and the easy translation of local abbreviations to the full form before performing a normal Automap search.
Table 1:
Examples of clinical terms
| Symptom Category | Examples of Possible descriptors | SNOMED Concept |
|---|---|---|
| Chest Pain | C/P, pleuritic pain, chest tightness, angina | 29857009 | Chest pain (finding) | |
| Abdominal Pain | Abdo pain, epigastric pain, flank pain | 21522001|Abdominal pain (finding) | |
| Dysponea | SOB, Shortness of breath, DIB, wheeze | 267036007 | Dysponea (finding) | |
| Trauma | Injury, fall, fell, laceration | 19130008 | Traumatic abnormality | |
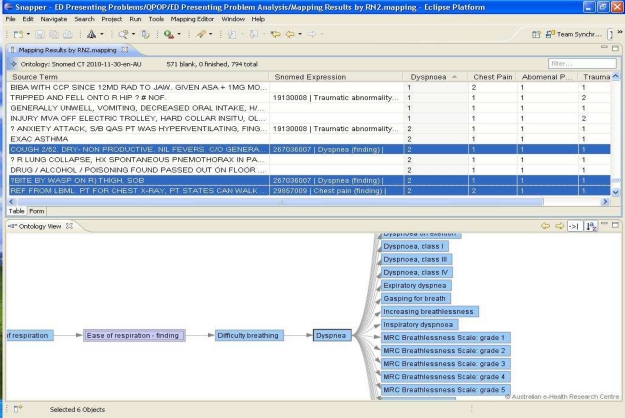
The functionality of our system after executing Automap with Script feature is shown in Figure 2 above. The source term shows the patient’s free-text presenting complaint narrative sourced from the hospital EDIS; one row for each ED presentation. The automatic identification of the symptom group is shown by the SNOMED expression. The relationship between the source term and SNOMED expression is set to “equivalent”. This column is not shown in the above figure for simplicity purpose. The columns chest pain, dysponea, etc shows the identification of symptom groups by Nurse2. The value 2 in the columns indicates that the nurse had identified the specific symptom group shown in the column heading and the value 1 indicates that the nurse did not identify the specific symptom group.
Figure 2.
Mapping of free-text ED presenting complaints to SNOMED concepts
Results
The system outcomes were compared with the manual identification of symptom groups by Nurse1 and Nurse2. The “chest pain” symptom group identified by Nurse2 was randomly selected as the Gold Standard for validating the system performance. The identification of all symptom groups by Nurse1 and Nurse2 individually was considered as the silver standard. The comparison of the system results against individual results by Nurse1 and Nurse2 is shown in Table 2 and Table 3 respectively. The results shown are within 95% confidence interval.
Table 2:
Comparison of Snapper results with Nurse1 results
| Symptom Category | Specificity | Accuracy | Precision/ +ve predictive value | Recall/Sensitivity | F-score |
|---|---|---|---|---|---|
| Chest Pain | 98.5% | 96.6% | 73.8% | 80.4% | 0.8 |
| Abdominal Pain | 91.3% | 90.8% | 81.6% | 32.0% | 0.5 |
| Dysponea | 99.9% | 96.9% | 96. 0% | 50.0% | 0.7 |
| Trauma | 96.4% | 80.7% | 72.8% | 31.1% | 0.4 |
Table 3:
Comparison of Snapper results with Nurse2 results
| Symptom Category | Specificity | Accuracy | Precision/+ve predictive value | Recall/Sensitivity | F-score |
|---|---|---|---|---|---|
| Chest Pain | 99.3% | 97.7% | 91.8% | 80.0% | 0.9 |
| Abdominal Pain | 99.3% | 89.9% | 86.8% | 30.6% | 0.5 |
| Dysponea | 99.7% | 96.5% | 92.0% | 46.9% | 0.6 |
| Trauma | 91.2% | 86.9% | 20.7% | 30.4% | 0.2 |
The system results were also analysed using the confusion matrices between the system results and results by the individual nurses. These matrices for the comparative results are shown below in Table 4 and Table 5 below.
Table 4:
Confusion Matrix Nurse1 Vs System Results
| System /Predicted | |||||||
|---|---|---|---|---|---|---|---|
| Chest Pain | Abdominal Pain | Dysponea | Trauma | Other | Total | ||
| Nurse1/Actual | Chest Pain | 45 | 0 | 0 | 0 | 11 | 56 |
| Abdominal Pain | 3 | 31 | 0 | 0 | 62 | 128 | |
| Dysponea | 13 | 0 | 24 | 0 | 8 | 72 | |
| Trauma | 8 | 0 | 0 | 59 | 117 | 184 | |
Table 5:
Confusion Matrix Nurse2 Vs System Results
| System /Predicted | |||||||
|---|---|---|---|---|---|---|---|
| Chest Pain | Abdominal Pain | Dysponea | Trauma | Other | Total | ||
| Nurse2/Actual | Chest Pain | 56 | 0 | 0 | 0 | 15 | 71 |
| Abdominal Pain | 4 | 33 | 0 | 0 | 69 | 106 | |
| Dysponea | 13 | 0 | 23 | 0 | 11 | 47 | |
| Trauma | 4 | 0 | 0 | 17 | 34 | 55 | |
The confusion matrix between two nurses is shown in Table 6 below. The symptom groups identified by Nurse2 were considered as the actual symptom groups present in the presenting complaint free-text. The symptom groups identified by Nurse2 were considered as the predicted outcomes. The comparison showed difference in symptom group classification by both nurses. The interpretation of the free-text narrative by one clinician may differ from the interpretation of the same by other clinician. This experiment confirms the need of a standard terminology to describe patient’s presenting complaint description.
Table 6:
Confusion Matrix Nurse2 Vs Nurse1
| Nurse1 /Predicted | |||||||
|---|---|---|---|---|---|---|---|
| Chest Pain | Abdominal Pain | Dysponea | Trauma | Other | Total | ||
| Nurse2/Actual | Chest Pain | 52 | 3 | 15 | 8 | 17 | 95 |
| Abdominal Pain | 2 | 93 | 2 | 4 | 13 | 114 | |
| Dysponea | 11 | 1 | 43 | 4 | 34 | 93 | |
| Trauma | 4 | 3 | 0 | 49 | 6 | 62 | |
The Cohen Kappa statistics [18] was used to identify level of agreement between the raters. The formula used to identify the Kappa coefficient is shown below.
Where Pr(a) is the relative observed agreement among two clinical research nurses, and Pr(e) is the hypothetical probability of chance agreement between the two nurses. The Kappa coefficient κ = 1 indicates complete agreement and κ= 0 indicates no agreement. The Cohen Kappa measures for each symptom category are shown in Table 7 below.
Table 7:
Agreement between two clinical nurses – Cohen’s Kappa Measure
| Symptom Category | Pr(a) | Pr(e) | Kappa coefficient (κ) |
|---|---|---|---|
| Chest Pain | 0.986 | 0.879 | 0.9 |
| Abdominal Pain | 0.997 | 0.914 | 0.9 |
| Dysponea | 0.999 | 0.941 | 0.9 |
| Trauma | 0.943 | 0.900 | 0.4 |
The Kappa Coefficient indicates high level of agreement (i.e. κ > 0.8) between the two nurses for detecting chest pain, abdominal pain and dysponea. The Kappa Coefficient for Trauma indicated low level of agreement between the two clinical nurses. A possible reason for such exception is the difference in subjective interpretation of the data due to the non standardized terminology. It can be observed that the system performance was strong when compared with the gold standard with chest pain F-score=0.9; Precision=91.8%; Recall or Sensitivity=80.0%; Specificity=99.3%. The confusion matrices shown in Table 4, Table 5 and Table 6 also indicate that the system performance was stronger in detecting “chest pain”. The system performance can be maximized for detecting other symptom categories.
Discussion
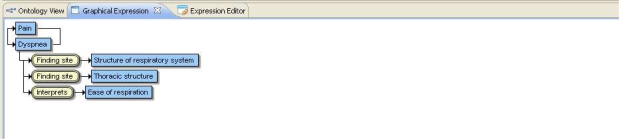
The system performance was higher for detecting “Chest Pain” than other symptom categories with accuracy greater than 96% and 80.4% recall or sensitivity. The system performance when tested separately with each nurse’s findings showed similar outcomes. For example, Abdominal Pain: F-score=0.5 for Nurse1 and F-score=0.5 for Nurse2. Dysponea: F-score=0.7 for Nurse1 and F-score=0.6 for Nurse2. The system was also tested to identify more symptom categories such as Headache, Urinary Symptoms, and Seizures. The results showed accuracy greater than 93%. The comparison of system results with the manual results showed that the system performed better when the free-text involved a single clinical concept per patient narrative. The system performance was weak in automatic identification of multiple symptoms per patient narrative. The system performance, especially the recall or sensitivity was low when multiple clinical concepts were present in the free-text. The system has the capability to manually manage the occurrence of multiple symptoms through the post-coordinated SNOMED expression. The system provides an ability to modify the SNOMED expression determined through the Automap feature. The edited SNOMED expression shows the group of symptoms identified in the presenting problem free text. The system also provides the ability to explore the symptom group in a graphical expression as shown in figure 3 below.
Figure 3.
Graphical Expression for Mapping Group of Symptoms
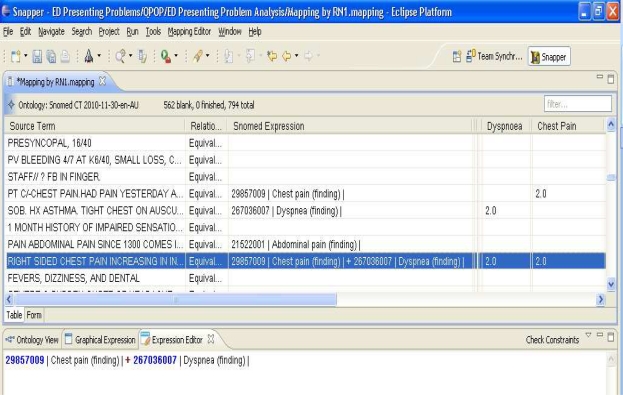
In addition to the Ontology view, mapping of multiple symptoms through SNOMED expression editor is shown in Figure 4 above. The mapping by RN1 is shown in “Dysponea” and “Chest Pain” columns. The SNOMED expression is manually edited to map group of symptoms. The system’s Automap capability can be improved in future to capture multiple symptoms automatically. The identification of patient symptom groups by the clinician is based on the subjective interpretation of the presenting complaints descriptions. The proposed system has identified patient sub-groups using the rule-base and SNOMED CT ontology. The results showed that SNOMED CT ontology combined with a rule-based system can improve the efficiency in analysing unstructured ED data. The identification of symptom groups can be used for variety of purposes. For example, impact of pathology ordering associated with a particular symptom group can be evaluated using our system results [19].
Figure 4.
Mapping group of symptoms to SNOMED concepts
The system currently does not have any machine learning ability. The machine learning approaches have shown advantages in text categorization [20]. However, the issues in realizing potential benefits of machine learning methods for biomedical domain are reported [21]. The rule-based approach demonstrated in this research provides an effective mechanism to model local or institution-specific knowledge that can improve the efficiency in ED clinical triage practices. This research showed that knowledge engineering approaches can be a pragmatic approach for specialist clinical application domain such as emergency medicine. The need to include machine learning component will be investigated in future.
Conclusion and Future Research
This research has demonstrated a prototype system for analysis of ED patient presenting complaints. The system outcomes show promising initial results for categorizing patient presentations into “chest pain” symptom group. The system performance can be further enhanced using SNOMED post-coordinated expressions to map group of symptoms. This paper also suggests the application of the research findings for the evaluation of pathology ordering associated with specific symptom groups. The reduction of excessive pathology ordering is a significant problem due to high ordering costs. Our system was further applied to a larger data set involving 1.5 years of data with 525,883 patient presentations from six hospital emergency departments. The association of the symptom classification results with corresponding pathology ordering will be undertaken in future.
Acknowledgments
This research was supported by the Queensland Emergency Medicine Research Foundation Grant, QEMRF-PROJ-2009-016-CHU. This research is a collaborative work between Department of Emergency Medicine at Royal Brisbane & Women’s Hospital and the Australian e-Health Research Centre.
References
- 1.Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, Olszewski RT. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med. 2005;33:31–40. doi: 10.1016/j.artmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Harkema H, Gaizauskas R, Hepple M, Roberts A, Roberts I, Davis N, Guo Y. A large-scale terminology resource for biomedical text processing. Proceedings of BioLINK; 2004. pp. 53–60. [Google Scholar]
- 3.Bramley M. NEHTA terminology analysts. Health Information Management Journal. 2009;38(3):59–63. doi: 10.1177/183335830903800310. [DOI] [PubMed] [Google Scholar]
- 4.International Health Terminology Standards Development Organisation (IHTSDO) SNOMED clinical terms user guide. 2010. pp. 1–99. [Google Scholar]
- 5.Bowman S. Coordinating SNOMED-CT and ICD-10: Getting the most out of electronic health record systems. Journal of AHIMA. 2005;76(7):60–61. [PubMed] [Google Scholar]
- 6.Brouch K. AHIMA project offers insights into SNOMED, ICD-9-CM mapping process. Journal of AHIMA. 2003;74(7):52–55. [PubMed] [Google Scholar]
- 7.Garde S, Knaup P, Hovenga EJS, Heard S. Towards semantic interoperability for electronic health records, Methods of Information in Medicine. 2007;46(2):332–343. doi: 10.1160/ME5001. [DOI] [PubMed] [Google Scholar]
- 8.Maulden S, Greim P, Bouhaddou O, Warnekar P, Megas L, Parrish F, Pharm D, Lincoln MJ. Using SNOMED CT as a mediation terminology: mapping Issues, lessons learned, and next steps toward achieving semantic interoperability. Proceedings of the Third International Conference on Knowledge Representation in Medicine; Phoenix, Arizona, USA. May 31st – June 2nd, 2008. [Google Scholar]
- 9.Truran D, Saad P, Zhang M, Innes K, Kemp M, Huckson S, Bennetts S. Using SNOMED CT - enabled data collections in a national clinical research program; primary care data can be used in secondary studies. Proceedings of Health Informatics Conference; Canberra, Australia. Aug, 2009. pp. 182–187. [Google Scholar]
- 10.Nguyen AN, Lawley MJ, Hansen DP, Colquist S. A simple pipeline application for identifying and negating SNOMED clinical terminology in free text. Proceedings of the Health Informatics Conference; Canberra, Australia. 2009. Aug, 2009. pp. 188–193. [Google Scholar]
- 11.Patrick J, Wang Y, Budd P. Australasian Workshop on Health Knowledge management and Discovery (HKMD) 2007. An automated system for conversion of clinical notes into SNOMED clinical terminology; pp. 219–226. [Google Scholar]
- 12.Chapman WW, Chu D, Dowling J, Harkema H. Identifying respiratory-related clinical conditions from ED reports with Topaz, Advances in Disease Surveillance. 2008;5:12. [Google Scholar]
- 13.Savova GK, Masanz JJ, Orgen PV, Zheng J, Sunghwan S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. JAMIA. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy S, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Medical Informatics and Decision Making. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesarus: the MetaMap program. Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 16.Tuttle MS, Blois MS, Erlbaum MS, Nelson SJ, Sherertz DD. Toward a Bio-Medical Thesaurus: Building the Foundation of the UMLS. Proc Annu Symp Comput Apl Med Care; 1988 November 9; pp. 191–195. [Google Scholar]
- 17.Vickers DM, Lawley MJ. Mapping existing medical terminologies to SNOMED CT: An investigation of the novice user’s experience. Proceedings of Health Informatics Conference; 2009. pp. 46–51. [Google Scholar]
- 18.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20(1):37–46. [Google Scholar]
- 19.Wagholikar A, Chu K, Hansen D, O’Dwyer J. An approach for analysing pathology ordering in emergency departments using HDI technology. Proceeding of Health Informatics Conference; 2010. pp. 158–164. [Google Scholar]
- 20.Sebastiani F. Machine learning in automated text categorization. ACM Computing Surveys. 2002;34(1):1–47. [Google Scholar]
- 21.Liu K, Hogan WR, Crowley RS. Methodological Review: Natural Language Processing methods and systems for biomedical ontology learning. Journal of Biomedical Informatics. 2011;44(1):163–179. doi: 10.1016/j.jbi.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]