Abstract
Increasingly, researchers use simulation to generate realistic population health data to evaluate surveillance and disease control methods. This evaluation approach is attractive because real data are often not available to describe the full range of population health trajectories that may occur. Simulation models, especially agent-based models, tend to have many parameters and it is often difficult for researchers to evaluate the effect of the multiple parameter values on model outcomes. In this paper, we describe Simulation Analysis Platform (SnAP) - a software infrastructure for automatically deploying and analyzing multiple runs of a simulation model in a manner that efficiently explores the influence of parameter uncertainty and random error on model outcomes. SnAP is designed to be efficient, scalable, extensible, and portable. We describe the design decisions taken to meet these requirements, present the design of the platform, and describe results from an example application of SnAP.
Introduction
Increasingly, researchers use simulation to generate realistic population health data to evaluate surveillance and disease control methods. This evaluation approach is attractive because real data are often not available to describe the full range of population health trajectories that may occur.
As more data have become available and computing power has increased, simulation models have tended to become more complicated. Agent-based models in particular, where a distinct software object is used to represent each person in the model, tend to have many parameters. While parsimonious models are generally preferred, many parameters may be needed to generate realistic outcome data. Complex interaction patterns, often modeled as networks, can also be used to define the interaction of agents. As a result, in addition to requiring many parameters, these simulation models can be computationally intensive. A challenging problem results, therefore, where researchers should perform many simulation runs to evaluate the sensitivity of their findings to the multiple parameters in their model, but encoding many parameter sets and running the corresponding models is cumbersome and time consuming.
In this paper, we describe Simulation Analysis Platform (SnAP) - a software infrastructure for automatically deploying and analyzing multiple runs of a simulation model in a manner that efficiently explores the influence of parameter uncertainty and random error on model outcomes. Although the initial development of this platform was motivated by a particular project, we designed SnAP to be suitable for a range of applications in public health research and policy-making. We begin by presenting a motivating research problem and then from this specific problem, we develop the general problem and system requirements. We then present our design for the platform, describe results from an example application of SnAP, and close the paper with a discussion of this work and identification of future directions.
Motivating Research Problem
Unfortunately, there are many examples of public health surveillance systems failing to detect massive infectious disease outbreaks (1; 2). In response to this reality, public health agencies have introduced new surveillance methods (3), but it is difficult to evaluate rigorously whether these new methods are effective (4). Such evaluation is critical to guide the appropriate adoption and use of new technologies for surveillance. As a specific example of this problem, we consider surveillance in an urban area to detect waterborne outbreaks due to the failure of a water treatment plant (5–8). Once such an outbreak is detected, the public health intervention is to issue a boil water advisory, which is maintained in place until water quality is returned to within normal limits (9; 10).
Laboratory-based surveillance is the standard approach used by most public health agencies to detect waterborne disease outbreaks in urban areas (11). In this type of surveillance, lab directors and physicians report to public health suspected and confirmed infections for diseases that are named in legislation. Lab surveillance is very specific, but it is neither sensitive nor timely (12–14). To augment this approach to surveillance, many public health departments have adopted syndromic surveillance (15), which follows healthcare utilization patterns and presenting symptoms of patients (16). This approach to surveillance is sensitive and timely, but not specific. The introduction of syndromic surveillance in this context raises questions of practical importance. For example, does syndromic surveillance offer any advantage over laboratory-based surveillance in this context? If so, how should these two approaches be configured to work together?
To help answer these and other related questions, we developed a simulation model to represent water distribution, human mobility, exposure to drinking water, infection, disease progression, healthcare utilization, laboratory testing, and reporting to public health (17). This model first creates a synthetic population from census data and then uses approximately 30 parameters to define the progression of individuals through the model. We want to explore questions related to the effectiveness of boil water advisories under different outbreak scenarios and to examine the role of different surveillance approaches in detecting waterborne disease outbreaks. Additionally, the parameter values in this model are not known with certainty, and we want to incorporate this uncertainty into our simulation results.
General Problem and Requirements
To address problems similar to what we described above, many other researchers have developed simulation software that can be used to evaluate the ability of public health authorities to detect and control disease outbreaks (18). Conducting this research, however, requires completion of multiple complex and coordinated computational tasks, such as generating simulated data representing realistic outbreak scenarios, applying detection algorithms with different configurations to the generated data, estimating detection performance metrics such as sensitivity and timeliness, comparing the outcomes of the outbreaks with or without an intervention from public health agencies, and quantifying the costs associated with different scenarios. Moreover, due to a large number of factors possibly affecting the outcomes of interest, these tasks must be repeated many times with different inputs to the simulation model. Given the complexity of this research design, a software infrastructure that can support parameterization and execution of all the tasks in an efficient and convenient manner, is necessary to make such research feasible and reproducible.
This problem is similar in some respect to the one faced by researchers in bioinformatics who must repeatedly perform a sequence of data manipulation and analysis on large data sets generated from sequencing, structure and expression experiments. To meet that need, analytical “pipeline” software was created, allowing researchers to assemble and connect visual analysis icons using a graphical interface, and then deploy the steps corresponding to the visual representation (19; 20). This pipeline software, however, is not flexible enough to support the types of simulation models and analytical methods required to evaluate public health surveillance. To our knowledge, no analytical pipeline exists to address our general problem.
The purpose of SnAP is to provide a computational environment for running large-scale experiments using simulated data, with a primary focus on studies to evaluate the effectiveness of public health surveillance systems and disease control strategies. To meet this objective, the system must fulfill the following requirements:
Efficiency
Simulations of population health, especially those that model individuals as software agents, tend to be computationally expensive. At the same time, in order to assess the effects of random error and parameter uncertainty on key outcomes, each simulation scenario must be repeated hundreds, or event thousands of times. Even more simulations are often required to explore systematically the effects of several configurable simulation parameters in one experiment. These factors make efficiency a primary requirement.
Scalability
High-performance computing capacity continues to advance and the system should be able to take advantage of additional computational resources to decrease the overall run-time.
Extensibility
Experimental analysis using simulated data has many applications. The platform should allow for application-specific extensions to be easily integrated with the rest of the system.
Portability
In order to be practically useful for public health applications, the platform should be portable and support multiple hardware configurations and operating systems.
In the following section we present the design decisions made to develop a software system that meets these requirements.
Design Considerations
To meet both the efficiency and scalability requirements, an obvious strategy is parallel computing. Even though individual simulation runs can take significant time, a single simulation run is usually completed within hours or days. The real computational challenge stems from the necessity to repeat each run many times with different random number generator seeds or parameter values. This type of problem is known as embarrassingly parallel in that it does not require parallelization deeper than the level of individual runs (21).
To facilitate task-level parallelization, we chose to use a modular design, where individual tasks or analytical steps are implemented independently, so that they impose no constraints on each other except for the format of input and output data. This choice allows the software for a task to be swapped with an alternative implementation or off-the-shelf software. To allow moving and processing vast quantities of data, the platform must also provide flexible communication and data flow facilities. We decided to use of a text file interface with a standard well known syntax to ensure extensibility and portability. For portability considerations, we are using well-established, freely-available (primarily open-source) platform-independent tools and languages as a choice for implementation.
Instead of using fixed values for uncertain simulation parameters, we find it informative to explicitly incorporate parameter uncertainty in the simulation results. A number of sampling methods have been proposed for this purpose (22). These methods define an N-dimensional parameter space, where N is the number of parameters and each parameter follows a known probability distribution. Parameter values are then sampled from their respective distributions to ensure unbiased coverage of the parameter space. The simulation is run multiple times using a different sample of parameter values each run, generating a distribution of outcome variables that reflects parameter value uncertainty. Among different sampling methods, Latin Hypercube Sampling (LHS) has the advantage of producing an unbiased representation of the parameter space with a small number of samples compared to other methods, such as random sampling (23; 24). To produce M samples from an N-dimensional parameter space using this technique, each of the N parameter distributions is partitioned into M equally probable bins, and the value of each parameter is uniformly drawn from one of these bins, so that bin numbers do not repeat in any given sample. A reduced number of parameter samples entails fewer simulation runs, which is critically important to ensure efficiency. We have therefore chosen LHS as a technique to account for simulation parameter uncertainty.
System Design
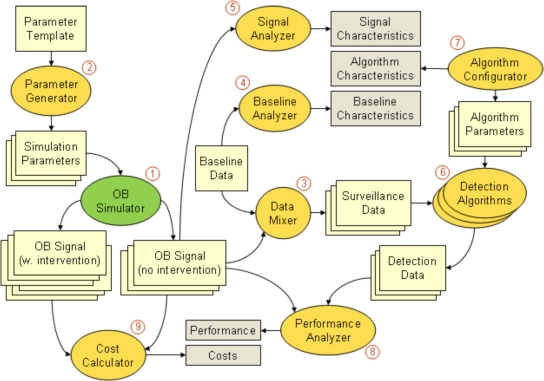
Figure 1 displays a schematic overview of the simulation analysis platform that we have designed and we give below a detailed description of the system components and interfaces.
Figure 1.
Schematic overview of SnAP. Numbers correspond to system components described in the article.
(1) The outbreak simulator is the central component of the system responsible for generating the outbreak data that will form the basis of the intended analysis. As described above, we have developed an agent-based simulation model for generating realistic multivariate outbreak signals similar to historical waterborne outbreaks of gastrointestinal disease (17). The model takes as input an outbreak scenario and simulation parameters as described below. As output, the outbreak simulator generates spatially distributed counts of cases (infected, symptomatic, seeking care at ED or a physician, reported to local public health department, etc.) over time. The model was written in Java using Mason, a low-level discrete event simulation library allowing for flexibility and efficiency at the same time.
(2) The outbreak simulator operates with a set of simulation parameters, which determine the shape of the generated outbreak signals. Simulation parameters can be categorized in two broad types: 1) a configurable parameter, which has an exact value defined in advance that can be varied experimentally, and 2) an uncertain parameter, which is represented by a probability distribution. Configurable parameters define an outbreak scenario and can be adjusted to conduct “what if” analyses, for example, examining the proportion of a population that will be infected when a certain level of pathogen concentration is maintained in drinking water for a particular duration of time. Uncertain parameters in our system are sampled from their respective distributions using LHS. Any specific analysis using the platform therefore involves a sizable set of simulation runs, in which both types of parameters can be varied. The results from this set of simulations represent a distribution of possible outbreak signals occurring under one or more outbreak scenarios and accounting for both random variation and parameter value uncertainty.
To implement the process of generating this complex mix of variously typed simulation parameters, we have devised a simple description language based on the JavaScript Object Notation (JSON). JSON is a lightweight data-interchange format that encodes data as name/value pairs and supports lists and nested objects (25). We create a parameter template, that defines a) the possible values of the configurable parameters in the simulation, and b) the distributions for all uncertain parameters. Consider an experiment (described in more detail later in the text) that examines several what-if scenarios by systematically varying two configurable parameters: duration of water contamination (p1) and the concentration of pathogen in the water (p2). Other parameters required in the simulation are uncertain parameters (i.e. they are governed by a probability distribution, and we want to sample their values with LHS). Figure 2 displays a fragment of a parameter template file that defines such an experiment. The template is used by the parameter generator – a component in charge of creating a set of specific parameter configurations to be fed to a corresponding set of simulators deployed in parallel. The parameter generator derives the required number of parameter configuration instances, in the following way:
- Generate a Cartesian product of possible values of all configurable parameters defining outbreak scenarios. The total number of scenarios (experimental conditions in the analysis) is:
where V is the total number of configurable parameters, and Ki is the number of possible values of the i-th parameter. In the example above V=2, K1=2, K2=3, and therefore S=6. Generate M samples from an N-dimensional parameter space using LHS, where N is the total number of uncertain parameters in the simulation. Each sample will contain unique values for all N parameters. For our example, we use M=1000 (note that the number of samples is independent of N).
From these, assemble the required set of parameter configurations, in which each outbreak scenario is combined with each of LHS samples.
Figure 2.
Parameter template specification example.
This would yield S × M parameter configurations in total (6000 in the example above) that can be fed to a corresponding set of outbreak simulations.
(3) Public health surveillance systems never operate directly on the outbreak signal, but have to detect the signal from baseline or endemic disease activity. To represent this situation in our analysis, the outbreak signals in SnAP are superimposed on baseline data by the data mixer component, which produces the synthetic surveillance data that can be used for evaluation of detection algorithms. Depending on the nature of the outbreak signal, different baseline data streams should be used. For example, the counts of people seeking medical help at the ED generated by our simulation model are superimposed on baseline ED utilization data for gastro-intestinal diseases. In each outbreak, we randomly select the start of the signal relative to the baseline.
(4) To determine which aspects of the baseline data affect detection performance, we must characterize the raw data. The baseline analyzer component computes a number of summary statistics on the baseline data, such as baseline mean and variance, trend, presence of weekly pattern, magnitude of yearly seasonality, and autocorrelation.
(5) Similarly to baseline analyzer, the signal analyzer summarizes the features of the OB Signal that may affect detection performance, such as peak size (number of standard deviations above the baseline mean), overall duration, time from onset to peak.
(6) For the purpose of the illustrative analysis, about 5 different detection algorithms used routinely in public health surveillance systems will be applied to the synthetic surveillance data. This part of SnAP is currently being implemented. The algorithms applied to surveillance data streams generate time series of binary alarms or outbreak probability that constitute detection data.
(7) Each Algorithm has a number of parameters that can be adjusted to affect detection performance. To facilitate comparison, we have created a list of algorithm characteristics that are not specific to any particular algorithm, but can be used to describe all algorithms involved in the analysis (e.g., adaptive vs. non-adaptive, whether or not the day-of-week effect is modeled). The algorithm configurator component generates specific parameter configurations for the detection algorithms (similarly to LHS simulation parameter generator) and annotates each configuration using these general algorithms characteristics.
(8) The initial outbreak signal is used to evaluate the performance of detection algorithms. Performance analyzer compares the detection data to the timing of the outbreak to determine if the outbreak was detected by the algorithm, and if so, on which day of the outbreak it was detected.
(9) To quantify the effects of timely outbreak detection, we have implemented public health intervention in the form of a boil-water advisory in our simulation model. It is thus possible to compare the outcomes of interventions applied at different time points during the course of an outbreak with no-intervention scenario.
Application of the System
Study design
As an example application of SnAP, we present a simple simulation study, in which we evaluate how the health burden of a waterborne cryptosporidiosis outbreak depends on the duration of water contamination and the concentration of the cryptosporidium pathogen in the source water. Qualitatively, it is clear how both factors are likely to affect the infection rate. Quantitatively, however, the effect of these factors on the likely health burden from a waterborne outbreak in a particular geographical setting would be very challenging to estimate without an infrastructure like SnAP. The challenging nature of this analysis may explain why, to our knowledge, no researchers have yet conducted such a study.
In our study, we simulated the failure of one of the water treatment facilities on the island of Montreal for durations of 4 hours and 72 hours. We used three concentration levels: 0.1, 1, and 10 oocysts per litre. These conditions yield a total of six water contamination scenarios, and we performed 1000 runs of the simulation model for each scenario to explore the contribution to the results of uncertainty in parameter values, as described in the System Design section.
Results
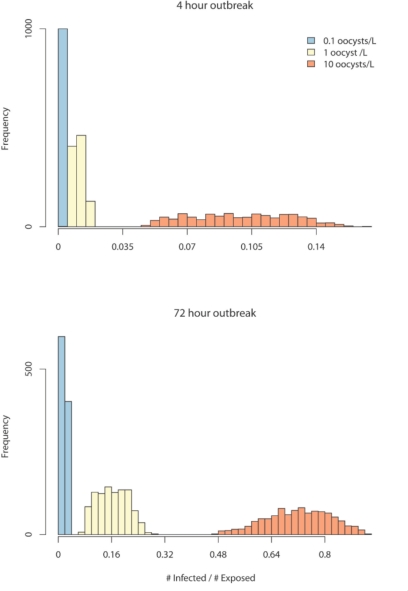
Application of SnAP for the scenario described above produced a range of results; we present those related to the LHS sampling. In Table 1, we display the mean of selected outbreak characteristics for each of the six outbreak scenarios. The 95% confidence interval for the mean of these characteristics summarizes the distribution over the 1,000 simulation runs. In Figure 3, we explicitly display the distribution of the proportion of exposed that become infected for each of the six scenarios. These distributions allow assessment of the variation in these outcomes when both random variation and parameter uncertainty are considered in the model. Although we do not explicitly decompose the variation in this example in variation attributable to randomness and parameter uncertainty, we have demonstrated previously that the total variation is driven mainly by parameter uncertainty with random variation contributing a relatively small amount.
Table 1.
Outbreak characteristics by duration of treatment failure and concentration of contamination in the source water. All estimates are followed by 95% confidence intervals that incorporate random error and parameter uncertainty.
| Duration of Treatment Failure | Concentration of Contamination in Source Water | |||
|---|---|---|---|---|
| 10 oocysts /L | 1 oocyst /L | 0.1 oocysts /L | ||
| Average Number Infected | 4 hours | 137,958 (135,473 – 140,442) | 15,317 (15,019 – 15,615) | 1,547 (1,517 – 1,578) |
| 72 hours | 1,136,345 (1,125,356 – 1,147,333) | 258,678 (254,062 – 263,294) | 28,981 (28,418 – 29,544) | |
| Average Number Symptomatic | 4 hours | 82,793 (81,212 – 84,373) | 9,193 (9,005 – 9,381) | 928 (909 – 948) |
| 72 hours | 681,863 (674,068 – 689,657) | 155,236 (152,298 – 158,175) | 17,397 (17,041 – 17,753) | |
| Average Number Deaths | 4 hours | 11 (10 – 11) | 1.2 (1.0 – 1.3) | 0.1 (0.1 – 0.1) |
| 72 hours | 93 (86 – 100) | 20 (19 – 22) | 2.2 (2.0 – 2.3) | |
| Infection Rate (Infection / Exposed) | 4 hours | 0.099 (0.097 – 0.101) | 0.011 (0.011 – 0.011) | 0.001 (0.001 – 0.001) |
| 72 hours | 0.719 (0.713 – 0.725) | 0.164 (0.161 – 0.166) | 0.018 (0.018 – 0.019) | |
| Attack Rate (Symptomatic / Exposed) | 4 hours | 0.059 (0.058 – 0.061) | 0.007 (0.006 – 0.007) | 0.001 (0.001 – 0.001) |
| 72 hours | 0.431 (0.427 – 0.436) | 0.098 (0.096 – 0.1) | 0.011 (0.011 – 0.011) | |
| Mortality Rate (Deaths / Exposed) | 4 hours | 7.55×10−6 (6.99×10−6 – 8.10×10−6) | 8.27×10−7 (7.52×10−7 – 9.01×10−7) | 8.13×10−8 (6.51×10−8 – 9.75×10−8) |
| 72 hours | 5.88×10−5 (5.45×10−5 – 6.30×10−5) | 1.26×10−5 (1.17×10−5 – 1.35×10−5) | 1.36×10−6 (1.25×10−6 – 1.46×10−6) | |
Figure 3.
Results from application of SnAP to the example scenario, highlighting the incorporation of random error and parameter uncertainty into simulation results. The variation in attack rates (#infected/#exposed) over 1,000 simulations is shown under two treatment failure scenarios and three concentrations of oocysts in the source water.
Performance
SnAP is currently implemented on a Top 500 supercomputer that forms part of the CLUMEQ network in Quebec. Its 960 nodes are highly optimized for the kind of intensive numerical analysis that our tasks require. A large number of simulations implied by the generated parameter configurations can be easily run in parallel via the standard queuing system, which we have slightly adapted to our particular needs with a simple Python script. To give an idea of our current level of computing performance, we have been able to complete 9000 simulation run in under 8 hours (although this figure is highly dependent on the number of available nodes in a certain time period, as this system is used concurrently by many research teams). By way of comparison, a single simulation run takes around 30 minutes on a normal desktop machine, and requires between 2 and 4 GB of memory. Given the simplicity and reliance of this setup, we are confident that it will scale reasonably well (i.e. almost linearly).
Discussion
We have described a platform for efficiently deploying multiple simulation runs in a manner that examines the sensitivity of model results to parameter uncertainty. The platform was designed to be efficient, scalable, extensible, and portable. We believe that our design meets these requirements, and we have used SnAP successfully to conduct many experiments, including evaluating the effectiveness of a boil water advisory to control waterborne outbreaks. Given the complexity of the research design required to evaluate public health outbreak detection and disease control through simulation, SnAP should make research more feasible, less error-prone, and more reproducible.
Although our file-based communication protocol between the components of the system has the advantage of simplicity and readability, it could possibly be improved by using a more flexible and centralized system. We have considered the use of a new kind of database derived from the so-called NoSQL paradigm, being currently developed very actively, especially in large-scale web environments. Databases of this type can be best understood as flexible and highly scalable key-value stores where the need for prior data type and schema definition is reduced to the minimum, and for which parallelization is one of the core architectural consideration. We think that such characteristics would fit well in our current architecture, and could potentially improve its overall performance and power.
In addition to using SnAP to evaluate surveillance strategies and public health interventions, we also anticipate that this platform will support the systematic evaluation of surveillance algorithms and that the SnAP may even be able to help guide decision-making in near real-time. In terms of evaluating algorithms, SnAP can help to automate the application of algorithms to a range of outbreak signals and the evaluation of the performance of those algorithms. Moreover, the platform can characterize the baseline data and signal characteristics in a consistent manner. All of these characteristics of an outbreak, the algorithms, and the data, can then be analyzed using data mining methods to identify the most appropriate algorithms to use for different types of surveillance (26; 27). In terms of guiding decision-making in near real-time, the efficiency of SnAP may allow public health decision-makers to evaluate the likely effect of different disease control options prior to initiating an intervention.
Conclusion
We have developed SnAP, a scalable, extensible, portable, and easily configurable platform for running high-throughput simulation experiments.
References
- 1.GAO Biosurveillance: Efforts to Develop a National Biosurveillance Capability Need a National Strategy and a Designated Leader (GAO-10-645) [Internet] 2010. Available from: http://www.gao.gov/products/GAO-10-645.
- 2.IOM . Washington, DC: 2010. BioWatch and Public Health Surveillance: Evaluating Systems for the Early Detection of Biological Threats [Internet] Available from: http://www.iom.edu/Reports/2010/BioWatch-Public-Health-Surveillance-Evaluating-Systems-Early-Detection-Biological-Threats.aspx. [PubMed] [Google Scholar]
- 3.Mandl KD, Overhage JM, Wagner MM, Lober WB, Sebastiani P, Mostashari F, et al. Implementing syndromic surveillance: a practical guide informed by the early experience. [Internet] Journal of the American Medical Informatics Association : JAMIA. 2004;11(2):141–50. doi: 10.1197/jamia.M1356. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14633933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckeridge DL. Outbreak detection through automated surveillance: a review of the determinants of detection. [Internet] Journal of biomedical informatics. 2007 Aug;40(4):370–9. doi: 10.1016/j.jbi.2006.09.003. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17095301. [DOI] [PubMed] [Google Scholar]
- 5.Semenza JC, Nichols G. Cryptosporidiosis surveillance and water-borne outbreaks in Europe [Internet] Euro Surveill. 2007;12(5):E13–4. doi: 10.2807/esm.12.05.00711-en. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17991392. [DOI] [PubMed] [Google Scholar]
- 6.Schuster CJ, Ellis AG, Robertson WJ, Charron DF, Aramini JJ, Marshall BJ, et al. Infectious disease outbreaks related to drinking water in Canada, 1974–2001 [Internet] Can J Public Health. 2005;96(4):254–258. doi: 10.1007/BF03405157. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16625790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laursen E, Mygind O, Rasmussen B, Ronne T. Gastroenteritis: a waterborne outbreak affecting 1600 people in a small Danish town [Internet] J Epidemiol Community Health. 1994;48(5):453–458. doi: 10.1136/jech.48.5.453. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7964354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuusi M, Nuorti JP, Hanninen ML, Koskela M, Jussila V, Kela E, et al. A large outbreak of campylobacteriosis associated with a municipal water supply in Finland [Internet] Epidemiol Infect. 2005;133(4):593–601. doi: 10.1017/s0950268805003808. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallis PM, Matson D, Jones M, Jamieson J. Application of monitoring data for Giardia and Cryptosporidium to boil water advisories [Internet] Risk Anal. 2001;21(6):1077–1085. doi: 10.1111/0272-4332.216176. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11824683. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds KA. Public reaction to boil water notices in the united states [Internet] Water conditioning and purification magazine. 2008;50(7) Available from: http://www.wcponline.com/pdf/0807On_Tap.pdf. [Google Scholar]
- 11.Benden CA, van, Lynfield R. Public Health Surveillance for Infectious Diseases. In: Lee LM, Teutsch SM, Thacker SB, St Louis ME, editors. Principles and Practice of Public Health Surveillance. Oxford University Press; 2010. pp. 236–254. [Google Scholar]
- 12.Jajosky RA, Groseclose SL. Evaluation of reporting timeliness of public health surveillance systems for infectious diseases. [Internet] BMC public health. 2004:429. doi: 10.1186/1471-2458-4-29. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15274746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Effler P, Ching-Lee M, Bogard A, Ieong MC, Nekomoto T, Jernigan D. Statewide system of electronic notifiable disease reporting from clinical laboratories: comparing automated reporting with conventional methods [Internet] JAMA : the journal of the American Medical Association. 1999 Nov;282(19):1845–50. doi: 10.1001/jama.282.19.1845. [cited 2010 Sep 15] Available from: http://www.ncbi.nlm.nih.gov/pubmed/10573276. [DOI] [PubMed] [Google Scholar]
- 14.Overhage JM, Grannis S, McDonald CJ. A comparison of the completeness and timeliness of automated electronic laboratory reporting and spontaneous reporting of notifiable conditions. [Internet] American journal of public health. 2008 Feb;98(2):344–50. doi: 10.2105/AJPH.2006.092700. [cited 2010 Sep 15] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2376898&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buehler JW, Sonricker A, Paladini M, Soper P, Mostashari F. Syndromic Surveillance Practice in the United States: Findings from a Survey of State, Territorial, and Selected Local Health Departments [Internet] Advances in Disease Surveillance. 2008. p. 6. Available from: http://www.isdsjournal.org/article/view/2618/2517.
- 16.Mandl KD, Overhage JM, Wagner MM, Lober WB, Sebastiani P, Mostashari F, et al. Implementing syndromic surveillance: a practical guide informed by the early experience. [Internet] Journal of the American Medical Informatics Association : JAMIA. 2004;11(2):141–50. doi: 10.1197/jamia.M1356. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14633933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okhmatovskaia A, Verma AD, Barbeau B, Carriere A, Pasquet R, Buckeridge DL. A simulation model of waterborne gastro-intestinal disease outbreaks: description and initial evaluation. [Internet]. AMIA ... Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2010. Jan, pp. 557–61. 2010. [cited 2011 Jul 8] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3041429&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 18.Modeling of Infectious Disease Agent Study (MIDAS) MIDAS Software Survey Results [Internet] 2011. [cited 2011 Jul 8] Available from: https://mission.midas.psc.edu/index.php?option=com_content&view=article&id=84&Itemid=113.
- 19.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, et al. A guided tour of the Trans-Proteomic Pipeline. [Internet] Proteomics. 2010 Mar;10(6):1150–9. doi: 10.1002/pmic.200900375. [cited 2011 Jul 8] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3017125&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerquin M, McDermott J, Frazier Z, Samudrala R. The Bioverse API and web application. [Internet] Methods in molecular biology (Clifton, N.J.) 2009. Jan, pp. 541511–34. [cited 2011 Jul 8] Available from: http://www.ncbi.nlm.nih.gov/pubmed/19381533. [DOI] [PubMed]
- 21.Foster I. Parallel Algorithm Examples In: Designing and Building Parallel Programs. Addison-Wesley; 1995. [Google Scholar]
- 22.Helton J, Johnson J, Sallaberry C, Storlie C. Survey of sampling-based methods for uncertainty and sensitivity analysis [Internet] Reliability Engineering & System Safety. 2006 Oct;91(10–11):1175–1209. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0951832005002292. [Google Scholar]
- 23.Helton J, Davis F, Johnson J. A comparison of uncertainty and sensitivity analysis results obtained with random and Latin hypercube sampling [Internet] Reliability Engineering & System Safety. 2005 Sep;89(3):305–330. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0951832004002340. [Google Scholar]
- 24.McKay MD, Beckman RJ, Conover WJ. A Comparison of Three Methods for Selecting Values of Input Variables in the Analysis of Output from a Computer Code. Technometrics. 1979;21(2):239–245. [Google Scholar]
- 25.2011. JavaScript Object Notation (JSON). JavaScript Object Notation (JSON)
- 26.Buckeridge DL, Okhmatovskaia A, Tu S, O’Connor M, Nyulas C, Musen MA. Understanding detection performance in public health surveillance: modeling aberrancy-detection algorithms. [Internet] Journal of the American Medical Informatics Association : JAMIA. 2008;15(6):760–9. doi: 10.1197/jamia.M2799. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18755992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckeridge DL, Okhmatovskaia A, Tu S, O’Connor M, Nyulas C, Musen MA. Predicting outbreak detection in public health surveillance: quantitative analysis to enable evidence-based method selection [Internet] AMIA Annual Symposium Proceedings. 2008. pp. 76–80. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18999264. [PMC free article] [PubMed]