Abstract
As part of a large-scale project to use DNA biorepositories linked with electronic medical record (EMR) data for research, we developed and validated an algorithm to identify type 2 diabetes cases in the EMR. Though the algorithm was originally created to support clinical research, we have subsequently re-applied it to determine if it could also be used to identify problem list gaps. We examined the problem lists of the cases that the algorithm identified in order to determine if a structured code for diabetes was present. We found that only just over half of patients identified by the algorithm had a corresponding structured code entered in their problem list. We analyze characteristics of this patient population and identify possible reasons for the problem list omissions. We conclude that application of such algorithms to the EMR can improve the quality of the problem list, thereby supporting satisfaction of Meaningful Use guidelines.
Introduction
The electronic medical record (EMR) provides a rich source of information that can be leveraged to improve the quality of patient care and for performing clinical research.1–3 While EMR adoption rates are currently low,4,5 this situation is likely to change with the recent publication of Meaningful Use guidelines by the U.S. Centers for Medicare and Medicaid Services.6,7 These guidelines, coupled with financial incentives and penalties, provide health care providers with specific and concrete objectives against which to gauge the effective use of an EMR system. The objectives of the guidelines range widely, from incorporating lab results as structured values in the EMR, to providing patients with electronic access to their health information.
We focus in this paper on the Meaningful Use objective to maintain an up-to-date problem list of current and active diagnoses, using structured codes from the ICD-9-CM or SNOMED CT® vocabularies.6 The maintenance of an up-to-date problem list has been found to facilitate quality care and effective communication among health care providers, yet studies have shown that problem lists can be inaccurate and poorly maintained.8–10 One option to improve problem list quality is to ease the burden of maintaining it by automatically detecting potential diagnosis omissions, using algorithms that leverage information in the rest of a patient’s EMR.10,11 This is the avenue of investigation we pursue in this paper.
As part of the electronic Medical Records and Genomics (eMERGE) Network,12 we have developed an algorithm to identify type 2 diabetes patients in the EMR.13 To develop our algorithm, we utilized EMR data contained within the Northwestern Medicine Enterprise Data Warehouse (EDW).14 The EDW is an integrated repository of over 10 terabytes of clinical and biomedical research data from 36 source systems. The EDW collects and aggregates data from Northwestern’s Feinberg School of Medicine (FSM), Northwestern Memorial Hospital (NMH), and Northwestern Medical Faculty Foundation (NMFF). The core data sources include Cerner Millennium (PowerChart, RadNet, etc.), EpicCare Ambulatory EMR, GE Centricity (outpatient billing and scheduling for NMFF), and PRIMES (inpatient billing). Other systems in the EDW include smaller specialized clinical and research databases. Most EDW data is synchronized nightly with its source systems.
Methods
The type 2 diabetes algorithm utilized in this study has been successfully ported and validated at multiple eMERGE Network sites.13 Here we provide an overview of the algorithm and methods used for validation. We also describe our methods for determining whether a patient does or does not have a problem list entry for type 2 diabetes, and our methods for examining selected characteristics of the patient population of interest.
Type 2 diabetes algorithm and validation
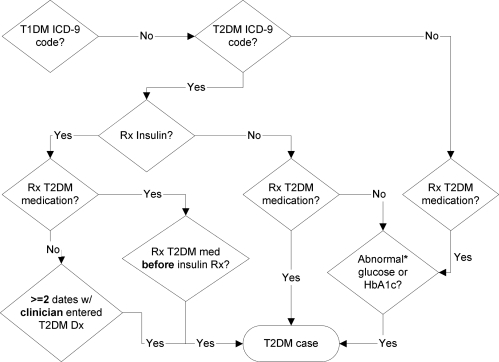
The type 2 diabetes algorithm (see Figure 1) utilizes a variety of types of information found in the EDW, including ICD-9 codes, medication orders, and laboratory results.13 The algorithm was designed to avoid confounding by inclusion of cases with type 1 diabetes (T1DM); hence, we start by excluding those who have a T1DM ICD-9 diagnosis code (250.x1 or 250.x3, where x is any number). Then we look for patients diagnosed with type 2 diabetes mellitus (T2DM) by an ICD-9 code (250.x0 or 250.x2, excluding 250.1x as it is indicative of T2DM with ketoacidosis, a condition also closely associated with T1DM). If the patient is not diagnosed with T2DM, we still select the patient if they were prescribed a medication for T2DM and have an abnormal lab indicating diabetes (see Figure 1 for what constitutes an abnormal lab). This is done to include patients lacking a structured diagnoses code in the EDW.
Figure 1:
Algorithm for identifying T2DM patients in the EMR. Abnormal glucose or HbA1c = random glucose > 200mg/dl, fasting glucose ≥ 125 mg/dl, or hemoglobin A1c ≥ 6.5%.
For those patients with a T2DM diagnosis, we then look separately at those only prescribed insulin (or Symlin®) as some of these patients could represent individuals with T1DM which has been misclassified as T2DM because of age of onset or other reasons. We only identify patients in this subset as a T2DM case if they have been on a T2DM medication in the past (see Table 1 for our list of medications), or have at least two visits on different dates with a clinician with the T2DM diagnosis in the problem list or as an encounter diagnosis. Lastly, if a patient has a T2DM diagnosis, and is prescribed a T2DM medication or has an abnormal lab indicating diabetes then they are also selected as a T2DM case. This captures patients who are able to control their diabetes through diet or exercise, as they would presumably have had at least one abnormal glucose test at the time of onset. In this way our algorithm ensures that each patient meets at least two criteria in order to be considered a T2DM case. This feature of the algorithm is designed to increase its precision by eliminating potential classes of false positives.
Table 1:
Prescribed T2DM medications meeting patient inclusion criteria. RxCUI = RxNorm ingredient level concept unique identifer
| RxCUI | Generic Name | Brand Names | Drug Class |
|---|---|---|---|
| 173 | acetohexamide | Dymelor | Sulfonylureas |
| 10633 | tolazamide | Tolinase | Sulfonylureas |
| 2404 | chlorpropamide | Diabinese | Sulfonylureas |
| 4821 | glipizide | Glucotrol | Sulfonylureas |
| 217360 | glipizide | Glucotrol XL | Sulfonylureas |
| 4815 | glyburide | Micronase, Glynase, Diabeta | Sulfonylureas |
| 25789 | glimepiride | Amaryl | Sulfonylureas |
| 73044 | repaglinide | Prandin | Meglitinides |
| 274332 | nateglinide | Starlix | Meglitinides |
| 6809 | metformin | Glucophage | Biguanides |
| 84108 | rosiglitazone | Avandia | Thiazoldinediones |
| 33738 | pioglitazone | ACTOS | Thiazoldinediones |
| 72610 | troglitazone | Rezulin | Thiazoldinediones |
| 16681 | acarbose | Precose | Alpha-glucosidase inhibitors |
| 30009 | miglitol | Glyset | Alpha-glucosidase inhibitors |
| 593411 | sitagliptin | Januvia | DPPIV inhibitor |
| 60548 | exenatide | Byetta | Injectables |
To validate our algorithm, two physicians (including one of the authors) reviewed 57 randomly selected cases, which were mixed with 43 randomly selected controls (patients without diabetes who had at least 1 normal glucose lab and at least 2 clinician visits among other criteria to ensure they did not have diabetes but had enough data). The physicians were blinded as to which patients were cases or controls. Two other eMERGE sites with different EMR systems (Marshfield Clinic and Vanderbilt University) similarly validated the algorithm via chart abstraction and clinician review of randomly selected charts (150 at Marshfield and 100 at Vanderbilt).
Unless otherwise noted in Figure 1, the diagnoses we use in our algorithm are ICD-9 codes in encounter diagnoses and problem lists which are entered by clinicians, and medical history diagnoses which could be entered by a clinician or a nurse.
Comparison with the Problem List
Using the EDW, we queried the problem lists of each of the T2DM cases that were identified by the algorithm. Because NMFF and NMH use separate EMRs, we searched two problem lists when they existed for each patient: the NMFF problem list in Epic (which contains ICD-9 codes only), and the NMH problem list in Cerner (which contains ICD-9 codes, SNOMED codes, and free text). We ran two queries: the first one looking for T2DM specific codes (ICD-9 codes 250.x0 or 250.x2), and the second one to find any diabetes diagnosis code: type 1, type 2, or unspecified type (i.e., any ICD-9 codes starting with 250 and any SNOMED code for diabetes). We ran the second query because we suspected that codes specific to type 2 diabetes may have been incorrectly omitted or incorrectly included in the problem list. This suspicion was based on initial reviews of results from the first query as well as from doing earlier chart reviews. In addition, as a converse study, we ran a query to see if there were any patients who had a T2DM diagnosis in their problem list, but were not identified by our T2DM algorithm.
To validate our queries, a physician (one of the authors) reviewed the charts of 45 randomly selected patients. As our initial queries indicated that patients did not have any T2DM codes in one or both of their two potential problem lists, we selected the 45 patients for review as follows: 25 who did not have codes in either problem list, 10 who did not have codes in the NMFF problem list but did in their NMH problem list, and 10 who did not have codes in the NMH problem list but did in their NMFF problem list. We chose to use the result set from first query to select patients for review, because our algorithm looks for T2DM specifically and our first query only looks for T2DM codes. During the review process, the reviewer noted if diabetes was mentioned in the problem list at all, if a similar diagnosis was noted, if the patient actually had T2DM, and if the patient was seen by a primary care physician (PCP), defined in this study as internal medicine or OB/GYN clinicians. If the patient was seen by a PCP, the reviewer noted where they were seen (NMFF, NMH or outside our institutions).
Patient Characteristics
To characterize and compare the diabetes patients including both those with and without a code in their problem list, we looked at several pieces of information: who saw the patients, where they were seen, and when they were seen. Specifically, we looked at the number and dates of in-person encounters with a clinician, at which institution these encounters occurred, and if any of these encounters were with a PCP. We also computed the approximate year of first T2DM diagnosis for the patients by capturing the earliest of the following: the first date a diagnosis code for T2DM was assigned or the first date a T2DM medication was prescribed.
Results and Discussion
In this section we present and discuss the results of running the algorithm and problem list comparison on the entire data set available in the EDW, including both inpatient and outpatient populations. The Northwestern EDW contains data extending as far back as as the source EMR systems have data, so for the results reported below we are looking at the entire longitudinal record that patients have in the source systems at the time the algorithm was executed.
Type 2 diabetes algorithm and validation
Using our algorithm we found 23,988 T2DM patients, out of approximately 2 million patients in our EDW. Only 1 patient out of the 57 T2DM cases identified by the electronic algorithm and chart reviewed for the study was found to not be a T2DM case, resulting in a 98% (56/57) positive predictive value (PPV). Marshfield and Vanderbilt had 99% and 100% PPV, respectively. Additionally, Marshfield and Northwestern both achieved 100% sensitivity. The number of charts we reviewed and our PPV is consistent with other algorithms validated as part of the eMERGE project.15
Comparison with the Problem List
Out of 23,988 patients in our EMR with T2DM according to our eMERGE algorithm, only 64% have either Epic or Cerner problem list codes for any type of diabetes (type 1 or type 2), and only 53% have T2DM codes in either Epic or Cerner problem lists. Thus, our algorithm has revealed there to be many probable instances of erroneous omissions of T2DM codes from the problem list.
The chart reviews of the 45 patients who had T2DM diagnosis codes missing from their problem list revealed that more than half (58%) did have other diabetes diagnosis codes (either type 1 or unspecified type of diabetes mellitus), and a few had related diagnoses such as impaired glucose tolerance, and diagnoses indicating complications of diabetes. These latter diagnoses, such as diabetic peripheral vascular disease, clearly implied that the patient had diabetes even though they lacked an explicit code for it in their problem list. The reviewer was able to confirm the accuracy of the results of the second query: whenever the query found there to be no diabetes diagnosis code in one of a patient’s problem lists, the reviewer confirmed there was no such code actually present. Whenever the query found there to be a diagnosis code in one of a patient’s problem lists, the reviewer was able to locate it.
Of those reviewed, 91% were confirmed to have T2DM. Of the rest: 1 had MODY (Mature Onset Diabetes of the Young), 2 had the pancreas removed, and 1 had a high dose of steroids which subsequently caused elevated blood glucose levels. In addition, the reviewer found both of the following types of errors in problem lists: codes specific to type 2 diabetes that were incorrectly omitted or incorrectly included in the problem list.
Lastly, when we queried to see if there were any patients who had T2DM in their problem list but did not meet our algorithm criteria, we found 5249 patients. Of these patients, 412 are probably T1DM patients because they are both on insulin and have T1DM diagnoses. Another 1,074 are likely to be T1DM patients because they had less than 2 encounters with a T2DM diagnosis entered by a clinician, were treated with insulin only (never on T2DM medications), and had T1DM diagnoses. Many of the remaining patients were not classified as T2DM positive by the algorithm because they only satisfy 1 criterion, not 2 as required. These are patients whom we might flag for verification by a clinician.
Patient Characteristics
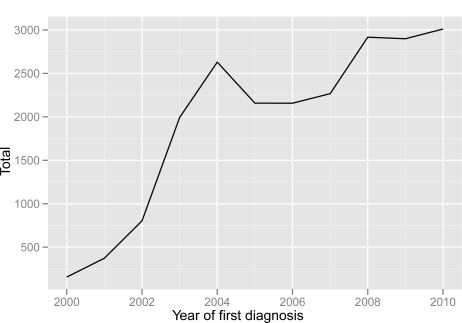
Figure 2 shows the total number of T2DM cases identified by the algorithm. The number of cases is graphed by year when records indicate that the case was first diagnosed. We do this in order to give a sense of the rate at which new T2DM cases can be expected to appear in the EMR. The graph starts at the year 2000, because this is near the beginning of the time period at which the EMR systems were widely deployed at Northwestern; data in the system before this date is relatively sparse. It can be seen from this graph that there is a steady increase in the number of cases discovered by the algorithm. Again, this increase can be at least partially explained by the fact that there was a phased roll-out of the EMR system to clinics over the time-span indicated.
Figure 2:
Type 2 diabetes mellitus patients identified by the eMERGE algorithm. Arranged by year when EDW records indicate first diagnosis. Year of first diagnosis was either the first date a diagnosis code was assigned or a T2DM medication was prescribed.
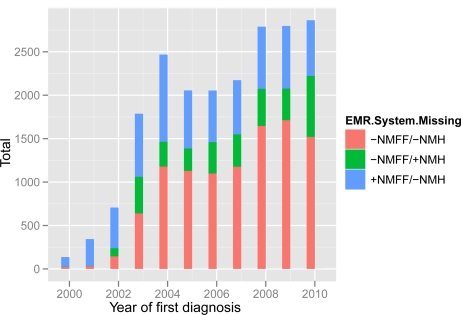
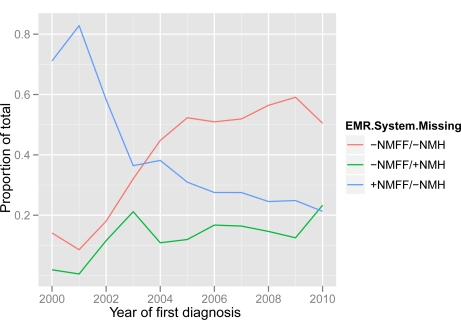
Figure 3 shows the distribution of T2DM patients without a problem list code, arranged by year when first diagnosed, and broken down by the EMR system from which the code is missing. The general shape of the distribution generally reflects the one found in Figure 2. This indicates that the relative rate of problem list omissions has been relatively stable over time. This situation is depicted more explicitly in Figure 4, which shows problem list omissions as a proportion of the total number of T2DM cases identified, broken down by EMR system. Viewing the trend lines after about 2004, there appears to be a rather steady rate of problem list omissions across the EMR systems.
Figure 3:
Type 2 diabetes mellitus patients without a problem list code. Arranged by year when EDW records indicate first diagnosis, and broken down by the EMR system from which the code is missing. Year of first diagnosis was either the first date a diagnosis code was assigned or a T2DM medication was prescribed.
Figure 4:
Type 2 diabetes mellitus patients without a problem list code, as a proportion of the total number of patients identified by our algorithm for a given year. Arranged by year when EDW records indicate first diagnosis, and broken down by the EMR system from which the code is missing. Year of first diagnosis was either the first date a diagnosis code was assigned or a T2DM medication was prescribed.
In the chart review of patients missing a T2DM diagnosis in the problem list, 74% of patients who did not have any diabetes diagnosis code in their problem list(s) (N=19) also did not have a PCP at our institutions (N=14). A subsequent query of those without any diabetes diagnoses in either problem list revealed that overall 26% had not seen a PCP at our institutions ever. This is in contrast to 47% of all T2DM diabetes patients being seen by a PCP at our institutions for comparison. Furthermore, 16% of patients with missing codes from the NMFF problem list were seen in-person at least once at NMFF, and 30% of patients with missing codes from the NMH problem list were seen in-person at least once at NMH. It is also interesting to note that 90% of patients in the chart review who had diabetes diagnoses codes in their NMH problem list but not their NMFF problem list had an empty NMFF problem list. Lastly, half (48–52%, either T2DM or any DM diagnosis) of T2DM patients had a diagnosis in one problem list, but had no diagnosis in the other problem list. This analysis therefore identifies a potential issue regarding the transmission of patient care information across institutional boundaries.
Conclusions and Future Work
From our results, the percentage of T2DM patients with diabetes in their problem list(s) failed to meet the 80% threshold required by the Meaningful Use Regulations. This was not surprising to us and confirmed that we need to take further actions to ensure that the problem list is appropriately populated. Patients without a primary care physician were particularly likely to have inadequate problem list maintenance; approximately one-quarter of patients without a diagnosis in the problem list did not have a PCP at our institutions. Given the projected shortage of primary care providers nationwide, it is likely impractical to rely on improved primary care availability to close this shortfall. Rather, this finding supports the development of improved methods for the capture and maintenance of problem lists adapted for a wide range of providers, and integrated into an EMR-enabled workflow.
We note a number of limitations in our study. Our study benefited from access to data from two separate EMRs merged and normalized into an Enterprise Data Warehouse, which may limit the generalizability of our findings. Our EMRs captured a significant proportion of data as structured elements; whereas other sites may require additional pre-processing with natural language processing (NLP). Although our algorithm performed well across a number of eMERGE sites, it is unclear if our findings would translate to care settings without the resources available at tertiary care research institutions.
Our future work is focused on further improvements to our algorithm, such as attempting to capture diet or exercise controlled diabetics, patients with similar diagnoses such as T1D, and patients having impaired glucose tolerance. We also intend to work on translating study findings into actionable components within our EMRs, such as automatically generated alerts for patients with near certain T2DM. Further refinements to the algorithm will likely require additional work to characterize patients with T2DM in their problem list who may need to have T2DM removed from their problem list, or are patients who are not being recognized by our current algorithm.
We identify NLP as a particularly useful tool in order to improve the sensitivity of our type 2 diabetes algorithm.16 For the phenotypes studied as part of the eMERGE network, use of NLP increased the sensitivity of many of the EMR-based algorithms, at times dramatically.15 Problem lists may be supplemented by identification of diagnosis concepts from the unstructured information in physician notes, which can be mined to create corresponding structured diagnoses.10,11
In summary, our study revealed numerous instances of potentially erroneous omissions of T2DM codes from the problem list. We conclude that T2DM algorithms similar to the one developed for the eMERGE Network could be applied to EMRs to fill in gaps and increase the overall accuracy of problem lists, and consequently improve compliance with Meaningful Use regulations.
Acknowledgments
The eMERGE Network was initiated and funded by the National Human Genome Research Institute, with additional funding from the National Institute of General Medical Sciences through the following grant for Northwestern University: U01-HG-004609. Funding for the Northwestern Medicine EDW is provided by Northwestern University Clinical and Translational Sciences Institute (NUCATS) grant UL1RR025741. We would also like to thank Marshfield Clinic and Vanderbilt University for their T2DM algorithm validation results.
References
- 1.Stead WW. Rethinking electronic health records to better achieve quality and safety goals. Annu Rev Med. 2007;58:35–47. doi: 10.1146/annurev.med.58.061705.144942. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhry B, Wang J, Wu S, Maglione M, Mojica W, Roth E, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med. 2006;144(10):742–52. doi: 10.7326/0003-4819-144-10-200605160-00125. [DOI] [PubMed] [Google Scholar]
- 3.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86(4):560–72. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon EG, Jha AK, Christino M, Honour MM, Fernandopulle R, Middleton B, et al. Assessing the level of health-care information technology adoption in the United States: a snapshot. BMC Med Inform Decis Mak. 2006;6:1. doi: 10.1186/1472-6947-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jha AK, DesRoches CM, Campbell EG, Donelan K, Rao SR, Ferris TG, et al. Use of electronic health records in U.S. hospitals. N Engl J Med. 2009;360(16):1628–38. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 6.2010. Electronic Health Record Incentive Program; Final Rule;
- 7.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363(6):501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 8.Szeto HC, Coleman RK, Gholami P, Hoffman BB, Goldstein MK. Accuracy of computerized outpatient diagnoses in a Veterans Affairs general medicine clinic. Am J Manag Care. 2002;8(1):37–43. [PubMed] [Google Scholar]
- 9.Campbell JR. Strategies for problem list implementation in a complex clinical enterprise. AMIA Annu Symp Proc; 1998. pp. 285–289. [PMC free article] [PubMed] [Google Scholar]
- 10.Meystre S, Haug PJ. Automation of a problem list using natural language processing. BMC Med Inform Decis Mak. 2005;5:30. doi: 10.1186/1472-6947-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui AA, Taira RK, El-Saden S, Dordoni A, Aberle DR. Automated medical problem list generation: towards a patient timeline. Stud Health Technol Inform. 2004;107(Pt 1):587–91. [PubMed] [Google Scholar]
- 12.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Armstrong LL, Denny JC, et al. Use of Diverse Electronic Medical Record Systems to Identify Genetic Risk for Type 2 Diabetes within a Genome Wide Association Study. JAMIA. doi: 10.1136/amiajnl-2011-000439. under review;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northwestern Medical Enterprise Data Warehouse Available from: https://edw.northwestern.edu.
- 15.Kho A, Pacheco J, Peissig P, Rasmussen L, Newton K, Weston N, et al. Electronic Medical Records for Genetic Research: Results of the eMERGE Consortium. Science Translational Medicine. 2011;3(79re1) doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turchin A, Kohane IS, Pendergrass ML. Identification of patients with diabetes from the text of physician notes in the electronic medical record. Diabetes Care. 2005;28(7):1794–5. doi: 10.2337/diacare.28.7.1794. [DOI] [PubMed] [Google Scholar]