Abstract
SHARPn is a collaboration among 16 academic and industry partners committed to the production and distribution of high-quality software artifacts that support the secondary use of EMR data. Areas of emphasis are data normalization, natural language processing, high-throughput phenotyping, and data quality metrics. Our work avails the industrial scalability afforded by the Unstructured Information Management Architecture (UIMA) from IBM Watson Research labs, the same framework which underpins the Watson Jeopardy demonstration. This descriptive paper outlines our present work and achievements, and presages our trajectory for the remainder of the funding period. The project is one of the four Strategic Health IT Advanced Research Projects (SHARP) projects funded by the Office of the National Coordinator in 2010.
Introduction
In December of 2010, the Office of the National Coordinator (ONC) announced the Strategic Health IT Advanced Research Projects (SHARP)[1] as part of the federal stimulus project. This report outlines the scope and trajectory of one of the four awarded projects which focuses upon secondary data use of information arising from Electronic Medical Records (EMRs). SHARPn[2] (for normalization) is a collaboration of 16 academic and industry partners to develop tools and resources that influence and extend secondary uses of clinical data. The program assembles modular services and agents from existing open-source software to improve the utilization of EHR data for a spectrum of use-cases and focus on three themes: Normalization, Phenotypes, and Data Quality/Evaluation. The program was assembled into six projects that span one or more of these themes, though together constitute a coherent ensemble of related research and development. The six projects are strongly intertwined and mutually dependent, specifically: (1) Semantic and Syntactic Data Normalization, (2) Natural Language Processing (NLP), (3) Phenotyping Applications, (4) Performance Optimizations and Scalability, (5) Data Quality Metrics, and (6) Evaluation Frameworks. All of these services are developing open-source deployments as well as commercially supported implementations.
The secondary use of EHR sourced data is a broad domain. It includes patient safety and clinical quality metrics and development programs as the most obvious, but other clinical applications range from clinical decision support to practice variation monitoring. The entire categories of clinical and translational research are fundamentally dependent on effective secondary use of clinical information, including clinical trials, observational cohorts, outcomes research, comparative effectiveness, and best evidence discovery.
Our vision is to develop and foster a federated informatics research community committed to open-source resources that can industrially scale to address barriers to the broad-based, facile, and ethical use of EHR data for secondary purposes. Within this collaborative community, we seek to create, evaluate, and refine informatics artifacts that contribute to EHR data use for improving care, generating new knowledge, and addressing population needs. We are committed to making all artifacts available as open-source tools, services, and scalable software. However, we partner with industry developers for commercial deployment and support of our software artifacts, recognizing that many adopters will eschew exclusively open-source resources that may lack reliable, commercial support.
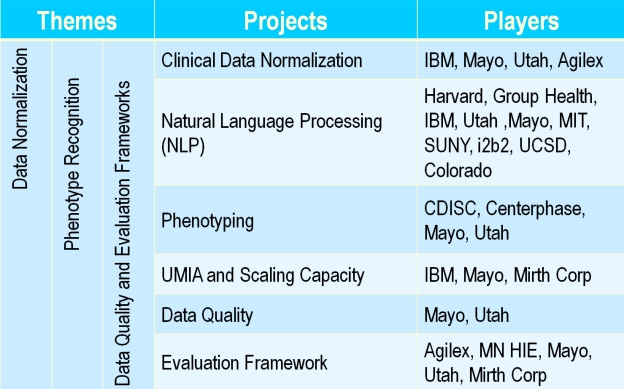
The consortium of organizations funded and participating in SHARPn include: Mayo Clinic, University of Utah, Intermountain Healthcare, Agilex Technologies, CDISC (Clinical Data Interchange Standards Consortium), Centerphase Solutions, Deloitte, Group Health Seattle, IBM Watson Research Labs, Harvard University & i2b2, Minnesota HIE (MNHIE), MIT, Mirth Corporation, State University of New York at Buffalo, University of California San Diego, University of Pittsburgh, University of Colorado. As is evident in Figure 1, the consortium is a blend of academic communities, large and small businesses, an HIT standards development organization, and a consulting group. Such multidisciplinary talent is crucial to achieve the ambitious goals of the consortium.
Figure 1.
SHARPn Project Organization
We structure our presentation around each project, although the interdependencies are substantial. For example, the phenotyping algorithms rely heavily upon the pre-existing normalization of clinical data, to assure comparability and consistency of cohort retrieval among medical centers with differing EHR environments. Our closing discussion paints the spectrum of development and application which we target.
Semantic and Syntactic Normalization
An underlying principle for secondary data use is that all applications are more efficient and reliable if the data on which they are premised is comparable and consistent[3]. One way to make heterogeneous data from disparate EHR systems comparable and consistent is post-hoc normalization. This is precisely what we engineer with our first set of software appliances.
As with most of our software developed in SHARPn, we invoke the Unstructured Information Management Architecture[4] (UIMA) as our framework; this open-source artifact from IBM Watson Labs – the framework on which the Watson Jeopardy engine was built – is described in more detail in the Performance Optimization project below. We provide two dimensions of normalization, essentially syntactic and semantic.
Syntactic normalization relies heavily on the 20-year history of Health Information Exchange (HIE) and disparate data source normalization undertaken by the Regenstrief Institute and the Indiana HIE. The Regenstrief team has developed a normalization pipeline, Health Open Source Software[5] (HOSS), which will soon be formalized as open-source. HOSS excels at making ill-formed HL7 message contents into well-formed structures. Since these algorithms have accreted over decades, a SHARPn task will be to modularize and simplify normalization task elements, and render them in a high-throughput UIMA-AS[6] pipeline. SHARPn will publish these normalization algorithms and a reference implementation of their deployment in UIMA.
Semantic normalization invokes the mature technologies emergent from Mayo’s open-source LexGrid project[7] for terminology services, and in particular the caBIG supported LexEVS implementation[8]. Semantic normalization is not magic, and ultimately depends upon the creation or availability of mapping files, for example local laboratory codes into LOINC. However, Pareto optimization pertains, where in practice 99% of the cases are handled by 2% of the codes, making practical map creation scalable and practical. The standards-based generalization of these terminology services will soon be manifest in the CTS2 (Common Terminology Services) specification being finalized by HL7 and the Object Management Group. Our final semantic normalization functionality will wrap CTS2 services as UIMA artifacts, which are callable by a variety of SHARPn use cases, including NLP.
Both syntactic and semantic normalization require that a “common” form, or canonical representation, is specified as a target for normalization activities. While the HOSS pipeline has worked well using HL7 message syntax as a de facto normalization scheme, we believe that additional specificity is needed to address the spectrum of use-cases in secondary data use, and the clinical granularity of information being generated in today’s EHRs. SHARPn has chosen the Clinical Element Models[9] (CEMs), historically developed by Intermountain Healthcare, as our canonical representation. They are described as: “the basis for retaining computable meaning when data is exchanged between heterogeneous computer systems as well as the basis for shared computable meaning when clinical data is referenced in decision support logic.” Presently, over 4,000 XML schema for CEMs, such as blood pressure measurement or specific laboratory tests, are defined. SHARPn, and a growing consortium of informatics users, are contributing to the CEM library, which is an open-source artifact.
Natural Language Processing (NLP)
Within the NLP project, our goal is the development of enabling technologies for high-throughput phenotype extraction from clinical free text. In parallel, we are exploring the ability to hybridize clinical data extracted from medical reports with the already-structured data existing in data repositories to facilitate a complete phenotype. Our focus is NLP and Information Extraction (IE), defined as the transformation of unstructured free text into structured representations. Thus our goal is to research and implement general purpose modular solutions for the discovery of key components to be used in a wide variety of biomedical use cases. Specifically, our efforts are on methodologies for clinical event discovery and semantic relations between these events. Subsequently, the discovered entities and relations will populate normalization targets, in our case the CEMs where each CEM is invoked through the concept SNOMED CT or RxNORM code.
Labeling atomic events with their arguments facilitates more complex processing of the textual data such as identifying temporal information facilitates causal reasoning. Therefore, the discovery of clinical events and entities is the building block of a relation-extraction system for deep language understanding. Our methods use as features the local and global linguistic and domain context through multi-layered linguistic annotations generated by clinical Text Analysis and Knowledge Extraction System (cTAKES)[10][11], University of Colorado’s CLEAR TK[12], and the tools provided by the participating co-investigators. The extracted features are then fed to train a machine learner to distinguish events from non-events and to label each event. Our relation-extraction methodology relies on the syntactic structure of the sentence and each constituent’s semantic role in that sentence (e.g., “patient,” “agent,” “theme,” “location”) and employs machine learning. Concept graphs[13, 14] are then used as the knowledge representation for the discovered relations across the sentences in the given document. Events and entities are the nodes with the edges between the nodes being derived from the semantic relations. The information in the concept graphs is used to populate the CEM-based templates for entities and relations. In addition, we are exploring active learning techniques[15] to minimize the amount of training data. Each module will be evaluated in a standard 10-fold evaluation (9 folds for training, 1 fold for testing) for its task against the gold standard.
The engineering framework within which the NLP project functions is UIMA AS. Core technologies such as cTAKES and CLEAR TK are built within UIMA providing a solid basis for expandability and software development. One of the main UIMA concepts is that of a type system, which defines the annotations and their structure as generated by the pipeline. An agreed-upon basic type system ensures a software development foundation. Among our goals is to release to the community such a basic type system around which new modules can be contributed or existing modules can be wrapped within cTAKES.
The NLP project has set forth an aggressive release schedule. The core system, cTAKES, has been expanded with a number of modules. The dependency parser (released September, 2010) uses the CLEAR dependency parser[16], a state-of-the-art transition based parser developed by the CLEAR computational semantics group at the University of Colorado at Boulder. The Drug profile module (released December, 2010) discovers medication mentions in the patient’s clinical notes, normalizes them to an RxNORM code and populates a template with relevant attributes such as dosage, duration, start date, end date, form, frequency, route, strength. The Smoking status classifier (released March, 2011) assigns each patient one of four smoking status labels: current smoker, past smoker, non-smoker and unknown[17]. Among our 2011 releases are a full cycle prototype for processing text through cTAKES and populating OrderMedAbm CEMs (of note, OrderMedAmb represent medications). Thus, by fall of 2011, the phenotype applications project will be enabled to start directly consuming output from the NLP artifacts,
Phenotype Applications
The term phenotyping is significantly overloaded in our application of meaning. Our use implies the algorithmic recognition of any cohort within EHR for a defined purpose. These purposes were inspired by the algorithmic identification of research phenotypes[18] in the NHGRI funded eMERGE (electronic MEdical Records and GEnomics) cooperative agreement consortium[19] in which Mayo is a founding member; these phenotype algorithms are used to define case and control cohorts for genome-wide association studies (GWAS). However, the SHARPn view of phenotyping includes inclusion and exclusion criteria for clinical trials, numerator and denominator criteria for clinical quality metrics, epidemiologic criteria for outcomes research or observational studies, and trigger criteria for clinical decision support rules, among others. Nevertheless, the underlying principles of using well-defined algorithms across diagnostics fields, laboratory values, medication use, and NLP-derived observations adheres to the practices demonstrated in eMERGE[20]. In the following paragraphs, we introduce and discuss a range of research topics and issues addressed by this sub-project within the SHARPn program.
A key aspect of the EHR-based phenotyping algorithms is the ability to implement and execute them across multiple institutional boundaries and EHR system settings. While we were highly successful in eMERGE to achieve this goal, one of the limitations was the lack of a structured representation that can be leveraged for algorithm design and specification. Consequently, all the phenotyping algorithms developed by the eMERGE consortia resided in Microsoft® Word and Excel files without necessarily following any particular template and structure, making them primarily amenable to human consumption and interpretation. There are two main issues with such an approach: first, due to the lack of any structure or template used for algorithm representation, the algorithms themselves are not computable and machine processable, even semi-automatically, and second, the unstructured nature of the algorithms introduces the risk for misinterpretation and ambiguity in cohort eligibility criteria specification. Figure 2, for example, shows a graphical representation of the hypothyroidism algorithm developed within eMERGE. To address this limitation, in SHARPn we are investigating structured eligibility criteria representation models, in particular CDSIC’s Protocol Representation Model (PRM[21]). Based on the BRIDG model[22], PRM provides a standardized template and artifacts for specifying different aspects of a cohort eligibility criteria, thereby enabling case/control definitions for the phenotyping algorithms to be represented in an XML-based representation. While one has to still interpret, implement and execute information stored the XML files for the algorithms (that is, the XML files cannot be executed directly within an EHR system), PRM nonetheless provides a structured representation that can be queried and parsed via software applications. In fact, our goal is to implement a publicly available Web-based library of phenotyping algorithms that can be accessed and searched by a range of clients.
Figure 2.
Hypothyroidism algorithm (courtesy of the eMERGE consortia)
Related to the topic of cross-institutional implementation and execution of phenotyping algorithms, is understanding and addressing issues related to the variation and heterogeneity of EHR data across multiple institutions for a given disease or phenotype that arise due to a multitude of factors including modalities of data recording, storage, and organization. For example, the usage of a particular or set of ICD-9 codes or prescription of a specific brand of drug may vary across institutions, which arguably would result in the variability of algorithm query results, as well as, potentially cause issues in multi-site data integration and analysis. In SHARPn, one of our goals is to evaluate the extent to which such variability can result into issues for a comparable case/control definition, at least based on data from Mayo Clinic and Intermountain Healthcare.
Another key aspect relevant to EHR-based phenotyping algorithms is a cost-benefit analysis from the perspective of time and effort spent in algorithm design, execution and validation. Specifically, the historic norm for cohort identification has been to rely on manual chart/nurse abstraction and/or leverage billing and diagnoses codes (ICD-9, and CPT-4 primarily). The former is arguably non-scalable, expensive and time-consuming, while the latter is error-prone due to biases in coding practices. As evidenced by Figure 2 above, the EHR-based phenotyping algorithms on the other hand, apply a range of information typologies, including laboratory measurements and medications, which typically require development of sophisticated natural language processing techniques. This obviously raises the “cost-benefit” question for requiring the additional effort and resources for such an elaborate process. Led by Centerphase, within SHARPn we are currently implementing a pilot study to understand such issues more coherently for Type 2 Diabetes cohort identification. Specifically, our goal is to compare and document the pros and cons, primarily from a time, effort, resource, and cost perspective, in EHR-based phenotyping algorithm design and development.
The data flow for phenotyping has demonstrated convincingly that a persistence layer for clinical data is required. This is because phenotyping constitutes a question/answer process, where within an algorithm one ascertains whether a particular patient does or does not manifest definitional criteria. This persistence layer can be in any manner forms, including: tag data elements, RDF triple stores, or conventional modeled databases. For convenience in our development phases, we opt for SQL database technology although we recognize that decision is arbitrary. Our ultimate goal is to decouple the algorithms from any physical implementation of a persistence layer.
The clinical data persistence layer also comprises storage target for normalization services, including NLP. Phenotyping algorithms, to ensure reproducibility across EHR environments, are dependent on well-normalized data. Thus, one can regard the persistence layer as a data store for normalized representations. Because phenotyping is question and answer by nature, it would seem efficient to normalize data in batches, and pose questions against a normalized persistence layer. However, one can envision posing question across an HIE environment, doing just-in-time normalizations on the answers, thereby supporting phenotyping over a denormalized, distributed collection of clinical data.
Performance Optimization
UIMA[4] is a freely available, Apache Software Foundation open-source project, intended to bring interoperability and reusability in the arena of unstructured information analytics. It has been widely adopted by both academic and commercial developers, and is helping to create a community of unstructured information analytics developers that build upon each other’s work. Mayo Clinic was the alpha user of UIMA in 2003, using it as the backbone for our cTAKES NLP pipeline. UIMA includes an asynchronous scaleout capability, UIMA-AS[6] (AS = Asynchronous Scaleout), that is very effective in exploiting both multi-core architectures and clusters of networked machines to achieve high throughput. This capability was recently demonstrated by IBM’s Watson when it competed on the TV show Jeopardy!®[23] Watson used UIMA-AS to scale out the processing needed to compete on Jeopardy! to thousands of compute cores. UIMA-AS forms the core framework for SHARPn software artifacts.
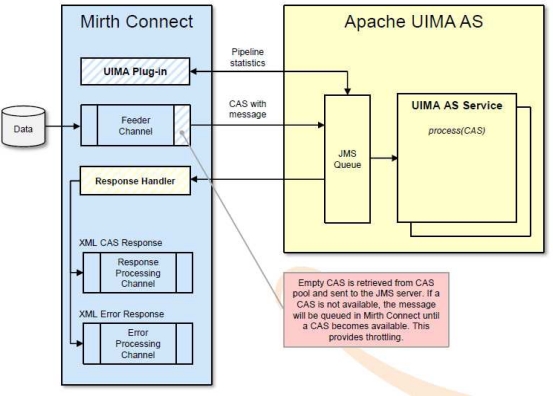
The SHARPn infrastructure approach attempts to span both immediate program needs, supporting the researchers in their quest for new and improved algorithms, while at the same time yielding a framework immediately available for evaluation and production implementations. SHARPn partnered with Mirth Corp. to develop a new adapter to their MIRTH CONNECT[24] open source interface engine, already supporting various protocols (HL7 2.x, HL7 3.0, SOAP, XML, etc.), the new adapter enables any number of UIMA based algorithms developed and published by the project to be invoked from clinical data flows. The diagram below depicts the design of the new adapter to be used to host UIMA processing.
UIMA represents a extremely useful infrastructure for high performance processing, and its overall utility within the SHARPn project will be investigated, however another infrastructure to also be utilized within the project is WSBPEL, specifically those services provided via Apache ODE. By utilizing MIRTH CONNECT’s existing SOAP capabilities, Service Oriented Architectures will be evaluated and contrasted with the UIMA pipelines capabilities to determine the relative benefits of each to the goals of the project.
It is interesting to note that the entire software stack used by the project is open source, components utilized were: Ubuntu for its cloud computing, NHIN CONNECT for its secure internet exchange support, MIRTH CONNECT as the hosting hub for UIMA-AS, and WS-BPEL, as well as JBOSS to host the ODE web services orchestration engine and MySql for persistence services for both test data sets and results output. The diagram below depicts the flow enabled via the use of these open source packages.
This means that any group wanting to evaluate the solutions developed by the SHARPn program can do so with minimal costs, those groups interested in accessing the fitness of the NLP, Data Normalization or Phenotyping algorithms developed are immediately enabled to carry out experiments and those wanting to utilize the algorithms in production can do so by installing the software within their local environment.
Evaluation Framework and Data Quality Metrics
The initial evaluation strategy is to exchange normalized data (as produced in the normalization pipeline) as the payload in NHIN Connect services communicating via the internet between Intermountain Healthcare in Salt Lake City and the Mayo Clinic in Rochester. A first working version of this communication will transmit de-identified laboratory data from Intermountain to a normalized data store that exists within the cloud at the Mayo clinic. Data in the form of HL7 Version 2 messages will be sent from Intermountain to an instance of the Mirth interface engine installed within the Intermountain network. The interface engine will interact with LexEVS terminology services and with UIMA components to convert the HL7 Version 2 message into a normalized Clinical Element (CE) instance. Coded data within the CE instance will be represented using LOINC and SNOMED CT codes. Normalization of the CE instance will be guided by the definition of the LOINC code, SNOMED CT value set, allowed units of measure, numeric constraints, and other information contained in the definition of the specific Clinical Element Model for the kind of lab data being sent. When the normalized CE instance has been created, the Mirth interface engine will initiate communication through a standard NHIN Connect Gateway that has been installed in the Intermountain network. The normalized CE instance will be placed as the payload within the NHIN Connect message, and the gateway will transfer the data to another instance of an NHIN Gateway that has been installed at the Mayo Clinic in Rochester. The NHIN gateway will forward the data to an instance of a Mirth interface engine installed in the Mayo cloud. The Mirth interface engine will then persist the data into a de-identified patient data store in the Mayo cloud. If the laboratory data includes elements that are free text, the Mirth engine will invoke an NLP pipeline to extract coded data from the free text. The newly extracted coded data would then also be stored into the patient data store.
Once we have the communication link working from end to end, we will evaluate the availability, stability, and performance of the system, as the primary framework for system throughput evaluation and benchmarking for continuing improvement strategies.
An important issue in secondary use of EHRs, is the variation across health care delivery institutions in the way medical information is recorded and coded. For example, the rate at which a given ICD-9 code is used may vary across institutions, even though the rate at which the condition exists may not vary. Such variations have potentially large implications both for the goal of generalizing findings from EHR analysis from one institution to a broader population, as well as for combining EHR-based results across institutions. It must be acknowledged that true variations across institutions in disease prevalence also exist, due to ethnic and cultural differences, behavioral and climate differences, genetic differences, etc. Nevertheless it is important to understand how variations in data collection and representation affect the ability to use EHR in a generalizable way.
In order to develop valid methods for combining information across health care delivery institutions, it is important to understand the variation in the EHR data relative to any given disease or phenotype. An example of a SHARP test case for automated determination of phenotype for subjects within a health system is that of the phenotype “type 2 Diabetes Mellitus”. Northwestern University, as a member of the eMERGE consortium, developed an algorithm for identifying cases with this phenotype based on routine EHR documentation. It would be important to understand to what extent the application of this algorithm, or any other algorithm, yields comparable case definition across Mayo Institute and Intermountain Health Care. At the end of this test case, we should be able to characterize differences between these 2 institutions, in terms of recording and coding of diagnoses, billing codes, lab results, medications, etc., that would have an impact on the generalizability of algorithms for phenotyping Type 2 DM.
Finally, we hope that this study will pave the way for a broader study on data heterogeneity issues relevant to inter-institutional mining of the EHR for secondary purposes.
Discussion
The challenges confronting all users of EMR data, with respect to comparability and consistency, are formidable. These challenges are magnified for secondary use cases, where data aggregation, integration, inferencing, and synthesis are made more complex in the face of heterogeneous data from disparate sources. SHARPn seeks first to facilitate data normalization through sharable software resources, including the generation of structured information from unstructured data through NLP. Drawing from virtual, distributed, or locally resident persistence layers of normalized clinical data, the execution of standardized phenotyping algorithms, for a spectrum of cohort identification use-cases, from libraries of well-curated and validated phenotyping standards will facilitate the speed and reduce the effort levels associated with reliable secondary use applications. Finally, establishing metrics for reliability and establishing frameworks for incremental improvements will continuously drive the quality and consistency of our software.
Substantial limitations persist, not the least being the unavoidable requirement for well-curated semantic mapping tables among native data representations and their target standards. This impediment remains restricted by human authoring and review, though some promising techniques for semi-automated mapping algorithms are beginning to appear. Fortunately, surprising large fractions of real data appear to be addressed by a modest proportion of all possible mapping, making this labor intensive step reasonably scalable, if one is willing to trade off 100% completeness for semantic transformation.
More interestingly, SHARPn has created a community which not only interacts remarkably well within itself, but is creating strategic links with related projects such as many of the Nation Centers for Biomedial Computing, including the National Center for Biomedical Ontology (NCBO), Informatics for Integrating Biology and the Bedside (i2b2), and Integrating Data for Analysis, Anonymization and SHaring (iDASH). The modular, open-source nature of SHARPn deliverables enables innovative adoption by academic groups and developers. However, as SHARPn matures, our commercial partners will lead the path toward commercially support implementations of SHARPn technologies which should expand our adoption community to a much broader audience. Nevertheless, its status as “middle-ware” will always limit any highly visible manifestations of the tools to end-users of secondary use applications.
Figure 3.
UIMA integration into MIRTH translation platform
Acknowledgments
This work was funded primarily by a SHARP award (90TR0002) to Mayo Clinic from HHS and the Office of the National Coordinator. Additional funding from the Mayo Clinic eMERGE (U01-HG04599) project.
References
- 1.Office_of_the_National_Coordinator Strategic Health IT Advanced Research Projects (SHARP) Program. 2011. [cited 13 March 2011]; Available from: http://healthit.hhs.gov/portal/server.pt/community/healthit_hhs_gov__sharp_program/1806.
- 2.SHARPn: Secondary Data Use and Normalization. 2011. [cited 13 March 2011]; Available from: http://sharpn.org.
- 3.Chute CG, Cohn SP, Campbell JR. A framework for comprehensive health terminology systems in the United States: development guidelines, criteria for selection, and public policy implications. ANSI Healthcare Informatics Standards Board Vocabulary Working Group and the Computer-Based Patient Records Institute Working Group on Codes and Structures. J Am Med Inform Assoc. 1998;5(6):503–10. doi: 10.1136/jamia.1998.0050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apache UIMA. http://incubator.apache.org/uima/. [cited 2010 January 20]; Available from: http://incubator.apache.org/uima/.
- 5.Health Open Source Software Collaborative. 2009. Regenstrief. [cited 14 March 2011]; Available from: https://tools.regenstrief.org/wiki/display/hoss/Health+Open+Source+Software+Collaborative. [Google Scholar]
- 6.Getting Started: UIMA Asynchronous Scaleout. http://incubator.apache.org/uima/doc-uimaas-what.html. [cited 2010 January 20]; Available from: http://incubator.apache.org/uima/doc-uimaas-what.html.
- 7.Pathak J, et al. LexGrid: A Framework for Representing, Storing, and Querying Biomedical Terminologies from Simple to Sublime. Journal of Americal Medical Informatics Association. 2009;16(3):305–15. doi: 10.1197/jamia.M3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.OBO: Open Biomedical Ontologies. [cited; Available from: http://obo.sourceforge.net.
- 9.Clinical Element Model (CEM) www.clinicalelement.com [cited 2010 January 20]; Available from: www.clinicalelement.com.
- 10.Savova GK, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayo_Clinic. clinical Text Analysis and Knowledge Extraction System. 2011. [cited 16 March 2011]; Available from: http://sourceforge.net/projects/ohnlp/files/cTAKES/ or www.ohnlp.org.
- 12.ClearTK toolkit for statistical NLP. 2011. [cited 16 March 2011]; Available from: http://code.google.com/p/cleartk/.
- 13.Sowa J. Conceptual graphs for a database inference. IBM Journal of Research and Development. 1976;20:336–357. [Google Scholar]
- 14.Sowa JF. Conceptual structures : information processing in mind and machine. xiv. Reading, Mass: Addison-Wesley; 1984. p. 481. [Google Scholar]
- 15.Chen J, et al. Human Language Technology conference - North American chapter of the Association for Computational Linguistics annual meeting (HLT-NAACL) New York, NY: 2006. An Empirical Study of the Behavior of Active Learning for Word Sense Disambiguation. [Google Scholar]
- 16.Choi J, Nicolov N. K-best, Locally Pruned, Transition-based Dependency Parsing Using Robust Risk Minimization. Collections of Recent Advances in Natural Language Processing. 2011;5:205–216. [Google Scholar]
- 17.Sohn S, Savova GK. Mayo clinic smoking status classification system: extensions and improvements. AMIA Annu Symp Proc; 2009. pp. 619–23. [PMC free article] [PubMed] [Google Scholar]
- 18.eMERGE . eMERGE Network Phenotype Library. 2011. [cited 13 March 2011]; Available from: https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Library_of_Phenotype_Algorithms. [Google Scholar]
- 19.McCarty CA, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4(1):13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kullo IJ, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CDSIC Protocol Representation Model. 2011. [cited 2011 March 15. Available from: http://www.cdisc.org/protocol.
- 22.Biomedical Research Integrated Domain Group (BRIDG) Model. 2011. [cited 2011 March 15. Available from: http://www.bridgmodel.org/.
- 23.IBM What is Watson? 2011. [cited 16 March 2011]; Available from: http://www.ibm.com/innovation/us/watson/index.html.
- 24.Mirth Mirth Connect. 2011. [cited 16 March 2011]; Available from: http://www.mirthcorp.com/community/mirth-connect.