Abstract
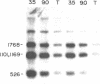
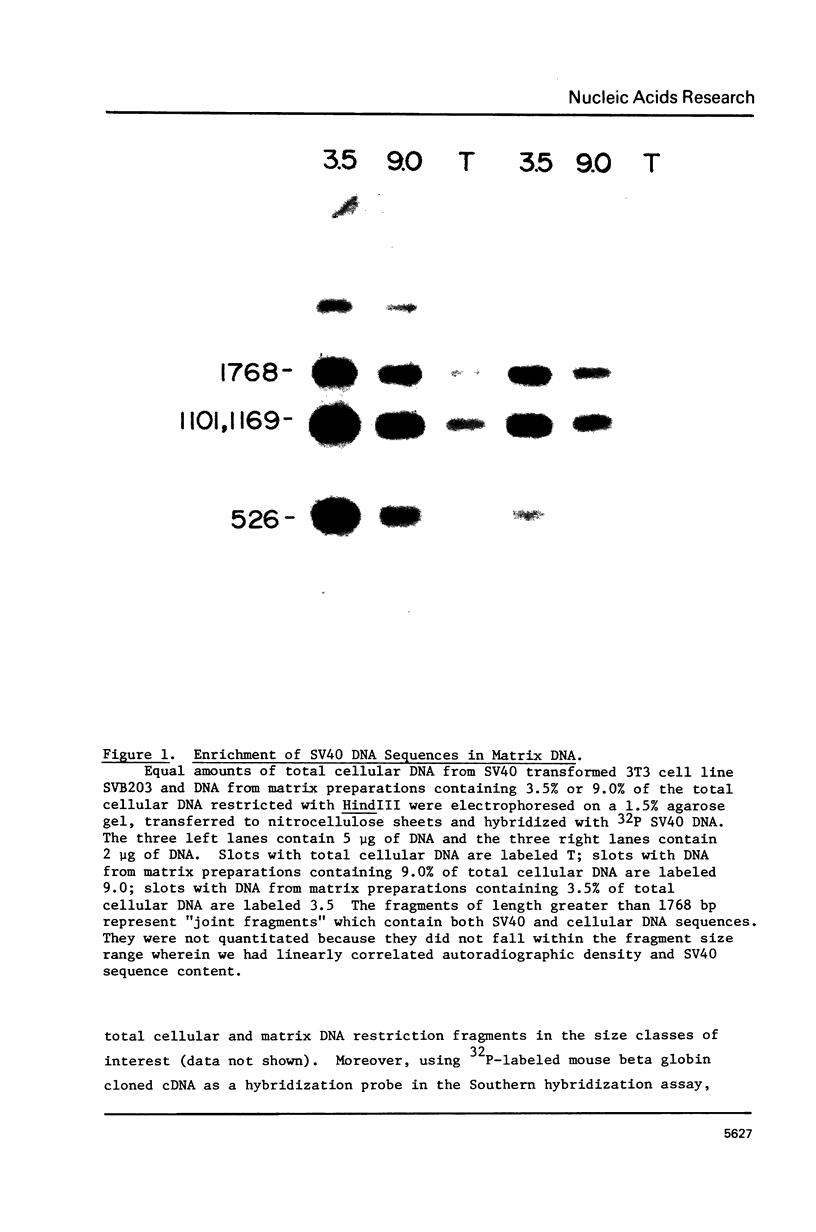
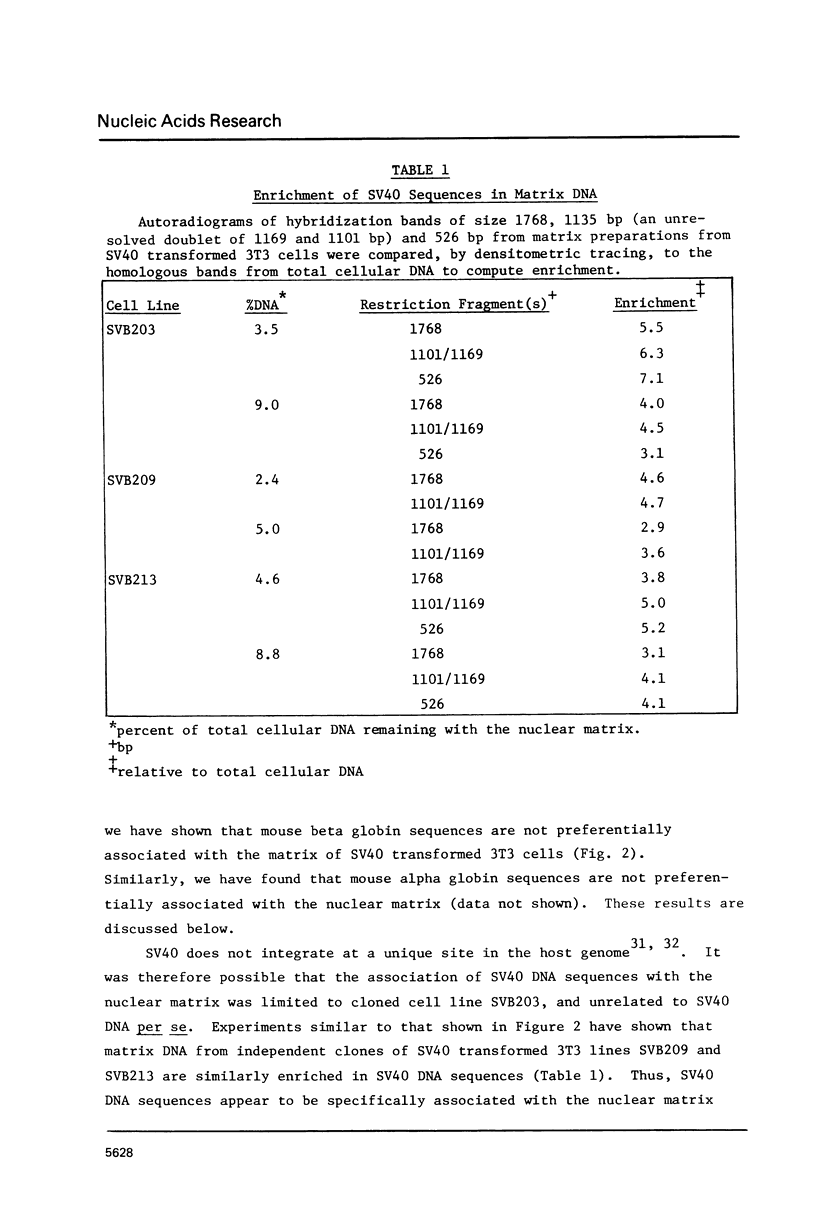
Recent studies indicate that eukaryotic DNA is organized into supercoiled loop domains. These loops appear to be anchored at their bases to an insoluble nuclear skeleton or matrix. Most of the DNA in the loops can be released from the matrix by nuclease digestion; the residual DNA remaining with the nuclear matrix represents sequences at the base of the loops, and possibly other sequences which are intimately associated with the nuclear matrix for other reasons. Using a quantitative application of the Southern blotting technique, we have found this residual DNA from SV40 infected 3T3 cells to be enriched in SV40 sequences, indicating that they reside near matrix-DNA attachment points. An enrichment of 3-7 fold relative to total cellular DNA, was found in each of three different lines of SV40 infected 3T3 cells. Control experiments with globin genes showed no such enrichment in this residual matrix DNA. This sequence specificity suggests that the spatial organization of DNA sequences within loops may be related to the functionality of these sequences within the cell.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrack E. R., Coffey D. S. The specific binding of estrogens and androgens to the nuclear matrix of sex hormone responsive tissues. J Biol Chem. 1980 Aug 10;255(15):7265–7275. [PubMed] [Google Scholar]
- Barrack E. R., Hawkins E. F., Allen S. L., Hicks L. L., Coffey D. S. Concepts related to salt resistant estradiol receptors in rat uterine nuclei: nuclear matrix. Biochem Biophys Res Commun. 1977 Dec 7;79(3):829–836. doi: 10.1016/0006-291x(77)91186-x. [DOI] [PubMed] [Google Scholar]
- Bekhor I., Mirell C. J. Simple isolation of DNA hydrophobically complexed with presumed gene regulatory proteins (M3). Biochemistry. 1979 Feb 20;18(4):609–616. doi: 10.1021/bi00571a010. [DOI] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977 Jun;73(3):616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Nuclear protein matrix: association with newly synthesized DNA. Science. 1975 Jul 25;189(4199):291–293. doi: 10.1126/science.1145202. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Chipchase M., Speirs J. The ribosomal RNA cistrons. Prog Nucleic Acid Res Mol Biol. 1971;11:351–389. doi: 10.1016/s0079-6603(08)60332-3. [DOI] [PubMed] [Google Scholar]
- Boer G. J. A simplified microassay of DNA and RNA using ethidium bromide. Anal Biochem. 1975 May 12;65(1-2):225–231. doi: 10.1016/0003-2697(75)90507-2. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- CALLAN H. G. THE NATURE OF LAMPBRUSH CHROMOSOMES. Int Rev Cytol. 1963;15:1–34. doi: 10.1016/s0074-7696(08)61114-6. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Mapping sequences in loops of nuclear DNA by their progressive detachment from the nuclear cage. Nucleic Acids Res. 1980 Jul 11;8(13):2895–2906. doi: 10.1093/nar/8.13.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Deppert W. Simian virus 40 (SV40)-specific proteins associated with the nuclear matrix isolated from adenovirus type 2-SV40 hybrid virus-infected HeLa cells carry SV40 U-antigen determinants. J Virol. 1978 Apr;26(1):165–178. doi: 10.1128/jvi.26.1.165-178.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel P. A., Mullenders L. H., Wanka F. Analysis of the attachment of replicating DNA to a nuclear matrix in mammalian interphase nuclei. Nucleic Acids Res. 1979 Jan;6(1):219–230. doi: 10.1093/nar/6.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiferman I., Pogo A. O. Isolation of a nuclear ribonucleoprotein network that contains heterogeneous RNA and is bound to the nuclear envelope. Biochemistry. 1975 Aug 26;14(17):3808–3816. doi: 10.1021/bi00688a013. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates D. M., Bekhor I. Distribution of active gene sequences: a subset associated with tightly bound chromosomal proteins. Science. 1980 Feb 8;207(4431):661–662. doi: 10.1126/science.7352280. [DOI] [PubMed] [Google Scholar]
- Gazit B., Panet A., Cedar H. Reconstitution of a deoxyribonuclease I-sensitive structure on active genes. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1787–1790. doi: 10.1073/pnas.77.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hartwig M. Organization of mammalian chromosomal DNA: supercoiled and folded circular DNA subunits from interphase cell nuclei. Acta Biol Med Ger. 1978;37(3):421–432. [PubMed] [Google Scholar]
- Herman R., Weymouth L., Penman S. Heterogeneous nuclear RNA-protein fibers in chromatin-depleted nuclei. J Cell Biol. 1978 Sep;78(3):663–674. doi: 10.1083/jcb.78.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Ide T., Nakane M., Anzai K., Ando T. Supercoiled DNA folded by non-histone proteins in cultured mammalian cells. Nature. 1975 Dec 4;258(5534):445–447. doi: 10.1038/258445a0. [DOI] [PubMed] [Google Scholar]
- Igó-Kemenes T., Zachau H. G. Domains in chromatin structure. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Ketner G., Kelly T. J., Jr Integrated simian virus 40 sequences in transformed cell DNA: analysis using restriction endonucleases. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1102–1106. doi: 10.1073/pnas.73.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Pardon J. F. Structure and function of chromatin. Annu Rev Genet. 1979;13:197–233. doi: 10.1146/annurev.ge.13.120179.001213. [DOI] [PubMed] [Google Scholar]
- Miller T. E., Huang C. Y., Pogo A. O. Rat liver nuclear skeleton and ribonucleoprotein complexes containing HnRNA. J Cell Biol. 1978 Mar;76(3):675–691. doi: 10.1083/jcb.76.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. E., Huang C. Y., Pogo A. O. Rat liver nuclear skeleton and small molecular weight RNA species. J Cell Biol. 1978 Mar;76(3):692–704. doi: 10.1083/jcb.76.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B., Coffey D. S. A fixed site of DNA replication in eucaryotic cells. Cell. 1980 Feb;19(2):527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B. Sequence analysis of nuclear matrix associated DNA from rat liver. Exp Cell Res. 1980 Aug;128(2):466–470. doi: 10.1016/0014-4827(80)90083-x. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Piñon R., Salts Y. Isolation of folded chromosomes from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2850–2854. doi: 10.1073/pnas.74.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Goutorbe F., Strauss F., Bernardi G. Yield of restriction fragments from yeast mitochondrial DNA. J Mol Biol. 1977 Feb 15;110(1):47–52. doi: 10.1016/s0022-2836(77)80097-1. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Mantieva V. L., Georgiev G. P. The similarity of DNA sequences remaining bound to scaffold upon nuclease treatment of interphase nuclei and metaphase chromosomes. Nucleic Acids Res. 1979 Nov 24;7(6):1713–1735. doi: 10.1093/nar/7.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Ferguson J., Davis R. W., Stark G. R. T antigen binds to simian virus 40 DNA at the origin of replication. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1605–1609. doi: 10.1073/pnas.72.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Scheer U., Franke W. W., Trendelenburg M. F., Spring H. Classification of loops of lampbrush chromosomes according to the arrangement of transcriptional complexes. J Cell Sci. 1976 Dec;22(3):503–519. doi: 10.1242/jcs.22.3.503. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Shaper J. H., Pardoll D. M., Kaufmann S. H., Barrack E. R., Vogelstein B., Coffey D. S. The relationship of the nuclear matrix to cellular structure and function. Adv Enzyme Regul. 1978;17:213–248. doi: 10.1016/0065-2571(79)90015-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Wolf S. F., Mareni C. E., Migeon B. R. Isolation and characterization of cloned DNA sequences that hybridize to the human X chromosome. Cell. 1980 Aug;21(1):95–102. doi: 10.1016/0092-8674(80)90117-8. [DOI] [PubMed] [Google Scholar]