1.0 INTRODUCTION
Luscious cherries, sweet peaches, creamy avocados, and tropical papayas are just a few of the tasty treats that come to mind when we think of fruit. Indeed, fruit come in all shapes and sizes, from gigantic pumpkins to the tiny fruit of the duckweed Wolffia angusta, which are as small as a grain of salt. Fruit range in texture from soft and fleshy to dry and papery with each design optimized for a different seed dispersal strategy. Fleshy fruit are often sweet, brightly colored, and are generally adapted to be eaten by vertebrates, which carry the seeds to a new location before depositing them in a pile of fertilizer. In contrast, wind, water, and the force generated by the opening of the seedpod commonly distribute the seeds of dry fruit. Of course there are many exceptions, such as the spiked, barbed, dry fruit that snag a ride by adhering to the fur of passing animals. Dry fruit are classified as either dehiscent, in which the walls of the ovary open to release the seeds into the environment, or indehiscent, in which the seeds remain enclosed in the fruit and the fruit is shed from the plant. Many important crops including peas, beans, lentils, soybeans and canola have dehiscent fruit.
Both crops with fleshy fruit and with dehiscent fruit are of such importance to agriculture and the human diet that fruit have been the focus of extensive research in recent years. Research on fleshy fruit has focused primarily on tomato and great progress has been made in understanding the genes that control the size and ripening of tomato fruit (for reviews see Giovannoni, 2004; Tanksley, 2004; Adams-Phillips, et al., 2004). Research on dehiscent fruit has focused on Arabidopsis thaliana, which will be the focus of this chapter (for additional reviews see Dinneny and Yanofsky, 2004; Ferrándiz, et al., 1999; Bowman et al., 1999).
In this chapter, we will first discuss wild-type fruit development and then turn to the genes and hormones that are known to regulate fruit formation in Arabidopsis. Specifically, we will examine the genes that are involved in specifying the development of the different tissue types within the fruit, the genes that control the formation of axes within the fruit, and the processes that regulate fruit development after fertilization (see Table 1 for a list of genes involved in fruit development). The fruit is arguably the most complex plant organ and its development is just beginning to be understood, making fruit development a ripe field for many years to come.
1.1 Wild-type Fruit Structure
The fruit is defined as the mature ovary (and, in some types of fruit, additional floral tissues) that forms a specialized structure designed to protect the seeds while they develop and disperse them at maturity. The fruit develops from the gynoecium after fertilization. The gynoecium is the female reproductive structure including the ovary and is usually formed from one or more fused carpels at the center of the flower. A carpel is a single ovule bearing structural unit of the gynoecium and is thought to have originated from a modified bract or leaf (Bowman et al., 1999).
The Arabidopsis gynoecium is composed of two fused carpels, each of which consists of a seedpod wall and surrounding tissues. The fusion of the carpels is congenital, meaning that the gynoecium arises as a single primordium. The Arabidopsis fruit develops from the fertilized gynoecium to form a silique, or seedpod, which dries and dehisces at maturity, releasing the seeds. The Arabidopsis fruit consists of many distinct cell types, which are derived from the gynoecium. From top to bottom there are four different regions of the gynoecium and subsequently the fruit: the stigma, style, ovary, and gynophore (Figure 1; see Figure 2 for definitions of axes in the fruit and other terms).
Figure 1.
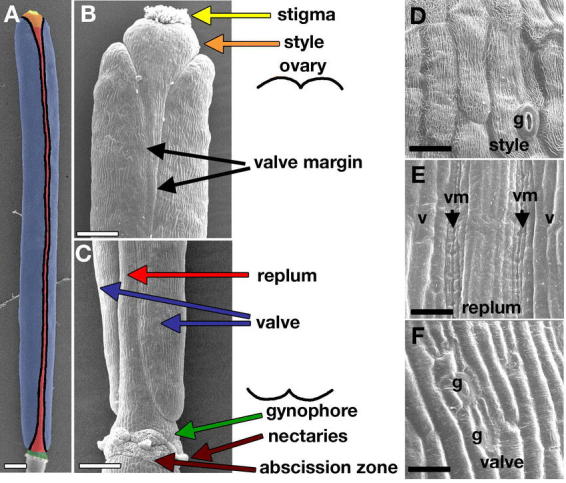
Structure of the wild-type fruit. (A) Scanning electron micrograph (SEM) of a Landsberg erecta (Ler) fruit at stage 17. The fruit has been false colored to distinguish the different parts and this color code has been used throughout the review. At the top of the fruit the stigma is colored yellow and the style is orange. The large central part of the fruit is the ovary, which consists of the valves (blue) or seedpod walls, the replum (red) or central ridge that remains attached to the plant after dehiscence, and the valve margins (black) where the valves connect to the replum. The gynophore (green) is the small internode at the base of the fruit above the floral organ abscission zone and the nectaries. (B) Close up SEM of the top of a Ler fruit at early stage 17 where the different parts of the fruit are indicated with arrows. (C) Close up SEM of the bottom of a Ler fruit at early stage 17 where the different parts are indicated with arrows. (D) SEM showing morphology of the epidermal style cells with the characteristic cuticular ridges and the interspersed stomata (g) at stage 17. (E) SEM showing the epidermis of the valves (v), valve margins (vm), and replum in the middle of the fruit. All of the cells are highly elongated, but the valve margin cells are very narrow, while the replum cells are of moderate width. (F) SEM showing the epidermal valve cells at stage 17. The valve cells are highly elongated and are interspersed with stomata (g). The scale bar in A represents 500 µm, the scale bars in B and C represent 200 µm, and the scale bars in D-F represent 20 µm.
Figure 2.
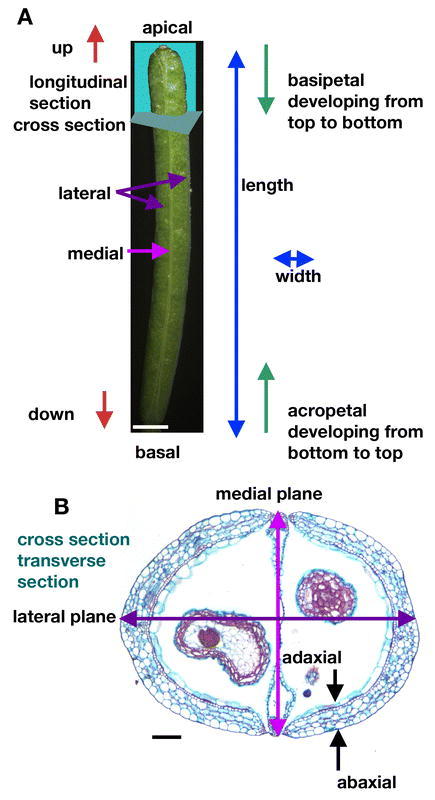
Terminology. (A) Picture of a stage 17 fruit with axes labeled. (B) Cross section of a stage 17 fruit with axes labeled. The scale bar in A represents 1 mm and the scale bar in B represents 100 µm.
Stigma
At the top of the gynoecium, the stigma is comprised of a single layer of elongated papillar cells specialized for the germination of pollen (Figure 1B). The stigma is the first component of the transmitting tract, a set of cells that secrete a polysaccharide-rich extracellular matrix, which-forms a pathway for the growth and guidance of pollen tubes (Sessions and Zambryski, 1995; for reviews of pollen tube guidance see Lord and Russell, 2002 and Palanivelu and Preuss, 2000).
Style
Below the stigma, the style forms a solid cylinder around the central transmitting tract cells that guide the pollen tubes from the stigma to the ovary. In the style, a ring of vascular tissue surrounds the transmitting tract cells. The outer epidermis of the style consists of large rectangular cells with characteristic wax ridges (or crenulations) interspersed with stomata (Figure 1D) (Sessions and Zambryski, 1995).
Ovary
Below the style, the ovary, which houses the developing seeds, forms the majority of the Arabidopsis fruit. The ovary takes the form of a silique with two separate locules or compartments. The ovary consists of several distinct tissues including the valves (seedpod walls), replum (middle ridge), septum, and valve margins.
Valves
The seedpod walls, or valves, lie on the lateral sides of the ovary surrounding and protecting the developing seeds (Figure 3A). The outer epidermal layer of the valves, or exo-carp, consists of long rectangular cells interspersed with stomata (Figure 1F). Inside the exocarp, three layers of thin walled cells containing chloroplasts (chlorenchyma cells) establish the mesocarp (Figure 3). The lateral vascular strands run through the center of the valve and branch to form the secondary vasculature in the valve (Alvarez and Smyth, 2002). Two layers of endocarp cells form on the interior of the valves (Figure 3). The inner epidermis, the endocarpa (ena), consists of enlarged cells (Figure 3 and 8F), which break down later as the fruit matures (Figure 8E). The second endocarp layer, enb (or lignified valve layer), consists of very narrow highly elongated cells, which become lignified late in development (Figure 3 and 8D to 8F). Lignins are phenolic polymers that increase the strength and thickness of the cell walls in which they are deposited. The lignified enb layer is thought to be involved in the spring-loaded shattering mechanism of seed dispersal employed by Arabidopsis and its important crop relative, canola. During dehiscence, the valves separate from the replum and fall from the fruit so that the seeds can be dispersed (Figure 9B and 9G). It has been proposed that as the seedpod dries, the outer layers of thin walled valve cells contract, creating tension against this rigid lignified enb layer which helps to pop the valves off the fruit (Figure 9H; Spence et al., 1996).
Figure 3.
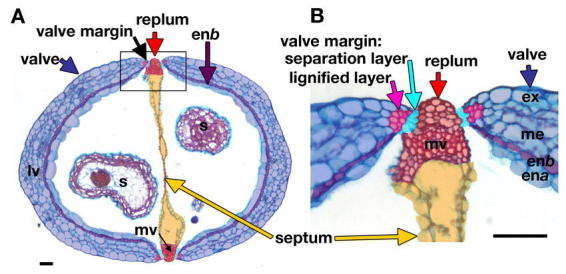
Cross section of a wild-type fruit. (A) Cross section of the ovary of a stage 17 wild-type (Ler) fruit. The fruit has been false colored to distinguish the parts as described in Figure 1. The septum (light orange) divides the fruit into two locules. The valve contains the enb layer (purple), which is lignified. The box encloses the region shown at higher magnification in B. The medial vascular bundle (mv) in the replum and the lateral vascular bundle (lv) in the valve are indicated, as are the developing seeds (s). (B) Close up of a cross section of the replum and valve margin. The valve margin consists of a lignified layer (pink) and the separation layer (aquamarine blue). The valve usually contains 6 cell layers. The outer epidermis is termed the exocarp (ex). The next three cell layers are the mesocarp (me). The inner two cell layers form the endocarp consisting of the lignified enb layer (purple) and the large cells of the ena layer. The scale bars in A and B represent 50 µm.
Figure 8.
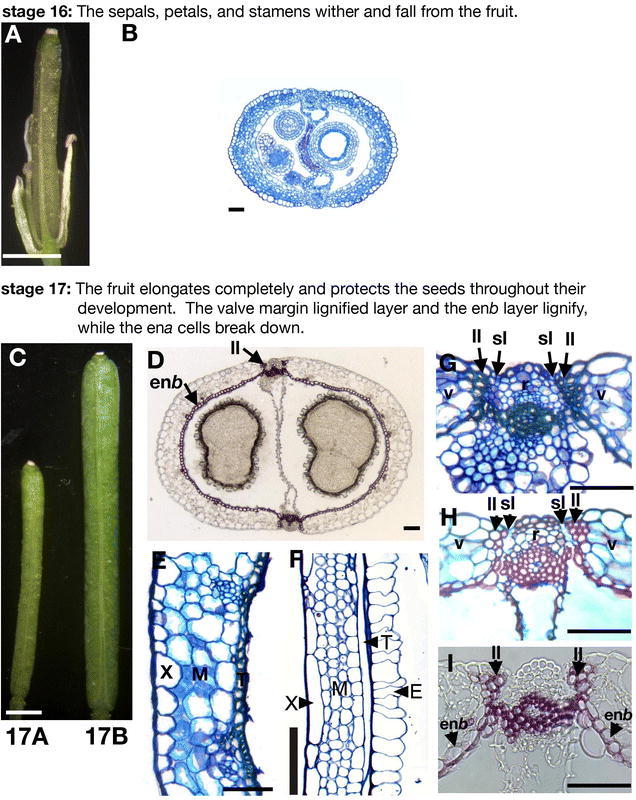
Fruit development: stages 16–17. (A) Stage 16 fruit showing the floral organs withering. The floral organs in front of the fruit have been removed. (B) Cross-section of a stage 16 fruit. (C) Picture of a stage 17A and a stage 17B fruit. (D) Cross-section of a late stage 17 fruit in which lignified cells have been stained with phloroglucinol. The enb layer and the lignified layer (ll) of the valve margin are indicated. (E) Cross-section of the valve wall of a mid stage 17 fruit showing the exocarp (X), mesocarp (M), and enb (T) cell layers. The ena layer has disintegrated. (F) Longitudinal section of the valve wall of a stage 17 fruit 5 days after pollination. The enlarged ena (E) cells are present. (G) Cross-section of the replum region stained with toluidine blue. The valve cells (v), valve margin lignified layer (ll), valve margin separation layer (sl), and replum (r) cells are labeled. (H) Cross-section of the replum region stained with safranin O and alcian blue. The thin walled separation layer (sl) cells stain light blue. (I) Cross-section of the replum region stained with the lignin specific stain phloroglucinol. The lignified layer (ll) and enb layer are stained. The scale bars in A and C represent 1 mm and the scale bars in B, D, E, G–I represent 50 µm. F: from Vivian-Smith and Koltunow, 1999.
Figure 9.
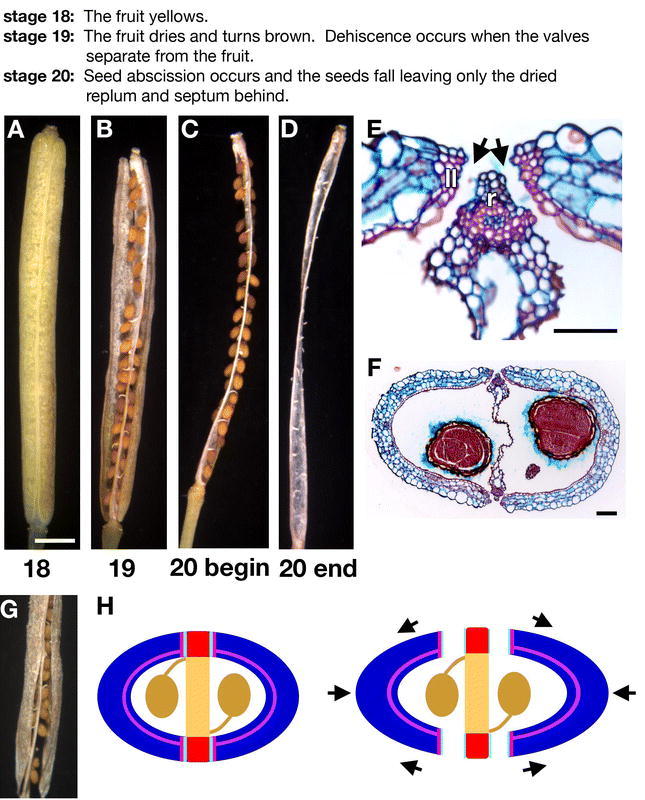
Fruit development: stages 18–20. (A) Picture of a stage 18 fruit. (B) Picture of a stage 19 fruit. (C) Picture of a stage 20 fruit at the beginning before the seeds have fallen. (D) Picture of a stage 20 fruit after the seeds have fallen. (E) Cross-section of the replum region of a stage 18 fruit stained with safranin O and alcian blue. The separation layer cells (arrow) have separated and disintegrated. The replum (r) and the lignified layer (ll) of the valve margin are labeled. (F) Cross-section of a stage 18 fruit stained with safranin O and alcian blue. (G) Close up of the bottom of a stage 19 fruit where the left valve is separating from the valve margin at the replum. (H) Diagram of a cross section of the fruit depicting the tensions that are thought to help pop the valves off the replum. As the outer thin walled valve cells (blue) dry, they are thought to contract creating tension against the rigid enb (purple) and lignified valve margin (pink) layers. The color scheme is valves (blue), replum (red), septum (light orange), seeds (brown), enb layer (purple), valve margin lignified layer (pink), and separation layer (aquamarine blue). Scale bar in A represents 1 mm (for A–D, G). Scale bar in E represents 50 µm and in F represents 100 µm.
Replum and septum
The ovary is divided into halves at the septum and the replum (Figures 1E and 3). The replum was originally defined as the structure that remains attached to the plant after fruit dehiscence, which includes the septum and the abaxial replum (Weberling, 1989). More recently, however, the term replum has come to refer to only the outer or abaxial portion and does not include the septum (Alvarez and Smyth, 2002). Each replum contains one of the medial vascular bundles. The ovules and funiculi arise from the placentae, which lie along each of the inner sides of each replum. The septum divides the fruit, stretching from the inner side of one replum to the other replum. In the middle of the septum, transmitting tract cells connect to the style forming a continuous tract for pollen tube growth.
Valve margins
The valve margins, the zones where the fruit opens, differentiate at the borders between the valves and the replum allowing the valve to separate from the replum in the mature fruit to disperse the seeds (Figure 9B and 9G). The valve margins constrict, forming an indentation, because the cells in the valve margins are smaller than the surrounding cells in the valve and the replum (Figure 1A to 1C, 1E, and 3). Each valve margin consists of two layers-a separation layer and a lignified layer (Figure 3B and Figure 8G to 8I). In the separation layer, or dehiscence zone, hydrolytic enzymes are secreted to break down the middle lamella between adjacent cells, allowing the cells to separate (Figure 9E; Meakin and Roberts, 1990). The lignified layer of the valve margin is thought to act in conjunction with the enb layer of the valve, creating tensions to assist in the seed dispersal process (Figure 9H; Spence et al., 1996).
Gynophore
At the base of the fruit there is a short stalk called the gynophore, which is also known as the internode or stipe (Figure 1A and 1C).
1.2 Wild-type Gynoecium Development
Each of these structures of the fruit is formed in the developing gynoecium during flower development, which is where we will begin with a detailed sequential account. Flower and fruit development have been divided into stages to provide landmarks and a framework within which to discuss the developmental events (Smyth et al., 1990). The development of wild-type Landsberg erecta (Ler) gynoecia is described below.
Stages 1–5
The flower first appears as a small bulge on the side of the shoot apical inflorescence meristem at stage 1 (Figure 4A). At stage 2, the floral primordium enlarges and becomes separated from the inflorescence meristem by a small groove (Figure 4A). The sepal primordia arise on the flanks of the floral meristem at stage 3 (Figure 4A). The sepals grow so that by stage 4 they partially cover the floral meristem (Figure 4A). At stage 5, the petal and stamen primordia are initiated. Late in stage 5, the floral meristem expands laterally to form an elliptical flattened platform where the gynoecium will develop (Figure 4B; Sessions, 1997).
Figure 4.
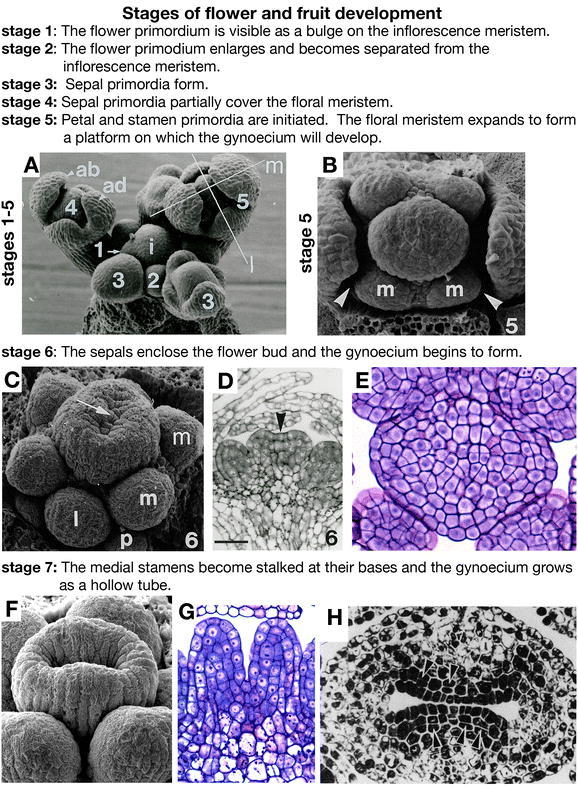
Flower development: stages 1–7. (A) SEM image of the inflorescence meristem (i) and floral stages 1–5. The floral stages are labeled with the corresponding number. The medial (m) and lateral (l) axes are labeled on a stage 5 flower. The abaxial (ab) and adaxial (ad) sides of a stage 4 flower are labeled relative to the inflorescence meristem. (B) SEM of a late stage 5 floral meristem that has formed a flattened oval where the gynoecium will arise. Arrowheads point to the petal primordia and two of the medial stamens are labeled m. (C) SEM of a stage 6 flower showing the beginning of formation of the gynoecium as a ridge of raised cells around a central cleft (arrow). A lateral stamen is labeled l. (D) Longitudinal section of a stage 6 gynoecium. The arrow points to the central cleft. (E) Cross section of a stage 6 gynoecium. (F) SEM of a stage 7 gynoecium showing the vertical growth of the tube. (G) Longitudinal section of a stage 7 gynoecium. (H) Transverse section of a late-stage 7 or early-stage 8 gynoecium. Scale bar in D represents 22 µm. A–D: from Sessions, 1997. E–G: Reprinted from Current Topics in Developmental Biology, 45, Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J., Molecular genetics of gynoecium development in Arabidopsis, 155–205, Copyright (1999), with permission from Elsevier. H: from Hill and Lord, 1989.
Stage 6
When the sepals enclose the bud at stage 6, the gynoecium begins to develop as a raised ridge around a central cleft (Figure 4C to 4E; Hill and Lord, 1989; Sessions, 1997; Smyth et al., 1990). The cleft forms because the cells in the center of the meristem do not divide. The gynoecium consists of two congenitally fused carpels, which arise as a single cylinder because they are joined at the margins in the medial region (Okada et al., 1989). Based on the analysis of sectors, eight progenitor cells in the floral meristem give rise to each carpel (Bossinger and Smyth, 1996). The gynophore is formed from the layers of cells above the stamen primordia and below the tube of the gynoecium (Sessions, 1997). Because the gynoecium arises as a tube, both the exocarp and ena layers are derived from the epidermal layer of the meristem and thus can be described as the outer epidermis and the inner epidermis respectively.
Stage 7
Stage 7 begins when the medial, or long, stamens become stalked at their bases. The gynoecium continues to grow upward to form a continuous hollow cylinder (Figure 4F to 4H). At this stage the inner surfaces of the tube almost touch and the top of the tube is not level (Alvarez and Smyth, 2002).
Stage 8
The gynoecium continues to grow taller and wider during stage 8, which is marked by the formation of locules in the anthers. At the beginning of stage 8, the Ler gynoecium is about 80 µm tall, but by the end it is about 180 µm tall (Figure 5A and 5B; Sessions, 1997). The carpel walls are about 5 to 6 cell layers thick, suggesting that all of the cell layers of the gynoecium are already present (Figure 5B and 5C). The medial vascular bundles begin to develop, but there is no sign of the differentiation of style, replum, or valve cells in the epidermis. The septum is initiated during stage 8 when the inner medial surfaces form ridges. By the end of stage 8, the medial ridges become flanked by placentae (Figure 5C; Sessions, 1997).
Figure 5.
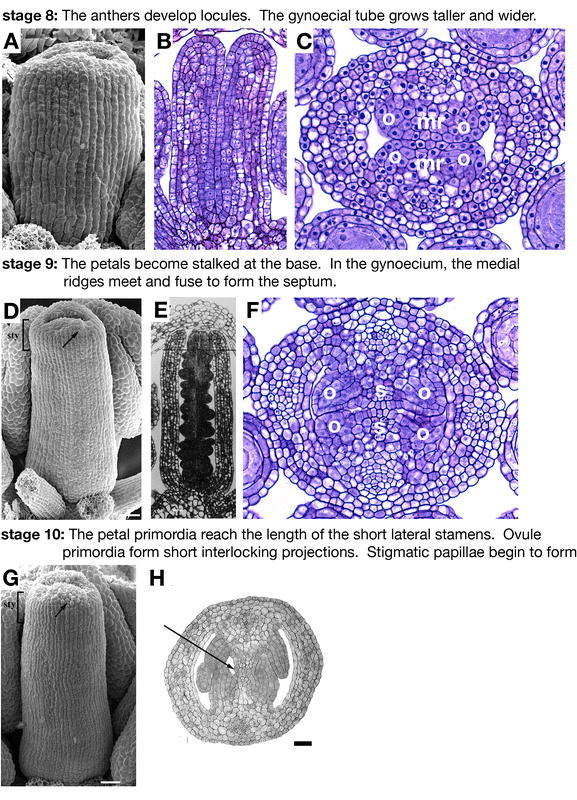
Flower development: stages 8–10. (A) SEM of a stage 8 gynoecium. (B) Longitudinal section of a stage 8 gynoecium. (C) Transverse section of a late-stage 8 gynoecium. Placentas (o) form on both sides of the medial ridges (mr). (D) SEM of a stage 9 gynoecium. The style region (bracketed) can be identified as morphologically distinct from the ovary. A few of the stigmatic papillae become visible as rounded cells on the top of the gynoecium (arrow). (E) Longitudinal section of a stage 9 gynoecium along the lateral axis. (F) Cross section of a stage 9 gynoecium. The three different classes of cells relating to the future exocarp, mesocarp, and endocarp can be distinguished in the valves. The ovule primordia (op) arise from the placentas flanking the medial ridges. The medial ridges meet in the center of the fruit to form the septum (s). (G) SEM of a stage 10 gynoecium. More stigmatic papillae (arrow) are evident at the top of the gynoecium, which has closed over. (H) Cross section of a stage 10 gynoecium. Transmitting tract precursors (arrow), smaller darkly staining cells, are present in the middle of the septum. Scale bars in D and G represent 25 µm. A–C and F: Reprinted from Current Topics in Developmental Biology, 45, Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J., Molecular genetics of gynoecium development in Arabidopsis, 155–205, Copyright (1999), with permission from Elsevier. D, G, and H: from Alvarez et al., 2002. E: from Sessions 1997.
Stage 9
Stage 9 is marked by the petal primordia becoming stalked at the base. The gynoecium continues to grow and by the end of stage 9, the apex starts to close (Figure 5D and 5E). Late in stage 9 the appearance of the first rounded cells at the top of the gynoecium suggest the development of the stigmatic papillae (Alvarez and Smyth, 2002). The style begins to become morphologically distinct from the ovary and the lateral vascular bundles start to differentiate (Ferrándiz et al., 1999). The septum forms when the leading edges of each medial ridge meet and fuse. The placenta produces ovule primordia (Figure 5F; Robinson-Beers et al., 1992).
Stage 10
Stage 10 begins when the petal primordia reach the length of the short stamens (Smyth et al., 1990). The upper end of the gynoecium closes and begins to produce immature stigmatic papillae (Figure 5G). The gynoecium continues to grow and reaches about 400 µm (Sessions, 1997). The ovule primordia grow to form finger-like projections, which mesh with those from the opposing placenta in an interlocking pattern (Figure 6C*; Robinson-Beers et al., 1992). The septa continue to grow out from the middle of the medial ridge and the cells at the middle begin to stain more darkly, suggesting that they are starting to differentiate as transmitting tract cells (Figure 5H; Alvarez and Smyth, 2002).
Figure 6.
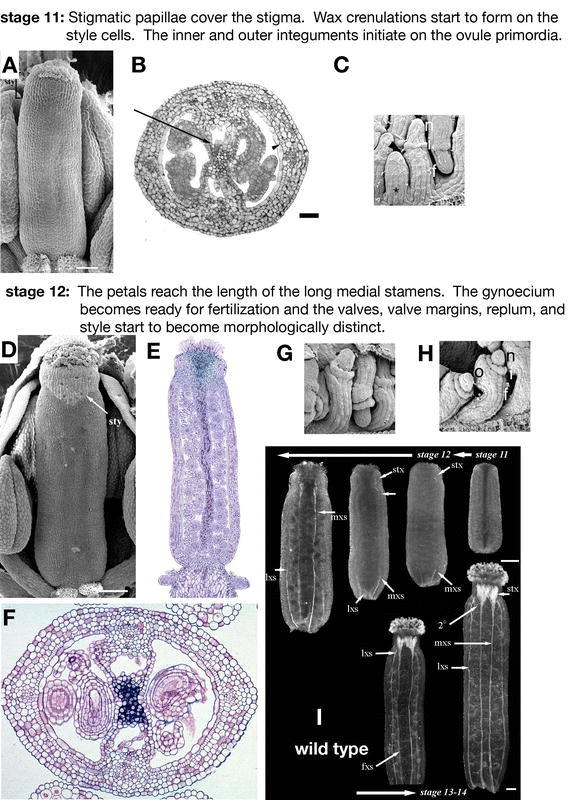
Flower development: stages 11–12. (A) SEM of a stage 11 gynoecium in which the stigma is covered with many papillae. (B) Cross-section of a stage 11 gynoecium. In the septum (arrow), small darkly staining cells near the middle of the gynoecium will form the transmitting tract. At the edges of the septum, large air filled pockets form with only a few loosely spaced cells. The cells in the enb layer have started dividing anticlinally to form long narrow cells (arrow head). (C) SEM of the ovule primordia in an early stage 11 gynoecium. The valve has been removed to expose the ovule primordia in one side of the gynoecium. The younger ovule primordium (asterisk) that forms a finger-like projection is typical of stage 10 while the slightly older ovule in which the inner integument (i) and outer integument (arrowhead) are initiating between the nucellus (n), and funiculus (f) is characteristic of the beginning of stage 11. (D) SEM of a mid stage 12 gynoecium where the stylar epidermal cells are clearly distinct from the rest of the gynoecium. (E) Longitudinal section of a stage 12 gynoecium. Note that the transmitting tract stains darkly. (F) Cross-section of a stage 12 gynoecium. The transmitting tract in the middle of the septum stains darkly. (G) SEM of the developing ovules early in stage 12, which have initiated inner and outer integuments. (H) SEM of the ovules inside the gynoecium midway through stage 12. The integuments have grown and start to cover the nucellus. (I) Cleared whole mount gynoecia viewed under dark field optics to visualize the thickening of the xylem cell walls. The progression of stages starts from the upper right, continues to the left, and curves around in the second row from left to right. At stage 11, no thickening of the xylem cell walls is detected. Early in stage 12, thickening begins at the base of the fruit in the medial xylem strands (mxs) and progresses apically. Then thickening begins in the stylar xylem (stx) and progresses toward the base in the medial vascular bundle. Later in stage 12, thickening of the lateral xylem strands (lxs) begins at the base of the fruit and develops toward the apex. Throughout stage 12 more xylem elements in the style differentiate. Additionally, a single strand of xylem differentiates in the funiculus (fxs). During stages 13 and 14, secondary (2°) vascular tissue forms in the valves branching from the lateral vascular bundle and differentiating toward the base of the gynoecium. Scale bars in A represent 50 µm, B 25 µm, and D and I 100 µm. A, B, D, and I: from Alvarez et al., 2002. C, G, and H: from Meister et al., 2002. E and F: Reprinted from Current Topics in Developmental Biology, 45, Bowman, J.L., Baum, S.F., Eshed, Y., Putterill, J., and Alvarez, J., Molecular genetics of gynoecium development in Arabidopsis, 155–205, Copyright (1999), with permission from Elsevier.
Stage 11
The stigmatic papillae cover the entire surface of the stigma during stage 11 (Figure 6A). The gynoecium reaches a length of about 600 µm (Sessions, 1997). The external surface of the style, characterized by its wax crenulations, begins to develop and continues to differentiate through stage 12 (Alvarez and Smyth, 2002). Cells in the enb layer begin to divide anticlinally to produce very narrow cells (Figure 6B). Two regions form in the septum: a pre-transmitting tract region with small cells that stain darkly lies in the center (first visible at stage 10) and a pocket with a few loosely spaced cells and ample air space forms closer to each edge of the fruit (Alvarez and Smyth, 2002). In the ovules, the funiculus, inner integument, and outer integument all start to differentiate (Figure 6C; Robinson-Beers et al., 1992).
Stage 12
At the beginning of stage 12, the petals reach the length of the long medial stamens. The gynoecium continues to grow and reaches about 800 µm (Figure 6D and 6E). Differentiation of the tissues starts to become apparent as the valves expand laterally to become clearly distinct from the narrow apical style. The valve margins and replum also become morphologically distinct. The gynoecium becomes ready for pollination when the stigmatic papillae elongate and fully differentiate. The transmitting tract simultaneously differentiates to guide the pollen tubes to the ovules (Figure 6E and 6F; Bowman et al., 1999). The integuments of the ovules grow and almost cover the nucellus (Figure 6G and 6H; Robinson-Beers et al., 1992). The secondary walls of the xylem thicken during stage 12 (Figure 6I; Alvarez and Smyth, 2002). Thickening of the medial vascular bundle cell walls begins at the base of the fruit and extends upward. Slightly later, cell wall thickening begins in the style and continues downward to meet with the upward-growing medial vasculature in the middle of the fruit. The lateral vascular bundle cell walls begin to thicken later than the medial bundles and the lateral vascular bundle cell walls thicken entirely from the bottom up. More xylem elements mature in the style and connect to the medial vascular bundles. A single strand of xylem in the funiculus thickens but does not connect to the medial xylem until stage 13 or 14.
Stage 13
Stage 13 is defined by anthesis, which is when the flower opens and self-pollinates (Figure 7A). After the pollen lands on the stigma and germinates, the pollen tube grows down the papilla cells between the inner and outer layers of the cell walls. The pollen tube takes 45 to 50 minutes to reach the extracellular matrix of the transmitting tract, where it grows between the transmitting tract cells until it reaches an ovule during stage 14 (Figure 7H; Kandasamy et al., 1994). By stage 13, the ovule is completely developed and ready for the pollen tube to reach it (Figure 7D; Robinson-Beers et al., 1992). The cells of the valve (the exocarp, mesocarp, endocarpb, and endocarpa) are small relative to the dramatic expansion they will undergo after fertilization as the fruit elongates to accommodate the developing seeds (Figure 7B and 7C, compare to 8F). Second order xylem strands begin to differentiate in the valves originating from the top of the lateral xylem (Figure 6I; Alvarez and Smyth, 2002).
Figure 7.

Gynoecium development: stages 13–15. (A) SEM of a stage 13 gynoecium showing the pollination of the stigma (stg). The style (sty), valves (va), replum (rep), and gynophore (gyn) are all clearly distinguishable by this stage. (B) Transverse section of a stage 13 gynoecium showing all the parts of the mature gynoecium: replum (rp), medial vascular bundle (mv), septum (sp), exocarp (ex), mesocarp (ms), transmitting tract (tt), ovule (o), lateral vascular bundle (lv), and locule (lc). (C) Longitudinal section through a stage 13 valve showing differentiation of the exocarp (X), enb (T), and ena (E). (D) SEM of stage 13 ovules, which finish development and become ready for fertilization. (E) SEM of a stage 14 flower where the medial stamens extend above the top of the stigma (reproduced with permission from Nature Publishing Group (http://www.nature.com/). (F) Cross-section of a stage 14 gynoecium. The replum (r), valve (v), valve margins (arrowheads vm), mesocarp (me), endocarpa (ena), and endocarpb (enb) are labeled. (G) SEM of a stage 14 ovule. (H) Paths of the pollen tubes growing through the stigma and transmitting tract to fertilize the ovules viewed with aniline blue fluorescence in a cleared stage 14 gynoecium. (I) Picture of a stage 15 fruit, which has extended above the top of the anthers. The floral organs in front of the fruit have been removed. (J) Cross section of a stage 15 gynoecium. Scale bars in A and C represent 100 µm, in B, F, and J represent 50 µm, and in E and I represent 500 µm. A and H: from Alvarez et al., 2002. B: from Ferrándiz et al., 1999. C: from Vivian-Smith and Koltunow, 1999. D and G: from Meister et al., 2002. E: from Liljegren et al., 2000.Note:Section E is reproduced with permission from Nature Publishing Group (http://www.nature.com/).
1.3 Wild-type Fruit Development
Fruit development is defined to begin after fertilization of the egg cell and endosperm during stage 14.
Stage 14
At stage 14, the anthers extend above the top of the stigma (Figure 7E; Smyth et al., 1990). The exocarp cells of the valves and the replum divide and expand primarily along the longitudinal axis, contributing to the lengthening of the fruit. The mesocarp cells divide and elongate primarily longitudinally. Many chloroplasts develop in the mesocarp cells of the valves, whereas the mesocarp cells in the valve margin do not develop chloroplasts and do not divide or expand as much as their neighbors. This causes the valve margins to constrict, forming a small groove along the edge of the replum (Figure 7E and 7F). The enb cells become long and narrow as they expand longitudinally (Spence et al., 1996). The ena layer appears swollen as the cells expand in all directions (Figure 7F). As in the mesocarp, endocarp cells in the valve margins do not expand as much as their neighbors. The pollen tubes grow through the transmitting tract and along the funiculus to enter each ovule and fertilize the egg cell and endosperm (Figure 7G and 7H; Robinson-Beers et al., 1992). Second order vascular strands continue to develop in the valves and the vascular strand in each funiculus connects to the medial vasculature (Figure 6I; Alvarez and Smyth, 2002).
Stage 15
By the beginning of stage 15, the gynoecium has elongated such that the stigma extends above the top of the anthers (Figure 7I). The fruit continues to grow and the valve cells divide and expand (Figure 7J). Second order vasculature continues to mature in the valves in a reticulate pattern and has completed development by the end of stage 15 (Alvarez and Smyth, 2002).
Stage 16
The sepals, petals, and stamens start to wither during stage 16 (Smyth et al., 1990), and these organs fall from the fruit by the end of the stage. Concurrently, the fruit continues to elongate through cell division and cell expansion (Figure 8A and 8B).
Stage 17
Stage 17 is a long stage, which starts when the floral organs fall from the fruit and ends when the fruit start to yellow. Stage 17 has been divided into two sub-stages, 17A and 17B (Figure 8C). In stage 17A the fruit elongates and increases in width to its mature form. From anthesis onward, growth in width of the fruit is almost entirely due to cell expansion in all of the cell layers of the valves (Vivian-Smith and Koltunow, 1999). In contrast, the total elongation of the fruit is achieved through both cell expansion and cell division (Figure 8F, compare with Figure 7C). Cell division is especially prevalent in the mesocarp where the number of cells increases about threefold, while cell expansion is more pronounced in both the exocarp and ena (Vivian-Smith and Koltunow, 1999). Cell expansion is limited at the valve margin where cells remain relatively small, forming an indentation between the valve and the replum (Figure 8G).
Stage 17B starts when the fruit reaches its full length and ends when the fruit yellows before it dries. The valve margin begins to differentiate into the dehiscence zone, preparing the fruit to open at maturity. The cell walls of the valve margin lignified layer start to thicken and later lignify (Figure 8I). These cells form a connection with the enb layer of the valve, which also undergoes lignification (Figure 8D; Spence et al., 1996). The middle lamella between the cells in the valve margin separation layer breaks down (Figure 8H). In addition, the cells of the ena layer and the cells on the epidermis of the septum break down, leaving behind their thickened inner cell wall (Figure 8E; Spence et al., 1996).
Stage 18
Stage 18 begins about 12 days after pollination when the siliques turn yellow from top to bottom and start to dry (Figure 9A; Vivian-Smith and Koltunow, 1999). The separation layer cells disintegrate, leaving the valves free to detach from the replum at stage 19 (Figure 9E and 9F).
Stages 19 and 20
The valves separate from the replum and fall from the siliques during stage 19 (Figure 9B and 9G) and the seeds fall from the replum in stage 20 (Figure 9C and 9D; Smyth et al., 1990).
2.0 SPECIFICATION OF CARPEL IDENTITY
Since the fruit develops from the gynoecium, much of the fruit structure is already determined before fertilization. Therefore, understanding fruit development is impossible without examining the patterning of the gynoecium. The first question is how the organs in the center of the flower are specified to form two fused carpels rather than sepals, petals, stamens, or even leaves.
2.1 AGAMOUS (AG) Specifies Carpel Identity
There is perhaps no gene with a more pivotal role in plant sexual reproduction than AGAMOUS (AG). Reproductive organs are completely lacking in ag single mutants, which consequently never produce fruit (Figure 10A and 10B; Bowman et al., 1989). The AG gene is necessary for determining stamen and carpel identity and provides the C function of the ABC model of floral organ identity (Coen and Meyerowitz, 1991; Bowman et al., 1991a). Close inspection of ag flowers reveals that while sepals and petals in whorls one and two develop normally, petals replace whorl three stamens. In addition, carpels, which normally occupy the central fourth whorl, are replaced with a new flower. Consequently, ag flowers are considered to have a “double flower” or “flower within a flower” phenotype and display the overall pattern of (sepal, petal, petal)n. Thus, in addition to its organ identity function, AG is needed to prevent the indeterminate growth of the floral meristem.
Figure 10.
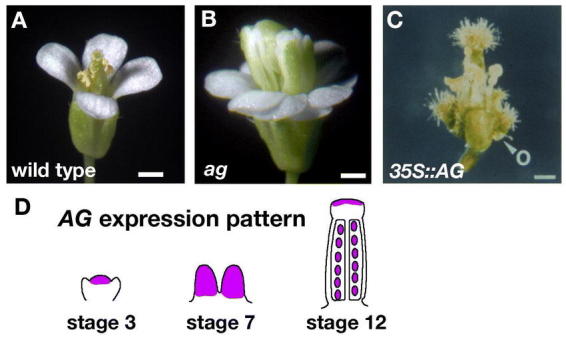
AGAMOUS specifies carpel development. (A) Wild-type flower. There are four whorls of floral organs: sepals, petals, stamens, and carpels. (B) ag flower. The ag flower consists of sepals, petals, petals, and new flower repeating this pattern. (C) 35S::AG flower. The sepals are converted to carpels and the petals are missing or stamenoid. The first whorl carpels produce ovules (O). (D) Diagram representing the expression pattern of AG. AG is expressed in the inner two whorls of the floral meristem starting at stage 3. AG continues to be expressed throughout the gynoecium through stage 7. Later, AG expression is limited to the developing ovules and the stigma. Scale bars in A–C represent 0.5 mm. C: Reprinted from Cell, 71, Mizukami, Y., and Ma, H., Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity, 119–131, Copyright (1992), with permission from Elsevier.
AGAMOUS encodes a MADS domain transcription factor
AGAMOUS was the first MADS-box gene cloned from Arabidopsis, and since that time dozens of related genes have been isolated and characterized (Yanofsky et al., 1990). Plant MADS-box genes typically have a canonical MIKC structure, where the M refers to the highly conserved DNA-binding MADS-domain at or near the amino terminus of the protein. The I refers to an intervening region that helps give each MADS protein its specificity and may contribute to dimer formation. The K-domain, which derives its name from the weak sequence similarity with keratin, likely forms a coiled coil domain that is important in protein-protein interactions. The C-terminus of plant MADS proteins is referred to as the C-domain, and although its precise functions have yet to be fully explored, it appears to play roles in protein-protein interactions as well as in transcriptional activation (for reviews of plant MADS-box genes see Reichmann and Meyerowitz, 1997 and Theissen et al., 2000).
Consistent with its role in stamen and carpel identity, the flower-specific AG RNA is first detected in the central domain of stage 3 flowers in cells that will later give rise to stamens and carpels (Figure 10D; Drews et al., 1991). AG RNA accumulates uniformly throughout stamen and carpel primordia during stages 4 through 7 (Figure 10D), but in more mature flowers it becomes localized to specific cell types within these organs (Bowman et al., 1991b). During stage 9, only low a level of AG RNA is detected in the valves of developing carpels, whereas a high level of AG RNA accumulates in ovule primordia. During stage 12, AG RNA is detected in all cells of developing ovules, in the septum, and in the stigmatic papillae, and no expression can be detected in the style and valves (Figure 10D). The specific localization of AG to these additional tissues within the carpels suggests that AG might have additional roles later in gynoecium development, and in fact AG is involved in specifying ovule identity (Pinyopich et al., 2003).
Ectopic expression of AG promotes carpel identity
Loss-of-function studies have shown that AG is required for stamen and carpel development. To determine if AG is sufficient to specify reproductive organ development in ectopic positions, gain-of-function transgenic plants were generated that ectopically expressed the AG gene from the constitutive CaMV 35S promoter (Mizukami and Ma, 1992; Mandel et al., 1992). 35S::AG plants produced flowers in which the first whorl sepals were converted into carpelloid organs and the second whorl petals were converted into staminoid organs (Figure 10C). Thus, within the context of the flower, ectopic AG expression is sufficient to promote reproductive organ development. It is also important to note that ectopic AG expression was unable to convert vegetative leaves into carpelloid organs, suggesting that additional factors are required. These factors include the SEPALLATA (SEP) genes, which are required for AG function (Honma and Goto, 2001; Pelaz et al., 2001). The SEP1,2,3 genes are also required for gynoecium formation and in the sep1 sep2 sep3 triple-mutant no gynoecium is formed (Pelaz et al., 2000).
2.2 Carpel Development in the Absence of AGAMOUS
The ABC model of flower organ identity predicts that the activities of the A-function genes, which are normally restricted to the outer two whorls, expand to include all floral whorls in ag-mutant flowers. This raises an interesting question regarding AG activity: is AG absolutely required for carpel development, as the ag single-mutant phenotype might suggest, or is there an AG-independent carpel development pathway that remains hidden in ag mutants because the ectopic expression of A function genes changes the fate of these organs?
In A function mutants such as apetala2 (ap2), carpels develop in both the first and fourth whorls (Figure 11A; Bowman et al., 1991b). Because AG is ectopically expressed in the first whorl carpels of ap2 mutants (Drews et al., 1991), the role of AG in carpel development can be examined in the first whorl organs of the ag ap2 double mutant.
Figure 11.
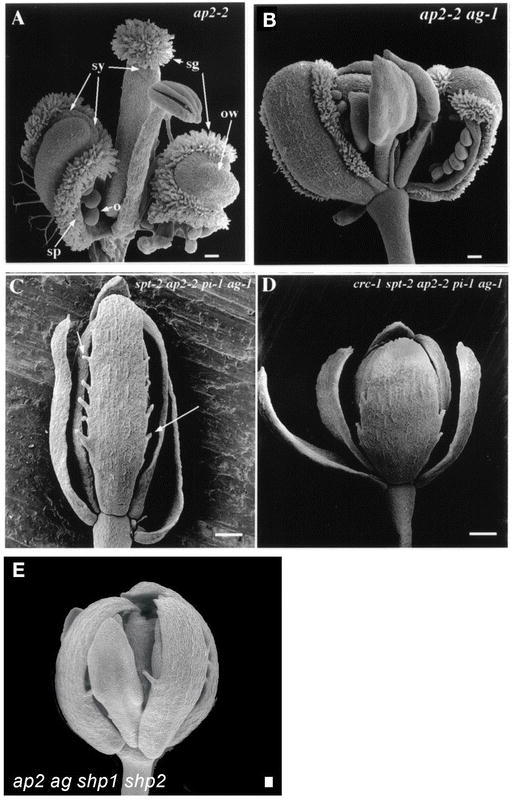
Carpel development can occur independently of AG. (A) ap2-2-mutant flower. The first whorl carpels have all of the characteristics of carpels: ovules (o), stigmatic tissue (sg), style tissue (sy), valve cells (ow), and septum pri-mordia (sp). (B) The ap2-2 ag-1 first whorl carpels still retain many carpel specific tissues despite the absence of AG. Ovules and stigmatic tissue are clearly present, but the valve cells are absent. (C) The spt-2 ap2-2 pi-1 ag-1 quadruple-mutant flower shows a dramatic reduction in carpelloid properties of the outer whorl carpels. The stigma and style cells are completely absent. The only carpelloid tissue remaining is the ovule projections from the margins (arrows). The pi-1 mutation was included to remove the stamenoid organs still present in ap2-2 ag-1 mutants and does not affect the carpels of the first whorl. (D) The crc-1 spt-2 ap2-2 pi-1 ag-1 quintuple-mutant flower. Removal of CRC removes most of the rest of the carpel-loid properties of the outer whorl organs. (E) The ag ap2 shp1 shp2 quadruple-mutant flower. SHP acts partially redundantly with AG in the specification of carpels in the first whorl of ap2 mutants. Scale bars in A and B are 100 µm, C and D are 250 um, and E is 50 µm. A–D: from Alvarez and Smyth, 1999. E: From Pinyopich et al., 2003. Reproduced with permission from Nature Publishing Group (http://www.nature.com/).
Surprisingly, many features of carpels (stigmatic tissue, marginal cell types, and ovules) can still be observed in the first-whorl organs of ag ap2 double mutants (Figure 11B; Bowman et al., 1991b; Alvarez and Smyth, 1999). These studies indicate that an AG-independent pathway can promote development of most carpel tissues and that this second pathway is negatively regulated by AP2 activity. In fact, the only features of carpels that are entirely lacking in ag ap2 double mutants are the specialized cells of valves including the enb layer, indicating that these cell types are AG-dependent.
Multiple factors promote carpel development in the absence of AG activity. The SHATTERPROOF (SHP) MADS-box genes, which are closely related to AG, are largely responsible for the carpelloid features seen in ag ap2 double mutants (Pinyopich et al. 2003; see also section 5.1.1 SHATTERPROOF (SHP) specifies valve margin identity). In the ag ap2 shp1 shp2 quadruple mutant, the first whorl organs are completely devoid of carpelloid features (Figure 11E). Conversely, expression of SHP2 throughout the flower can rescue carpel and stamen development in ag mutants. Therefore, AG and SHP proteins can act almost interchangeably in promoting carpel development. Their distinct roles seem to be the result of their unique expression patterns rather than a significant difference in the protein structures (Pinyopich et al. 2003).
Another factor involved in the AG-independent carpel development pathway is SPATULA (SPT), which also has a role in the development of transmitting tract and medial tissues (see section 5.6.5 SPATULA (SPT) is required for transmitting tract development). Remarkably, nearly all carpelloid features, including stigmatic papillae, style tissue, septum, and transmitting tract are absent from the ap2 ag spt triple mutant (Figure 11C; Alvarez and Smyth, 1999). These results indicate that SPT is largely responsible for promoting carpel development in the ag ap2 double mutant. SPT plays a role in promoting development of most of the different tissues of carpels and this role is redundant with AG.
The observation that ovule-like carpelloid outgrowths are still present in spt ag ap2 triple mutants indicates that additional carpel-promoting factors exist. The CRABS CLAW (CRC) gene encodes one such factor (see section 5.6.6 Fruit shaped like a CRABS CLAW (CRC)). The further reduction of carpelloid outgrowths in crc spt ag ap2 quadruple mutants demonstrates that CRC is one of the factors promoting carpel development in spt ag ap2 triple mutants (Figure 11D; Alvarez and Smyth, 1999). CRC and SPT may act downstream of SHP in promoting the AG-independent carpel development pathway.
3.0 CONTROL OF THE NUMBER OF CARPELS
While carpel identity is specified by AG and associated factors, these factors do not dictate how many carpels are produced. What determines the number of carpels in the gynoecium? The wild-type gynoecium consists of two congenitally fused carpels, each containing one valve flanked with valve margin, replum and septum tissues. Several mutants have been identified that produce either too many or too few carpels. Analysis of these mutants suggests that the number of carpels is determined by the amount of tissue available in the center of the flower, which is influenced both by the size of the floral meristem and the amount of tissue allocated to each of the floral whorls.
Mutants affecting the size of the floral meristem influence the number of carpels in the gynoecium. For example, in the clavata (clv) mutants, the size of the floral meristem is increased and consequently more carpels are formed. Strong alleles of clv1 and clv3 often have 3 to 8 carpels (Figure 12A and 12B; Clark et al., 1995). The name clavata comes from the club-shaped siliques formed by the fusion of numerous carpels. Although the increase in the carpel number is not as extreme in clv2 mutants, the gynoecia often have additional phenotypes including elongated gynophores and reduced valves, suggesting that CLV2 has additional roles in fruit development (Figure 12C; Kayes and Clark, 1998).
Figure 12.
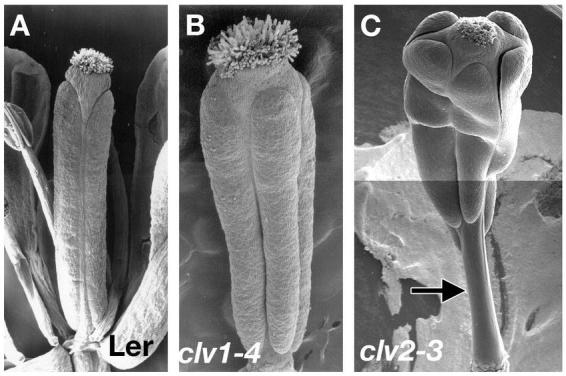
Control of carpel number by CLAVATA genes. (A) Ler stage 15 fruit with two carpels. (B) clv1 fruit with 4 carpels at stage 15. (C) clv2-3 fruit with 5 carpels and the characteristic elongated gynophore. A–B: from Ferrándiz et al., 1999. C: from Kayes and Clark, 1998.
Other genes that modify meristem size can affect the number of carpels. SHOOT MERISTEMLESS (STM) encodes a homeodomain protein required for the establishment and maintenance of meristems. Plants heterozygous for stm can partially suppress the increased carpel number phenotype of clv3 mutants (Clark et al., 1996). Likewise, POLTERGEIST (POL) can suppress the clv phenotype returning the meristem size to normal and reducing the number of extra carpels (Yu et al., 2000).
The number of carpels can also be affected by changing the proportion of the floral meristem allotted to carpel development. For example, in superman (sup) mutants, the number of stamens is increased at the expense of the carpels. In sup mutants the gynoecium contains 0 to 2 carpels, which are often mosaics of carpel and stamen tissue (Bowman et al., 1992). SUP encodes a putative transcription factor with a zinc finger and a basic leucine zipper motif (Sakai et al., 1995). SUP is expressed at the boundary between the third and fourth whorls and limits the expression of the B class floral homeotic genes to the third whorl. Thus, in sup mutants, the third whorl encroaches on the fourth whorl snatching more than its fair share of the floral meristem.
4.0 CONTROL OF FRUIT SHAPE BY ERECTA (ER)
While the number of carpels dramatically affects the overall shape of the fruit, the shape of each carpel also contributes to the fruit structure. One of the most well known modifications of fruit shape is caused by the erecta (er) mutation, which is found in the commonly used Landsberg erecta (Ler) background. In fact, many of the other fruit mutations described here have been isolated in the presence of the er mutation and the er fruit is often considered the “wild type.” For example, Ler fruit have been described in the section on wild-type development (above). The er fruit are blunt as well as shorter and broader than wild type (e.g. Columbia which does not harbor the er mutation; Figure 13A to 13C; Torii et al., 1996). The pedicels on er plants are shorter, the inflorescences are more compact, and the stems are both thicker and shorter causing the plants to stand upright. It has been hypothesized that ER acts to coordinate organ initiation with cell division and expansion.
Figure 13.
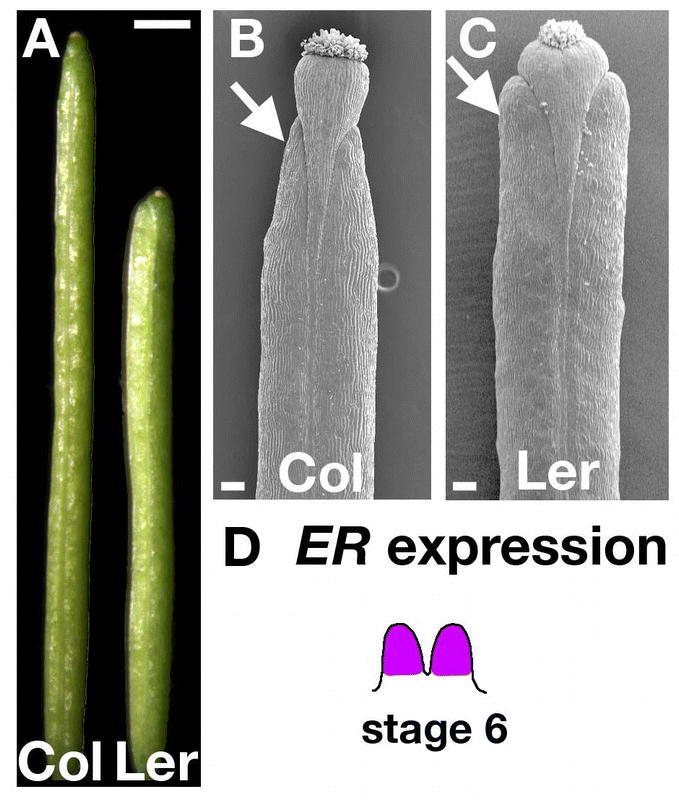
ERECTA controls carpel shape. (A) Wild-type Columbia (Col) fruit not carrying the er mutation. Note that the fruit is longer and narrower. Landsberg erecta (Ler) fruit carrying the er mutation. Note that the fruit is shorter and broader. (B) SEM of the apex of a Col fruit at stage 17 showing that the valves are tapered (arrow). (C) SEM of the apex of a Ler fruit at stage 17 showing the blunted end of the valves (arrow). (D) ER is expressed throughout the gynoecium at stage 6. Scale bar in A represents 1 mm. Scale bars in B–C represent 100 µm.
ERECTA encodes a leucine rich repeat receptor-like kinase
ERECTA encodes a leucine rich repeat receptor protein kinase (Torii et al., 1996). Two hydrophobic domains in the ER protein are thought to form a signal peptide and a transmembrane domain. The C-terminal intracellular domain encodes the catalytic domain of a putative serine threonine kinase, while the extracellular domain contains twenty leucine-rich repeats (LRRs). ER is expressed strongly in the shoot apical meristem (Yokoyama et al., 1998). In addition, ER is expressed throughout the floral meristem from stages 1–3. In stages 4–6, expression is limited to the developing stamens and carpels (Figure 13D). By stage 8, expression is reduced throughout the flower. It is likely that ER acts to receive extracellular signals to promote cell division and expansion and thus controls the fruit shape.
5.0 GENES PATTERNING THE FRUIT
So far we have discussed the genes that are broadly involved in setting up the overall structure of the gynoecium through the specification of carpel identity, controlling the number of carpels, and shaping the carpels. However this is just the beginning of our analysis of fruit development. The gynoecium is a complex structure requiring the action of many additional genes to control the development of the many distinct tissues and cell types within the carpels. The correct development of these tissues is essential for fertilization, protection of the developing seeds, and dispersal of the mature seeds. Specification of carpel identity launches a program of gynoecium development, which induces the transcription of genes involved in the specification of each of the cell types of the fruit. Stefan de Folter and colleagues have examined the expression profiles of 1100 transcription factors during fruit development and found that most of these genes are expressed either in the fruit or in the seed (2004). Now we will examine each tissue of the fruit in turn focusing on the genes that control the development of these specific cell types.
5.1 Valve Margin Identity Genes
We will begin our examination of the specific cell types in the fruit with the valve margin because the development of the valve margin is crucial for seed dispersal and is therefore carefully regulated by a multitude of genes (for a review see Ferrándiz, 2002). Dehiscence, or fruit opening, occurs at the valve margins where the valves separate from the replum (Figure 9H). The valve margin consists of the separation layer and the lignified layer. The separation layer contains thin-walled cells that secrete hydrolytic enzymes to break down the middle lamella, allowing the cells to physically separate (Figure 8H and Figure 9E). The lignified layer of the valve margin acts in conjunction with the lignified enb layer of the valve to create tension, which causes the valves to pop off (Figure 8D and I and Figure 9H). Several transcription factors have been identified that are involved in the specification of valve margin identity.
5.1.1 SHATTERPROOF (SHP) Specifies Valve Margin Identity
The SHATTERPROOF genes (SHP1 and SHP2) are redundantly required for the differentiation of both the lignified layer and separation layer of the valve margin (Liljegren et al., 2000). The shp1 shp2 double-mutant fruit fails to dehisce when it matures and consequently traps the seeds inside, preventing their dispersal. In contrast to wild-type fruit, which open easily upon contact, shp1 shp2 valves remain firmly attached and cannot be removed when rubbed between fingers. In fact, when force is applied, the shp-mutant pods often tear across the valves instead of splitting at the valve margins. The defect in shp fruit is first observed after fertilization when the constriction in the valve margin is reduced, particularly near the base of the fruit. The absence of a defined valve margin becomes increasingly prominent as the fruit matures (Figure 14A and 14E). Furthermore, molecular markers for the valve margin are absent in shp fruit and lignification of the valve margin is reduced in shp fruit (Figure 14B and 14F).
Figure 14.

Indehiscent fruit result from a failure of valve margin development. (A) SEM of the apex of a wild-type stage 17 fruit with a distinct valve margin (arrowhead). (B) Cross-section of the wild-type replum region stained with lignin specific stain phloroglucinol. The lignified cells are present in the lignified layer of the valve margin (arrowhead). (C) In alc fruit the valve margin is also distinct despite the failure of the fruit to dehisce. (D) The lignified layers of the valve margins are present in alc fruit. In alc mutants the development of the separation layer is affected and a lignified bridge (lb) connects the vasculature of the replum to the lignified layer and the enb layer blocking the dehiscence of the fruit. (E) In the apex of shp1 shp2 fruit, the valve margin is still distinct whereas in the base of shp1 shp2 fruit, the valve margin is not distinct (not shown). (F) The valve margin lignified layer fails to differentiate near the base of shp1 shp2 fruit. However lignified layer cells can be detected near the apex of the fruit (not shown). (G) In ind-mutant fruit the valve margin is not distinct in the apex and the base. (H) The lignified layer fails to differentiate throughout ind fruit. (I) The ind alc double-mutant fruit have a phenotype very similar to ind single-mutant fruit. (J) The ind alc lignified layer is absent similar to ind single mutants. (K) Valve margin definition is further reduced in ind shp1 shp2 triple-mutant fruit indicating that SHP1 and SHP2 have roles in valve margin development that are separate from those of IND. (L) The valve margin lignified layer is absent in ind shp1 shp2 fruit. Furthermore, the lignified enb layer, which normally abuts the valve margin (see B), retreats a few cells away from the replum (bracket). (M) Valve margin development is further reduced in ind alc shp1 shp2 quadruple-mutant fruit. This suggests that ALC has some roles in valve margin development that are independent of IND and of SHP. (N) The enb layer retreats several cells into the valve of ind alc shp1 shp2 fruit. (See also Figure 21). (O) Ectopic expression of FUL under the 35S promoter in 35S::FUL fruit prevents differentiation of the valve margin and the entire circumference of the fruit appears to be covered with valve cells. (P) 35S::FUL blocks the formation of the valve margin lignified layer. The outer cells of the replum region appear to have differentiated as valve cells. However, the inner part of the replum including the vascular bundle is present. Scale bars in SEMs are 100 µm and scale bars in sections are 50 µm. A, C, E, G, I, K, and M: Reprinted from Cell, 116, Liljegren, S.J., Roeder, A.H.K., Kempin, S.A., Gremski, K., Østergaard, L., Guimil, S., Reyes, K.D., and Yanofsky, M.F., Control of fruit patterning in Arabidopsis by INDEHISCENT, 843–853, Copyright (2004), with permission from Elsevier. O and P: from Ferrándiz et al., 2000b.
The SHATTERPROOF genes encode redundant MADS box transcription factors
The SHATTERPROOF genes encode two very closely related members of the MADS-domain family of transcription factors (Liljegren et al., 2000). The 87% identical SHP proteins are functionally redundant because neither single mutant has any abnormal phenotype. Likewise, the SHP genes have almost identical expression patterns. Expression of the SHP genes is initially detected broadly in the developing gynoecium, but soon thereafter (stage 12) expression becomes limited to the valve margins and ovules (Figure 15A and 15B; Flanagan et al., 1996; Savidge et al., 1995; Roeder et al., 2003). This localized expression pattern coincides with the stage at which valve margins become distinct (Sessions, 1997). Expression of the SHP genes at the valve margins continues after fertilization through early stage 17 (Figure 15C). The SHP genes are also expressed in the nectaries, ovules, septum, and style, although no defect in any of these tissues is observed in the shp double mutant. The role of the SHP genes in these locations may be obscured by redundancy with other genes. In fact, the SHP genes have redundant roles in ovule development with the related SEEDSTICK (STK) MADS-box gene (Pinyopich et al. 2003). The observations that the SHP genes are not expressed in ag single-mutants and that AG can bind to a site in the SHP2 promoter in vitro suggests that the AG carpel identity gene plays a role in promoting SHP gene expression (Flanagan et al., 1996; Savidge et al., 1995). SHP in turn activates the expression of other genes involved in valve margin development, including ALCATRAZ and INDEHISCENT.
Figure 15.
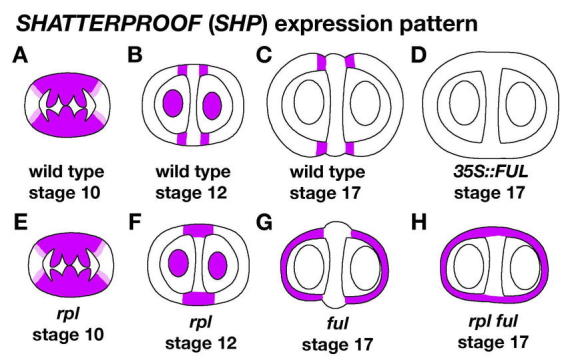
SHP expression is negatively regulated by FUL and RPL. (A) Early in development the SHP genes are broadly expressed in the gynoecium. At stage 10, their expression extends throughout the valve margins, replum, septum, and developing ovules. Weak expression is also seen extending into the edges of the valves. (B) At stage 12, SHP expression is limited specifically to the valve margins. SHP also continues to be expressed in the developing ovules. (C) SHP continues to be expressed in the valve margins through stage 17. (D) Ectopic expression of FUL in 35S::FUL fruit is sufficient to block expression of SHP in the valve margins. (E) In rpl mutants, SHP expression is similar to wild type in early stages. (F) At stage 12 in rpl mutants, SHP continues to be expressed in the replum indicating that RPL is required to negatively regulate SHP expression in the replum. SHP is ectopically expressed in the replum region of rpl mutants through stage 17 (not shown). (G) In ful mutants, SHP is ectopically expressed throughout the valves indicating that FUL is required to negatively regulate SHP expression in the valves. (H) In rpl ful double mutants SHP expression completely surrounds the fruit.
5.1.2 alcatraz (alc) Mutants Keep the Seeds Imprisoned
Dehiscence is also blocked in alcatraz (alc) mutants (Rajani and Sundaresan, 2001). The indehiscent phenotype of alc mutants is less severe than that of shp1 shp2 because the alc fruit will open under pressure. The external definition of the valve margin is not affected in alc fruit (Figure 14A and 14C). Likewise, the valve margin lignified layer is present in alc fruit (Figure 14B and 14D). What causes indehiscence in the alc fruit? The separation layer of the valve margin is affected in alc mutants. Wild-type fruit open at the separation layer when the cells secrete enzymes to break down the middle lamella between the cell walls. In wild-type plants, this process leaves a clean boundary with even edges. In alc, these cells appear torn as if cell separation did not occur. Furthermore, in wild type none of the cells of the separation layer are lignified. A lignified bridge forms in alc mutants connecting the lignified valve margin and enb layer with the lignified vascular tissue of the replum (Figure 14D). This lignified bridge, together with a failure to specify separation layer cells, apparently accounts for the indehiscent phenotype of alc mutants. Therefore, alc is specifically required for the differentiation of the separation layer of the valve margin.
ALCATRAZ encodes a basic helix loop helix transcription factor
ALC encodes a basic helix loop helix (bHLH) transcription factor (Rajani and Sundaresan, 2001). Although ALC is broadly expressed in the gynoecium prior to fertilization, it becomes restricted to the dehiscence zone at stage 16 (Figure 16A and 16B; Liljegren, et al., 2004; Rajani and Sundaresan, 2001). At stage 17, ALC continues to be expressed in the valve margin, but expression expands into the outer replum cells (Figure 16C). When the fruit is starting to turn yellow at stage 18, ALC expression can be seen in the inner cells of the valve margin where it prevents the ectopic lignification of these separation layer cells (Figure 16D). ALC is also expressed in the stigma, ovules, nectaries, newly emerging leaves, lateral root primordia, and the fruit pedicel branch point, although it has no known function in these tissues.
Figure 16.
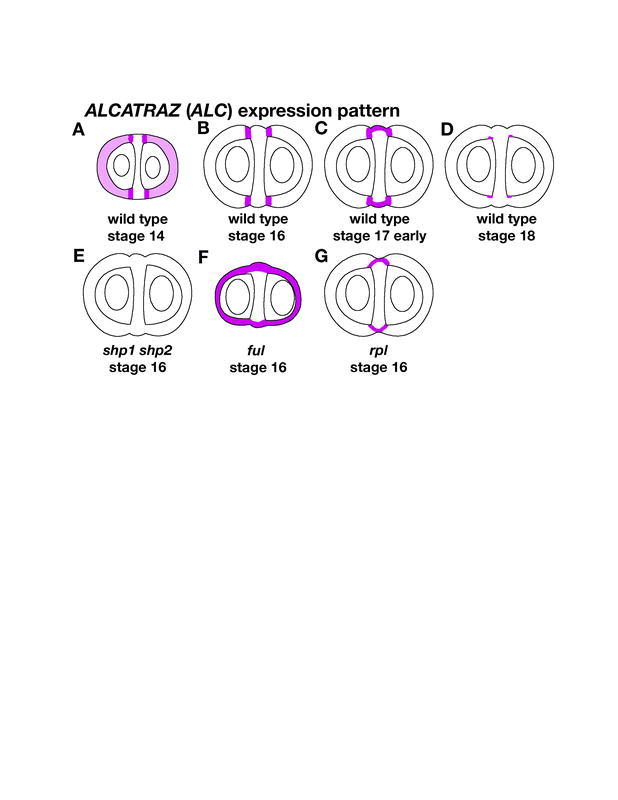
ALC is positively regulated by SHP and negatively regulated by FUL and RPL. (A) In early stages, ALC is expressed in the valves as well as the valve margin as shown here at stage 14. (B) At stage 16, ALC expression becomes limited specifically to the valve margins. (C) At stage 17, ALC is expressed in the outer cells of the replum as well as in the valve margins. (D) At stage 18, ALC is expressed in the inner few cells of the separation layer. In alc mutants these cells ectopically lignify to form the lignified bridge joining the valves to the replum. (E) ALC expression is not detected in the valve margins of shp1 shp2 fruit at stage 16. (F) ALC is ectopically expressed in the valves of ful-mutant fruit. ALC expression is also observed in the replum of ful mutants. (G) ALC is ectopically expressed in the outer cells of the rpl-mutant replum at stage 16 when ALC expression is valve margin specific in wild-type fruit.
5.1.3 INDEHISCENT (IND) Is Required for Valve Margin Specification
The loss of valve margin development in indehiscent (ind) mutants is more severe than either shp or alc mutants (Liljegren et al., 2004). The loss of valve margin definition in ind mutants can be seen throughout the fruit instead of primarily at the base as in shp1 shp2 mutants (Figure 14G). IND is a key factor controlling the specification of the valve margin separation and lignified layers, which are absent throughout ind-mutant fruit (Figure 14H).
INDEHISCENT also encodes a basic helix loop helix transcription factor
Like ALC, IND encodes a bHLH transcription factor. IND falls into an atypical class of bHLH proteins with an unusual amino acid sequence in the basic domain. Most bHLH proteins contain a critical glutamate residue in the basic domain that contacts the CA bases of the DNA binding site (Toledo-Ortiz et al., 2003), but in the wild-type IND protein this glutamate is replaced by an alanine (Liljegren et al., 2004). It would be interesting to investigate how this affects the DNA binding properties of the IND protein and whether it binds to a different consensus site or completely fails to bind DNA as has been suggested for this class of bHLH proteins (Toledo-Ortiz et al., 2003).
The helix loop helix domain of bHLH proteins is often involved in homo- or herterodimerization. In yeast, IND can interact with ALC, suggesting that these two proteins may heterodimerize to specify the separation layer (Liljegren et al., 2004).
IND is expressed in the valve margin. The GT140 molecular marker reports the expression pattern of IND in the valve margin from before fertilization (at stage 12) to early stage 17 (Figure 17A and 17B).
Figure 17.

IND is positively regulated by SHP and negatively regulated by FUL and RPL. (A) IND expression is first detected at stage 12 in the valve margins. This is the same stage at which SHP expression becomes limited to the valve margins. (B) IND continues to be expressed in the valve margins through early stage 17. (C) IND expression is not detected in shp1 shp2 mutants indicating that SHP1 and SHP2 positively regulate IND. (D) IND is ectopically expressed throughout the valves when SHP1 and SHP2 are ectopically expressed in 35S::SHP1 35S::SHP2 plants. (E) IND is ectopically expressed in the replum in rpl mutants. (F) IND is ectopically expressed throughout the valves of ful-mutant fruit. (G) IND is detected at low levels throughout the valves of shp1 shp2 ful mutants indicating that unknown factors in addition to SHP are involved in the activation of IND expression. (H) IND is not detected in the valve margins of 35S::FUL fruit.
5.1.4 Indehiscent Canola Plants
Every year canola farmers lose an average of 20% of the crop when the seedpods open before they can be harvested (Child et al., 1998; MacLeod, 1981). Not only are the farmers losing profit, but the escaped seeds also contaminate the field, becoming weeds as other crop plants are grown in the same location. Farmers can go to great lengths to trap the seeds within the pods, including gluing the pods closed. Since canola (Brassica napus and Brassica rapa) is closely related to Arabidopsis it may be possible to apply our understanding of Arabidopsis fruit development to inhibiting pod shatter in canola. Two canola orthologs of IND, BIND1 and BIND2, have been isolated. Reducing expression of BIND in canola plants has resulted in indehiscent canola seedpods (Guy Vancanneyt, personal communication). However, the strong nature of the indehiscent phenotype in these plants inhibited seed recovery, suggesting that the production of lines with less severe phenotypes will be necessary to apply this technology.
5.1.5 The Transcription Factor Network Specifying Valve Margin Identity
IND, SHP, and ALC all encode transcription factors that have overlapping roles in specifying the valve margin. Liljegren, et al., undertook an extensive genetic analysis to understand the network of action and interaction between these genes (2004). SHP positively regulates both IND and ALC, since in the shp1 shp2 mutant neither IND nor ALC expression is detected at the valve margin (Figure 17C, 16E, and 18; Liljegren et al., 2000; Liljegren et al., 2004). Furthermore, since IND and ALC proteins can interact in yeast, it is likely that IND and ALC heterodimerize to specify the separation layer of the valve margin together (Figure 18). IND appears to be primarily responsible for development of the lignified layer of the valve margin.
Figure 18.
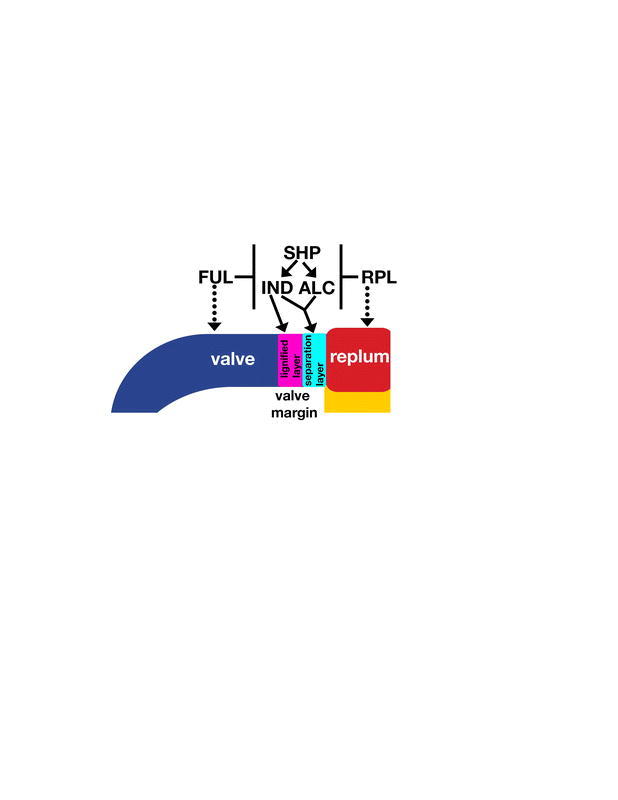
Model for formation of the valve margin at the border between the valve and the replum. SHP, IND, and ALC together form a nonlinear network to specify valve margin formation. SHP positively regulates both IND and ALC. IND and ALC are likely to heterodimerize to specify the separation layer, while IND specifies the lignified layer. SHP, IND, and ALC are all negatively regulated by FUL in the valves limiting valve margin formation to the edge of the valves. Likewise, SHP, IND, and ALC are negatively regulated by RPL in the replum limiting valve margin formation to the edge of the replum. FUL is not directly required for most aspects of valve formation since most aspects of valve development occur in the absence of FUL activity in ind alc shp1shp2 ful quintuple mutants. Likewise, RPL is not directly required for replum formation because the replum differentiates in shp1 shp2 rpl triple mutants. RPL and FUL primarily function to limit the differentiation of the valve margin precisely to a band at the border between the valve and replum ensuring that the fruit opens properly.
Although IND and ALC primarily act downstream of SHP, IND and ALC also have roles independent of SHP. For example, the ind-mutant phenotype is more severe than the shp1 shp2-mutant phenotype (Figure 14E to 14H). Likewise, the loss of valve margin specification is more extreme in the ind shp1 shp2 triple mutant than in either ind or shp1 shp2, showing that there is a slight additive effect and that SHP also has roles that are separate from IND (Figure 14K and 14L). Most of the functions of ALC are thought to be a subset of those of IND because the two proteins are likely to act as a heterodimer, requiring both partners to specify the formation of the separation layer. As expected, the ind alc double-mutant phenotype is similar to the ind single-mutant phenotype (Figure 14G to 14J). However, the ind alc shp1 shp2 quadruple mutant shows an even more severe loss of valve margin development than the ind shp1 shp2 triple mutant, suggesting that ALC has further roles in valve margin development separate from both IND and SHP (Figure 14K to 14N). In conclusion, these genes act together in a nonlinear network to specify the valve margin.
5.2 Valve Development
Unlike the valve margins, which disperse the seeds, the role of the valves is to enclose and protect the seeds as they develop. The valves are derived from the ovary walls and must expand dramatically as the fruit elongates to allow for the growth of the seeds. When the fruit matures, the valves separate at the valve margins and fall from the fruit to release the seeds.
Currently, the genes directly involved in specifying valve development largely remain a mystery. As mentioned above, AG is a good candidate for promoting valve development because valve cells are absent in the carpels of ag ap2 double mutants (see section 2.2 Carpel development in the absence of AG). Due to the difficulty of working with ag ap2 double mutants, the role of AG in the valves has not been extensively investigated. Another MADS-box gene FRUITFULL was initially thought to specify valve cell development, but later its role in valve development was shown to be largely indirect.
5.2.1 fruitfull (ful) Valves Fail to Expand and Differentiate Correctly
Initially the FRUITFULL (FUL) gene was thought to specify valve identity because the valves fail to differentiate correctly in ful mutants. Mutations in the FUL gene result in small, compact fruit that fail to elongate after fertilization (Figure 19A; Gu et al., 1998). A typical wild-type fruit will elongate at a steady rate, increasing in length eightfold only 5 days post-fertilization. In contrast, ful-mutant fruit elongate only twofold even 20 days post-fertilization. Close inspection of the surface of ful fruit by scanning electron microscopy and of the internal valve cells through cross sections reveals that the fruit fail to elongate because valve cell development is dramatically altered and the valve cells fail to expand (Figure 20A to 20B and 19F to 19G; Gu et al., 1998; Ferrándiz et al., 2000a). The role of FUL in valve development is not limited to cell expansion because ful fruit contain more than twice as many cells in the ena layer as wild type, which suggests that FUL also has a role in regulating valve cell division. Additional evidence that valve development is altered in ful is the absence of stomatal complexes, which are normally present in wild type valves (Figure 20A and 20B). Based on the ful single-mutant phenotypes, the FUL gene appeared to be required to promote the normal growth and differentiation of valve cells.
Figure 19.
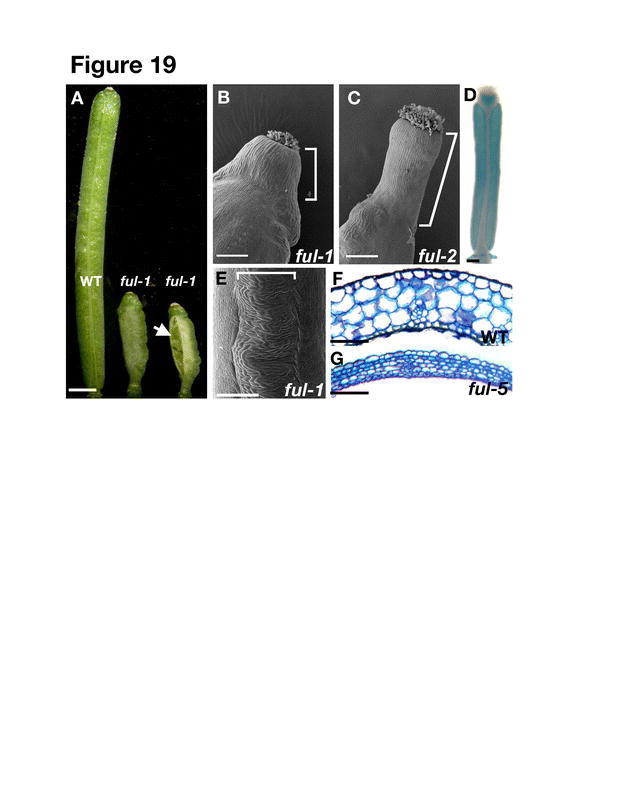
fruitfull valves fail to expand and differentiate correctly. (A) Picture comparing a fully elongated wild-type fruit at stage 17 to two comparable ful-1 fruit. In the second ful-1 fruit, the valve has split open. (B) SEM of the style (bracketed) of a ful-1 fruit, which is a strong allele in the Landsberg erecta ecotype. The erecta mutation suppresses the abnormal elongation of the style in ful mutants. (C) SEM of the style (bracketed) of a ful-2 fruit, which is a moderate allele in the Columbia ecotype showing abnormal elongation of the style in ful mutants. (D) Expression pattern of FUL in the valves, style, and nectaries of a stage 16 fruit. Expression of FUL is detected in plants heterozygous for the ful-1 allele (ful-1/+), which carries an enhancer trap DS insertion that reflects the expression pattern of FUL. See also Figure 21G. (E) The replum (bracketed) of ful mutants is enlarged and twisted. (F) Cross-section of a wild-type valve. (G) Cross-section of a ful valve showing that the cells fail to expand after fertilization as they do in wild type. Scale bar in A represents 1 mm, scale bars in B-E represent 200 µm, and scale bars in F-G represent 100 µm. B-C: from Ferrándiz et al., 2000a. F: Reprinted from Cell, 116, Liljegren, S.J., Roeder, A.H.K., Kempin, S.A., Gremski, K., Østergaard, L., Guimil, S., Reyes, K.D., and Yanofsky, M.F., Control of fruit patterning in Arabidopsis by INDEHISCENT, 843–853, Copyright (2004), with permission from Elsevier.
Figure 20.

Valve cell differentiation is restored in ful mutants by removal of valve margin identity. (A) SEM of the epidermal cells of wild-type (Ler) valves at stage 17. The cells are elongated (one is outlined) and interspersed with stomata (arrowheads). (B) ful valve cells remain small and rounded. No stomata differentiate in ful valves. (C) Valve cells of shp1 shp2 ful do not elongate much more than ful valve cells. However, a few stomata are present in shp1 shp2 ful-mutant valves (not shown). (D) Elongation is partially restored in alc ful valve cells although stomata are not present. (E) Elongation is restored slightly more in alc shp1 shp2 ful valve cells. (F)‘Removal of IND in ind ful fruit greatly restores both valve cell elongation and formation of stomata. (G)‘The valve cells of ind alc ful appear similar to ind ful valve cells. (H) Cell elongation is similarly restored in ind shp1 shp2 ful valves. (I) The epidermal valve cells in ind alc shp1 shp2 ful appear very similar to wild-type valve cells. Scale bars represent 25 µm. B and F: Reprinted from Cell, 116, Liljegren, S.J., Roeder, A.H.K., Kempin, S.A., Gremski, K., Østergaard, L., Guimil, S., Reyes, K.D., and Yanofsky, M.F., Control of fruit patterning in Arabidopsis by INDEHISCENT, 843–853, Copyright (2004), with permission from Elsevier.
Another striking indication that ful valve cell fate is altered is that all of the internal mesocarp cells are ectopically lignified in ful valves, indicating that FUL is normally required to prevent valve cell lignification (Figure 21A and 21D; Ferrándiz et al., 2000b). In wild-type fruit, the mesocarp cells are only lignified at the valve margin. Therefore, the ectopic lignification of the ful valve mesocarp cells is the first hint that ful valves have adopted a valve margin identity (see section 5.2.3 FRUITFULL negatively regulates SHATTERPROOF, INDEHISCENT, and ALCATRAZ).
Figure 21.

Control of lignification in the valves. (A) Cross-section of a wild-type fruit stained with the lignin specific stain phloroglucinol. The lignified layer of the valve margin (lm) and the enb layer (lv) are lignified, but the mesocarp cells (m) are not lignified. (B) Lignification of the enb layer retracts a few cells (asterisk) from the replum in ind alc shp1 shp2 mutants. The lignified layer of the valve margin also fails to differentiate. (C) The enb layer fails to lignify in ind alc shp1 shp2 ful quintuple mutants. (D) The mesocarp cells are ectopically lignified in ful mutants. (E) The mesocarp cells of 35S::SHP1 35S::SHP2 fruit are also ectopically lignified. (F) Removal of IND activity from ful-mutant valves in ind ful restores the lignification pattern in the valves. Only the enb layer of ind ful mutants is lignified and not the mesocarp cells. (G) Cross-section showing the expression pattern of FUL in the valves, but not the replum. The expression pattern shown is derived from plants heterozygous for the ful-1 enhancer trap insertion. (H) In ind alc shp1 shp2 fruit, the domain of FUL expression in the valves retracts several cell widths (asterisks) from the replum. The retraction of FUL correlates with the retraction of the lignification of enb cells seen in B. (I) Model for lignification of the enb layer. FUL, IND, ALC, and SHP all contribute to lignification of the enb layer. IND, ALC, and SHP negatively regulate a hypothetical replum factor, which in turn negatively regulates FUL. Scale bars represent 100 µm. A-C and G-H: Reprinted from Cell, 116, Liljegren, S.J., Roeder, A.H.K., Kempin, S.A., Gremski, K., Østergaard, L., Guimil, S., Reyes, K.D., and Yanofsky, M.F., Control of fruit patterning in Arabidopsis by INDEHISCENT, 843–853, Copyright (2004), with permission from Elsevier. E: From Liljegren et al., 2000. Reprinted with permission from Nature Publishing Group (http://www.nature.com/). F: courtesy of Sarah Liljegren.
One consequence of failure of the ful fruit to elongate is that the seeds become densely packed inside. This contrasts with the wild-type fruit, whose continued growth provides ample room to accommodate the growing seeds. In fact, the developing seeds of the ful mutant apply pressure on the valve walls, which often tear (Figure 19A). This pressure also apparently accounts for the fact that ful-mutant seeds are smaller than those of the wild type. In contrast to ful valve cells, ful replum cells continue to expand, creating a highly twisted structure (Figure 19E). In addition, medial and lateral vascular bundles are poorly differentiated in ful fruit, although the precise defect is unknown. The style of ful fruit undergoes a dramatic elongation, indicating that FUL is normally required to prevent elongation of the style (Figure 19C; Ferrándiz et al., 2000a). This elongated-style phenotype of ful mutants is not observed when the ful mutation is in the Landsberg erecta (Ler) background, which is capable of suppressing this phenotype (Figure 19B; Ferrándiz et al., 2000a). In addition to the changes in fruit development, ful plants show abnormal development of the cauline leaves, which appear wider than wild type and have defects in the vasculature.
FRUITFULL encodes a MADS domain transcription factor
The FUL gene is a member of the extended family of MADS-box genes and is most closely related to the APETALA1 and CAULIFLOWER flower meristem identity genes. Interestingly, ap1 cal ful triple mutants show a dramatic non-flowering phenotype, indicating that these three genes share redundant roles in promoting flower meristem identity (Ferrándiz et al., 2000a).
The FUL gene is strongly expressed in the fruit valves, consistent with the location of the defects in ful mutants (Figure 19D and 21G; Mandel and Yanofsky, 1995; Gu et al., 1998). During flower development, FUL expression is first detected during stage 3 in a central region of the flower meristem in cells that will later form the carpels. As these carpels develop, FUL becomes restricted to the valves. FUL expression can also be detected in the style, consistent with the greatly elongated style that occurs in ful mutants. FUL is also expressed in the inflorescence meristem, where it presumably acts to promote flowering, and in leaves, which show developmental abnormalities in ful mutants.
5.2.2 Ectopic Expression of SHATTERPROOF or INDEHISCENT Produces Fruit Resembling fruitfull Mutants
Is ectopic expression of the valve margin genes SHP or IND in other tissues sufficient to convert those cells into valve margin cells? Overexpression of the SHP genes with the CaMV35S promoter (35S::SHP1 35S::SHP2) produces fruit with a phenotype resembling weak alleles of ful (Liljegren et al., 2000). The valves are much narrower than wild type and often split due to the pressure of the developing seeds, similar to ful valves, but the 35S::SHP fruit are longer than ful fruit. Molecular markers for the valve margin including IND are expressed in the valves of 35S::SHP fruit (Figure 17D) and the valve cells become ectopically lignified, indicating that valve cells have been partially converted into valve margin cells (Figure 21E). The effect of ectopic expression of IND (35S::IND) is even stronger than that seen for 35S::SHP, and the resulting fruit even more closely resemble ful fruit (Liljegren et al., 2004). The resemblance of both 35S::SHP and 35S::IND fruit to ful fruit raised the question of whether ful-mutant valve cells have a partial valve margin identity.
5.2.3 FRUITFULL Negatively Regulates SHATTERPROOF, INDEHISCENT, and ALCATRAZ
In the ful mutant, SHP, IND, and ALC are all ectopically expressed throughout the valves of the fruit and the valve cells appear to adopt a partial valve margin identity (Figure 15G, 17F, and 16F; Ferrándiz et al., 2000b; Liljegren et al., 2004). As mentioned above, the ectopic lignification of the ful mesocarp cells is one sign that the valve cells have adopted a valve margin identity because in wild-type fruit only the mesocarp cells of the valve margin are lignified (Figure 21A and 21D; see section 5.2.1 fruitfull (ful) valves fail to expand and differentiate correctly). Likewise, the expression domains of downstream molecular markers for the valve margin expand throughout the valves in ful mutants. These loss-of-function studies showed that the FUL gene is required to negatively regulate the expression of valve margin identity genes in the valves, thereby limiting SHP, IND, and ALC expression to the valve margin (Figure 18).
5.2.4 35S::FUL Fruit Are Indehiscent
To determine if FUL is sufficient to negatively regulate valve margin development and/or promote a valve cell fate in ectopic positions, transgenic plants were constructed in which the FUL gene was constitutively expressed from the CaMV35S promoter (35S::FUL). The most striking phenotype of 35S::FUL fruit is that cells of the outer replum and valve margin appear to adopt a valve cell fate (Figure 14O; Ferrándiz et al., 2000b). Thus, the cells on the entire surface of 35S::FUL fruit have the appearance of valve cells. SHP and IND expression is not detected in the valve margins of 35S::FUL fruit, which are converted into valve tissue, and consequently the fruit fail to dehisce (Figure 15D and 17H). Moreover, the lignified cells of the valve margin cannot be detected in 35S::FUL fruit, indicating that ectopic FUL expression is sufficient to prevent lignification of these cells (Figure 14P). These studies indicate that within the context of the fruit, FUL expression is sufficient to repress the valve margin identity genes and convert the valve margin and outer replum cells into valve cells.
5.2.4 Restoration of Valve Development in fruitfull Mutants by Removal of Valve Margin Identity
The conversion of ful valve cells into valve margin cells raises the question of whether FUL really specifies valve cell fate or alternatively just represses valve margin fate. The fact that ectopic FUL expression can convert the outer replum cells to valve cells suggests that FUL has some role in valve identity. How much of the ful-mutant fruit phenotype can be attributed to the ectopic expression of SHP, ALC, and IND in the valves? This question can be addressed by removing the ectopic valve margin identity. When SHP activity is removed from ful fruit in the shp1 shp2 ful triple mutant, the ful phenotype is partially rescued (Ferrándiz et al., 2000b). The shp1 shp2 ful triple-mutant fruit are slightly longer than ful single-mutant fruit (Figure 22). Also a few guard cells, which are entirely absent from the ful valves, differentiate in the shp1 shp2 ful valves (Figure 20C). However, the valves of shp1 shp2 ful fruit are still ectopically lignified, indicating that rescue of the ful phenotype by removal of SHP activity is only partial. Removal of ALC activity from the ful valves restores fruit elongation to a greater extent than removal of SHP, since alc ful fruit reach 40% the length of wild-type fruit (Figure 22; Liljegren et al., 2004). Likewise, the epidermal valve cells in alc ful fruit elongate slightly more than ful valve cells, although the formation of stomata is not restored (Figure 20D). In contrast, removal of IND activity from ful fruit dramatically restores fruit elongation to 67% the length of wild type (Figure 22). Furthermore, removal of IND eliminates the ectopic lignification of the valves suggesting that the ectopic IND expression is the primary cause of ectopic valve lignification in ful mutants. In ind ful fruit, as in wild type, only the enb layer of the valves is lignified (Figure 21A, 21D, and 21F). Since IND controls the lignification of the valve margin, the ectopic lignification of the ful valves can be interpreted as an expansion of this lignified layer throughout the valves. Furthermore, stomata are present in the valves of ind ful fruit and the epidermal valve cells elongate almost as much as wild type (Figure 20F).
Figure 22.
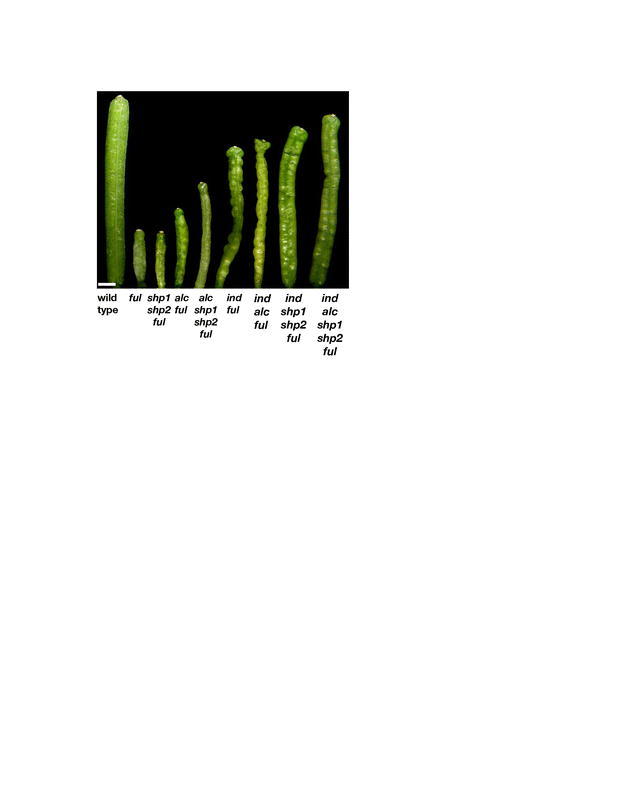
Fruit elongation is restored in ful mutants by removal of valve margin identity. Picture of wild type (Ler), ful, shp1 shp2 ful, alc ful, alc shp1 shp2 ful, ind ful, ind alc ful, ind shp1 shp2 ful, ind alc shp1 shp2 ful fruit (from left to right) at stage 17 after the fruit have fully elongated. Removal of valve margin identity progressively restores fruit elongation. Removal of IND activity in ind ful mutants contributes most to the restoration of fruit elongation. Scale bar represents 1 mm. Reprinted from Cell, 116, Liljegren, S.J., Roeder, A.H.K., Kempin, S.A., Gremski, K., Østergaard, L., Guimil, S., Reyes, K.D., and Yanofsky, M.F., Control of fruit patterning in Arabidopsis by INDEHISCENT, 843–853, Copyright (2004), with permission from Elsevier.
At first, the striking rescue of ful by removal of IND activity seems enigmatic. Since IND acts downstream of SHP, how can removal of IND rescue valve development to a greater degree than removal of SHP from ful mutants? If IND were to act exclusively downstream of SHP, we would not expect IND to be expressed in shp1 shp2 ful valves and the rescue of ful fruit by shp1 shp2 should be at least as good as rescue by ind. The answer to this dilemma seems to be that other factors contribute to the activation of IND in a SHP-independent manner. In fact, a low level of IND expression can be detected throughout shp1 shp2 ful valves (Figure 17G; Ferrándiz, et al., 2000b). Although IND expression is not detected in the valve margin region of shp1shp2 mutants, indicating that SHP is one of the main factors contributing to the activation of IND expression, the increased loss of valve margin definition in ind shp1 shp2 compared to either shp1 shp2 or ind mutants alone suggests that a low level of IND is present in shp1 shp2 mutants (see section 5.1.5 The transcription factor network specifying valve margin identity).
Although removal of IND activity dramatically rescues ful, the restoration of valve identity is not complete. Valve elongation and differentiation is more fully restored in ind shp1 shp2 ful mutants than in either ind ful or shp1 shp2 ful mutants (Figure 22 and 20H). Likewise, genetic analysis with alc indicates that ALC also has SHP-independent and IND-independent functions and removal of ALC also restores valve development. The progressive rescue of fruit elongation and valve cell development in ful by the progressive removal of valve margin identity (Figure 22 and 20A to 20I) indicates that these transcription factors act in a nonlinear network instead of a strictly linear pathway. The most complete rescue of valve development occurs in the ind alc shp1 shp2 ful quintuple mutant, in which fruit elongation is restored to 90% of the wild-type length (Figure 22). The outer epidermal cells of the valve appear almost identical to wild-type valve cells in that they are elongated and interspersed with stomata (Figure 20I). This rescue of valve development in the absence of FUL activity indicates that FUL is not directly required for the majority of valve cell differentiation, but instead acts to prevent these cells from adopting a valve margin cell fate. However, the ind alc shp1 shp2 ful fruit are bumpy and the cells in the inner ena layer of the valve do not fully enlarge, indicating that valve development is not completely restored. These defects may reflect a requirement for FUL activity in valve differentiation or may indicate that other factors, which have not yet been identified, also play a role in the ful phenotype. FUL does play a redundant role in the lignification of the enb cell layer of the valve, as we shall see next.
5.2.4 Endocarp b (enb): The Lignified Layer of the Valve
The enb layer, the second innermost cell layer of the valves, becomes rigid when it lignifies during the middle of stage 17 (Figure 21A). The enb layer is one of three cell types thought to be involved in dehiscence, acting in conjunction with the separation and lignified layers of the valve margin. When the valves desiccate upon fruit maturation, the outer thin-walled valve cells are thought to contract, putting tension on the inflexible enb layer, which causes the shattering of seedpods (Figure 9H; Spence, et al., 1996). AG is involved in the specification of the enb layer, which is absent in the first whorl carpels of ag ap2 double mutants (see section 2.2 Carpel development in the absence of AGAMOUS; Alvarez and Smyth, 1999). IND, ALC, SHP1, SHP2, and FUL are also all redundantly involved in the lignification of the enb layer, which fails to lignify throughout the ind alc shp1 shp2 ful fruit except for a few cells at the base (Figure 21A and 21C; Liljegren, et al. 2004). A less penetrant reduction of enb lignification is also observed in ind shp1 shp2 ful fruit. However, the enb layer lignifies in the ind ful double mutant (Figure 21F). Since lignification of the enb layer is only absent when all five genes are mutated, it appears all these genes function redundantly in this process.
In wild-type fruit the enb layer extends throughout the valves and joins the valve margin lignified layer. In contrast, a gap of a few cells appears between the replum and the enb layer in ind alc shp1 shp2 quadruple-mutant fruit (Figure 21A and 21B; Liljegren, et al., 2004). The cells in the gap are larger and appear similar to the valve mesocarp cells instead of the small enb cells. This gap correlates with a retraction of the domain of FUL expression into the valve and away from the replum (Figure 21G and 21H). Again these results indicate that in the absence of IND, ALC, SHP, and FUL activity, the development of the enb layer is affected. It has been proposed that the domain of a hypothetical replum factor expands in the absence of the valve margin genes and causes the retraction of FUL (Figure 21I; Liljegren, et al. 2004; Ferrándiz et al., 2000). In the future, it would be interesting to identify this putative replum factor.
5.3 Replum Development
The replum is the central ridge in the gynoecium that remains attached to the fruit after the seedpod shatters. Each replum contains an outer layer of cells and the internal medial vascular bundle. The replum connects to the septum, which divides the fruit into two compartments. As in the valves, genes that directly control replum development, including the putative replum factor (see section 5.2.4 endocarp b (enb)-the lignified layer of the valve), remain elusive.
5.3.1 REPLUMLESS (RPL) Limits Valve Margin Development to the Edge of the Replum
Mutants affecting the replum have been difficult to isolate in traditional screens because the replum is such a narrow stripe of tissue that it is difficult to see. The ful mutant has an enlarged and twisted replum that is more apparent. In a genetic screen in the ful-mutant background, a mutant in which the replum fails to differentiate was identified and named replumless (rpl) (Roeder et al., 2003). The rpl ful double mutant was crossed to wild type to isolate the single mutant and determine whether it also had a defect in replum development.
In the rpl single mutant the outer cell layers of the replum fail to differentiate as replum cells (Figure 23A to 23D) and instead adopt characteristics of valve margin cells (Roeder et al., 2003). For example, in rpl fruit the cells in the replum region lignify like the valve margin lignified layer or stain similarly to cells of the valve margin separation layer (Figure 23G to 23J). Only the outer cell layers of the replum are affected in the rpl mutant and the inner parts of the replum including the vascular bundle are present (Figure 23B and 23D).
Figure 23.
Loss of replum development in replumless mutants is restored by removal of valve margin identity. (A) SEM close up of the replum (bracketed r), valve margins (vm arrowhead), and edges of the valves (v) of a wild-type (Ler) stage 17 fruit. (B) Cross section of the replum (arrow), valve margins (arrowheads) and edges of the valves (v) of a wild-type (Ler) stage 17 fruit. The medial vascular bundle (vb) runs through the inner part of the replum. (C) In rpl mutants, the replum fails to differentiate (bracket) and in its place the cell take on the characteristics of the valve margin cells (arrowheads). (D) In rpl mutants, the outer cells of the replum region are small and similar to valve margin cells. However, the inner parts of the replum including the vascular bundle are present in rpl mutants. (E) Removal of SHP activity from rpl mutants in shp1 shp2 rpl triple mutants rescues replum development. (F) The cells in replum of shp1 shp2 rpl fruit appear similar to wild-type replum cells. (G) Cross section of a wild-type replum region stained with safranin O and alcian blue. The separation layer (SL arrowhead) stains light blue, which is distinct from the lignified layer (LL arrowhead) and replum (R arrow). (H)‘In a moderate allele rpl-1, the replum region is covered with light blue staining cells that appear similar to the separation layer. (I) Cross section of a wild-type replum region stained with lignin specific stain phloroglucinol. The lignified layers (LL arrowhead) of the valve margins stain. (J) In a strong allele rpl-3, the valve margin lignified layers can connect across the replum. (K) RPL::RPL-GUS is expressed (pink) in the replum of a cross section of the gynoecium at stage 6. (L) RPL::RPL-GUS continues to be expressed strongly in the replum of a cross section of a stage 12 gynoecium. (M) RPL::RPL-GUS expression pattern in the replum and style of a stage 12 gynoecium. Scale bars in A-F represent 20 µm, scale bars in G-J represent 25 µm, and scale bars in K-M represent 50 µm. A-M: Reprinted from Current Biology, 13, Roeder, A.H.K, Ferrándiz, C., and Yanofsky, M.F., The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit, 1630–1635, Copyright (2003), with permission from Elsevier.
The cells in the rpl replum region ectopically express the valve margin identity genes SHP, IND, and ALC (Figure 15F, 17E, and 16G). The rpl-mutant phenotype is evident through alterations in the expression pattern of SHP2 in the gynoecium at stage 12 when the first morphological differences are seen between replum, valve, and valve margin in wild-type gynoecia. Early in the development of the gynoecium, SHP is expressed broadly throughout the replum, valve margins, and even the edges of the valves (Figure 15A and 15E). At stage 12, SHP expression becomes limited specifically to the valve margins in wild type, but in rpl mutants, SHP continues to be ectopically expressed in the replum (Figure 15B and 15F). Therefore RPL is likely to act at stage 12 to negatively regulate SHP, restricting it to the valve margin.
Replum development can be rescued in rpl mutants by the removal of SHP activity in the shp1 shp2 rpl triple mutant (Figure 23E and 23F). Therefore, RPL is not directly required for replum development and instead is required to negatively regulate SHP, preventing the replum from differentiating as valve margin (Figure 18).
REPLUMLESS encodes a homeodomain protein
RPL encodes a homeodomain protein in the BELL1 family (Roeder et al., 2003). RPL is also known as PENNYWISE (PNY), BELLRINGER (BLR), and VAAMANA (VAN) for its additional roles in meristem function and internode elongation (Smith and Hake, 2003; Byrne et al., 2003; and Bhatt et al., 2004).
RPL is expressed in the replum beginning very early in gynoecium development at stage 6 (Figure 23K). RPL::RPL-GUS is expressed in the replum at stage 12 when RPL negatively regulates SHP (Figure 23L to 23M). RPL is also expressed in the ovules, stem, leaves, roots, and shoot apical meristem.
Homeodomain transcription factors in the BELL family often heterodimerize with members of the KNOX family (Bellaoui et al., 2001; Smith et al., 2002). Smith and Hake, Byrne et al., and Bhatt et al. have found that the RPL protein interacts with certain members of the KNOX family including SHOOT MERISTEMLESS (STM), KNAT1, and KNAT6, but no interaction was observed with KNAT2, KNAT3, KNAT4, or KNAT5 (2003; 2003; 2004). Furthermore, Bhatt et al. have found that RPL becomes localized to the nucleus when it interacts with a KNOX partner (2004). Currently it is unknown whether these interactions have a role in fruit development.
Interestingly, Bao et al. have found that RPL also negatively regulates AG expression (2004). They have shown that the RPL protein can bind to AG regulatory elements in vitro, suggesting that the regulation of AG may be direct. Since AG is closely related to SHP, it would be interesting to determine whether the role of RPL in negatively regulating SHP is also direct.
5.4 Model for the Formation of a Stripe of Valve Margin Tissue
The function of RPL in the replum closely parallels the function of FUL in the valves. The SHP genes specifying valve margin identity are initially broadly expressed and are subsequently confined to a narrow stripe at the junction between the valve and the replum by FUL and RPL (Figure 18). In this way the valve margin is guaranteed to differentiate precisely at the border between the valves and the replum so that the fruit will shatter and disperse the seeds properly.
The restoration of valve and replum development in ful and rpl mutants respectively by the removal of valve margin identity indicates that for the most part FUL and RPL are not directly required for the differentiation of these tissues. Instead FUL and RPL are required to negatively regulate the valve margin identity genes, giving the valves and replum the opportunity to differentiate. Thus, FUL and RPL appear to be involved in an elaborate mechanism for accurately positioning the stripes of valve margin tissue. This valve margin patterning mechanism must be superimposed on an underlying initial patterning of valve and replum and these underlying factors specifying replum and valve identity have yet to be discovered.
What approaches can we take to identify upstream patterning factors? Since floral organs including carpels are derived from modified leaves, it is likely that some of the genes more generally involved in lateral organ development will also be involved in carpel development. Genes that pattern the meristem have been shown to activate specific organ identity genes. For example, the WUSCHEL (WUS) homeobox gene, which controls meristem maintenance, also activates AGAMOUS expression in the center of the floral meristem (Lohmann et al, 2001). Other meristematic genes may contribute to patterning at later stages in carpel development as well.
5.5 Meristematic Role of the Replum?
It is intriguing that RPL has roles in both meristem function and development of the replum. Various genes functioning in the meristem such as SHOOT MERISTEMLESS (STM) are also expressed in the replum (Long et al., 1996). It is tempting to think that the replum and medial ridge may act as meristematic tissues producing the placenta and the septum. However, the roles of these other meristematic genes in replum development remain hidden partially because mutations in these genes tend to block development long before the formation of gynoecia. Nevertheless, the role of one set of genes has been uncovered.
5.5.1 CUP-SHAPED COTYLEDONS (CUC) Function in Replum and Septum Formation
The CUP-SHAPED COTYLEDONS (CUC) genes are thought to function in the separation of organs from neighboring organs and from the meristem. The cuc1 cuc2 double mutant is best known for its embryonic phenotype in which the cotyledons fuse together into a cup shape and the shoot meristem fails to form (Aida et al., 1997). Since the double mutant is normally lethal at the seedling stage, the role of the CUC genes in replum and septum development remained hidden until Ishida et al. regenerated the cuc-mutant shoots from calli (2000). The septa of cuc1 cuc2 double-mutant gynoecia fail to fuse and about half of these gynoecia lack the outer replum tissue in their upper halves. Early in gynoecium development, the medial ridge of tissue is present in cuc1 cuc2 double mutants, but the medial ridge fails to expand and fuse to generate the septum. Additionally, in the cuc-1/+ cuc2/cuc2 plants, about 50% of the septa do not fuse completely, leaving large holes. Therefore CUC1 and CUC2 are redundantly required for septum and replum formation. It is unclear whether this role in replum and septum development is related to the functions of the CUC genes in establishing boundaries between organs.
The CUP-SHAPED COTYLEDON genes encode NAC domain transcription factors
CUC1 and CUC2 encode two closely related members of the plant-specific family of NAC domain transcription factors (Aida et al., 1997; Takada et al., 2001, Duval et al., 2002). CUC1 and CUC2 also act redundantly with CUC3, a third closely related member of the NAC family (Vroemen et al., 2003), although the role of CUC3 in gynoecium development has yet to be investigated.
As expected, the CUC genes are expressed in the boundaries between organs. In addition, CUC1 and CUC2 are expressed in the gynoecium during stage 7 at the inner edge of the medial ridge where the septum will form (Ishida et al., 2000; Takada et al., 2001). As the septum primordia grow, CUC1 and CUC2 continue to be expressed on the inner ridge and in the middle of the septum until stage 10 or 11. Since the CUC genes are not expressed in the replum, it is surprising that the replum is also affected in the cuc1 cuc2 double mutant.
5.6 Fusion and the Formation of the Stigma, Style, Septum, and Transmitting Tract
The replum is one of the medial tissues of the fruit. Now we will turn from our focus on the replum to examine the other medial tissues including the septum and transmitting tract and their involvement in the fusion of the gynoecium. The gynoecium undergoes two fusion events (Bowman et al., 1999). The first is the congenital fusion of the two carpels, meaning that the gynoecium is composed of two fused carpels despite its formation as a single primordium. Although it is not visible in wild-type gynoecia, the fusion of carpels occurs in the medial tissues, including the replum and septum primordia. The second fusion event in gynoecium development is the postgenital fusion of the two medial ridges to form the septum and the fusion of the style to form a solid cylinder. This fusion is evident during development when the two medial ridges, which are initially separated, grow inward until they meet in the center of the gynoecium. After fusion the transmitting tract differentiates in the center of the style and septum, providing the pathway through which the pollen tube travels from the stigma to reach the ovule. The transmitting tract cells secrete a rich extracellular matrix supporting pollen tube growth. As we will see, several genes including LEUNIG, TOUSLED, STYLISH, SPATULA, and CRABS CLAW are involved in the formation of the stigma, style, septum, and transmitting tract and are also required for the fusion of the gynoecium. Furthermore, mutants in many of these genes have synergistic interactions, suggesting that they act in the same genetic pathway.
5.6.1 LEUNIG (LUG) and Fusion
The leunig (lug) mutants are defective in both congenital fusion of the carpels at the tips and postgenital fusion of the septum. The apex of the lug gynoecium is characteristically split into four unfused protrusions (Figure 24A and 24B; Liu and Meyerowitz, 1995; Liu et al., 2000; Chen et al., 2000). Two of these protrusions are medial and composed of style tissue topped by stigmatic papillae. The other two lateral protrusions are horns of valve tissue. The apical defects can be detected as early as stage 7 when the apex of the gynoecium already has an irregular shape. The lug septum grows outward but the two medial ridges fail to fuse (Figure 24D). In addition, the number of carpels is affected in lug mutants and ranges from 1 to 3 (Liu et al., 2000).
Figure 24.
LEUNIG is involved in fusion of the carpels. (A) lug fruit, showing horn like protrusions of the valves (h) above the stigma (s). (B) Close up of the unfused apex of a lug gynoecium. There are two medial stigmatic surfaces (s) and two later horn shaped protrusions from the valves (h). (C) lug ant double mutant showing the two completely unfused horn shaped carpels of the fourth whorl (4). Because of the homeotic affects of lug on the other floral organs, the medial first whorl sepals have also been converted to horn shaped unfused carpels (1). (D) Cross-section of a lug gynoecium with two carpels at stage 10 showing the failure of the septum to fuse. (E) LUG is expressed throughout the carpels at stage 7. Carpel expression is reduced in stage 9, but strong LUG expression is detected in the developing ovule primordia. At stage 12 LUG expression is restricted to the ovules. A-D: from Liu et al., 2000.
lug mutants are best known for other floral phenotypes, including the occasional conversion of sepals to carpels or stamens and the conversion of some petals to stamens (Liu and Meyerowitz, 1995). These homeotic transformations are caused by ectopic expression of AGAMOUS (AG) (see section 2.1 AGAMOUS (AG) specifies carpel identity) in the outer whorls of the flower. LUG negatively regulates AG in the first two whorls of the flower. One question that remains is whether the defects in lug gynoecia are also caused by misregulation of AG expression.
LEUNIG (LUG) encodes a transcriptional corepressor
LUG encodes a putative transcriptional corepressor related to the yeast protein Tup1 and the Drosophila protein Groucho with two glutamine rich regions and seven WD repeats (Conner and Liu, 2000). It is likely that LUG interacts with other transcription factors because it does not contain a DNA binding domain. The LUG protein is localized to the nucleus, consistent with its proposed role in transcription.
LUG is highly expressed in stage 1 to 2 floral primordia and subsequently in each of the floral organ primordia, including the carpels as they arise (Figure 24E; Conner and Liu, 2000). LUG is weakly expressed in the carpel valves, but it is strongly expressed in the vasculature and the developing placenta and ovules (Figure 24E).
5.6.2 leunig (lug) aintegumenta (ant) Double Mutants Lack Medial Tissues
The AINTEGUMENTA (ANT) gene promotes cell proliferation in newly arising organ primordia. The ant mutant has phenotypes weakly resembling those of lug mutants including defects in fusion of the carpels and narrower floral organs. The similar phenotypes of lug and ant prompted Liu et al. to construct the lug ant double mutant to determine if the two genes have redundant roles during development (2000).
Amazingly, the lug ant double-mutant gynoecia are completely unfused and consist entirely of two horn-shaped valves (Figure 24C; Liu et al., 2000). Medial tissues including ovules, placenta, septum, and replum are completely absent from these unfused horn-shaped carpels, which consist of valve tissue, including lateral vascular bundles and valve epidermal cells. Small rectangular cells resembling valve margin cells can still be found at the edge of these valves. These unfused valves develop relatively normally without medial tissues, suggesting that the development of the valves and the medial tissues are largely independent. This synergistic phenotype of lug ant gynoecia indicates that LUG and ANT function together to regulate the formation of medial tissues. Mutations in LUG also interact synergistically with tousled mutants producing gynoecia with severely reduced stigma and style tissues (see section tousled leunig double mutants have reduced apical tissues).
5.6.3 tousled (tsl) Mutants Have Reduced Apical Tissues, which Fail to Fuse
Gynoecium formation in the tousled (tsl) mutant is characterized by reduced differentiation of apical tissues, including the stigma and style, and failure of the style and septum to fuse (Figure 25A to 25F; Roe et al., 1997b). Stigmatic papillae are reduced and form in variable patches at the top edge of the carpels (Figure 25D). Molecular markers show that stigmatic papillae and inner style tissues are present, but the number of cells in each is reduced. The number of carpels is somewhat variable, ranging from one to four, although the majority of tsl gynoecia have two carpels. The defects in tsl gynoecia are seen early at stage 7 when the two carpels are partly unfused (Figure 25G). In stages 10 and 11 the top of the gynoecium remains uneven and only patches of style and stigmatic tissue differentiate. TSL appears to function in the proliferation of apical tissues including stigma and style (Roe et al., 1997b). In addition, tsl mutants have pleiotropic effects on the rest of the plant.
Figure 25.
tousled gynoecia have reduced apical tissues. (A) Wild-type (Ws) gynoecium at stage 13. The regions of the stigma (sg), style (st) and ovary (ov) are delineated between arrowheads. (B) Close up of wild-type style (st) at stage 13. (C) tsl-1 gynoecium at stage 13 showing that the apex is unfused and the number of stigmatic papillae is reduced. (D) Close up of the top of a tsl-1 gynoecium at stage 13. The apex is unfused and stigmatic papillae are absent from portions of the apex. Abaxial style cells are present at the apex and are present on the inner rim of the gynoecium. (E) Cross-section of a wild-type (Ws) gynoecium showing the fused septum. (F) Cross-section of a tsl-1 gynoecium revealing the failure of the septum to fuse. (G) Stage 9 tsl-1 gynoecium where the apex is already uneven. (H) TSL::GUS reporter line showing expression in the style and the upper valves at stage 11. (I) TSL::GUS reporter line at stage 13 showing expression limited to the style. Scale bars A and C represent 300 µm. Scale bars in B, D, and G represent 50 µm. A-I: from Roe et al., 1997b.
The TOUSLED kinase
The TSL protein belongs to a family of a nuclear localized serine threonine kinase that is conserved in plants and animals (Roe et al., 1993; Roe et al., 1997a; Ehsan et al., 2004). Studies of TOUSLED-like kinases in animal systems suggest that they are involved in chromatin metabolism, particularly in chromatin assembly, DNA repair, and the regulation of gene expression (Ehsan et al., 2004). The Arabidopsis TSL protein has been shown to transautophosphorylate and to phosphorylate histone H3 and Asf1b/SGA1 (an Arabidopsis relative of yeast Asf1 which recruits histone H3 and H4 during nucleosome assembly) in vitro, suggesting that the Arabidopsis TSL may also have a role in regulating chromatin metabolism (Roe et al., 1997a; Ehsan et al., 2004). Based on northern blot analysis, TSL is expressed strongly in inflorescences and expression continues at moderate levels in flowers through anthesis stage to the beginnings of fruit development. TSL transcript is also present in shoots and leaves although it is absent in stems (Roe et al., 1993). Within the gynoecium, TSL::GUS is expressed in the apical half of the gynoecium at stage 11 (Figure 25H). During stage 13, TSL::GUS expression becomes restricted to the style (Figure 25I). The expression of TSL in the style has been confirmed by in situ hybridization (Roe et al., 1997b). The broad pattern of TSL expression and the multiple phenotypes of tsl mutants as well as the phenotypes caused by mutations in different chromatin regulatory genes including gymnos (see section GYMNOS encodes a chromatin-remodeling enzyme) show the importance of chromatin remodeling in plant development.
tousled leunig double mutants have reduced apical tissues
Strong synergistic affects are seen when tsl mutants are combined with leunig (lug) mutants. As in tsl mutants, floral organ number is reduced in lug mutants and lug gynoecia are unfused at the top (Figure 26A; see section 5.6.1 LEUNIG (LUG) and fusion), suggesting that lug might have a role in the same genetic pathway as tsl. Most tsl lug double-mutant gynoecia have only one carpel with extensions of tissue occasionally topped with stigmatic papillae (Figure 26B). The placenta tissue in these single carpel gynoecia is greatly reduced. When two carpels are formed in tsl lug double mutants, they are largely unfused with long extensions devoid of stigmatic tissue (Figure 26C). The extreme severity of both of these phenotypes suggests that the developmental pathways in which TSL and LUG function overlap.
Figure 26.
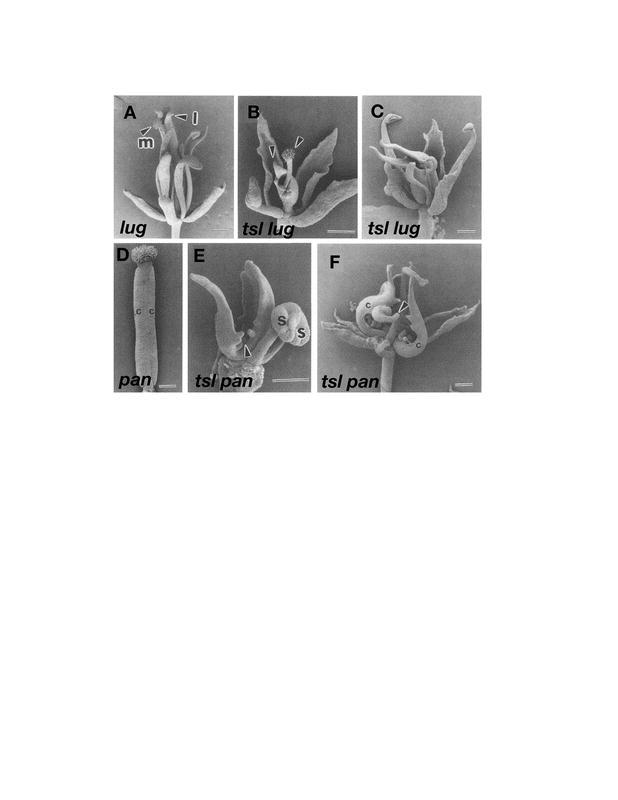
Synergistic interactions of tsl with lug and pan mutants. (A) The unfused apex of a lug gynoecium with four protrusions: two medial (m) and two lateral (l). (B) tsl lug double mutant with only one carpel. There are two projections: one lateral and one medial (arrow heads). (C) tsl lug double mutant with two carpels, which are fused at the base of the gynoecium, but become unfused horns lacking stigmatic tissue at the apex. (D) Stage 13 bicarpellate pan gynoecium. Mutations in pan have minor effects on gynoecium development. (E) Stage 10 tsl pan double mutant showing unfused carpels with ovule primordia (arrowhead). (F) Mature tsl pan flower with two unfused carpels (c). Scale bars in A-E represent 300 µm. Scale bar in F represents 150 µm. A-F: from Roe et al., 1997b.
tousled perianthia double mutants form unfused gynoecia
Strong synergistic affects are also seen in tsl perianthia (pan) double mutants. Gynoecium development is only minimally affected in pan single mutants, which occasionally have three carpels and/or an incompletely fused septum (Figure 26D). However, in the tsl pan double mutant, the gynoecium degenerates into two or three completely unfused carpelloid organs with very little style, stigma, or placenta (Figure 26E and 26F). TSL and PAN act redundantly in gynoecium development. Only in the tsl mutant is the role of PAN in the gynoecia uncovered. tsl mutants also interact synergistically with ett mutants (see section ETTIN interacts synergistically with TOUSLED).
5.6.4 STYLISH (STY) Genes Specify Style Development
The STYLISH (STY1 and STY2) genes redundantly control the development of the stigma and style, which are reduced and develop abnormally in the sty1 sty2 double mutant (Kuusk et al., 2002). In sty1 sty2 fruit, a depression forms in the middle of the stigma while protrusions of stylar tissue grow around the edges and, consequently, stigmatic papillae point in all directions (Figure 27A and 27C). Likewise, the xylem fans typical of wild-type styles are greatly reduced in sty1 sty2 mutants (Figure 27D and 27E). In addition, the septum is often reduced or absent in the apical parts of the sty1 sty2 gynoecium. The sty1 single mutant has similar but less severe defects in stigma and style development (Figure 27B). The sty2 single mutant shows no defects in gynoecium development.
Figure 27.

STYLISH specifies style development. (A) SEM of the apex of a wild-type gynoecium at stage 13. The stigma, style, valves and replum have been marked. (B) SEM of the apex of a sty1 gynoecium at stage 13 showing the presence of style cells on the top of the gynoecium (arrowhead) and reduced disorganized stigmatic papillae. (C) SEM of the apex of a sty1 sty2 double mutant at stage 13 showing that the style has formed short horn-like protrusions (arrowhead) and clustered stigmatic papillae. The amount of style and stigmatic tissue is reduced in comparison to wild type. (D) Vascular pattern of a cleared wild-type gynoecium at stage 13. Note that the medial vascular bundle bifurcates (arrowhead) immediately below the style where large xylem fans form. (E) The vascular pattern of sty1 sty2 double mutants showing basalized bifurcation of the medial vasculature (arrowheads) and reduced or absent xylem fans in the style. (F) SEM of a 35S::STY1 gynoecium at stage 13. Regions with ectopic style are bordered with a white line. (G) 35S::STY1 fruit dehiscing. The ectopic style cells block dehiscence (arrowhead and arrow). (H) Transmission electron micrograph (TEM) of a transverse section of wild-type valve epidermal cells showing smooth cell walls in the absence of wax crenulations. (I) TEM of wild-type style epidermal cells showing the wax crenulations characteristic of style cells. (J) TEM of 35S::STY1 valve epidermal cells in the region of ectopic style development showing that these cells form the wax crenulations characteristic of style cells. (K) STY1::GUS reporter line showing expression in the stigma and style of stage 12 gynoecium. (L) Inflorescence showing the expression of the STY2::GUS reporter in the style, stigma, and anthers of the flowers. (M) Representation of the STY1 expression pattern as seen through in situ hybridization. STY1 is expressed throughout the top of the gynoecium at stage 8. Expression is stronger in the medial regions than in the lateral regions. Scale bars in A-G represent 100 µm. Pictures H-J are taken at magnification 5200x. A-L: from Kuusk et al., 2002.
Ectopic expression of either STY1 or STY2 throughout the plant (35S::STY1 or 35S::STY2) is sufficient to cause the differentiation of ectopic style cells in the valves of the fruit (Figure 27F). These ectopic style cells in the valves of 35S::STY1 fruit form the cuticular ridges characteristic of style cells (Figure 27H to 27J). Likewise, the vascular tissue forms xylem fans near the ectopic style cells, which is another indication of ectopic style development. The patches of ectopic style block the formation of the dehiscence zone in the apex of the valves and inhibit the opening of the fruit (Figure 27G).
The STYLISH genes encode RING finger proteins
STY1 and STY2 encode closely related C3HC3H zinc finger proteins (Kuusk et al., 2002). The zinc finger domains are similar to RING finger motifs, which are involved in protein-protein interactions. The STY proteins also have nuclear localization signals and IGGH domains. The STY genes belong to a family of ten genes in Arabidopsis including SHORT INTERNODES (SHI) and LATERAL ROOT PRIMORDIUM1 (LPR1).
Both STY1 and STY2 are expressed in the apex of the developing gynoecium where the stigma and style form (Figure 27K to 27M; Kuusk et al., 2002). STY1 expression begins in the gynoecium at stage 6, while STY2 is not observed in apex of the gynoecium until stage 9. The absence of STY2 expression in the earliest stages of gynoecium development may account for its incomplete redundancy with STY1.
5.6.5 SPATULA (SPT) Is Required for Transmitting Tract Development
The transmitting tract is absent from the septum of spatula (spt) mutants and the development of medial tissues is also affected (Figure 28A to 28D; Alvarez and Smyth, 1999; Alvarez and Smyth, 2002). The clusters of cells at the tip of each septum that normally form the transmitting tract are missing in spt gynoecia (Figure 28C and 28D). The absence of transmitting tract cells, results in poor pollen tube growth, and consequently fewer than a quarter of the ovules are fertilized. The spt carpels are often unfused at the top because the growth of the style, the stigma, and the septum is reduced. At anthesis, spt gynoecia are narrow in the stylar region and have fewer stigmatic papillae at the tip (Figure 28B). As spt fruit develop, they become flatter than normal due to increased lateral expansion, resulting in a spatula-shaped structure (Figure 28E and 28F). Fusion of the septum often fails to occur in the upper-most regions allowing the gynoecium to flatten into the spatula shape.
Figure 28.
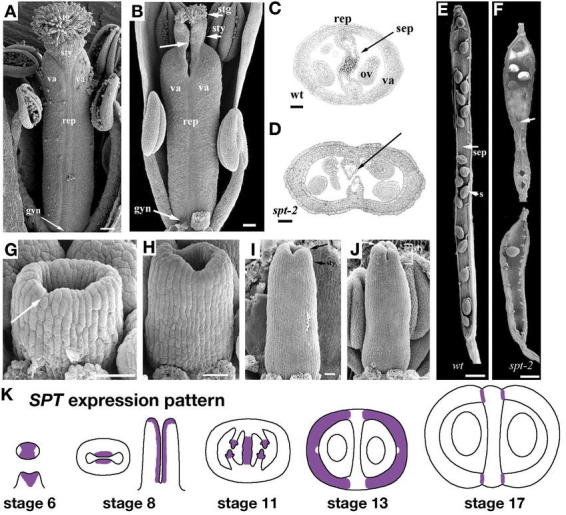
SPATULA is required for transmitting tract formation and apical fusion. (A) Wild-type gynoecium at stage 13. Note the numerous stigmatic papillae. The stigma (stg), style (sty), valves (va), replum (rep), and gynophore (gyn) are labeled. (B) Stage 13 spt gynoecium showing the characteristic split in the style and reduced numbers of stigmatic papillae. (C) Cross-section of a wild-type gynoecium. Note the fused septum (sep) filled with transmitting tract, which stains darkly. The ovule (ov), replum (rep), and valve (va) are labeled. (D) Cross-section of a spt gynoecium at stage 13 showing that the septum has not fused and transmitting tract cells are absent. (E) Wild-type silique at stage 20 with one valve removed showing the complete septum and seeds. (F) Mature spt-2 siliques at stage 20 with one wall removed showing the characteristic spatula shape. The septum is completely unfused or fused only at the base. (G) Stage 7 spt gynoecium where the medial region is already retarded in development (arrow). (H) A cleft deepens in the medial regions of spt gynoecia during stage 9. (I) At early stage 11, the spt gynoecium is prematurely starting to close over. In contrast, no stigmatic papillae are evident because the development of papillae is both reduced and delayed in spt. (J) At late stage 11, the spt style (bracketed) becomes distinct from the ovary and the cleft where the style failed to fuse is obvious. Stigmatic papillae start to develop. (K) At stage 6, SPT is expressed in a cone of cells in the gynoecium primordium. SPT expression is localized primarily to the medial regions. At stage 8, SPT is expressed in the inner medial regions where the medial ridges are growing to form the septum. SPT is also expressed at the top of the gynoecium where the stigma and style will develop. At stage 11, SPT is expressed in the septum including the developing transmitting tract. SPT is also expressed in the ovule primordia. At stage 13, SPT is expressed throughout the valves excluding the exocarp cells. By stage 17, SPT expression becomes restricted to the valve margins although SPT has no known function in fruit dehiscence. Scale bars in A-B represent 100 µm, C-D and J represent 50 µm, E-F represent 500 µm, and G-I represent 25 µm. A-J: from Alvarez and Smyth, 2002.
The defect in spt gynoecia is first detected at stage 7 when the growth of the medial region begins to lag behind the lateral regions (Figure 28G; Alvarez and Smyth, 2002). Growth of the medial region continues to be retarded through stages 8 and 9 and by stage 10 a cleft has formed (Figure 28H). Also at stage 10, the apex of the gynoecium narrows earlier than wild type. However the stigmatic papillar cells that form on wild-type gynoecia at stages 9 and 10 are not found on spt gynoecia until late stage 11 and even then, their numbers are reduced (Figure 28I and 28J).
SPATULA encodes a bHLH transcription factor
The SPATULA gene encodes a basic helix-loop-helix (bHLH) transcription factor (Heisler et al., 2001). SPT is expressed in a dynamic pattern within the developing gynoecium paralleling its role in the development of medial tissues. Within stage-4 floral meristems, SPT RNA is first detected in a pie-wedge-shaped domain that appears to mark the cells that will later form the gynoecium. By stage 6, SPT transcript is strongest in the medial regions of the gynoecium (Figure 28K) and by stage 7, SPT expression is limited to the inner medial regions of the gynoecium where the septum will form. At the apex of the gynoecium, SPT expression spreads where the future stigma will develop (Figure 28K). SPT expression is further restricted to only a few cells at the leading edge of the medial ridge during stage 8 (Figure 28K). From stage 9 to 11, SPT is expressed throughout the septum and the stigmatic papillae, but this expression ceases in stage 12 (Figure 28K). This early expression pattern correlates directly with the abnormal phenotypes observed in spt mutants. Beginning in stage 12, SPT is expressed throughout the valves except the epidermis and the vascular bundles (Figure 28K). After fertilization as the fruit expands, SPT is restricted to the valve margins (Figure 28K). Currently, no role for SPT in the valves or valve margins has been uncovered.
SPT is also expressed in petals, stamens, ovules, the seed attachment site, young leaves, stipules, maturing pith cells in the stem, differentiating vascular cells, and lateral root caps although no abnormal phenotype is observed in these tissues in spt mutants.
The expression of SPT in many tissues where no mutant phenotype has been observed raises the possibility that additional factors may act redundantly with SPT. Specifically, bHLH proteins often heterodimerize, so other interacting bHLH proteins may redundantly mediate these functions. For example, SPT is specifically expressed in areas where cell separation will occur, such as the valve margin, the stomium of the anther, and the seed abscission zone, although currently this expression has no known function. It will be interesting to test whether SPT heterodimerizes with the ALCATRAZ or INDEHISCENT bHLH proteins, which are also expressed in the valve margin (see sections 5.1.2 alcatraz (alc) mutants keep the seeds imprisoned and 5.1.3 INDEHISCENT (IND) is required for valve margin specification).
5.6.6 Fruit Shaped Like a CRABS CLAW (CRC)
Mutations in the CRC gene prevent the fusion of the style and upper part of the ovary. Each side of the reduced style curves inward creating a notch reminiscent of a crab's claw (Figure 29A and 29B). Additionally, crc mutants fail to produce nectaries and crc fruit are shorter and wider than wild type with fewer seeds (Figure 29A to 29D; Alvarez and Smyth, 1999). Occasionally, an ovule or an ovule-like primordium projects outward from the replum hinting that crc fruit have a defect in abaxial/adaxial polarity (Figure 29G; see section 6.1 Abaxial/adaxial axis of the fruit). The defects in crc first become evident when the crc gynoecial platform is already broader than wild type at the initiation of the gynoecium during stage 6 (Alvarez and Smyth, 2002). By stage 8, as the crc gynoecium elongates, two clefts start to form in the top of the medial regions (Figure 29E). The clefts deepen in stage 9 and the upper ridges where the stigma and style will form begin to turn inward at stage 10 (Figure 29F). During stage 12, the stigmatic papillae forming on the top of the style halves commingle (Figure 29G). At the cleft, the septum often fails to fuse (Alvarez and Smyth 2002).
Figure 29.
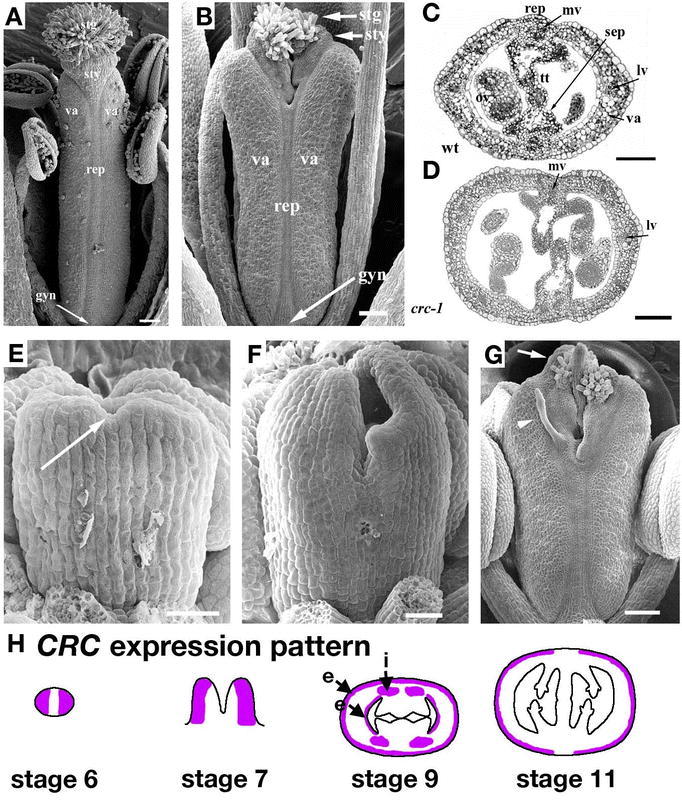
CRABS CLAW is necessary for apical fusion of the carpels. (A) Wild-type gynoecium at stage 13. (B) crc mutant at stage 13. Note the unfused style and upper ovary. (C) Cross-section through a wild-type gynoecium at stage 13. (D) Cross-section through a crc gynoecium at stage 13 showing the unfused septum. (E) A cleft (arrow) begins to form in the medial regions of a stage 6 crc gynoecium. (F) Stage 10 crc gynoecium where the cleft has deepened and the tips of the carpels where the stigma will form are starting to turn inward. (G) Late stage 12 crc gynoecium where the two halves of the stigmatic surface are curved inward and the stigmatic papillae are interlocking. An aberrant ovule primordium has arisen on the outside of the replum (arrowhead). (H) Based on in situ hybridization results, CRC is expressed in the lateral sides of the gynoecium at stage 6 and stage 7. At stage 8, CRC is expressed in two domains. The epidermal (e) domain includes both the outer and inner epidermis of the carpels. The internal (i) domain includes 4 spots near the placentas. By stage 11, CRC is expressed only in the outer epidermis of the valves and not the replum or the internal domains. Scale bars on A-D represent 100 µm, E-F represent 25 µm, and G represents 50 µm. A-G: from Alvarez and Smyth, 2002.
CRABS CLAW encodes a YABBY transcription factor
CRC is the founding member of the YABBY gene family, a family of six transcription factors each with a zinc finger and a helix loop helix domain (Bowman and Smyth, 1999). CRC is expressed in the developing nectaries and gynoecium paralleling the disruptions in the mutant. CRC is expressed in the two lateral sides of the gynoecium primordium at stage 6 (Figure 29H). During stages 7 and 8, two domains of expression become apparent: epidermal and internal. The epidermal expression is primarily in the outer epidermis although weak expression can be seen in the ena layer during stages 7 through 9. Expression in the outer epidermis completely encircles the fruit including the replum until stage 10 when expression in the replum begins to decline (Figure 29H). By stage 12, expression in the outer epidermis is not detected. Simultaneously, during stages 7 through 10, CRC is expressed internally in four stripes adjacent to the cells that will form the placentas (Figure 29H). Both the epidermal and internal expression domains run the whole length of the gynoecium, including the future style.
Constitutive expression of CRC (35S::CRC) produces solid cylindrical gynoecia composed primarily of style tissue topped with stigmatic tissue, which is consistent with the role of CRC in the development of the style (Eshed et al., 1999). In addition carpelloid tissues including stigma and ovules develop on the sepal margins supporting the role of CRC in the carpel specification pathway (see section 2.2 Carpel development in the absence of AGAMOUS).
5.6.7 Synergistic Interactions between CRABS CLAW, STYLISH, and SPATULA
Because of the similarity in the failure of apical fusion in crc and spt, crc spt double mutants were constructed to test whether these genes act in the same pathway. The phenotype of spt crc double mutants is far more severe than either single-mutant phenotype (Figure 30A and 30B; Alvarez and Smyth, 1999; Alvarez and Smyth, 2002). The spt crc gynoecia are almost completely unfused except at the base. Furthermore, marginal tissues including stigmatic papillae, style, ovules, and septum are severely reduced (Figure 30A and 30B). Only the valve tissue remains fairly normal. The synergistic phenotype of the crc spt double mutant can be identified early in development at stages 7 and 8 when large clefts arise in the medial regions of the gynoecial tube (Figure 30C). The interaction between SPT and CRC must be indirect because the expression domains of these two genes do not overlap. SPT expression is not affected in crc gynoecia, but CRC transcript is increased in spt gynoecia (Heisler et al., 2001). The synergistic phenotype reveals the roles of these genes in overlapping redundant pathways controlling the development of the medial tissues.
Figure 30.

The crc spt double mutant. (A) SEM of a crc spt double-mutant fruit showing that the carpels are largely unfused. The stigma (stg), ovules (ov) and replum (rep) are labeled. (B) Cross-section of a crc spt gynoecium of the lower part of a mature flower. The septum (sep), medial vascular bundle (mv), lateral vascular bundle (lv), and ovule (ov) are labeled. (C) SEM of a crc spt gynoecium at stage 8, showing that the carpels are largely unfused and curve inward. Scale bars in A-B represent 100 µm and the scale bar in C represents 25 µm. A-C: from Alvarez and Smyth 2002.
Similarly, mutations in STY interact synergistically with mutations in CRC and SPT. The phenotypes of both sty1 crc and sty1 spt double mutants are more severe than any of the single mutants (Kuusk et al., 2002). The style, septum, and stigmatic papillae are all greatly reduced in both sty1 crc and sty1 spt. In fact in sty1 spt, the stigmatic papillae are completely absent and the upper part of the gynoecium becomes unfused and leaf-like.
6.0 AXES OF POLARITY IN THE FRUIT
In the last section, we have discussed the factors involved in the development of specific tissues and cell types within the fruit. However, these factors generally control what cell type develops, but not where within the fruit that cell type is formed, as we saw in several ectopic expression studies. For example, the ectopic expression of STYLISH produces style cells in the valves (see section 5.6.4 STYLISH (STY) genes specify style development). How is each cell type correctly situated during wild-type fruit development? As we have seen there is a fairly elaborate mechanism involving FRUITFULL and REPLUMLESS, to ensure that the valve margin differentiates at the border between the valve and the replum (see section 5.4 Model for the formation of a stripe of valve margin tissue). But, how does the fruit ensure that the stigma develops only at the top and the ovules only form on the inside? Furthermore, how does the developing gynoecium determine how to proportion the style relative to the ovary and the gynophore?
The positions of different tissues in the fruit are arranged on three axes of polarity: the medial/lateral axis, the abaxial/adaxial axis, and the apical/basal axis (Figure 2). On the medial/lateral axis, the replum and septum are in the middle and the valves are at the edges. Little is known about the specification of the medial/lateral axis, although as we have seen there are many mutants that affect the formation of medial tissues (see section 5.6 fusion and the formation of the stigma, style, septum, and transmitting tract). The kanadi1 kanadi2 mutant, which primarily affects abaxial/adaxial patterning may also have a role in medial/lateral axis specification because the ovules, a medial tissue, spread throughout the lateral valves (see section 6.1.3 kanadi inside out gynoecia; Eshed, et al., 2001). More is known about the specification of the abaxial/adaxial axis and the apical/basal axis, so we will discuss these axes in detail.
6.1 Abaxial/adaxial Axis of the Fruit
All lateral organs, including fruit, exhibit differences between the adaxial side (the side facing the meristem) and the abaxial side (the side away form the meristem). These differences allow each organ to specialize with different functions in different regions. For example, the top of the leaf primarily captures light while the bottom of the leaf exchanges gasses with the environment. In the gynoecium, abaxial/adaxial polarity is manifested as the difference between the outer cell layers and the inner cell layers of the fruit. The inner side of the fruit including the placenta, ovules, and septum are adaxial tissues because they face the location of the former floral meristem (Figure 31A). Conversely, the replum and the outer valve epidermis are abaxial tissues. One of the most striking signs of a defect in abaxial/adaxial identity is the formation of ovules on the outside of the fruit.
Figure 31.
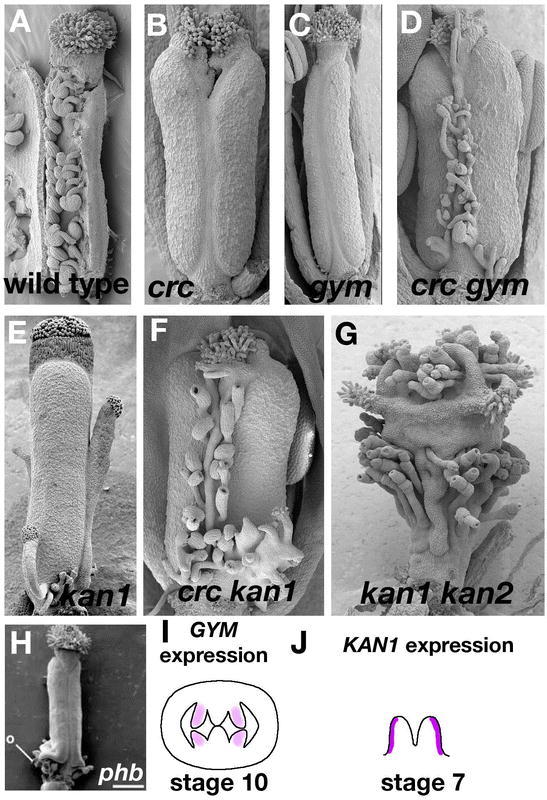
Abaxial/adaxial polarity in the gynoecium. (A) SEM of a wild-type gynoecium opened to show the ovules, which have developed inside. (B) crc gynoecium with no ectopic ovules on the exterior of the gynoecium. (C) gym gynoecium with no ectopic ovules. The gynoecium is narrower and shorter than wild type. (D) crc gym double mutant showing ectopic ovules developing along the replum. (E) kan1 gynoecium with ectopic tissue growing from the replum, but no ectopic ovules. (F)crc kan1 double-mutant gynoecium with two rows of ovules developing on the outside of the gynoecium. Septal tissue is also growing out where the replum should be. (G) kan1 kan2 gynoecium where the entire external surface of the gynoecium is producing ectopic ovules except for a septum that is growing out. (H) phb gynoecium showing a few ectopic ovules growing from the base of the gynoecium. (I) GYM is expressed in developing ovules at stage 10 based on in situ hybridization results. (J) KAN1 is expressed in the abaxial side of early stage 7 gynoecia. Scale bar in H represents 0.38 mm. A, E-G: Reprinted from Current Biology, 11, Eshed, Y., Baum, S.F., Perea, J. V., and Bowman, J.L. Establishment of polarity in lateral organs of plants, 1251–1260, Copyright (2001), with permission from Elsevier. B-D: from Eshed et al., 1999. H: from McConnell and Barton, 1998.
6.1.1 Role of CRABS CLAW in Abaxial/adaxial Polarity
Members of the YABBY gene family generally specify abaxial cell fate (Siegfried et al., 1999). Although CRABS CLAW (CRC) is a member of the YABBY family and is expressed in the abaxial side of the gynoecium (see section CRABS CLAW encodes a YABBY transcription factor), the crc-mutant phenotype shows little evidence of altering abaxial/adaxial polarity (Figure 31B). Does CRC have a hidden role in establishing polarity in the fruit? To uncover the role of CRC in the abaxial/adaxial polarity of the gynoecium, Eshed et al. screened for genetic enhancers of crc (1999).
6.1.2 Ovules Form on the Outside of gymnos (gym) Crabs Claw Gynoecia
Mutations in gymnos (gym) (also referred to as pickle (pkl) Ogas et al., 1999) enhance the crc phenotype producing twenty to thirty external ovules on the double-mutant gynoecia (Figure 31D; Eshed et al., 1999). During stage 8, these external ovule primordia are formed in a row on the edges of the replum mirroring the development of the ovule primordia from the placenta on the inside of the fruit. As the gynoecium continues to develop, secondary asynchronous waves of ovule development occur on the outside of the gynoecium. The loss of abaxial identity is incomplete in crc gym double mutants because an external septum does not develop and the abaxial valves remain. However, large cells of aberrant morphology are scattered throughout the valve epidermis.
Although gym single mutants show a number of abnormalities (Ogas et al., 1997), no defect in the abaxial/adaxial polarity of the fruit has been observed. The gym gynoecia are shorter and narrower than wild type and seem to mature slowly (Figure 31C; Eshed et al., 1999).
GYMNOS encodes a chromatin-remodeling enzyme
GYM, also known as PICKLE (PKL) (Ogas et al., 1999), encodes a CHD3/4 DNA-dependant ATPase in the family of SWI2/SNF2 chromatin-remodeling enzymes (Eshed et al., 1999). A PHD finger and a MYB DNA-binding domain are also predicted in the GYM protein. GYM is related to Drosophila Mi-2, which forms complexes with histone deacetylases to repress transcription of target genes. GYM, likewise, may form complexes with histone deacetylases to repress the transcription of genes involved in maintaining an undifferentiated state.
GYM is expressed in undifferentiated tissue including embryos, meristems and organ primordia. Also, GYM is expressed throughout gynoecia during stages 5–7, but becomes limited to the medial ridge and ovule primordia during stage 8 (Eshed et al., 1999). Finally GYM expression becomes restricted to the developing ovules (Figure 31I).
Why are ectopic ovules produced on the outside of gym crc double-mutant fruit? Eshed et al., hypothesize that in addition to the roles of CRC in establishing carpel identity and promoting style formation, CRC specifies abaxial cell fate within the gynoecium, while GYM mediates the repression of genes that promote an undifferentiated state (1999). It is possible that mutations in gym prolong the expression of meristematic genes in the gynoecium extending the period of indeterminacy and allowing placenta to develop. Simultaneously, mutations in CRC lift the normal restriction of placenta development to the inside (adaxial side) of the fruit. However, the reversal in abaxial/adaxial polarity appears to be limited to the medial regions in crc gym. One possible reason that the valves are not as affected in crc gym mutants is that other YABBY genes expressed in the valve may act redundantly with CRC. FILAMENTOUS FLOWER (FIL), YABBY2 (YAB2), and YABBY3 (YAB3) are all expressed in the abaxial side of the valves, but not in the medial regions (Siegfried et al., 1999). Other genes specifying abaxial cell fate could act redundantly with CRC as well. One such gene, KANADI1, was also isolated as an enhancer of crc.
6.1.3 Inside Out kanadi (kan) Gynoecia
Mutations in both KAN1 and KAN2 nearly abolish abaxial/adaxial polarity throughout the plant. Adaxial cell types replace abaxial ones suggesting that the KAN genes redundantly promote abaxial cell fate. The effect on the gynoecium is monstrous; ovules cover the outside of kan1 kan2 gynoecia replacing all of the abaxial valve tissue (Figure 31G; Eshed et al., 2001). It is interesting that the inside of the fruit is not merely reproduced on the outside; instead, the placenta expands to cover the surface suggesting that kan1 kan2 gynoecia also have a defect in medial/lateral patterning.
KAN1 and KAN2 act redundantly and kan2 mutants look completely normal. In kan1-mutant gynoecia, a ridge of tissue grows out from the replum and a few ectopic ovules are found on the exterior base of the gynoecium hinting at a slight alteration in abaxial/adaxial polarity (Figure 31E; Kerstetter et al., 2001; Eshed et al., 2001).
kanadi1 enhances crabs claw abaxial/adaxial defects
The kan1 mutant was identified in a screen to find enhancers of the crc mutant (see section 6.1.1 Role of CRC in abaxial/adaxial polarity; Eshed et al., 1999). The crc kan1 double mutant produces two complete rows of ovules on the exterior of the gynoecium and the replum grows out to produce an exterior septum including transmitting tract tissue (Figure 31F). The development of the external ovules and septum coincides with the development of the internal ones. Ectopic carpel tissue is also formed from the bottom of the gynoecium. In contrast to kan1 kan2, the defect in abaxial/adaxial polarity is largely limited to the medial regions and the base of the crc kan1 fruit.
KANADI genes encode GARP transcription factors
The closely related KANADI genes encode members of the GARP family of putative transcription factors (Kerstetter et al., 2001; Eshed et al., 2001). In Arabidopsis, the GARP family comprises more than fifty members including two additional genes closely related to KAN1/2. The expression pattern of KAN1 is consistent with its role in promoting abaxial identity (Kerstetter et al., 2001). KAN1 is expressed in the abaxial side of young leaves and floral organs. Within the gynoecium, KAN1 is first expressed on the abaxial side (Figure 31J).
The KANADI genes have a central role in specifying abaxial polarity as is demonstrated by the severity of the mutant phenotypes. It is likely that the KANADI and YABBY genes act in separate, but overlapping pathways in specifying abaxial fate (Eshed, et al., 2001).
6.1.4 PHABULOSA (PHB) Promotes Adaxial Fate
While the KANADI and YABBY genes promote abaxial fate, PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV), three members of the class III homeodomain leucine zipper (HD-ZIP) family, promote adaxial fate (Emery et al., 2003; McConnell et al., 2001). Gain-of-function alleles of PHB, PHV, or REV cause lateral organs to become adaxialized, whereas loss-of-function mutations in all three genes (phb phv rev) cause the cotyledons to become radialized and abaxialized and block formation of the shoot apical meristem. In the fruit, the affects of PHB gain-of-function alleles are rather minor causing only a few ovules to be produced on the outside of the fruit at the base (Figure 31H; McConnell and Barton, 1998).
The gain-of-function alleles of PHB, PHV, and REV block a microRNA-binding site thereby disrupting the spatial regulation of transcript accumulation (Emery et al., 2003; Rhoades et al., 2002). Consequently, in the gain-of-function alleles, PHB is ectopically expressed throughout developing leaves instead of being limited to the adaxial side as in wild type (McConnell et al., 2001). It has been proposed that PHB, PHV, and REV may also receive a signal from the meristem signaling adaxial localization since these proteins contain START domains which are putative sterol binding sites.
6.2 Formation of the Apical/basal Axis of the Fruit
The stigma, style, ovary and gynophore are arranged along the fruit from apex to base. Both the ETTIN gene and the plant hormone auxin play important roles in establishing the apical/basal axis.
6.2.1 ETTIN (ETT) Establishes Boundaries Along the Apical/basal Axis
Mutations in the ETTIN (ETT) gene cause spectacular alterations in gynoecium morphology, especially along the apical/basal axis (Sessions and Zambryski, 1995; Sessions, 1997). The ovary in the middle of the fruit is greatly reduced in size whereas the gynophore at the bottom of the fruit and the stigma and style at the top of the fruit increase in size (Figure 32A to 32H). In the ovary, the valves show the greatest reduction, but the placenta and septum are also reduced. Abaxial/adaxial polarity is also affected in ett gynoecia where an everted transmitting tract tissue develops in outgrowths on the exterior of the gynoecium (Figure 32H). The everted transmitting tract is stylar in origin and is evidence of the expansion of stylar tissues basally. Also, the ett stigma and style are split and the vascular tissue is affected (Sessions and Zambryski, 1995). Additionally, ett flowers tend to have one extra sepal and one extra petal, but tend to be missing a stamen.
Figure 32.
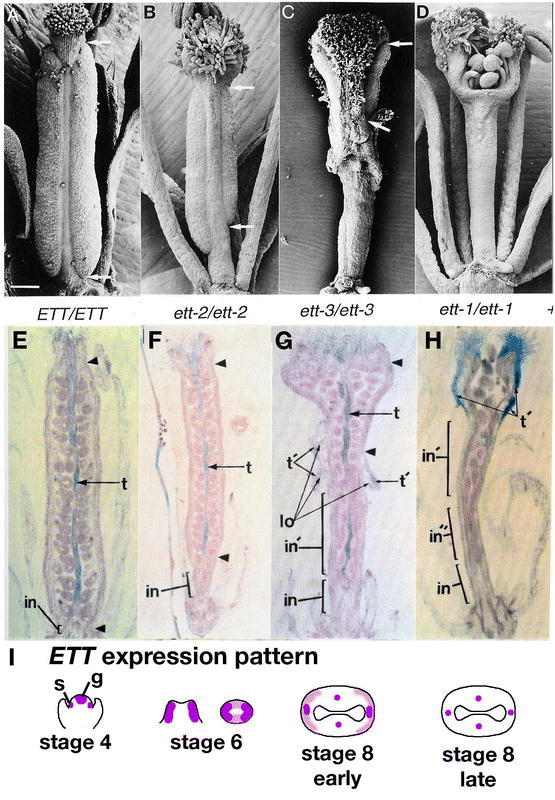
ETTIN and the apical/basal axis of the gynoecium. (A) Wild-type gynoecium post-pollination. The arrows mark the extent of valve elongation. (B) Gynoecium of the weak ett-2 allele. Note the uneven valves. (C) Gynoecium of the intermediate ett-3 allele. Note the small valves surrounded entirely by stigmatic and stylar tissue. (D) Gynoecium of the strong ett-1 allele. Valves are completely absent. (E) Longitudinal section of a wild-type gynoecium. The blue color indicates transmitting tract cells. t = transmitting tract; t′ = everted transmitting tract; in = internode with a solid stem; in’ = externally gynophore stalk, but contains ovules; in″ = solid gynophore stalk that has formed from a postgenital fusion event; lo = lateral outgrowths. (F) ett-2 with a slightly elongated gynophore (in). (G) ett-3 with some transmitting tract on the outward projections (t′). (H) ett-1 gynoecium with transmitting tract cells primarily on the outer apical surfaces. Abnormal ovules are present on part of the gynophore (in′). (I) Diagram depicting the expression pattern of ETT based on in situ hybridization (Sessions et al., 1997). At stage 4, ETT is expressed in the stamen primordia (s) and the gynoecium primordium (g). At stage 6, ETT is expressed abaxially in the gynoecium. ETT expression is stronger in the lateral regions of the gynoecium than the medial regions. By early stage 8, ETT is strongly expressed in the medial and lateral vascular bundles and weakly expressed abaxially the gynoecium. By late stage 8, ETT expression is only detected within the vascular bundles of the gynoecium. Scale bar in A represents 153 µm. A-H: from Sessions and Zambryski, 1995.
The severity of the ett phenotype ranges along an allelic series. In ett-2, a weak allele, the fruit is fertile and valves are present although they are reduced in size (Figure 32B and 32F). Often the valves are uneven, suggesting that the development of one valve is independent of the other. In ett-3, an intermediate allele, small valves develop that are completely surrounded by stylar and stigmatic tissue (Figure 32C). The everted transmitting tract occurs in both medial and lateral outgrowths (Figure 32G). The basal stalk is covered in style-like epidermal cells. In the strong ett-1 allele, valves are often completely absent and the gynoecia are sterile (Figure 32D). The base of the ett-1 gynoecia consists of a solid stock (Figure 32H). At the top of the stalk region, the center opens into a narrow space filled with abnormal ovules. Style-like epidermal cells cover the stalk. The top of the ett-1 gynoecium is covered with everted transmitting tract. Although these descriptions typify the gynoecia from each allele, the ett phenotype is somewhat variable and gynoecia from a single plant will vary in severity of the phenotype (Sessions and Zambryski, 1995; Sessions, 1997).
Defects in ett-mutant morphology appear early in flower development (Sessions, 1997). At stage 5, the meristem elongates more than wild type, producing extra layers of cells below the gynoecium that likely contribute to the elongated gynophore. As the ett gynoecium grows during stages 7 and 8 it forms a trumpet shape instead of a cylinder. Differentiation begins prematurely in stage 8 when stigmatic papillae become visible.
The ett phenotype has been interpreted as altering two hypothetical boundaries along the apical/basal axis of the fruit (Figure 33; see also section 6.2.4 Relationship of ETTIN and SPATULA to polar auxin transport; Sessions, 1997; Nemhauser et al., 2000). One boundary marks the apex of the gynoecium and the other lies at the base of the ovary delineating it from the gynophore. In ett mutants, the basal boundary is pulled upward leading to the elongated gynophore, while the apical boundary is pulled downward, increasing the stigma and style tissue. In ett-1, the basal boundary is interpreted as being above the apical boundary and so no valves are formed (Sessions, 1997). The basal boundary must be independent on each side of the fruit because the size of the valve on each side is independent. The ETT gene may be involved in establishing or interpreting these boundaries within the gynoecium (see section 6.2.4 Relationship of ETT and SPT to polar auxin transport).
Figure 33.
A model for the interaction of auxin and ETT in establishing apical/basal boundaries within the gynoecium. (A) Without treatment. The triangle represents the auxin gradient within the gynoecium. Auxin concentrations are high within the style and decrease toward the gynophore. The cylinder represents the gynoecium with border a between the style (dark green) and ovary (light green) and border b between the ovary and gynophore (yellow). (i) In wild-type gynoecia ETT interprets the concentration of auxin to establish an apical boundary and a basal boundary. (ii) In weak ett-2 mutants these boundaries are slightly destabilized. (iii) In strong ett-1 mutants the boundaries are further destabilized and the middle valve region is completely lost. (iv) In spt mutants, the apical boundary moves toward the top, so that apical tissue is reduced. (B) When treated with NPA, the auxin gradient is thought to becomes steeper. (i) In wild type, when the gradient becomes steeper, the valve regions are reduced. (ii) In ett-2 mutants the boundaries become further destabilized and the valves are lost. (iii) In strong ett-1 mutants, the boundaries are already destabilized, so the change in the auxin gradient has no effect. (iv) In spt, the increased auxin in the apex of the gynoecium caused by NPA treatment can rescue the defect. At the same time, the NPA has little affect on the valve length or gynophore length. The effects of NPA on wild type may act through SPT. From Nemhauser et al., 2000.
ETTIN encodes an auxin response factor
The ETTIN gene encodes a putative transcription factor with a DNA binding domain similar to that of Auxin Response Factors (ARF; Sessions et al., 1997). ARFs bind to auxin response elements (AuxREs) in the promoters of auxin-regulated genes to control their transcription. In addition to the DNA binding domain, ARFs contain a protein-protein interaction domain, which is not present in ETT. The ETT protein has a predicted nuclear localization sequence and two serine rich regions. The ETT protein probably functions to mediate an auxin response in the gynoecium (Sessions et al. 1997). Like other ARF transcription factors, ETT expression is not auxin inducible (Nemhauser et al., 2000).
ETT has a dynamic expression pattern that matches its many functions during flower development (Sessions et al., 1997). The ETT gene is first expressed in early floral meristems before stage 1. At stage 2 the ETT transcript becomes localized to an apical patch in the floral meristem and to the provascular tissue in the developing pedicel. ETT RNA is not detected in sepal primordia, but is found in both the developing petals and stamens during stages 3 to 9 (Figure 32I). ETT forms a thick ring around the terminal meristem preceding the formation of the gynoecium in stage 5. ETT RNA is localized abaxially in the gynoecium from stages 5 to 8 when it becomes restricted to the four vascular strands in cells between the phloem and the xylem during stages 8 to 12 (Figure 32I).
The expression pattern of ETT in the gynoecium coincides with the hypothesis that ETT acts to establish or interpret boundaries within the apical/basal axis of the gynoecium. At stage 5, the upper edge of the ETT transcript ring could establish the hypothetical apical boundary. Likewise the bottom of the ETT expression ring delineates the bottom of the ovary and could form the basal boundary. Alternatively ETT could act throughout the gynoecium at these early stages to interpret positional information (Sessions et al., 1997).
6.2.2 ETTIN Negatively Regulates SPATULA Expression
Mutations in spt suppress the ett phenotype by restoring valves and eliminating the overgrown gynophore and everted transmitting tract (Figure 34A to 34D; Alvarez and Smyth, 1998; Heisler et al., 2001). ETT negatively regulates SPT expression restricting SPT to the developing septum and transmitting tract. In ett mutants, SPT is ectopically expressed in the locations where ETT transcript would normally be present. At stage 6, SPT transcript is expanded into lateral regions of ett gynoecia where it is not found in wild type (Figure 34E). In stage 7, SPT is ectopically expressed in the abaxial cells of ett mutants. During stage 8, these abaxial cells undergo periclinal divisions similar to those that normally occur to form the septum and placenta. These periclinal divisions start to form the everted transmitting tract showing that the ectopic SPT expression is responsible for the formation of the everted transmitting tract. In summary, ETT negatively regulates the expression of SPT excluding it from the lateral and the abaxial zones of the gynoecium (Heisler et al., 2001). It will be interesting to determine whether ETT directly represses SPT since the SPT promoter contains several putative AuxRE sites that could serve as binding sites for ETT protein.
Figure 34.
ETT negatively regulates SPT. (A) Wild-type gynoecium at stage 12 with the parts labeled: stigma (sg), style (sy), replum (rep), valves (va), and gynophore (gyn). (B) spt gynoecium at stage 12 showing reduced stigmatic papillae and an unfused style. (C) ett gynoecium at stage 12 with an elongated gynophore and expanded stigma and style tissue. (D) ett spt double-mutant gynoecium in which valve development has been restored and looks fairly similar to spt single mutants. (E) Diagram of the expression pattern of SPT in ett mutants based on in situ hybridization results (Heisler et al., 2001). SPT is ectopically expressed in the abaxial regions of the ett gynoecium where the everted transmitting tract will develop. Scale bars in A-D represent 200 µm. A-D: from Heisler et al., 2001.
ETT patterns the gynoecium largely by restricting SPT activity. The ectopic expression of SPT in the abaxial domain clearly explains the defect in abaxial/adaxial polarity seen in ett mutants. However, ett spt double mutants also eliminate the apical/basal defects seen in ett; the apical and basal boundaries in the ett spt double mutant appear to form in the correct locations (Heisler et al., 2001).
ETTIN interacts synergistically with TOUSLED
In ett tsl double mutants, the gynoecium forms a fantastical structure composed only of placenta and ovules on a stalk (Figure 35A; Roe et al., 1997b). This extreme structure lacking valves, stigmatic tissue, and style results even when tsl is combined with the weak ett-2 allele, which itself has fairly large valves. This structure arises from a stalk with two laminar sheets producing ovule primordia at stage 10 (Figure 35B).
Figure 35.
ETT restricts TSL expression to the apex. (A) ett-1 tsl-1 double-mutant gynoecium at stage 13. The gynoecium consists entirely of an elongated gynophore topped with ovule bearing tissue. Arrowhead points to a spike of tissue emerging between the placentae. (B) ett-1 tsl-1 double-mutant gynoecium at stage 10 where ovule primordia (o) are just starting to develop. (C) TSL::GUS expression in ett-1 gynoecium at stage 13 is expanded down to the gynophore. (D) TSL::GUS expression in the weak ett-2 allele at stage 13 is also expanded down and partly into the valves. Scale bars in A-B represent 100 µm. A-D: from Roe et al., 1997b.
TSL is ectopically expressed in ett mutants stretching not just into the ectopic stylar medial tissue, but also into the valves of weak ett-2 mutants (Figure 35C to 35D compared to Figure 25I). Therefore, ETT limits the expression of TSL to the apex of the gynoecia establishing the apical boundary. The tsl ett double mutant would be expected to partially suppress the ett phenotype by eliminating the ectopic TSL. Instead, the phenotype is enhanced. One explanation is that TSL regulates other proteins involved in gynoecium development, which enhance the ett phenotype when they are misregulated in the double mutant. The observation that mutations in the spt gene can suppress the ett phenotype raises the question of whether spt mutants can also suppress the ett tsl double-mutant phenotype.
6.2.3 Auxin as a Morphogen in the Formation of the Apical/basal Axis of the Gynoecium
The plant hormone auxin is proposed to act as a morphogen forming a gradient that directs patterning in the Arabidopsis gynoecium (Nemhauser et al., 1998; Nemhauser et al., 2000). Generally auxin is thought to be synthesized in young apical tissues and transported basally throughout the plant. Members of the PIN family of proteins control the polarized transport of auxin and are thought to act as efflux carriers. The PIN proteins are dynamically localized to a particular side of the cell, so that transport has a net flow in the direction of their localization (Friml, 2003). This polarized transport of auxin is crucial for development of the plant. Application of polar auxin transport inhibitors is thought to disrupt the auxin gradient causing auxin to accumulate in high concentrations near the source and depriving the downstream cells of auxin. N-1-naphthylphthalamic acid (NPA) inhibits polar auxin transport and can be used to study the effects of disrupting auxin transport on development.
Application of auxin transport inhibitors throughout plant development has severe effects on the plant and greatly reduces the number of floral meristems produced (Okada et al., 1991). Therefore, to study the effects on gynoecium development, Nemhauser et al. (2000) devised a system of transient application of NPA to the inflorescence meristem. Even transient application of NPA caused the inflorescence meristem to become tapered and produce few floral meristems. The effect was temporary and after 13 days, resumption of normal flowering occurred. The flowers that developed within the period during which NPA affected development were studied in detail. In these flowers, the number of sepals and petals was variable and the number of stamens was reduced. The sepals and petals tended to be narrower than wild type. These phenotypes mimic those observed in pinformed (pin1) mutants confirming the disruption of polar auxin transport by NPA (see section 6.2.5 Mutations in PIN-FORMED (PIN1) disrupt polar auxin transport and the apical/basal axis of the gynoecium). In the gynoecium, application of low levels of NPA caused a slight increase in the stigma and style and a slight decrease in the ovary length (Figure 36A and 36B). Application of higher levels of NPA caused a more dramatic decrease in the valves while the stigma, style and gynophore increased in size (Figure 36C). In NPA treated gynoecia, stigmatic papillae developed earlier than normal and the replum protruded instead of being set in an indentation. These phenotypes caused by transient application of NPA to wild-type gynoecia mimicked weak ett phenotypes.
Figure 36.
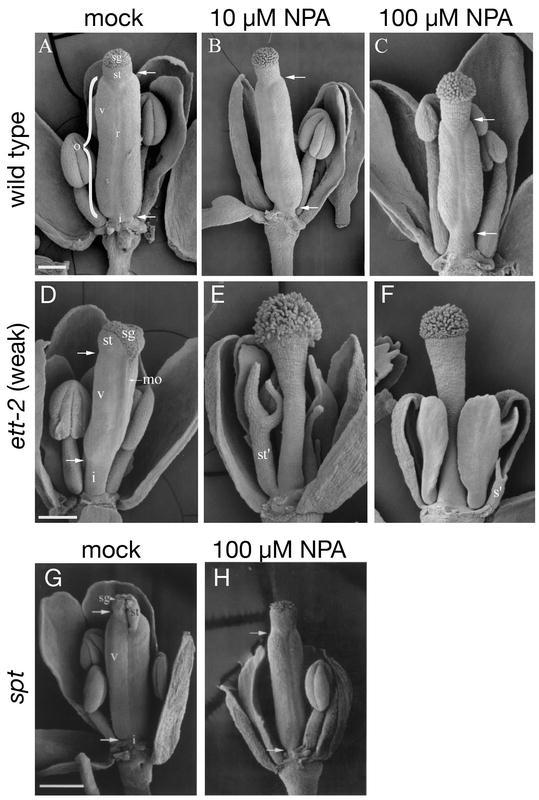
Inhibition of auxin polar transport produces phenotypes resembling ett mutants. (A) A mock treated wild-type gynoecium with the stigma (sg), style (st), ovary (o), vlaves (v), replum (r), and gynophore (i) labeled. (B) Wild-type gynoecium treated with 10µM NPA. The valves are slightly decreased whereas the stigma style and gynophore are slightly increased. (C) Wild-type gynoecium treated with 100µM NPA. The valves are further reduced whereas the stigma, style and gynophore are further increased. The replum also begins to protrude outward. (D) A mock treated ett-2 gynoecium displaying the weak ett phenotype where valves are still present. (E) Treatment of an ett-2 gynoecium with 10µM NPA dramatically increases the severity of the phenotype. Valve tissue is completely lost and the phenotype is comparable to the strong ett-1 phenotypes. The wild-type response to 10µM NPA is relatively minor. The dramatic change in the ett-2 phenotype highlights its increased susceptibility to NPA. (F) Treatment of ett-2 with 100µM NPA does not further increase the severity of the phenotype. (G) A mock treated spt-2 gynoecium. (H) Treatment of spt-2 gynoecia with 100µM NPA dramatically rescues the spt phenotype. The style fuses and more stigmatic papillae are produced. A-H: from Nemhauser et al., 2000.
6.2.4 Relationship of ETTIN and SPATULA to Polar Auxin Transport
Since NPA treatment promoted ett-like phenotypes, Nemhauser et al. examined the effect of NPA on ett mutants (2000). Weak ett alleles were sensitive to application of NPA, which dramatically enhanced the severity of the ett phenotype. Application of low levels of NPA, which had only a slight effect on wild-type gynoecia, completely eliminated the formation of valves in ett-2 gynoecia similar to the phenotype of strong ett-1 alleles (Figure 36D and 36E). Increasing the NPA dosage further did not increase the effect (Figure 36F). Application of NPA to ett-1 mutants, only slightly enhanced the already severe phenotype. For both ett-2 and ett-1, NPA treatment prevented the formation of ovules. NPA treatment seemed to completely disrupt the boundaries between style and ovary and the ovary and gynophore in weak ett alleles.
Based on the phenotypes of NPA treated wild type and ett gynoecia, Nemhauser et al. (2000) have proposed a model suggesting that auxin forms a morphogen gradient within the gynoecium and ETT interprets the gradient establishing the boundaries between regions of the gynoecium (Figure 33). The auxin gradient is thought to form with high concentrations at the apex of the developing gynoecium and decreasing levels toward the base. In normal gynoecia, these thresholds lie at the top and the bottom of the ovary. If NPA is applied, it is thought that the gradient becomes very steep with pools of auxin in the style and very low levels of auxin below. In this case, extra stigma and style tissue develop as the apical boundary moves downward and extra gynophore develops as the basal boundary moves upward, all at the expense of the valves. In ett mutants, the interpretation of the auxin gradient is affected and the boundaries become destabilized causing the apical boundary to move down while the basal boundary moves up. Application of NPA to ett mutants further destabilizes the boundaries increasing the severity of the phenotype.
In contrast to the enhancement of ett phenotypes with NPA, treatment of spt-mutant gynoecia with NPA rescued the spt phenotype allowing complete fusion of the style and increased development of stigmatic papillae (Figure 36G and 36H). According to the model, addition of NPA increases the concentrations of auxin at the top of the gynoecium rescuing this phenotype (Figure 33). In spt mutants treated with NPA, the valve and gynophore remained unaffected in contrast to wild type suggesting that the effects of auxin polar transport probably act through a pathway mediated by SPT (Nemhauser et al., 2000). Since ETT negatively regulates SPT, the enhanced sensitivity of ett to NPA is likely to result from increases in the ectopic SPT expression (Heisler et al., 2001).
6.2.5 Mutations in PIN-FORMED (PIN1) Disrupt Polar Auxin Transport and the Apical/basal Axis of the Gynoecium
Mutations in the PINFORMED (PIN1) gene lead to a decrease in polar auxin transport to less than 15% of that seen in wild-type plants (Okada et al., 1991). The pin inflorescence initially forms a barren stalk without any floral meristems, but later becomes fasciated and forms flowers. The number of sepals and petals tends to be increased, but stamens are almost absent in pin flowers. The petals tend to fuse at their bases. In pin flowers, a conversion of organs toward carpels is seen including carpelloid sepals, carpelloid petals, and carpelloid stamens. Flowers at the top of the fasciated inflorescence tend to have more than one gynoecium. The pin gynoecia display a range of phenotypes from almost normal structures with two valves, style and stigma, to stalk like gynoecia with an elongated gynophore topped with a style and stigma (Figure 37A; Goto et al., 1991). The few stamens that are present do not produce pollen and ovules fail to develop in the gynoecia, making the pin mutant completely sterile (Goto et al., 1991; Okada et al., 1991). The phenotypes of pin mutants can be mimicked by the application of polar auxin transport inhibitors throughout the life of the plant (Okada et al., 1991). The amount of auxin synthesized in pin plants is dramatically reduced which could be a secondary effect of the lack of transport either through a feedback mechanism or because the flower buds which normally synthesize a large part of the auxin are absent (Okada et al., 1991).
Figure 37.

Polar auxin transport in the gynoecium: PIN-FORMED and PINOID. (A) A pin-mutant flower showing that the gynoecium appears to be a stalk with no valves. (B) A pid gynoecium with no valves where the center has continued to grow through the stigmatic tissue (arrow). (C) A pid gynoecium with one small valve (arrow). Scale bars represent 250 µm.
PIN-FORMED encodes a transmembrane protein
The PIN1 gene is the founding member of the PIN family of putative auxin efflux carriers (Gälweiler, et al., 1998). PIN1 encodes a protein with 8 to 12 putative transmembrane domains surrounding a hydrophilic core, which has been proposed to function as a transmembrane carrier. PIN1 is expressed throughout the plant in cotyledons, flowers, roots, rosette leaves, seedlings, inflorescences, and siliques. In situ hybridization shows that the transcript accumulates in parenchymatous xylem and cambial cells. PIN1 protein is localized to one edge of the cell and this polar localization is often interpreted to indicate the direction of auxin transport (Reinhardt, et al., 2003; Benková, et al., 2003). Consistent with its role in basipetal auxin transport, PIN1 protein localizes to the basal cell membrane in the stem. Further characterization of the localization of PIN1 and the other PIN family members in the gynoecium should shed light on the flow of auxin in this tissue.
6.2.6 pinoid (pid) Gynoecia Are Missing the Central Ovary
Like pin1, the pinoid (pid) mutant forms very few flowers and the meristem forms a barren pin-like stalk. In the few flowers that arise, the pid gynoecia form a long gynophore capped by a style and stigma (Figure 37B and 37C). Therefore, the pid gynoecia appear similar to the most severe phenotypes seen in the pin gynoecia. About one third to one half of the pid gynoecia have small valves often only on one side of the gynoecium (Figure 37C). A small cone of growth occasionally continues up through the center of the stigma (Figure 37B). The defects in pid-mutant gynoecia are evident early in development when the pid gynoecia form as stalks with a small trumpet shaped opening at the top (Bennett et al., 1995). The pid gynoecium is similar to that of ett except that the abaxial/adaxial polarity is not affected (Alvarez and Smyth, 1998). Mutations in PID also affect the rest of the flower, reducing the number of stamens and sepals, but increasing the number of petals. The petals are often fused together forming a heart shape.
PINOID encodes a kinase
PID encodes a serine threonine protein kinase, which autophosphorylates in vitro (Christensen et al., 2000). In the developing gynoecium, PID is transiently expressed in the vascular tissues (Benjamins et al., 2001). PID expression is induced by the application of auxin and the promoter contains a single AuxRE. PID controls the polar localization of PIN protein within the cell and thus regulates polar auxin transport (Friml et al., 2004). High levels of PID expression (i.e. 35S::PID) lead to apical PIN localization while low levels of PID (i.e. in the pid mutant) lead to basal PIN localization. The targets of the PID kinase activity and the mechanism through which PID controls the polar localization of PIN remain elusive.
7.0 THE ROLE OF FERTILIZATION IN FRUIT DEVELOPMENT
The role of the fruit is to protect the developing seeds and ensure their dispersal upon maturation. After fertilization, the developing seeds communicate with the surrounding fruit to coordinate their simultaneous development. The initial decision to begin fruit development generally requires successful fertilization. In Arabidopsis, when the flower is not pollinated, the development of the gynoecium arrests and either the whole flower senesces and abscises or the petals, sepals, and stamens abscise leaving the immature gynoecium (Figure 38A; Vivian-Smith and Koltunow, 1999). The gynoecia of unfertilized flowers do not mature into fruit as is evident from their failure to dehisce and the failure of the enb layer to lignify (Vivian-Smith and Koltunow, 1999). The length of unpollinated gynoecia increases slightly due to cell elongation in the exocarp and endocarp and a little cell division in the mesocarp, but the increase falls far short of that seen in pollinated fruit (Figure 38B to 38D).
Figure 38.
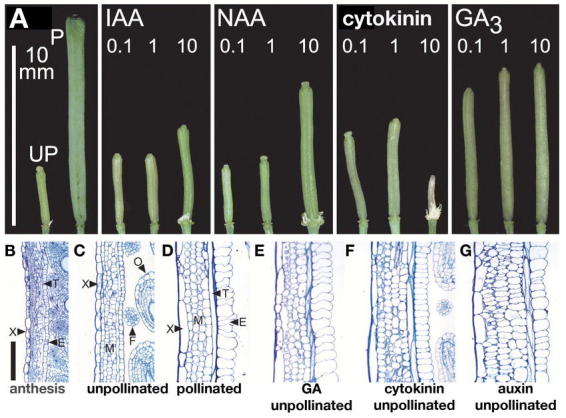
Hormone induced parthenocarpy. (A) Effects of hormone application to wild-type gynoecia at stage 13. 10 µmol of auxin (indoleacetic acid IAA or naphthylacetic acetic acid NAA), 1 µmol of cytokinin (benzyl adenine BA), or 0.1, 1, and 10 µmol of gibberellin (gibberellic acid GA3) induce elongation of the fruit without fertilization. 0.1, 1, or 10 µmol of each hormone were tested. (B) Longitudinal section of the valve of a stage 13 Ler gynoecium. (C) Longitudinal section of the valve of a mature unpollinated Ler gynoecium. Some cell expansion has occurred, but very little cell division. The enb layer has not lignified. (D) Stage 17 mature pollinated wild-type fruit. The exocarp cells have elongated. The mesocarp cells have divided. The enb cells have elongated extensively. The ena cells have expanded in all directions. (E) Longitudinal section of the valve of an unpollinated silique treated with GA. The valve cells have developed as they have in pollinated siliques. (F) Longitudinal section of cytokinin treated unpollinated silique. (G) Longitudinal section of an auxin treated unpollinated silique. The cells have expanded especially in width. Little cell division has occurred. Scale bar in A represents 1 cm. Scale bar in B represents 100 µm. A-G: from Vivian-Smith and Koltunow, 1999.
Even after fertilization, the development of the fruit is coordinated with seed development. In some species, lopsided fruit are produced when one portion of the fruit expands to develop normally around fertilized seeds while the growth of another unfertilized area of the same fruit is retarded (Gillaspy et al., 1993). The seeds are thought to signal to the surrounding fruit via growth hormones, which have been found at high levels in the developing seeds (Vivian Smith and Koltunow, 1999).
7.1 Gibberellins (GA) Promote Post-fertilization Fruit Elongation
In contrast to auxin, which appears to have a dramatic role in the early patterning of the gynoecium, gibberellins have a role later in the post-fertilization elongation of the fruit. Analysis of gibberellin synthesis and perception mutants has shown that gibberellins are required for silique development after fertilization. In ga1-3 mutants, which are severely deficient in GA synthesis, no silique formation is seen after pollination (Vivian-Smith and Koltunow, 1999; Barendse et al., 1986). In mutants with slightly higher levels of active GA, fruit development occurs, but elongation is abnormal. For example, the morphology of the valve cells is affected in ga5-1, which has greatly reduced levels of active endogenous gibberellins (Figure 39A and 39B). During wild-type fruit development after fertilization, the valve cells both divide and expand longitudinally. In contrast in ga5-1 siliques, cell division is reduced and cell expansion is increased especially in width (Vivian Smith and Koltunow, 1999).
Figure 39.

The affect of GA synthesis and perception mutants on valve development. (A) Longitudinal section through a stage 17 valve, 7 days after pollination. The exocarp (X), mesocarp (M), enb (T), and ena (E) are labeled. (B) Longitudinal section through a ga5 valve after pollination. The valve cells, especially the mesocarp cells, expand more laterally and have fewer divisions. (C) The gai valves look similar to the ga5 valves. (D) Valves of wild-type unfertilized parthenocarpic fruit induced with 10mM auxin, which appear similar to the gai and ga5. A-D: from Vivian-Smith and Koltunow, 1999.
Silique development is similarly affected in GA-insensitive gai mutants. GAI is a candidate transcriptional coactivator that is thought to repress growth in the absence of GA (Harberd et al., 1998). In the gai mutant, a 17 amino acid region has been deleted causing the mutant GAI protein to have a reduced sensitivity to GA. GA levels are elevated in gai mutants, but the plant has a phenotype similar to GA deficient mutants (Harberd et al., 1998). The gai valves have an abnormal morphology with reduced cell division and increased cell expansion similar to that found in GA deficient mutants (Figure 39C; Vivian-Smith and Koltunow, 1999). The valve cell morphologies of both gai and ga5-1 closely resemble the valve cells of unpollinated siliques treated with auxin (Figure 39D; see section 7.2.1 Hormone induced parthenocarpic fruit; Vivian-Smith and Koltunow, 1999). Auxin treatments of the silique in the absence of fertilization cause cell expansion especially in width, while cell division is reduced relative to pollinated siliques. These results suggest that when GA levels are low, an auxin-like effect dominates in the elongation of the fruit (Vivian-Smith and Koltunow, 1999).
Not all GA mutants are affected in fruit development; a threshold level of active GAs seems to exist above which fruit development is not affected. No effect on fruit development is seen in ga4-1 mutants, which are blocked in the final step of synthesis of the active GA1 and GA4. However, the concentration of active GAs in these plants is only three fold lower than wild type. The presence of active GAs may be explained by redundancy of other GA biosynthetic genes of the presence of other bioactive GAs. Apparently this three fold lower concentration of GAs present is sufficient to allow normal fruit formation (Vivian-Smith and Koltunow, 1999).
The roles of many hormones, especially GA, have also been studied in inducing parthenocarpy or fruit formation without seed formation.
7.2 Parthenocarpy–Fruit Development without Fertilization
Commercially, seedless fruit are often desired for their ease of consumption and extended shelf life. There are several classes of seedlessness in commercially available fruit ranging from the partially developed seeds of the “seedless” watermelon to the truly seedless Corinth grapes, which are parthenocarpic. Parthenocarpic fruit develop without fertilization of the ovules or formation of seeds. Parthenocarpy can be facultative meaning that parthenocarpic fruit form only when fertilization is actively blocked (Varoquaux et al., 2000). Examination of parthenocarpic fruit may lead to a better understanding of the processes that are normally activated by fertilization, since these processes have apparently been artificially activated, bypassing the fertilization step.
7.2.1 Hormone Induced Parthenocarpic Fruit
In Arabidopsis and many agricultural species, parthenocarpic fruit development can be triggered by application of gibberellins (GA), auxins, and cytokinins to unfertilized gynoecia (Figure 38A; Vivian Smith and Koltunow, 1999). These growth hormones applied to the entire gynoecium are thought to substitute for those normally provided by the seeds. Vivian-Smith and Koltunow (1999) have found that in Arabidopsis, gibberellic acid (GA3) applied to the gynoecium of an emasculated flower at stage 13 caused a significant increase in the length of the gynoecium (Figure 38A and 38E) and the fruit shattered upon desiccation indicating that the fruit had matured, not just expanded. Similarly, application of cytokinin (benzyl adenine BA) or auxin (indoleacetic acid IAA or naphthylacetic acetic acid NAA) also generated parthenocarpic fruit (Figure 38A, 38F and 38G; Vivian-Smith et al., 1999). Although none of these parthenocarpic fruit treated with single growth hormones came close to reaching the length of pollinated fruit, fruit treated with both GA and cytokinin or both GA and auxin elongated as much as pollinated fruit (Vivian-Smith et al., 1999). These studies suggest a complex set of hormonal interactions occur during normal fruit development.
Of the parthenocarpic fruit treated with single hormones, those treated with GA3 were the largest and the valve development most closely resembled that of pollinated fruit (Figure 38; Vivian-Smith and Koltunow, 1999). The number of cells and size of the cells in the valves of GA treated parthenocarpic fruit parallel those in wild type except that there are fewer cells in the mesocarp (Figure 38E compared with 38D). It is possible that normally GA acts fairly early after fertilization and is capable of inducing almost all of the downstream components for fruit expansion and maturation.
In contrast, the valve cells of auxin stimulated parthenocarpic siliques differed from those of pollinated siliques. The enlargement of the auxin treated gynoecia is primarily due to expansion of the valve cells especially in width. Cell division is reduced and only contributes a small amount to the total expansion of the silique (Figure 38G). The auxin treated siliques appear wider than pollinated siliques and the width of the valve wall is greater. The sclerenchymous cells of the enb layer wall do not thicken as much as in pollinated siliques although the dehiscence zone is functional and the fruit open. These differences between the auxin induced parthenocarpic siliques and pollinated siliques suggest that auxin is not sufficient to completely trigger normal fruit maturation. The expansion of the valve cells hints that auxin may primarily cause cell expansion during normal fertilized fruit development.
7.2.2 fruit without fertilization (fwf) Mutants Produce Parthenocarpic Fruit
A major advance to our understanding of parthenocarpy has come with the discovery of the fruit without fertilization (fwf) mutant of Arabidopsis. The fwf mutant is a facultative parthenocarp, setting seed like normal when pollinated, but also forming short seedless fruit when left unpollinated (Figure 40A; Vivian-Smith et al., 2001). The unpollinated fruit are 40% shorter than wild-type pollinated fruit. In the valves of fwf parthenocarpic fruit, greater cell expansion and reduced cell division are evident in the mesocarp layer than in pollinated wild-type fruit (Figure 40B to 40E). The vegetative fwf plant is indistinguishable from wild type, although a few other defects are seen in the flowers and the ovules.
Figure 40.
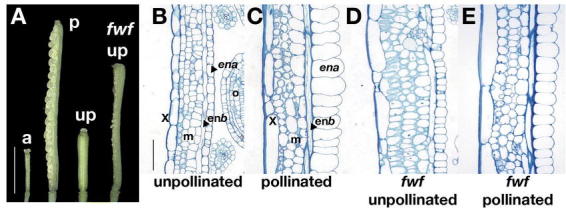
FRUIT WITHOUT FERTILIZATION restricts fruit development in the absence of fertilization. (A) Comparison of the lengths of a wild-type gynoecium at anthesis (a), a wild-type fruit seven days after pollination (p), an unpollinated wild-type fruit 7 days post-anthesis (up), and an unpollinated fwf parthenocarpic fruit (fwf up). (B) Longitudinal section of a wild-type valve at anthesis. The exocarp (x), mesocarp (m), enb, ena, and ovule (o), are labeled. (C) Longitudinal section of a pol-linated wild-type valve. (D) Longitudinal section of a fwf unpollinated valve. Cell expansion is increased and cell division is reduced relative to wild-type pollinated siliques. (E) Longitudinal section of an fwf pollinated valve, which has developed indistinguishably from wild type. Scale bar in A represents 3 mm. Scale bar in B-E represents 50 µm. A-E: from Vivian-Smith et al., 2001.
Vivian-Smith et al. made an interesting discovery when they tested different methods of blocking pollination in fwf mutants and found that parthenocarpic fruit of different lengths resulted (2001). Longer fruit were produced when all of the floral organs were removed from the fwf flower prior to anthesis than when fwf was combined with a male sterile mutant, which only blocked pollination. Therefore, the other floral organs may have an inhibitory role on fruit development. In normal fruit development, this inhibition is removed when the floral organs abscise during stage 16. Therefore Vivian-Smith et al., postulate that a signal inhibiting fruit development travels from the floral organs to the gynoecium.
It is likely that FWF acts as an inhibitor of fruit development and that this inhibition is released after fertilization. Better understanding of FWF function awaits cloning of the gene.
7.2.3 Overexpression of Cytochrome P450 CYP78A9 in an Activation Tagged Line Promotes Parthenocarpy
Another parthenocarpic mutant was isolated in an activation-tagging screen in which CaMV35S enhancers were randomly inserted into the genome to drive ectopic expression of the surrounding genes (Ito and Meyerowitz, 2000; Weigel et al., 2000). Serendipitously, a plant that failed to self-pollinate and produced parthenocarpic fruit was identified. This dominant activation tagged parthenocarpic mutant was designated 28-5. At anthesis, 28-5 gynoecia are already slightly longer and twice as wide as the wild-type gynoecium, suggesting that fruit development has already begun before fertilization can take place. Without fertilization, the 28-5 fruit elongate 2.5 times as long and 1.7 times as wide as emasculated wild-type gynoecia. When pollinated with wild-type pollen, 28-5 siliques elongate to become 10 to 20% longer than wild-type fruit.
The 28-5 phenotype is caused by the overexpression of CYP78A9, a cytochrome P450 gene (Ito and Meyerowitz, 2000). Cytochrome P450 proteins synthesize or degrade secondary products such as brassinosteroids, flavonoids, and lignin. The endogenous CYP78A9 gene is not expressed in vegetative tissue and is first detected in the funiculi of stage 14 ovules. Signals from the ovules are thought to stimulate fruit development after fertilization and CYP78A9 might be involved in production of a signal that activates fruit development (Ito and Meyerowitz, 2000).
7.2.4 knuckles Mutants Produce Indeterminate Parthenocarpic Fruit
Although knuckles (knu) mutants are male sterile (at 25°C), the fruit develop parthenocarpically and form a large bump, which appears similar to the knuckle of a finger, protruding from the silique (Payne et al., 2004). This knuckle consists of ectopic stamens and carpels that initiate from the placental tissue in the base of the fruit. The development of ectopic organs from the placenta occurs reiteratively such that multiple layers of carpel tissue can be present in the fruit. About half of the knu gynoecia produce knuckles and only the gynoecia with knuckles form weakly parthenocarpic fruit in the absence of fertilization (at 25°C; Payne et al., 2004). The correlation between parthenocarpic fruit development and production of ectopic organs within the gyneocium is also observed in tomato plants when TM29, a SEPALLATA homologue, is downregulated (Ampomah-Dwamena et al., 2002). Payne et al. speculate that the ectopic organs growing within the fruit might produce growth regulators or proliferative signals that substitute for those normally produced in the ovules causing fruit development in the absence of fertilization. Production of ectopic organs within gynoecia could be a strategy for the production of parthenocarpic fruit in diverse agriculturally important fruit crops.
In addition to the production of parthenocarpic fruit, knu mutants have defects in patterning the basal elements of the fruit (Payne et al., 2004). Elongated gynophores often form at the base of knu fruit. Furthermore, the knuckle structure arises from the basal placental tissue. The knu mutation is temperature sensitive and homozygous seeds can be produced when the plants are grown at 16°C.
KNUCKLES encodes a C2H2 zinc finger protein
KNU encodes a small C2H2 zinc finger protein with an EAR-like active repression domain and is thought to act as a transcriptional repressor (Payne et al., 2004). KNU is expressed in a large patch at the base of the developing gynoecium during stages 6–9 as well as the developing pollen and ovules. No expression of KNU is detected after stage 13, suggesting that KNU acts early in gynoecium formation before fertilization. Payne et al. propose that KNU acts as a transcriptional repressor of the proliferation of cells from the placenta into non-ovule floral organs.
8.0 MATURATION AND GLOBAL PROLIFERATIVE ARREST
At the end of fruit development, the mature fruit turns yellow and begins to dry in preparation for dehiscence. Constitutive expression of the AGAMOUS-like 15 (AGL15) MADS domain transcription factor from the 35S promoter delays this maturation of the fruit (Fernandez et al., 2000). In these 35S::AGL15 transgenic plants the fruit require about 24 to 26 days to mature in comparison to wild-type Wassilewskija (WS) siliques, which mature in about 17 to 18 days after pollination. Likewise, abscission of the floral organs is delayed so that many fully elongated siliques still have all of their sepals, petals and stamens attached (Figure 41A to 41C). The speed of embryo maturation within these siliques is not affected, suggesting that the delay of development is not universal. It has been hypothesized that AGL15 is involved in maintaining an immature state and that increased expression of AGL15 prolongs the immature state of these organs.
Figure 41.
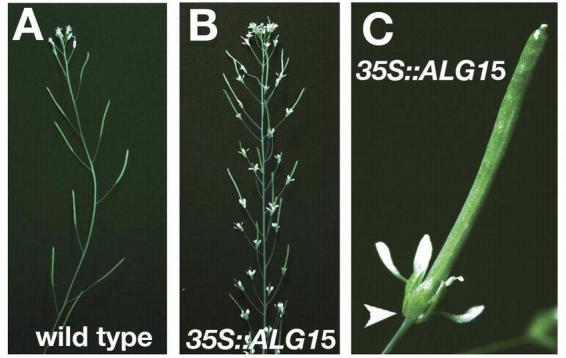
Maturation and 35S::AGL15 (A) WS wild-type inflorescence and siliques. (B) 35S::AGL15 causes a delay in the abscission of floral organs. (C) Close up of 35S::AGL15 silique stage 17B with floral organs still attached. A-C: from Fernandez et al., 2000.
Another effect of constitutive expression of AGL15 is an increase in the number of fruit produced along the main stem before the plant stops flowering (Fernandez et al., 2000). Under standard growth conditions, wild-type Ler plants produce about 28 to 38 fruit on the main stem of the plant before entering proliferative arrest in which the meristem becomes quiescent and stops producing new flowers (Hensel et al., 1994). The proliferative arrest of the inflorescence meristem becomes global spreading to the inflorescence meristems on all of the branches within two days of the arrest of the primary meristem. Proliferative arrest is not terminal and a resumption of growth occurs in about 10% to 30% of primary inflorescences about two weeks after arrest. Proliferative arrest depends on fruit development because male sterile plants do not undergo global proliferative arrest and instead produce about 63 to 71 flowers until the meristem finally terminally differentiates often in carpelloid structures (Hensel et al., 1994). Fruit development, not just fertilization, is required because plants in which the fruit were systematically removed after fertilization do not undergo global proliferative arrest. The number of seeds developing within the fruit may be the determining factor in signaling when global proliferative arrest should occur. In mutants with reduced fertility where the seed set was less than 50% that of wild type, global proliferative arrest does not occur although many developed fruit are present (Hensel et al., 1994). However, in plants heterozygous for an embryo lethal mutation in which one quarter of the ovules are aborted, no affect is seen on the timing of arrest. This evidence suggests that a signal emanates from the developing seeds out through the fruit to the whole plant directing the meristems to arrest. Therefore the development of the fruit has a direct affect on the development of the whole plant.
9.0 CONCLUSIONS AND PERSPECTIVES
The fruit is arguably the most complex plant organ and its development is an essential part of the reproductive success of angiosperms. Not only does the fruit protect the offspring through their early stages of development, the fruit also disperses the offspring into the environment so that they will have the best chance of settling in an advantageous place.
Recent genetic studies have made tremendous progress in unraveling some of the mysteries surrounding the development of fruit, but in spite of these advances, we have barely scratched the surface in terms of elucidating the overall genetic network controlling fruit development. One indication of the enormity of the task that lies ahead comes from the fact that only a handful of the genes required for the formation of different parts of the fruit have been identified and essentially nothing is known about how individual cell types within these structures acquire their specific identities. For example, the different cell layers of the valve including the exocarp, mesocarp, and endocarp layers a and b each have distinct cell morphologies. How do each of these, and other, cell types within the valves attain their specific identities?
It is perhaps not surprising that some of the first fruit development genes to be identified encode transcription factors that presumably regulate the cascade of gene activity that ultimately leads to the differentiation of specific cell types. It is also clear that, late in fruit development, enzymes involved in lignin biosynthesis as well as pectin degrading enzymes are induced in some of these cells. However, a large black box connects these early and late events and it will undoubtedly take years to fill in all the pieces. Perhaps even more confusing at present are the roles of plant hormones, where the complexities of their interactions obscure their roles. Furthermore, although it is clear that many of the cells must communicate with each other to ensure proper fruit development, almost nothing is known about the nature of these signals and their signaling pathways.
Rapid advances, with the aid of new innovations, will eventually provide a framework upon which to build a coherent picture of the mechanisms that underlie fruit development. As we begin to unravel this complex story, it will be interesting to see the extent to which these mechanisms have been conserved throughout angiosperm evolution, where the diversity of fruit structures is truly remarkable.
Acknowledgments
We would like to thank Sarah Liljegren, José Dinneny, Kristina Gremski, Lars Østergaard, Brian Crawford, Gary Ditta, and Eric Albert for comments on the manuscript. We thank Sarah Liljegren for allowing us to use unpublished pictures. We thank John Bowman, Donna Fernandez, Cristina Ferrándiz, Charles Gasser, Sangho Jeong, Anna Koltunow, Sandra Kuusk, Sarah Liljegren, Zongchi Liu, Hong Ma, Jennifer Nemhauser, Anusak Pinyopich, Judith Roe, Allen Sessions, David Smyth, and Eva Sundberg for providing figures and Academic Press Inc., American Association for the Advancement of Science, American Society of Plant Biologists, Elsevier, The Company of Biologists, Nature Publishing Group, NRC Research Press, and the University of Chicago Press for the permission to reproduce published photographs and figures. Our research on fruit development is funded by grants from the National Science Foundation, and A.H.K.R. is a Howard Hughes Medical Institute Predoctoral Fellow.
Footnotes
Citation: Roeder A.H.K., and Yanofsky M.F. (2006) Fruit Development in Arabidopsis. The Arabidopsis Book 4:e0075. doi:10.1199/tab.0075
elocation-id: e0075
Published on: February 22, 2006
Table 1.
Genes involved in fruit development
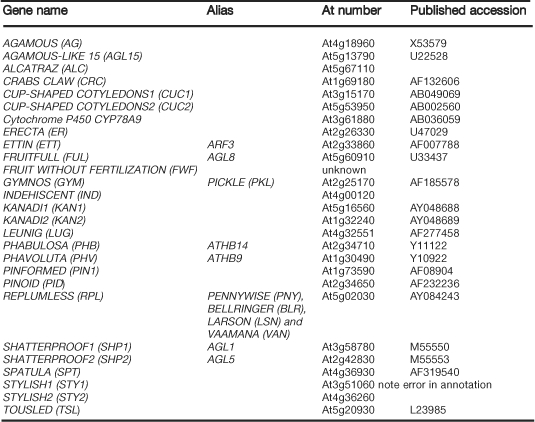
REFERENCES
- Adams-Phillips L., Barry C., Giovannoni J. Signal transduction systems regulating fruit ripening. Trends Plant Sci. 2004;94(1):331–338. doi: 10.1016/j.tplants.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;94(1):841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J., Smyth D. R. Genetic pathways controlling carpel development in Arabidopsis thaliana. J. Plant Res. 1998;1114(1):295–298. [Google Scholar]
- Alvarez J., Smyth D. R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development. 1999;1264(1):2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- Alvarez J., Smyth D. R. CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 2002;1634(1):17–41. [Google Scholar]
- Ampomah-Dwamena C., Morris B. A., Sutherland P., Veit B., Yao J. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002;1304(1):605–617. doi: 10.1104/pp.005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Franks R. G., Levin J. Z., Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;164(1):1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendse G. W. M., Kepczynski J., Karssen C. M., Kornneeef M. The role of endogenous gibberellins during fruit and seed development: studies on gibberellin-deficient genotypes of Arabidopsis thaliana. Physiol. Plant. 1986;674(1):315–319. [Google Scholar]
- Bellaoui M., Pidkowich M. S., Samach A., Kushalappa K., Kohalmi S. E., Mordusan Z., Crosby W. L., Haughn G. W. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell. 2001;124(1):2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykass P., Offringa R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development. 2001;1284(1):4057–4067. doi: 10.1242/dev.128.20.4057. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;1154(1):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bennett S. R. M., Alvarez J., Bossinger G., Smyth D. R. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 1995;84(1):505–520. [Google Scholar]
- Bhatt A. M., Etchells J. P., Canales C., Lagodienko A., Dickinson H. VAAMANA-a BEL1-like home-odomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene. 2004;3284(1):103–111. doi: 10.1016/j.gene.2003.12.033. [DOI] [PubMed] [Google Scholar]
- Bossinger G., Smyth D. R. Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development. 1996;1224(1):1093–1102. doi: 10.1242/dev.122.4.1093. [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Baum S. F., Eshed Y., Putterill J., Alvarez J. Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Develop. Biol. 1999;454(1):155–205. doi: 10.1016/s0070-2153(08)60316-6. [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Drews G. N., Meyerowitz E. M. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell. 1991a;34(1):749–758. doi: 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Sakai H., Jack T., Weigel D., Mayer U., Meyerowitz E. M. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development. 1992;1144(1):599–615. doi: 10.1242/dev.114.3.599. [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;1264(1):2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. Genes directing flower development in Arabidopsis. Plant Cell. 1989;14(1):37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991b;1124(1):1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Groover A. T., Fontana J. R., Martienssen R. A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development. 1304(1):3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- Chen C., Wang S., Huang H. LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis. 2000;264(1):42–54. doi: 10.1002/(sici)1526-968x(200001)26:1<42::aid-gene7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Christensen S. K., Dagenais N., Chory J., Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;1004(1):467–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Child R. D., Chauvaux N., John K., Ulvskov P., Onckelen H. A. Ethylene biosynthesis in oilseed rape pods in relation to pod shatter. J. Exp. Bot. 1998;494(1):829–838. [Google Scholar]
- Clark S. E., Jacobsen S. E., Levin J., Meyerowitz E. M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;1224(1):1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Clark S. E., Running M. P., Meyerowitz E. M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;1214(1):2057–2067. [Google Scholar]
- Coen E. S., Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;3534(1):31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Conner J., Liu Z. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA. 2000;974(1):12902–12907. doi: 10.1073/pnas.230352397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Busscher J., Colombo L., Losa A., Angenent G. C. Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol. Biol. 2004;564(1):351–366. doi: 10.1007/s11103-004-3473-z. [DOI] [PubMed] [Google Scholar]
- Dinneny J. R., Yanofsky M. F. Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. BioEssays. 2004;274(1):42–49. doi: 10.1002/bies.20165. [DOI] [PubMed] [Google Scholar]
- Drews G. N., Bowman J. L., Meyerowitz E. M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;654(1):991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Duval M., Hsieh T-F., Kim S. Y., Thomas T. L. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002;504(1):237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- Ehsan H., Reichheld J. P., Durfee T., Roe J. L. TOUSLED kinase activity oscillates during the xell cycle and interacts with chromatin regulators. Plant Physiol. 2004;1344(1):1488–1499. doi: 10.1104/pp.103.038117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J., Floyd S., Alvarez J., Eshed Y., Hawker N. P., Izhaki A., Baum S. F., Bowman J. L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;134(1):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S. F., Bowman J. L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;994(1):199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S. F., Perea J. V., Bowman J. L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001;114(1):1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- Fernandez D. E., Heck G. R., Perry S. E., Patterson S. E., Bleecker A. B., Fang S-C. The embryo MADS domain factor AGL15 acts postembryoically: inhibition of perianth senescence and abscission via constitutive expression. Plant Cell. 2000;124(1):183–197. doi: 10.1105/tpc.12.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C. Regulation of fruit dehiscence in Arabidopsis. J. Exp. Bot. 2002;534(1):2031–2038. doi: 10.1093/jxb/erf082. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Gu Q., Martienssen R., Yanofsky M. F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1, and CAULIFLOWER. Development. 2000a;1274(1):725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Liljegren S. J., Yanofsky M. F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000b;2894(1):436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Pelaz S., Yanofsky M. F. Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 1999;684(1):321–354. doi: 10.1146/annurev.biochem.68.1.321. [DOI] [PubMed] [Google Scholar]
- Flanagan C. A., Hu Y., Ma H. Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant Journal. 1996;104(1):343–353. doi: 10.1046/j.1365-313x.1996.10020343.x. [DOI] [PubMed] [Google Scholar]
- Friml J. Auxin transport - shaping the plant. Curr. Opin. Plant Biol. 2003;64(1):7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P. B. F., Ljung K., Sandberg G., Hooykaas P. J. J., Palme K., Ofringa R. A PINOID-dependant binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;3064(1):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;2824(1):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Gillaspy G., Ben-David H., Gruissem W. Fruits: A developmental perspective. Plant Cell. 1993;54(1):1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. J. Genetic regulation of fruit development and ripening. Plant Cell. 2004;164(1):S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N., Katoh N., Kranz A. R. Morphogenesis of floral organs in Arabidopsis: Predominant carpel formation of the pin-formed mutant. Jpn. J. Genet. 1991;664(1):551–567. [Google Scholar]
- Gu Q., Ferrándiz C., Yanofsky M. F., Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998;1254(1):1509–1517. doi: 10.1242/dev.125.8.1509. [DOI] [PubMed] [Google Scholar]
- Harberd N. P., King K. E., Carol P., Cowling R. J., Peng J., Richards D. E. Gibberellin: inhibitor of an inhibitor of…? BioEssays. 1998;204(1):1001–1008. doi: 10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Heisler M. G. B., Atkinson A., Bylstra Y. H., Walsh R., Smyth D. R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001;1284(1):1089–1098. doi: 10.1242/dev.128.7.1089. [DOI] [PubMed] [Google Scholar]
- Hensel L. L., Nelson M. A., Richmond T. A., Bleecker A. B. The fate of inflorescence meristem is controlled by developing fruits in Arabidopsis. Plant Physiol. 1994;1064(1):863–876. doi: 10.1104/pp.106.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. P., Lord E. M. Floral development in Arabidopsis thaliana: a comparison of the wild type and the homeotic pistillata mutant. Can. J. Bot. 1989;674(1):2922–2936. [Google Scholar]
- Honma T., Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;4094(1):525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Ishida T., Aida M., Takada S., Tasaka M. Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 2000;414(1):60–67. doi: 10.1093/pcp/41.1.60. [DOI] [PubMed] [Google Scholar]
- Ito T., Meyerowitz E. M. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large seedless fruit in Arabidopsis. Plant Cell. 2000;124(1):1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M. K., Nasrallah J. B., Nasrallah M. E. Pollen-pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development. 1994;1204(1):3405–3418. [Google Scholar]
- Kayes J. M., Clark S. E. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;1254(1):3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- Kerstetter R. A., Bollman K., Taylor R. A., Bomblies K., Poethig R. S. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;4114(1):706–712. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- Kuusk S., Sohlberg J. J., Long J. A., Fridborg I., Sundberg E. STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development. 2002;1294(1):4707–4717. doi: 10.1242/dev.129.20.4707. [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Ditta G. S., Eshed Y., Savidge B., Bowman J. L., Yanofsky M. F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;4044(1):766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Roeder A. H. K., Kempin S. A., Gremski K., Østergaard L., Guimil S., Reyes K. D., Yanofsky M. F. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;1164(1):843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- Liu Z., Franks R. G., Klink V. P. Regulation of gynoecium marginal tissue formation by LEUNIG and AIN-TEGUMENTA. Plant Cell. 2000;124(1):1879–1891. doi: 10.1105/tpc.12.10.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Meyerowitz E. M. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995;1214(1):975–991. doi: 10.1242/dev.121.4.975. [DOI] [PubMed] [Google Scholar]
- Lohmann J. U., Hong R. L., Hobe M., Busch M., Parcy F., Simon R., Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;1054(1):793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;3794(1):66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Lord E. M., Russell S. D. Mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 2002;184(1):81–105. doi: 10.1146/annurev.cellbio.18.012502.083438. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Bowman J. L., Kempin S. A., Ma H., Meyerowitz E. M., Yanofsky M. F. Manipulation of flower structure in transgenic tobacco. Cell. 1992;714(1):133–143. doi: 10.1016/0092-8674(92)90272-e. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Yanofsky M. F. The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell. 1995;74(1):1763–1771. doi: 10.1105/tpc.7.11.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod J. Oilseed Rape Book. 1981;4(1):107–119. Cambridge Agricultural, Cambridge. [Google Scholar]
- McConnell J. R., Barton M. K. Leaf polarity and mersitem formation in Arabidopsis. Development. 1998;1254(1):2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell J. R., Emery J., Eshed Y., Bao N., Bowman J. L., Barton M. K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;4114(1):709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Meakin J. P., Roberts J. A. Dehiscence of fruit in oilseed rape 1. Anatomy of pod dehiscence (Brassica napus L.). J. Exp. Bot. 1990;414(1):955–1002. [Google Scholar]
- Meister R. J., Kotow L. M., Gasser C. S. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development. 2002;1294(1):4281–4289. doi: 10.1242/dev.129.18.4281. [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;714(1):119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Feldman L. J., Zambryski P. C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development. 2000;1274(1):3877–3888. doi: 10.1242/dev.127.18.3877. [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Zambryski P. C., Roe J. L. Auxin signaling in Arabidopsis flower development? Curr. Opin. Plant Biol. 1998;14(1):531–535. doi: 10.1016/s1369-5266(98)80047-2. [DOI] [PubMed] [Google Scholar]
- Ogas J., Cheng J-C., Sung Z. R., Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;2774(1):91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. PNAS. 1999;964(1):13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Komaki M., Shimura Y. Mutational analysis of pistil structure and development of Arabidopsis thaliana. Cell Differ. Dev. 1989;284(1):27–38. doi: 10.1016/0922-3371(89)90020-8. [DOI] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M. K., Bell C. J., Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;34(1):677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R., Preuss D. Pollen tube targeting and axon guidance: parallels in tip growth mechanisms. Trends Cell Biol. 2000;104(1):517–524. doi: 10.1016/s0962-8924(00)01849-3. [DOI] [PubMed] [Google Scholar]
- Payne T., Johnson S. D., Koltunow A. M. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development. 2004;1314(1):3737–3749. doi: 10.1242/dev.01216. [DOI] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E., Yanofsky M. F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;4054(1):200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pelaz S., Tapia-López R., Alvarez-Buylla E. R., Yanofsky M. F. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2001;114(1):182–184. doi: 10.1016/s0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G. S., Savidge B., Liljegren S. J., Baumann E., Wisman E., Yanofsky M. F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;4244(1):85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Rajani S., Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 2001;114(1):1914–1922. doi: 10.1016/s0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E-R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;4264(1):255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., Bartel D. P. Prediction of plant microRNA targets. Cell. 2002;1104(1):513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Meyerowitz E. M. MADS domain proteins in plant development. Biol. Chem. 1997;3784(1):1079–1101. [PubMed] [Google Scholar]
- Robinson-Beers K., Pruitt R. E., Gasser C. S. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;44(1):1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe J. L., Rivin C. J., Sessions R. A., Feldman K. A., Zambryski P. C. The TOUSLED gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell. 1993;754(1):939–950. doi: 10.1016/0092-8674(93)90537-z. [DOI] [PubMed] [Google Scholar]
- Roe J. L., Durfee T., Zupan J. R., Repetti P. P., McLean B. G., Zambryski P. C. TOUSLED is a nuclear serine/threonin protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. J. Biol. Chem. 1997a;2724(1):5838–5845. doi: 10.1074/jbc.272.9.5838. [DOI] [PubMed] [Google Scholar]
- Roe J. L., Nemhauser J. L., Zambryski P. C. TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell. 1997b;94(1):335–353. doi: 10.1105/tpc.9.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A. H. K., Ferrándiz C., Yanofsky M. F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003;134(1):1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Sakai H., Medrano L. J., Meyerowitz E. M. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;3784(1):199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- Savidge B., Rounsley S. D., Yanofsky M. F. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral identity genes. Plant Cell. 1995;74(1):721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions R. A. Arabidopsis (Brassicaceae) flower development and gynoecium patterning in wild type and ettin mutants. Am. J. Bot. 1997;844(1):1179–1191. [PubMed] [Google Scholar]
- Sessions R. A., Nemhauser J. L., McCall A., Roe J. L., Feldman K. A., Zambryski P. C. ETTIN patterns the Arabidopsis floral mersitem and reproductive organs. Development. 1997;1244(1):4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Sessions R. A., Zambryski P. C. Arabidopsis gynoecium structure in the wild type and in ettin mutants. Development. 1995;1254(1):1519–1532. doi: 10.1242/dev.121.5.1519. [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Eshed Y., Baum S. F., Otsuga D., Drews G. N., Bowman J. L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;1264(1):4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Smith H. M. S., Boschke I., Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Nat. Acad. Sci. 2002;994(1):9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. M. S., Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell. 2003;154(1):1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. Early flower development in Arabidopsis. Plant Cell. 1990;24(1):755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J., Vercher Y., Gates P., Harris N. ‘Pod shatter’ in Arabidopsis thaliana, Brassica napus, and B. juncea. J. Microscopy. 1996;1814(1):195–203. [Google Scholar]
- Takada S., Hbara K., Ishida T., Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;1284(1):1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- Tanksley S. D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell. 2004;164(1):S181–S189. doi: 10.1105/tpc.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J. T., Münster T., Winter K-U., Saedler H. A short history of MADS-box genes in plant development. Plant Mol. Biol. 2000;424(1):115–149. [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P. H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;154(1):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K. U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., Whittier R. F., Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;84(1):735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux F., Blanvillain R., Delseny M., Gallois P. Less is better: new approaches for seedless fruit production. Trends Biotechnol. 2000;184(1):233–242. doi: 10.1016/s0167-7799(00)01448-7. [DOI] [PubMed] [Google Scholar]
- Vivian-Smith A., Koltunow A. M. Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 1999;1214(1):437–451. doi: 10.1104/pp.121.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A., Luo M., Chaudhury A., Koltunow A. Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development. 2001;1284(1):2321–2331. doi: 10.1242/dev.128.12.2321. [DOI] [PubMed] [Google Scholar]
- Vroemen C. W., Mordhorst A. P., Albrecht C., Kwaaitaal M. A. C. J., de Vries S. C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;154(1):1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberling F. Morphology of flowers and inflorescences. 1989;4(1):341. R. J. Pankhurst Trans. (Cambridge Univ. Press, Cambridge). p. [Google Scholar]
- Weigel D., Ahn J. H., Blázquez M. A., Borevitz J. O., Christensen S. K., Fankhauser C., Ferrándiz C., Kardailsky I., Malancharuvil E. J., Neff M. M., Nguyen J. T., Sato S., Wang Z-Y., Xia Y., Dixon R. A., Harrison M. J., Lamb C. J., Yanofsky M. F., Chory J. Activation tagging in Arabidopsis. Plant Physiol. 2000;1224(1):1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldman K. A., Meyerowitz E. M. The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature. 1990;3464(1):35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yokoyama R., Takahashi T., Kato A., Torii K., Komeda Y. The Arabidopsis ERECTA gene is expressed in the shoot apical mersitem and organ primordial. Plant J. 1998;154(1):301–310. doi: 10.1046/j.1365-313x.1998.00203.x. [DOI] [PubMed] [Google Scholar]
- Yu L. P., Simon E. J., Trotochaud A. E., Clark S. E. POLTERGEIST functions to regulated mersitem development downstream of the CLAVATA loci. Development. 2000;1274(1):1661–1670. doi: 10.1242/dev.127.8.1661. [DOI] [PubMed] [Google Scholar]


