Abstract
Plants accumulate storage substances such as starch, lipids and proteins in certain phases of development. Storage proteins accumulate in both vegetative and reproductive tissues and serve as a reservoir to be used in later stages of plant development. The accumulation of storage protein is thus beneficial for the survival of plants. Storage proteins are also an important source of dietary plant proteins. Here, we summarize the genome organization and regulation of gene expression of storage protein genes in Arabidopsis.
Introduction
Plant storage proteins can be classified into two classes; seed storage proteins (SSPs) and vegetative storage proteins (VSPs). SSPs are a set of proteins that accumulate to high levels in seeds during the late stages of seed development, whereas VSPs are proteins that accumulate in vegetative tissues such as leaves, stems and, depending on plant species, tubers. During seed germination, SSPs are degraded and the resulting amino acids are utilized by the developing seedlings as a nutritional source. SSPs are the major proteins in grains and, of the plant proteins, represent those that are the most abundantly consumed by humans. On the other hand, VSPs accumulate in the vegetative tissues when excess resources are available and serve as a temporal reservoir of amino acids for use in subsequent phases of growth and development.
SSP genes are classic targets for plant molecular biology. Their abundant expression in seeds allowed for easy detection of the gene transcripts and cDNA cloning during the dawn of plant molecular biology in late 70's to early 80's. Their expression is strictly limited to developing seeds during the mid- to late-stages of embryogenesis, a characteristic that makes them ideal for the study of tissue- and temporal-specific regulation of gene expression. Studies on the expression of SSP genes has contributed greatly to the development of plant molecular biology. Molecular genetic studies using Arabidopsis have led to a further understanding of expression and accumulation of SSPs and the recent genomic exploration of Arabidopsis has also begun to impact on our understanding of seed maturation processes associated with SSP accumulation.
In this review, we will summarize the studies on the regulation of SSP accumulation in Arabidopsis with reference to other seed-specific genes. The VSPs of Arabidopsis are also described in terms of genomic organization and regulation of gene expression.
Seed storage protein genes of Arabidopsis
The major SSPs of Arabidopsis are comprised of 12S globulins and 2S albumins (Figure 1). This classification of SSPs as albumins or globulins is based on whether the proteins are water-soluble or soluble in salt solution, respectively. In addition to this classification, the SSPs of crops also often have specific names. For example, glycinin, glutelin and zein represent specific SSP fractions from soybean, rice and maize, respectively. The Arabidopsis SSPs have also been named after those of Brassica napus. 12S globulin proteins are referred to as cruciferin, whereas the 2S albumins are referred to as either napin or arabin.
Figure 1.
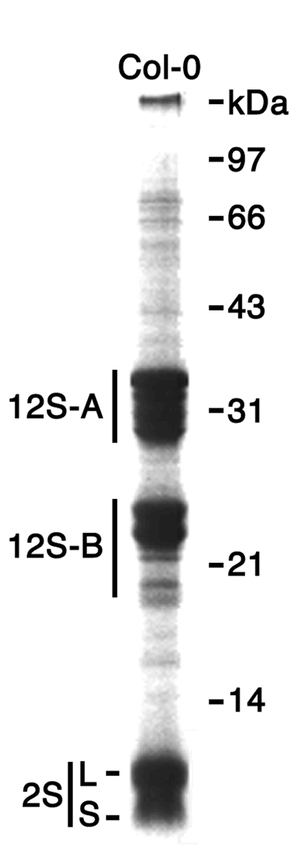
SSPs of Arabidospsis.Proteins were extracted from mature seeds of wild-type Col-0 strain and subjected to SDS-PAGE. Position of major SSPs, acidic (12S-A) and basic (12S-B) subunits of 12S globulin (cruciferin), and large (L) and small (S) subunits of 2S albumin (napin) are shown.
Both 12S globulins and 2S albumins are initially synthesized as a precursor and accumulate in protein bodies after processing of the preproteins. The 12S proteins are a member of the legumin-like 11S globulins and accumulate as hexameric complexes consisting of six a and six b polypeptides linked via disulfide bridges. The a and b polypeptides are generated from a single polypeptide-precursor following specific cleavages by processing enzymes. Cysteine residues involved in the crosslinking of the polypeptides and formation of the hexameric complexes have been identified (Inquello et al., 1993). In contrast, 2S albumin accumulates as a heterodimer consisting of two subunits linked by disulfide bridges. Similar to that for 12S globulin, these subunits are generated by cleavage of a precursor polypeptide (Krebbers et al., 1988). Synthesis of both 12S and 2S SSPs takes place on the rough ER, followed by sorting into protein bodies.
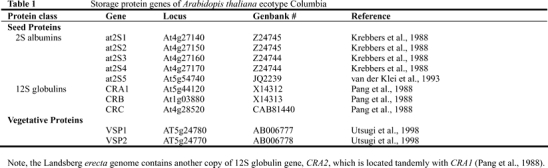
Similar to that in most other plant species, Arabidopsis SSPs are encoded by a small multi-gene family (Table 1). Five isoforms of genes encoding 2S albumin, referred to as at2S1 to at2S5, have been reported (Krebbers et al., 1988; van der Klei et al., 1993). Of these, four of the 2S albumin genes (at2S1 through at2S4) are present as a tandem cluster in the lower half of chromosome 4 (approx 12,575 kbp position). The fifth gene of 2S albumin (At5g54740; van der Klei et al., 1993) is located in the lower region of chromosome 5. Proteins corresponding to all of these genes have been detected, suggesting that all five genes are actively transcribed (van der Klei et al., 1993). Phylogenetic analysis of the amino acid and nucleotide sequences of 2S albumin genes in Brassicaceae suggests that duplication of the 2S albumin genes occurred prior to the Brassicaceae-Sysimbrieae split followed by concerted evolution (Boutilier et al., 1999).
Cloning of the 12S globulin genes of Arabidopsis was first reported by Pang et al. (1988). Three genes encoding 12S globulin are present in the genome of Columbia ecotype. These genes, designated as CRA1, CRB and CRC, are located on chromosome 5, 1 and 4, respectively (Table 1). The Landsberg erecta genome contains another copy of the 12S globulin gene, CRA2, which is located in tandem with CRA1 (Pang et al., 1988).
Other than the 12S and 2S proteins, 7S globulin proteins (vicilins) are also among the major SSPs of many plant species. Arabidopsis does not contain detectable 7S storage protein in mature seeds but a vicilin-like gene (At2g18540) is present towards the middle of chromosome 2 in a region covered by the BAC clone T17D12. A search of the Arabidopsis genome database has also revealed two other globulin-like genes, one in the upper region of chromosome 1 (At1g07750, BAC clone F24B9) and the other near the bottom of chromosome 4 (At4g36700, BAC clone AP22). A number of ESTs from siliques or seeds match At1g07750 or At4g36700, suggesting that these genes are expressed in seeds or siliques.
A number of genes encoding proteins other than SSPs are also expressed abundantly during late embryogenesis. These include oleosins and late embryogenesis abundant (LEA) proteins. Oleosins are relatively small hydrophobic proteins (15-25 kD) associated with oil bodies, a cellular organelle for storage of triacylglycerols. They are synthesized specifically in seeds and considered to be important for stability of oil bodies. LEA genes are a set of genes that are expressed in later stages of embryogenesis. LEA genes encode proteins that are highly hydrophilic and are proposed to play a role in the acquisition of desiccation tolerance. The expression of LEA genes is however not always strictly limited to late embryogenesis and many LEA genes are also expressed vegetatively in response to various stresses or in reproductive tissues, such as pollen. Two other genes, ATS1 and ATS3 (Arabidopsis thaliana seed genes), have also been identified in Arabidopsis as seed-specific genes that were isolated through a differential display method (Nuccio and Thomas, 1999). On the other hand, AtEm1 and AtEm6 are expressed in both late embryogenesis and vegetative tissues in response to exogenous application of abscisic acid (ABA) (Soderman et al., 2000). Recent progress in genomic studies using Arabidopsis has greatly increased the number of seed-specific genes identified (Girke et al., 2000).
Expression of seed storage protein genes — cis-acting factors
Similar to other SSP genes, the SSP genes of Arabidopsis are expressed specifically during the mid- to late-stages of seed development. However, the detailed patterns of expression during these developmental stages do differ between the genes.
The at2S1 gene is expressed essentially in the embryo axis, whereas at2S2 and at2S3 genes are highly expressed throughout the embryo (Guerche et al., 1990). A similar difference in expression levels and patterns has also been observed in corresponding promoter-reporter constructs, suggesting that the expression levels and tissue specificity are transcriptionally regulated (Conceicao et al., 1994). Studies using hybrid promoter constructs between at2S1 and at2S2 identified a region of the promoters required for 2S albumin expression in palisade parenchyma and specific epidermal cells (Conceicao and Krebbers, 1994). It was also demonstrated that some of the hybrid promoters exhibited altered temporal regulation (Conceicao and Krebbers, 1994).
5′-deletion analysis of the at2S1 promoter defined a region required for seed-specific expression (Conceicao and Krebbers, 1994), although no further analysis of cis-acting elements responsible for seed-specific expression of Arabidopsis SSPs has been reported so far. Detailed promoter analysis has been mostly carried out in legumes and cereals (Shewry et al., 1995; Wohlfarth et al., 1998 for review). These analyses identified the RY repeat motif (also refered to as the Sph element or legumin box; Baumlein et al., 1992; Chamberland et al., 1992; Fujiwara and Beachy, 1994; Sakata et al., 1997; Bobb et al., 1997) as a key cis-acting element for seed-specific gene expression. The RY repeat is a highly conserved cis-acting element acting as both an enhancer for seed-specific gene expression and a repressor of expression in vegetative tissue. A sequence corresponding to the RY repeat motif or a sequence related to it is also present in promoter regions of Arabidopsis SSP genes (Conceicao and Krebbers, 1994).
Analysis of cis-acting elements of the Arabidopsis oleosin promoter in transgenic B. napus revealed DNA fragments involved in seed-specific gene expression (Plant et al., 1994). Sequences present between −2500 bp and −1100 bp of the promoter were shown to be involved in modulating the levels of gene expression during the early stages of embryo development, whereas the −400 bp to −200 bp region contained sequences involved in osmotic induction. Similar promoter deletion analyses have also been reported for other Arabidopsis genes expressing in the seed, including LEA genes (Hull et al., 1996).
Regulation of seed storage protein gene expression — trans-acting factors
Genetic studies have been essential in elucidating the molecular mechanisms underlying regulation of SSP gene expression. Mutations causing reductions in SSP levels are powerful tools in the effort to understand the network of transcriptional regulation of SSP genes. There are two known examples demonstrating an interaction between a cis-acting element and its trans-acting factor in the regulation of SSP gene expression in Arabidopsis. These are the RY repeat/B3 domain proteins (ABI3 and FUS3) and the ABA-responsive element (ABRE)/bZIP protein (ABI5). Other loci affecting the expression of SSP genes appear to act either via another cis element(s) controlling SSP gene expression or indirectly by modulating “direct” trans-acting factors.
The abi3 (abscisic acid-insensitive3), fus3 (fusca3), and lec1 (leafy cotyledon1) mutants produce seeds that are intolerant to desiccation (Nambara et al., 1992; Keith et al., 1994; Meinke et al., 1994; West et al., 1994). When these mutant seeds are harvested and sown on medium prior to desiccation they show normal growth, although desiccation intolerant seeds are still produced in the next generation. Accumulation of SSPs and LEA proteins is severely reduced in these mutants (Nambara et al., 1992; Vicient et al., 2000) and the profile of the global pattern of gene expression is also altered during mid- to late-embryogenesis. During these stages, SSP and LEA genes are downregulated and genes normally expressed during or after germination are precociously activated (Nambara et al., 2000; Parcy et al., 1994, 1997; West et al., 1994). Therefore, ABI3, FUS3, and LEC1 appear to act as important regulatory components of mid- to late-stages of embryogenesis. Another class of mutants also exists that exhibit altered expression of LEA genes, although they can produce desiccation tolerant seeds. The abi4 (abscisic acid-insensitive4) and abi5 (abscisic acid-insensitive5) mutants are defective in gene expression of a subset of LEA genes during late-embryogenesis (Finkelstein, 1994). Although the monogenic mutants of these genes exhibit little or no phenotype in SSP gene expression, other data suggests ABI4 and ABI5 are involved in regulation of SSP gene expression. The ABI3, FUS3, LEC1 and ABI4 trans-acting factors are discussed individually below with respect to their role in regulation of seed storage proteins.
The ABI3 gene encodes a transcription factor, which is an ortholog of the maize VP1 gene (Giraudat et al., 1992). The phenotype of maize vp1 mutants is quite similar to that of abi3 mutants that produce desiccation intolerant ABA-insensitive seeds (McCarty et al., 1991). ABI3/VP1 orthologs from both monocots and dicots contain 4 conserved domains (A, B1, B2, and B3; Figure 2). The B3 domain of VP1 is able to bind specifically to the Sph element that contains a RY repeat in vitro (Suzuki et al., 1997) and it has been established that the RY repeat is required for transcriptional activation by ABI3 (Reidt et al., 2000). On the other hand, involvement of the B2 domain of ABI3 in regulation of At2S2, AtEm1, and AtEm6 gene expression is suggested (Bies-Etheve et al., 1999). Furthermore, ABI3 appears to act in concert with other transcription factors. Transcription factors interacting with ABI3 have been identified and are suggested to be a component of the transcriptional regulatory network for embryo development in Arabidopsis (Kurup et al., 2000).
Figure 2.
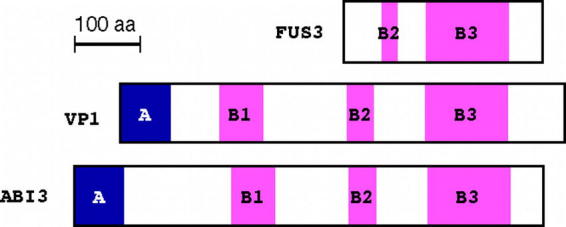
Domains of the ABI3/VP1 proteins.ABI3 and VP1 contain 4 conserved domains, designated as A, B1, B2 and B3. The N-terminal domain A is rich in acidic residues. One of the basic domains, B1, is necessary for physical interaction with bZIP transcription factors such as ABI5. The B2 domain contains a putative nuclear localization signal and is also suggested to function in other transcriptional regulation. The B3 domain is the DNA-binding domain that is highly conserved. The structure of FUS3 is a truncated version of ABI3/VP1. This contains only the B3 domain and an incomplete B2 domain.
The ABI3 gene is expressed abundantly during embryogenesis, which ceases after germination (Parcy et al., 1994). Recent reports have demonstrated that it is also expressed in dormant tissue, thus suggesting another function of ABI3 in tissues other than seeds (Rohde et al., 2000, for review). Transgenic Arabidopsis carrying a CaMV 35S promoter (35S):ABI3 construct expresses some of the SSP genes ectopically in response to exogenous ABA, demonstrating that ABI3 is one of the essential elements for seed-specific gene expression (Parcy et al., 1994).
The FUS3 gene encodes a B3 domain-containing transcription factor that lacks the N-terminal sequence, a conserved sequence in other orthologs of ABI3/VP1 (Luerssen et al., 1998) (Figure 2). The RY repeat has also been shown to be under the regulation of FUS3-mediated transcriptional activation (Wohlfarth et al., 1998, for review) and it was recently demonstrated that the RY repeat is the direct target of the B3 domain of FUS3 (Reidt et al., 2000). Recently it was demonstrated that the LEC2 gene, whose mutant shows similar phenotype as does the fus3 mutant but shows partial intolerance to desiccation, also encodes a B3 domain-containing protein (Stone et al. 2001). Other transcription factors containing the highly conserved B3 domain of VP1/ABI3/FUS3, designated as RAV1 and RAV2, are reported (Kagaya et al., 1999).
Although ABI3 and FUS3 act as transcriptional regulators cooperatively via the RY repeat, they also function independently to control SSP gene expression. This suggests that other domains are also important in defining their distinct functions. One of the important differences between ABI3 and FUS3 is the distinct role in the ABA response in seeds. The rice bZIP transcription factor TRAB1 that binds to ABRE interacts with the N-terminal region of OsVP1, a rice VP1 ortholog, in an ABA-dependent manner (Hobo et al., 1999). Interestingly, the Arabidopsis ABI5 gene encodes a bZIP transcription factor that is similar to Rice TRAB1 (Finkelstein and Lynch, 2000) and it also appears to bind to ABRE in vitro (Lopez-Molina et al., 2001). The N-terminal region of ABI3 (A and B1 domains), which is missing in FUS3, appears to be important for ABA-induced transcriptional activation by ABI3 (Reidt et al., 2000). Furthermore, ABI5 is required for the activation of SSP genes in vegetative tissues by 35S:ABI3 (Soderman et al., 2000). Recently Nakamura et al. (2001) reported that the B1 domain of ABI3 is necessary for interaction with ABI5 in a yeast two-hybrid assay.
The LEC1 gene is expressed in a seed-specific manner and encodes a putative transcription factor that is a homolog of the subunit of CAAT box-binding factor (Lotan et al., 1998), although the target DNA sequence of LEC1 is yet to be identified. Similar to that for the fus3 mutant, trichomes are present on leaf cotyledons of the lec1 mutant, suggesting a transformation of cell-type specificity (Figure 3). Ectopic expression of the LEC1 gene results in growth arrest after germination and occasionally produces embryo-like structures expressing SSP genes in these organs. This indicates that expression of the LEC1 gene is sufficient to initiate embryo development (Lotan et al., 1998). Interestingly, At2S1 and oleosin genes are ectopically expressed in roots of the pickle (pkl) mutant, which misexpresses the LEC1 gene in these tissues (Ogas et al., 1997, 1999). The PKL gene encodes a chromatin-remodelling factor (Ogas et al., 1999), suggesting that PKL-mediated repression of the LEC1 gene is important to repress seed-specific gene expression in non-embryonic tissues.
Figure 3.
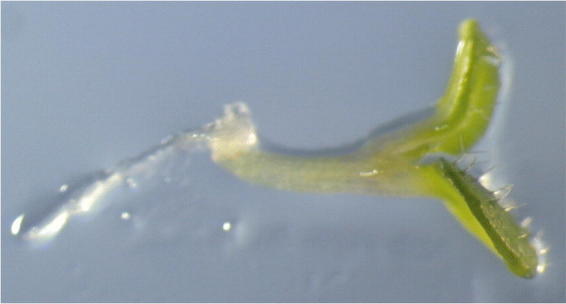
lec1 mutant seedlings possess trichomes on cotyledons.lec1 embryos rescued prior to desiccation can precociously germinate. Unlike wild type, trichomes appear on the adaxial surfaces of the cotyledons in the lec1 mutant.
Expression of 2S albumin genes is also dependent on genome organization. As mentioned above, four genes encoding 2S albumin are clustered within the Arabidopsis genome. Stable introduction of a transgene carrying a tandem repeat of 2S albumin genes into Arabidopsis resulted in unexpectedly high expression levels of the construct (Conceicao et al., 1994), suggesting an importance of gene organization in regulation of the 2S albumin genes. Chromatin structure has also been shown to be involved in seed-specific expression of the phaseolin gene in French bean (Li et al., 2001, for review). In vivo footprinting experiments of the phaseolin gene promoter revealed that nucleosome positioning is altered in seed to allow access of transcriptional regulators. The transcription factor PvALF, a French bean ortholog of ABI3/VP1, is intrinsically involved in nucleosome positioning, as are other non-histone proteins and ABA.
The ABI4 gene encodes a transcription factor containing an AP2-domain, a conserved DNA-binding domain in plants (Finkelstein et al., 1998). The target sequence of ABI4 remains unidentified, although the structure of the DNA binding domain is similar to that of ethylene responsive element binding protein or the cold-responsive element binding protein, CBP1. Expression of the ABI4 gene is abundant during embryogenesis, and it is also weakly expressed in vegetative tissues. Some of the genes encoding LEA proteins (AtEm1, AtEm6, and PAP140) have also been found to be downregulated in abi4 mutant seeds (Finkelstein et al., 1998). Although loss-of-function of the ABI4 gene does not significantly affect the expression of 12S globulin nor 2S albumin genes, transgenic plants carrying the 35S:ABI4 construct are able to ectopically express the SSP genes including At2S3 and CRC genes in response to exogenous ABA.
Genomic studies on seed-specific gene regulation
As part of a study on lipid metabolism in seeds, micro-arrays containing approximately 5,000 seed-expressed Arabidopsis genes were used to further understand seed-specific gene expression. Comparison with probes derived from seeds, leaves and roots revealed that approximately 10% of the genes were expressed at ratios greater than or equal to 10-fold higher in seeds than in leaves or roots. Included in this list are a large number of proteins of unknown function, and potential regulatory factors such as protein kinases, phosphatases and transcription factors (Ohlrogge et al., 2000). These factors may be involved in seed-specific regulation of gene expression. In another study using micro-arrays containing approximately 2,600 seed-expressed genes, approximately 25% of the genes were expressed at ratios greater than or equal to 2-fold higher in seeds than leaves or roots and 10% at ratios greater than or equal to 10 (Girke et al., 2000).
Effect of phloem transport and nutritional conditions on seed storage protein accumulation
Synthesis of SSPs depends on sugars and amino acids translocated from source leaves via the phloem. The accumulation of SSPs is therefore influenced by plant metabolic status and nutritional conditions. Expression of a soybean SSP was affected by sucrose supply (Bray and Beachy, 1985) and a potato tuber storage protein, patatin, has also been shown to respond to sucrose (Martin et al., 1997). Similarly, sugar transport to seed is likely to affect expression of SSP gene expression in Arabidopsis, although no direct evidence supporting this hypothesis has been reported. Recent analysis on sugar-induced signal transduction revealed an involvement of hexokinase, SNF1 kinase, and regulatory PRL1 protein (Smeekens, 2000, for review).
As described above, the Arabidopsis abi3 mutation affects SSP gene expression. In addition to this direct effect on SSPs, plants carrying the abi3 mutation also partitioned more resources into seed development than the wild type. These extra resources were available as a result of delayed senescence of the cauline leaves in the mutant. The change in distribution of photosynthate was confirmed by tracer analysis (Robinson and Hill, 1999).
Lipid metabolism is also important for expression of a class of SSPs. Transcription of the gene for oleosin during seed development was delayed and reduced in an Arabidopsis mutant carrying a lesion affecting diacylglycerol acyltransferase, without a reduction in the level of oleosin protein in seeds, suggesting the presence of a coordinated regulation of lipid biosynthesis-regulated gene expression (Zou et al., 1996). Expression of oleosin was also affected both in terms of timing and levels by other mutations affecting seed development such as fus3 (Kirik et al., 1996).
Amino acids play another critical role in the accumulation of SSPs. The broad specificity H+-amino acid co-transporter AAP1 is expressed in the endosperm and the cotyledons, whereas the similar transporter AAP2 is expressed in vascular strands of siliques and in funiculi. These transporters are implicated to be involved in long-distance transport of amino acids to seeds (Hirner et al., 1998). In addition, transgenic Arabidopsis plants overexpressing sense or antisense genes for a peptide transport gene, AtPTR2-B have been described. All four antisense lines and one sense line exhibited significant phenotypic changes, including late flowering and arrested seed development, suggesting a role of AtPTR2-B in nitrogen nutrition delivery to seeds (Song et al., 1997).
Sulfur nutrition has also been shown to affect SSP accumulation. In general, SSPs with a high content of sulfur-containing amino acids accumulate to a lower level under conditions of limited sulfur supply and, conversely, accumulation of SSPs with low contents of sulfur-containing amino acids are increased (Fujiwara et al., 1992). The SSPs of Arabidopsis have relatively high contents of sulfur-containing amino acids and the accumulation of 2S and 12S proteins under sulfur deficiency seem to be somewhat downregulated (Naito et al., 1994a). The b conglycinin SSP of soybean is a well studied example of SSPs responding to nutritional conditions. Under a limited supply of sulfur, accumulation of the b subunit of b-conglycinin is elevated and that of glycinin, another major SSP, is downregulated (Gayler and Sykes, 1985). Application of Met, on the other hand, results in an increase in the accumulation of glycinin and reduces or suppresses accumulation of the b subunit (Holowach et al., 1984). Transgenic Arabidopsis have been used effectively to examine the mechanisms of sulfur-regulated expression of soybean SSPs. Similar to that in soybean, expression of the b subunit gene of b-conglycinin is upregulated by sulfur deficiency (Hirai et al., 1995) and downregulated by Met application (Hirai et al., 1994) or mto1 mutations (Naito et al., 1994b; Naito et al., 1995, for review) causing an overaccumulation of soluble Met (Inaba et al., 1994; Chiba et al., 1999) in transgenic Arabidopsis. The gene promoter of the b subunit of b-conglycinin has been shown to be responsible for this regulation. Regulation of the b subunit promoter by the mto1 mutation is maternally controlled, i.e., expression of the b-subunit promoter is determined by the genotype of the female parent whereas the genotype of the male parent has little or no effect on the expression (Naito et al., 1994b). These findings suggest that Met, or one of its metabolites, transported from the maternal tissues affects SSP gene expression.
Expression of the b-subunit promoter in transgenic Arabidopsis was not only regulated according to sulfur nutrition, but also according to nitrogen nutrition (Kim et al., 1999). Sulfur and nitrogen assimilation pathways merge at the step of cysteine biosynthesis and a precursor for cysteine biosynthesis, O-acetyl-L-serine (OAS), has been shown to be accumulated depending on sulfur and nitrogen availability (Kim et al., 1999). Sulfur deficiency results in an increase in the accumulation of OAS, whereas nitrogen deficiency decreases OAS accumulation. This pattern was similar to that for the b-subunit gene regulation (Kim et al., 1999). Moreover, exogenous application of OAS in the presence of high sulfate in the media induced accumulation of the b subunit in cultured soybean cotyledons (Kim et al., 1999). These findings suggest that OAS acts as a key metabolite controlling the expression of SSP genes in response to sulfur and nitrogen availability in the environment. It is also intriguing that OAS contents were increased by ABA application in immature soybean cotyledons (Kim et al., 1997). ABA application to immature soybean cotyledon induces a similar response to that of sulfur deficiency, i.e., an increase in b subunit accumulation and a decrease in glycinin accumulation (Figure 4), suggesting a possible involvement of OAS in the ABA response.
Figure 4.
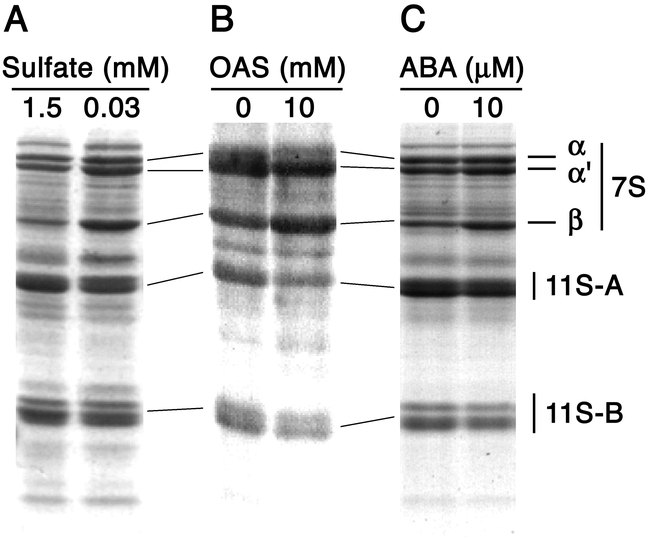
Effect of ABA application on accumulation of soybean seed storage proteins.A, Immature soybean cotyledons (about 20 mg in fresh weight) were cultured for six days in vitro in control (1.5 mM sulfate) or sulfur deficient (0.03 mM sulfate) medium. Proteins were extracted and subjected to SDS-PAGE analysis. Position of major seed storage proteins, a, a' and b subunits of 7S globulin (b-conglycinin), and acidic (11S-A) and basic (11S-B) subunits of 11S globulin (glycinin) are shown. Accumulation of the b subunit was increased, whereas those of glycinins are reduced. B and C, OAS or ABA was added to the control medium at concentrations shown in the figure. Accumulation of the b subunit was increased by OAS or ABA application, whereas glycinin accumulation was reduced.
Degradation during germination
Degradation of SSPs has been studied in legumes and cereals but there are little reports on similar studies in Arabidopsis. Proteases involved in SSP degradation include cysteine proteinase B (EPB) of barley (Mikkonen et al., 1996) and REP-1 of rice (Kato et al., 1999). Expression of the gene encoding EPB is regulated by both ABA and gibberellic acid, and cis- and trans-acting factors involved in this regulation have been described (Cercos et al., 1999).
Biotechnology
Seed storage proteins are the major source of plant proteins in the human diet. SSPs, however, do not necessarily have ideal compositions of essential amino acids for human needs. SSPs of legumes in general have limited contents of Met, whereas cereal SSPs do not contain sufficient amounts of other essential amino acids such as Lys, Trp and Tyr. One of the major goals for improvement of SSPs is to improve amino acid compositions. Given such a background, the alteration of SSP composition has been a major focus for biotechnology. Due to the fact that they are Met-rich proteins, 2S proteins have been expressed in transgenic plants to increase Met contents in seeds with moderate success (Altenbach et al., 1989).
Alteration of SSP composition has been reported in some crops including soybean (Kinney et al., 2001) and rice (Katsube et al., 1999). SSP gene promoters have also been used to drive expression of nutritionally useful genes including ferritin (Goto et al., 1999). As a model plant species, Arabidopsis has contributed in this field. It was demonstrated that antisense expression of 2S albumin gene in transgenic Arabidopsis carrying the Phaseolus vulgaris arcelin (arc) 5-I gene resulted in reduced accumulation of 2S albumin and an increase in accumulation of Arc5 proteins (Goossens et al., 1999). This was likely due to a diverging of resources for SSP synthesis to the foreign protein.
Vegetative storage proteins
VSPs were first described in leaves of depodded soybean plants (Wittenbach, 1982) and accumulated in soybean leaves to approximately 50% of the soluble proteins in sink-deprived plants (Wittenbach, 1983). Soybean has two types of VSPs, VSPa and VSPb (Staswick, 1988). Both VSPs are glycoproteins and share a high degree of sequence identity (about 80%) with a molecular mass of 27 kD and 29 kD, respectively, and carry phosphatase activity. These proteins are expressed in young seedlings, elongating hypocotyls, young leaves, stems, flowers and pods (Staswick, 1988). VSPs have also been described in a variety of other tissues, such as the tubers of potato and sweet potato (Wiltshire and Cobb, 1996, for review) and the bark of poplar (Lawrence et al., 1997; Zhu and Coleman 2001). These storage proteins accumulate in such tissues when excess photosynthates are produced.
Two genes have been described in Arabidopsis that are homologous to soybean VSPs (Berger et al., 1995; Utsugi et al., 1998). These genes are expressed in various tissues including the shoots, petioles, peduncles and siliques (Utsugi, 1998). It is yet to be clearly demonstrated if these homologous genes are expressed under high carbon and nitrogen conditions, however, it has been demonstrated that expression of the genes encoding Arabidopsis VSPs is regulated by jasmonic acid (JA) (Benedetti et al., 1995), similar to that of soybean VSPs. VSPs are also induced by wounding and phosphate starvation (Berger et al., 1995). Using JA-regulated expression of VSP1 promoter as a marker, the Arabidopsis mutant cev1 was identified in a study on the regulation of gene expression by JA (Ellis and Turner, 2001). Expression of VSP1 and VSP2 that code for Arabidopsis VSPs was enhanced in the cev1 (constitutive expression of VSP1) mutant and it was suggested that CEV1 functions in the early steps of the JA pathway and requires COI1 (coronatine insensitive 1, a F-box protein required for JA response) and ETR1 (ethylene response 1, a putative ethylene receptor) for its function (Ellis and Turner, 2001). Expression of VSP1 was also affected by the sos (salt overly sensitive) mutations (Gong et al., 2001).
Conclusions
In this review we have attempted to provide a summary of both the early and current work on storage proteins. Research using agricultural crop species such as legumes and cereals has provided us with a wealth of information on the composition and characteristics of storage proteins, however genetic analyses on the regulation of the genes encoding storage proteins has proven difficult in these species. As we have shown above, such studies now being carried out using Arabidopsis as a model are expected to further our understanding of the mechanisms behind storage protein accumulation at the genetic level.
Footnotes
Citation: Fujiwara T., Nambara E., Yamagishi K., Goto D.B., and Naito S. (2002) Storage Proteins. The Arabidopsis Book 1:e0020. doi:10.1199/tab.0020
elocation-id: e0020
Published on: September 30, 2002
Table 1.
Storage protein genes of Arabidopis thaliana ecotype Columbia
References
- Altenbach S. B., Ltenbach S. B., Pearson K. W., Meeker G., Staraci L. C., Sun S. S. M. Enhancement of the methionine content of seed proteins by the expression of a chimeric gene encoding a methionine-rich protein in transgenic plants. Plant Mol. Biol. 1989;131(1):513–522. doi: 10.1007/BF00027311. [DOI] [PubMed] [Google Scholar]
- Baumlein H., Nagy I., Villarroel R., Inze D., Wobus U. Cis-analysis of a seed protein gene promoter - the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992;21(1):233–239. [PubMed] [Google Scholar]
- Benedetti C. E., Xie D. X., Turner J. G. COI1-Dependent Expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 1995;1091(1):567–572. doi: 10.1104/pp.109.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Bell E., Sadka A., Mullet J. E. Arabidopsis thaliana Atvsp is homologous to soybean Vspa and Vspb, genes encoding vegetative storage protein acid-phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 1995;271(1):933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N., Conceicao A. D., Giraudat J., Koornneef M., Leon-Kloosterziel K., Valon C., Delseny M. Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol. Biol. 1999;401(1):1045–1054. doi: 10.1023/a:1006252512202. [DOI] [PubMed] [Google Scholar]
- Bobb A. J., Chern M. S., Bustos M. M. Conserved RY-repeats mediate transactivation of seed-specific promoters by the developmental regulator PvALF. Nucl. Acids Res. 1997;251(1):641–647. doi: 10.1093/nar/25.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K., Hattori J., Baum B. R., Miki B. L. Evolution of 2S albumin seed storage protein genes in the Brassicaceae. Biochem. System. Ecol. 1999;271(1):223–234. [Google Scholar]
- Bray E. A., Beachy R. N. Regulation by ABA of b-conglycinin expression in cultured developing soybean cotyledons. Plant Physiol. 1985;791(1):746–750. doi: 10.1104/pp.79.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercos M., Gomez-Cadenas A., Ho T. H. D. Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 1999;191(1):107–118. doi: 10.1046/j.1365-313x.1999.00499.x. [DOI] [PubMed] [Google Scholar]
- Chamberland S., Daigle N., Bernier F. The legumin boxes and the 3′ part of a soybean b-conglycinin promoter are involved in seed gene-expression in transgenic tobacco plants. Plant Mol. Biol. 1992;191(1):937–949. doi: 10.1007/BF00040526. [DOI] [PubMed] [Google Scholar]
- Chiba Y., Ishikawa M., Kijima F., Tyson R. H., Kim J., Yamamoto A., Nambara E., Leustek T., Wallsgrove R. M., Naito S. Evidence for autoregulation of cystathionine g-synthase mRNA stability in Arabidopsis. Science. 1999;2861(1):1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- Conceicao A. D., Vanvliet A., Krebbers E. Unexpectedly higher expression levels of a chimeric 2S albumin seed protein transgene from a tandem array construct. Plant Mol. Biol. 1994;261(1):1001–1005. doi: 10.1007/BF00028867. [DOI] [PubMed] [Google Scholar]
- Conceicao A. D., Krebbers E. A cotyledon regulatory region is responsible for the different spatial expression patterns of Arabidopsis 2S albumin genes. Plant J. 1994;51(1):493–505. doi: 10.1046/j.1365-313x.1994.05040493.x. [DOI] [PubMed] [Google Scholar]
- Ellis C., Turner J. G. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell. 2001;131(1):1025–1033. doi: 10.1105/tpc.13.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;51(1):765–771. [Google Scholar]
- Finkelstein R. R., Wang M. L., Lynch T. J., Rao S., Goodman H. M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;101(1):1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Lynch T. J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;121(1):599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M. Y., Chino M., Komeda Y., Naito S. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 1992;991(1):263–268. doi: 10.1104/pp.99.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Beachy R. N. Tissue-specific and temporal regulation of a b-conglycinin gene - Roles of the RY repeat and other cis-acting elements. Plant Mol. Biol. 1994;241(1):261–272. doi: 10.1007/BF00020166. [DOI] [PubMed] [Google Scholar]
- Gayler K. R., Sykes G. E. Effects of nutritional stress on the storage proteins of soybeans. Plant Physiol. 1985;781(1):582–585. doi: 10.1104/pp.78.3.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J., Hauge B. M., Valon C., Smalle J., Parcy F., Goodman H. M. isolation of the Arabidopsis-ABI3 gene by positional cloning. Plant Cell. 1992;41(1):1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T., Todd J., Ruuska S., White J., Benning C., Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;1241(1):1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z. Z., Koiwa H., Cushman M. A., Ray A., Bufford D., Kore-eda S., Matsumoto T. K., Zhu J. H., Cushman J. C., Bressan R. A., Hasegawa P. M. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;1261(1):363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens A., Van Montagu M., Angenon G. Co-introduction of an antisense gene for an endogenous seed storage protein can increase expression of a transgene in Arabidopsis thaliana seeds. FEBS Lett. 1999;4651(1):160–164. doi: 10.1016/s0014-5793(99)00943-6. [DOI] [PubMed] [Google Scholar]
- Goto F., Yoshihara T., Shigemoto N., Toki S., Takaiwa F. Iron fortification of rice seed by the soybean ferritin gene. Naturte Biotech. 1999;171(1):282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- Guerche P., Tire C., Grossi D. S. F., De Clercq A., Van Montagu M., Krebbers E. Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell. 1990;21(1):469–478. doi: 10.1105/tpc.2.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Fujiwara T., Goto K., Komeda Y., Chino M., Naito S. Differential regulation of soybean seed storage protein gene promoter-GUS fusions by exogenously applied methionine in transgenic Arabidopsis thaliana. Plant Cell Physiol. 1994;351(1):927–934. [Google Scholar]
- Hirai M., Fujiwara T., Chino M., Naito S. Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol. 1995;361(1):1331–1339. [PubMed] [Google Scholar]
- Hirner B., Fischer W. N., Rentsch D., Kwart M., Frommer W. B. Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J. 1998;141(1):535–544. doi: 10.1046/j.1365-313x.1998.00151.x. [DOI] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA. 1999;961(1):15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowach L. P., Thompson J. F., Madison J. T. Storage protein composition of soybean cotyledons grown in vitro in media of various sulfate concentrations in the presence and absence of exogenous L-methionine. Plant Physiol. 1984;741(1):584–589. doi: 10.1104/pp.74.3.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull G. A., Bies N., Twell D., Delseny M. Analysis of the promoter of an abscisic acid responsive late embryogenesis abundant gene of Arabidopsis thaliana. Plant Sci. 1996;1141(1):181–192. [Google Scholar]
- Inaba K., Fujiwara T., Hayashi H., Chino M., Komeda Y., Naito S. Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine: temporal and spatial patterns of soluble methionine accumulation. Plant Physiol. 1994;1041(1):881–887. doi: 10.1104/pp.104.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inquello V., Raymond J., Azanza J. L. Disulfide interchange reactions in 11S globulin subunits of Cruciferae seeds: Relationships to gene families. Eur. J. Biochem. 1993;2171(1):891–895. doi: 10.1111/j.1432-1033.1993.tb18318.x. [DOI] [PubMed] [Google Scholar]
- Kagaya Y., Ohmiya K., Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucl. Acids. Res. 1999;271(1):470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Shintani A., Minamikawa T. The structure and organization of two cysteine endopeptidase genes from rice. Plant Cell Physiol. 1999;401(1):462–467. doi: 10.1093/oxfordjournals.pcp.a029565. [DOI] [PubMed] [Google Scholar]
- Katsube T., Kurisaka N., Ogawa M., Maruyama N., Ohtsuka R., Utsumi S., Takaiwa F. Accumulation of soybean glycinin and its assembly with the glutelins in rice. Plant Physiol. 1999;1201(1):1063–1073. doi: 10.1104/pp.120.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K., Kraml M., Dengler N. G., McCourt P. fsca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;61(1):589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney A. J., Jung R., Herman E. M. Cosuppression of the alpha subunits of b-conglycinin in transgenic soybean seeds induces the formation of endoplasmic reticulum-derived protein bodies. Plant Cell. 2001;131(1):1165–1178. doi: 10.1105/tpc.13.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Hirai M. Y., Hayashi H., Chino M., Naito S., Fujiwara T. Role of O-acetyl-L-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta. 1999;2091(1):282–289. doi: 10.1007/s004250050634. [DOI] [PubMed] [Google Scholar]
- Kim H., Fujiwara T., Hayashi H., Chino M. Effects of exogenous ABA application on sulfate and OAS concentrations, and on composition of seed storage proteins in in vitro cultured soybean immature cotyledons. Soil Sci. Plant Nutr. 1997;431(1):1119–1123. [Google Scholar]
- Kirik V., Kolle K., Balzer H. J., Baumlein H. Two new oleosin isoforms with altered expression patterns in seeds of the Arabidopsis mutant fus3. Plant Mol. Biol. 1996;311(1):413–417. doi: 10.1007/BF00021803. [DOI] [PubMed] [Google Scholar]
- Krebbers E., Herdies L., De Clercq A., Seurinck J., Leemans J., Van Damme J., Segura M., Gheysen G., Van Montagu M., Vandekerckhove J. Determination of the processing sites of an Arabidopsis 2S albumin and characterization of the complete gene family. Plant Physiol. 1988;871(1):859–866. doi: 10.1104/pp.87.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup S., Jones H. D., Holdsworth M. J. Interactions of the developmental regulator AB13 with proteins identified from developing Arabidopsis seeds. Plant J. 2000;211(1):143–155. doi: 10.1046/j.1365-313x.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- Li G. F., Chandrasekharan M. B., Wolffe A. P., Hall T. C. Chromatin structure and phaseolin gene regulation. Plant Mol. Biol. 2001;461(1):121–129. doi: 10.1023/a:1010693703421. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2001;981(1):4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K. M., West M. A. L., Lo R., Kwong R. W., Yamagishi K., Fischer R. L., Goldberg R. B., Harada J. J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;931(1):1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Luerssen K., Kirik V., Herrmann P., Misera S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998;151(1):755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- Martin T., Hellmann H., Schmidt R., Willmitzer L., Frommer W. B. Identification of mutants in metabolically regulated gene expression. Plant J. 1997;111(1):53–62. doi: 10.1046/j.1365-313x.1997.11010053.x. [DOI] [PubMed] [Google Scholar]
- McCarty D. R., Hattori T., Carson C. B., Vasil V., Lazar M., Vasil I. K. The viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;661(1):895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- Meinke D. W., Franzmann L. H., Nickle T. C., Yeung E. C. Leafy cotyledon mutants of Arabidopsis. Plant Cell. 1994;61(1):1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkonen A., Porali I., Cercos M., Ho T. H. D. A major cysteine proteinase, EPB, in germinating barley seeds: Structure of two intronless genes and regulation of expression. Plant Mol. Biol. 1996;311(1):239–254. doi: 10.1007/BF00021787. [DOI] [PubMed] [Google Scholar]
- Naito S., Hirai M. Y., Chino M., Komeda Y. Expression of a soybean (Glycine max (L) Merr.) seed storage protein gene in transgenic Arabidopsis thaliana and its response to nutritional stress and to abscisic acid mutations. Plant Physiol. 1994a;1041(1):497–503. doi: 10.1104/pp.104.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S., Inaba-Higano K., Kumagai T., Kanno T., Nambara E., Fujiwara T., Chino M., Komeda Y. Maternal effects of mto1 mutation, that causes overaccumulation of soluble methionine, on the expression of a soybean b-conglycinin gene promoter-GUS fusion in transgenic Arabidopsis thaliana. Plant Cell Physiol. 1994b;351(1):1057–1063. [Google Scholar]
- Naito S., Hirai M. Y., Inaba H. K., Nambara E., Fujiwara T., Hayashi H., Komeda Y., Chino M. Expression of soybean seed storage protein genes in transgenic plants and their response to sulfur nutritional conditions. J. Plant Physiol. 1995;1451(1):614–619. [Google Scholar]
- Naito S., Inaba H. K., Kumagai T., Kanno T., Nambara E., Fujiwara T., Chino M., Komeda Y. Maternal effects of mto1 mutation, that causes overaccumulation of soluble methionine, on the expression of a soybean b-conglycinin gene promoter-GUS fusion in transgenic Arabidopsis thaliana. Plant Cell Physiol. 1994;351(1):1057–1063. [Google Scholar]
- Nakamura S., Lynch T. J., Finkelstein R. R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;261(1):627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Nambara E., Hayama Y., Tsuchiya Y., Nishimura M., Kawaide H., Kamiya Y., Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 2000;2291(1):412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- Nambara E., Naito S., McCourt P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 1992;21(1):435–441. [Google Scholar]
- Nuccio M. L., Thomas T. L. ATS1 and ATS3: Two novel embryo-specific genes in Arabidopsis thaliana. Plant Mol. Biol. 1999;391(1):1153–1163. doi: 10.1023/a:1006101404867. [DOI] [PubMed] [Google Scholar]
- Ogas J., Cheng J-C., Sung Z. R., Somerville C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science. 1997;2771(1):91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;961(1):13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J., Pollard M., Bao X., Focke M., Girke T., Ruuska S., Mekhedov S., Benning C. Fatty acid synthesis: from CO2 to functional genomics. Biochem Soc. Trans. 2000;281(1):567–574. [PubMed] [Google Scholar]
- Pang P. P., Pruitt R. E., Meyerowitz E. M. Molecular genomic organization expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 1988;111(1):805–820. doi: 10.1007/BF00019521. [DOI] [PubMed] [Google Scholar]
- Parcy F., Valon C., Raynal M., Gaubier-Comella P., Deleseny M., Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;61(1):1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F., Valon C., Kohara A., Misera S., Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;91(1):1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant A. L., Van Rooijen G. H. H., Anderson C. P., Moloney M. M. Regulation of an Arabidopsis oleosin gene promoter in transgenic Brassica napus. Plant Mol. Biol. 1994;251(1):193–205. doi: 10.1007/BF00023237. [DOI] [PubMed] [Google Scholar]
- Reidt W., Wohlfarth T., Ellerstroem M., Czihal A., Tewes A., Ezcurra I., Rask L., Baumlein H. Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000;211(1):401–408. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- Robinson C. K., Hill S. A. Altered resource allocation during seed development in Arabidopsis caused by the abi3 mutation. Plant Cell Environ. 1999;221(1):117–123. [Google Scholar]
- Rohde A., Kurup S., Holdswoth M. ABI3 emerges from the seed. Trends Plant Sci. 2000;51(1):418–419. doi: 10.1016/s1360-1385(00)01736-2. [DOI] [PubMed] [Google Scholar]
- Sakata Y., Chiba Y., Fukushima H., Matsubara N., Habu Y., Naito S., Ohno T. The RY sequence is necessary but not sufficient for the transcription activation of a winged bean chymotrypsin inhibitor gene in developing seeds. Plant Mol. Biol. 1997;341(1):191–197. doi: 10.1023/a:1005841125832. [DOI] [PubMed] [Google Scholar]
- Shewry P. R., Napier J. A., Tatham A. S. Seed storage proteins: structure and biosynthesis. Plant Cell. 1995;71(1):945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. Sugar-induced signal transduction in plants. Ann Rev, Plant Phys. Plant Mol. Biol. 2000;511(1):49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- Soderman E. M., Brocard I. M., Lynch T. J., Finkelstein R. R. Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 2000;1241(1):1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Koh S., Czako M., Marton L., Drenkard E., Becker J. M., Stacey G. Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol. 1997;1141(1):927–935. doi: 10.1104/pp.114.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E. Soybean vegetative storage protein structure and gene expression. Plant Physiol. 1988;891(1):309–315. doi: 10.1104/pp.87.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kao C. Y., McCarty D. R. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;91(1):799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L., Kwong L. W., Yee K. M., Pelletier J., Lepiniec L., Fischer R. L., Goldberg R. B., Harada J. J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA. 2001;981(1):11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi S., Sakamoto W., Murata M., Motoyoshi F. Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol. Biol. 1998;381(1):565–576. doi: 10.1023/a:1006072014605. [DOI] [PubMed] [Google Scholar]
- van der Klei H., Van Damme J., Casteels P., Krebbers E. A fifth 2S albumin isoform is present in Arabidopsis thaliana. Plant Physiol. 1993;1011(1):1415–1416. doi: 10.1104/pp.101.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient C. M., Bies-Etheve N., Delseny M. Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J. Exp. Bot. 2000;511(1):995–1003. doi: 10.1093/jexbot/51.347.995. [DOI] [PubMed] [Google Scholar]
- West M. A. L., Yee K. M., Danao J., Zimmerman L., Fischer R. L., Goldberg R. B., Harada J. J. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;61(1):1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire J. J. J., Cobb A. H. A review of the physiology of potato tuber dormancy Ann. Appl. Biol. 1996;1291(1):553–569. [Google Scholar]
- Wittenbach V. A. Effect of pod removal on leaf senescence in soybeans. Plant Physiol. 1982;701(1):1544–1548. doi: 10.1104/pp.70.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach V. A. Effect of pod removal on leaf photoshythesis and soluble protein composition of field-grown soybeans. Plant Physiol. 1983;731(1):121–124. doi: 10.1104/pp.73.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfarth T., Braun H., Kirik V., Koelle K., Czihal A., Tewes A., Luerssen H., Misera S., Shutov A., Baeumlein H. Regulation and evolution of seed globulin genes. J. Plant Physiol. 1998;1521(1):600–606. [Google Scholar]
- Zhu B., Coleman G. D. The poplar bark storage protein gene (Bspa promoter is responsive to photoperiod and nitrogen in transgenic poplar and active in floral tissues, immature seeds and germinating seeds of transgenic tobacco. Plant Mol. Biol. 2001;461(1):383–394. doi: 10.1023/a:1010600504740. [DOI] [PubMed] [Google Scholar]
- Zou J., Brokx S. J., Taylor D. C. Cloning of a cDNA encoding the 21.2 kDa oleosin isoform from Arabidopsis thaliana and a study of its expression in a mutant defective in diacylglycerol acyltransferase activity. Plant Mol. Biol. 1996;311(1):429–433. doi: 10.1007/BF00021805. [DOI] [PubMed] [Google Scholar]


