Abstract
Bioactive gibberellins (GAs) are diterpene phytohormones that modulate growth and development throughout the whole life cycle of the plant. Arabidopsis genes encoding most GA biosynthesis and catabolism enzymes, as well as GA receptors (GIBBERELLIN INSENSITIVE DWARF1, GID1) and early GA signaling components have been identified. Expression studies on the GA biosynthesis genes are beginning to reveal the potential sites of GA biosynthesis during plant development. Biochemical and genetic analyses demonstrate that GA de-represses its signaling pathway by binding to GID1s, which induce degradation of GA signaling repressors (DELLAs) via an ubiquitin-proteasome pathway. To modulate plant growth and development, the GA pathway is also regulated by endogenous signals (other hormones) and environmental cues (such as light, temperature and salt stress). In many cases, these internal and external cues directly affect GA metabolism and bioactive GA levels, and indirectly alter DELLA accumulation and GA responses. Importantly, direct negative interaction between DELLA and PIF3 and PIF4 (2 phytochrome interacting transcription factors) appears to integrate the effects of light and GA on hypocotyl elongation.
INTRODUCTION
Gibberellin (GA)-promoted stem growth was originally discovered by studies of the Bakanae (foolish seedling) disease in rice. Gibberella fujikuroi, a pathogenic fungus, produces chemicals that cause the infected rice plants to grow so tall that they fall over (Figure 1). In the 1930s, Japanese researchers obtained impure crystals of these compounds (named gibberellin A) from fungal filtrate. In 1950s, chemical structures of giberellin A were determined to be a mixture of GA1, GA2 and GA3 with GA3 being the major component by scientists in the US, UK and Japan (Takahashi et al., 1991). GA3 is also called gibberellic acid. Subsequent studies of dwarf mutants and analysis of GA content in wild-type (WT) and mutant plants revealed that bioactive GAs are endogenous plant hormones that modulate diverse developmental processes. Although 136 GAs have been identified in higher plants and fungi (MacMillan, 2002); http://www.plant-hormones.info/ga1info.htm), only a few are biologically active, e.g., GA1, GA3, GA4 and GA7 (Figure 2; Hedden and Phillips, 2000). Many of the other GAs are biosynthetic intermediates or catabolites of bioactive GAs. GA# is based on chronological order of its discovery, not based on its position in the biosynthetic pathway. Bioactive GAs regulate both vegetative and reproductive growth in plants. In Arabidopsis, GA is essential for seed germination, leaf and root growth, floral induction (in short-day conditions), inflorescence stem elongation (bolting), anther and petal development, and fruit and seed development. Severe GA-deficient mutants display pleiotropic phenotypes throughout their life cycle: non-germinating seeds (that can be rescued by seed coat removal), dark green compact rosette, extreme dwarf, late flowering (under long-day conditions) and male-sterile. This chapter will review GA biosynthesis and catabolism pathways, the physiological roles of GA and developmental regulation of GA metabolism, GA perception and its signaling pathway, and environmental and hormonal regulation of GA pathways in Arabidopsis.
Figure 1.

The Bakanae (Foolish Seedling) Disease in Rice Caused by Gibberella fujikuroi Infection (photo courtesy of Yuji Kamiya). The arrrow indicates the spindly seedling with the disease.
Figure 2.
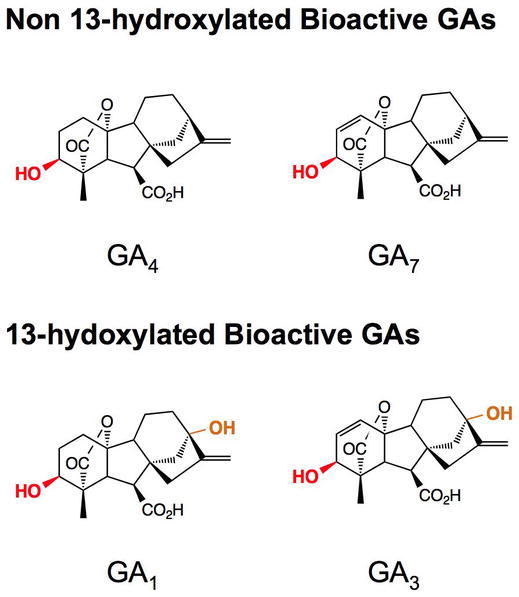
Major Bioactive GAs from Plants and Fungi.
GA4 is the major active GA in Arabidopsis. GA3 is the most abundant active GA made in fungi. The 3β-hydroxyl groups are highlighted in red and the 13-hydroxyl groups are highlighted in orange.
GA BIOSYNTHESIS AND CATABOLISM
GA biosynthesis and catabolism pathways have been studied extensively by gas chromatography-mass spectrometry analysis of GA content, purification of GA metabolism enzymes, isolation of GA-deficient mutants, and cloning of the corresponding genes (Hedden and Phillips, 2000; Yamaguchi, 2008). Biosynthesis of GA in higher plants can be divided into three stages: (i) biosynthesis of ent-kaurene from geranyl geranyl diphosphate (GGDP) in proplastids, (ii) conversion of ent-kaurene to GA12 via cytochrome P450 monooxygenases, and (iii) formation of C20- and C19-GAs in the cytoplasm (Figure 3). GGDP is a common precursor for GAs, carotenoids and chlorophylls. It is converted to ent-kaurene, the first committed intermediate in the GA pathway, in a two-step cyclization reaction, catalyzed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS). In the second stage of the pathway, stepwise oxidation followed by ring contraction is catalyzed by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) to produce GA12 (Figure 3A). In the third stage of GA biosynthesis, GA12 can be further converted to GA53 by 13-hydroxylation. GA12 and GA53 are then converted to various GA intermediates and bioactive GAs, including GA1 and GA4, by two parallel pathways that include a series of oxidation steps catalyzed by 2-oxoglutarate-dependent dioxygenases, GA 20-oxidases (GA20ox) and 3-oxidases (GA3ox; Figure 3B). The four major active GAs in plants all contain a 3β-hydroxyl group (Figures 2 and 3B).
Figure 3.
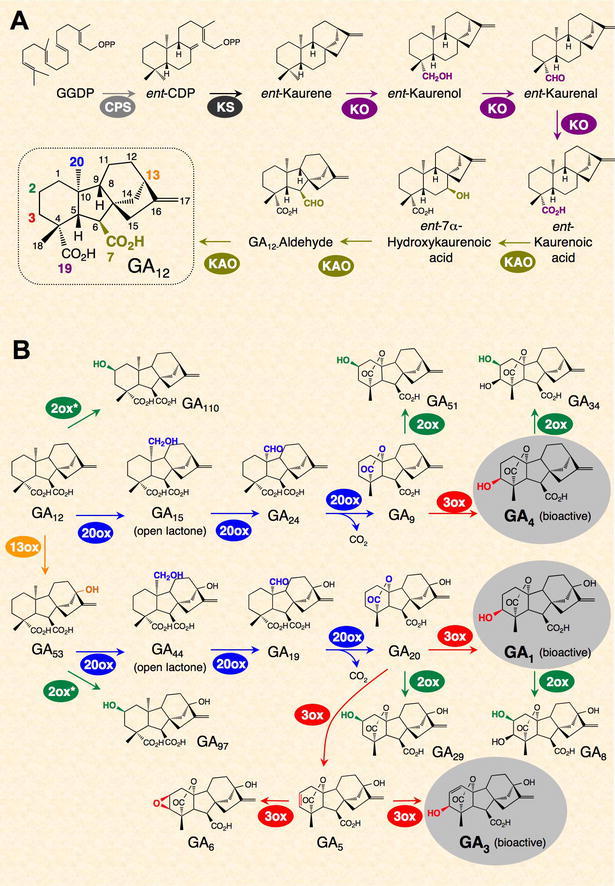
GA Biosynthetic and Catobolic Pathways in Plants. Adapted from Yamaguchi, 2008: reprinted, with permission, from the Annual Review of Plant Biology, Vol. 59 ©2008 by Annual Reviews, www.annualreviews.org.
(A) Synthesis of GA12 from GGDP.
(B) GA biosynthesis and deactivation (by GA2ox) pathways from GA12. The three active GAs are highlighted in grey circles. GA7 (13-nonhydroxy GA3), another active GA, is synthesized from GA9 (not shown).
GA Biosynthetic and Catobolic Pathways in Plants.
The amount of bioactive GAs can be affected by the rates of their synthesis and deactivation. It has been known for a long time that 2β-hydroxylation, which is catalyzed by GA 2-oxidases (GA2ox; Figure 3B), can convert active GAs (and/or their precursors) to inactive forms. Recently, two novel deactivation mechanisms were identified: epoxidation of non-13-hydroxylated GAs in rice (Zhu et al., 2006) and methylation of GAs in Arabidopsis (Varbanova et al., 2007) (Figure 4). The recessive rice mutant, elongated uppermost internode (eui), has elevated active GA levels only in the upper internodes. EUI encodes a P450 enzyme (CYP714D1) that catalyzes 16α,17-epoxidation of non-13-hydroxylated GAs (GA4, GA9 and GA12). 16a, 17-[OH]2-GAs are present in many species, suggesting that epoxidation of GAs is a common deactivation mechanism. Functional ortholog(s) of EUI in Arabidopsis remain to be characterized. The Arabidopsis GA methyltransferases (GAMT1 and GAMT2) are mainly expressed in developing and germinating seeds. Seeds of the double null mutant contain high levels of active GAs and are resistant to a GA biosynthesis inhibitor during germination. However, methylated GAs have not been detected in plant extracts. Future studies will be needed to determine whether and how these two new types of modification are widely used in different species to modulate active GA content.
Figure 4.
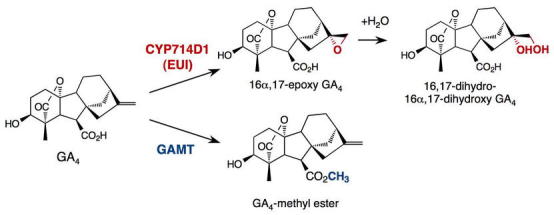
Epoxidation by EUI in Rice and Methylation by GAMT in Arabidopsis Are Alternative GA Deactivation Reactions. From Yamaguchi, 2008: reprinted, with permission, from the Annual Review of Plant Biology, Vol. 59 ©2008 by Annual Reviews, www.annualreviews.org.
GENES ENCODING ENZYMES FOR GA BIOSYNTHESIS AND CATABOLISM
Recent molecular genetic and biochemical studies helped to identify GA biosynthesis and catabolism genes in Arabidopsis and other species. A number of GA-deficient dwarf mutants in Arabidopsis were isolated by Maarten Koornneef and he named them ga1, ga2, ga3, ga4 and ga5 (Koornneef and van der Veen, 1980). After the cloning of corresponding genes, we now know that GA1 encodes CPS (Sun and Kamiya, 1994), GA2 encodes KS (Yamaguchi et al., 1998a), GA3 encodes KO (Helliwell et al., 1998), GA4 encodes GA3ox1 (Chiang et al., 1995), and GA5 encodes GA20ox1 (Phillips et al., 1995). In the last decade, genes encoding enzymes in most steps in the GA metabolic pathways (except for 13-hydroxylation) have been isolated from different species (Hedden and Phillips, 2000; Yamaguchi, 2008). CPS, KS and KO are each encoded by a single gene in Arabidopsis (Table 1), and also in most plant species examined. Synthesis of ent-kaurene, the first committed intermediate in the GA pathway, is catalyzed by CPS and KS. qRT-PCR analysis showed that extremely low amounts of CPS mRNA are present throughout development in Arabidopsis, and the expression pattern of this gene is cell-type specific (Silverstone et al., 1997b). Rapidly growing tissues, including shoot apices, root tips, developing anthers and seeds, contain higher amounts of CPS mRNA. The expression pattern of KS is similar to that of CPS, but the overall amount of KS mRNA is much higher than CPS (Yamaguchi et al., 1998a), suggesting that the synthesis of ent-kaurene is primarily determined by controlling the expression and location of CPS. In addition, overexpression of CPS resulted in a dramatic increase in ent-kaurene accumulation (Fleet et al., 2003), supporting previous speculation that CPS may act as a gatekeeper, controlling the location and the activity of the early stages of GA biosynthesis. Null alleles of the three early genes in Arabidopsis (ga1-3, ga2 and ga3) are non-germinating, male-sterile, severe dwarfs (Koornneef and van der Veen, 1980). Two KAO genes are present in Arabidopsis, and both are expressed in all tissues examined (Helliwell et al., 2001b).
Table 1.
GA Metabolism and Signaling Genes
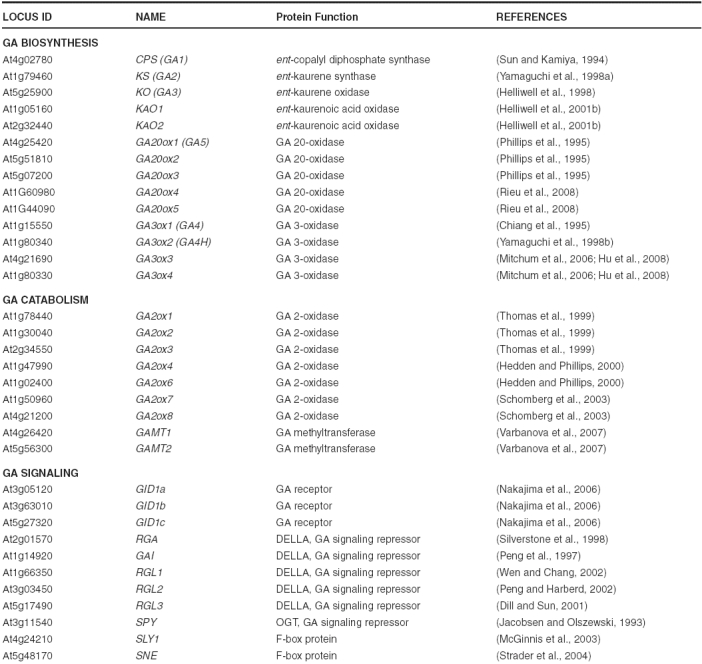
In contrast, GA20ox, GA3ox and GA2ox are each encoded by a small gene family (Table 1), and each gene shows a tissue-specific expression pattern (Thomas et al., 1999; Mitchum et al., 2006; Rieu et al., 2008). In the case of GA3ox genes, GA3ox1 is expressed throughout all stages of plant development, and GA3ox2 is mainly expressed in germinating seeds and vegetative tissues. In contrast, GA3ox3 and GA3ox4 are only expressed in developing flowers and siliques. As predicted by their expression patterns and levels, GA3ox1 has a predominant role in plant development: the ga3ox1 mutant displays a semi-dwarf phenotype (with normal fertility), whereas the other ga3ox single mutants have no obvious phenotypes because of functional redundancy. Consistent with their developmental expression profiles, the ga3ox1 ga3ox2 double mutant is more dwarfed than ga3ox1, and ga3ox1 ga3ox3 ga3ox4 displays floral defects (see details below). Among the GA20ox genes, GA20ox1, GA20ox2 and GA20ox3 have higher expression in most tissues examined (Rieu et al., 2008). GA20ox1 transcript is highly abundant in the inflorescent stem, consistent with the semi-dwarf phenotype of the ga20ox1 knockout mutant. The single and double mutant analysis indicates that GA20ox1 and GA20ox2 have overlapping roles throughout plant development. However, GA20ox1 plays a more prominent role in elongation of internodes and stamen filaments, whereas GA20ox2 contributes more in promoting floral initiation and silique growth. Overexpression of GA20ox and GA2ox genes in transgenic Arabidopsis alters the concentrations of bioactive GA (Huang et al., 1998; Coles et al., 1999; Schomberg et al., 2003). The GA20ox overexpression lines display early flowering and increased stem growth phenotypes, whereas GA2ox overexpression lines show late flowering and dwarf phenotypes. In contrast, overexpression of CPS, KS or KO did not affect bioactive GA levels or plant phenotype (Fleet et al., 2003; Swain et al., 2005), indicating that genes in the late stages of the pathway are crucial regulatory steps in modulating active GA content.
GA HOMEOSTASIS AND FEEDBACK MECHANISM
Accumulating evidence indicates that GA homeostasis is achieved by a feedback mechanism. Alterations in GA signaling activity due to chemical treatments (GA or GA biosynthesis inhibitors) or genetic mutations cause changes in expression of some of the GA biosynthesis genes (GA20ox and GA3ox) as well as GA catabolism genes GA2ox (reviewed in Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000; Olszewski et al., 2002; Sun and Gubler, 2004). Expression of early GA biosynthesis genes CPS, KS and KO is not affected by the feedback mechanism (Helliwell et al., 1998). In Arabidopsis, transcript levels of GA3ox1, GA20ox1, GA20ox2 and GA20ox3 are down-regulated by applied GA or by mutations that cause elevated GA signaling. In contrast, the GA catabolism genes GA2ox1 and GA2ox2 are up-regulated by GA treatment (Thomas et al., 1999). Conversely, GA-deficiency or mutations that reduce GA signaling result in up-regulation of GA3ox and GA20ox expression, and down-regulation of GA2ox expression. Interestingly, expression of the other GA3ox and GA20ox genes are not feedback regulated (Matsushita et al., 2007; Rieu et al., 2008), suggesting that they may promote elevated active GA levels that are essential for growth and development as will be discussed later in the case of the role of GA3ox2 in seed germination.
SUBCELLULAR COMPARTMENTALIZATION AND INTER-CELLULAR SEPARATION OF GA METABOLIC PATHWAY
The GA biosynthetic pathway is separated into distinct subcellular compartments (Figure 5). GGDP for GA biosynthesis is mainly derived from the methylerythritol phosphate pathway in the plastid, although the cytoplasmic mevalonate pathway also has a minor contribution (Kasahara et al., 2002). Biochemical studies showed that the two terpene synthases CPS and KS are localized in the stroma of plastids (Sun and Kamiya, 1994; Helliwell et al., 2001a). Transient expression studies of AtKO- and AtKAO-green fluorescent protein (GFP) fusions in tobacco leaves suggest that KO is located on the outer surface of the plastid envelope, whereas KAO is associated with the endoplasmic reticulum (Helliwell et al., 2001a). GA20ox, GA3ox and GA2ox are soluble enzymes and presumably localized in the cytoplasm. Therefore, the hydrocarbon ent-kaurene synthesized in the plastids (by CPS and KS) is somehow transported across the plastid envelope to be converted to ent-kaurenoic acid (by KO), and then to GA12 by the ER-associated KAO. GA12 is subsequently converted to the active GA4 by GA20ox and GA3ox in the cytoplasm. In addition to the subcellular compartmentalization, early (CPS) and later (KO and GA3ox) steps in GA biosynthetic pathway appears to occur in distinct cell types during seed germination (Yamaguchi et al., 2001), in elongating roots (Mitchum et al., 2006) and developing flowers (Hu et al., 2008). Such physical separation of the early and late steps in the GA biosynthetic pathway, which will be discussed in more detail in the next section, may add another layer of regulation to GA production.
Figure 5.
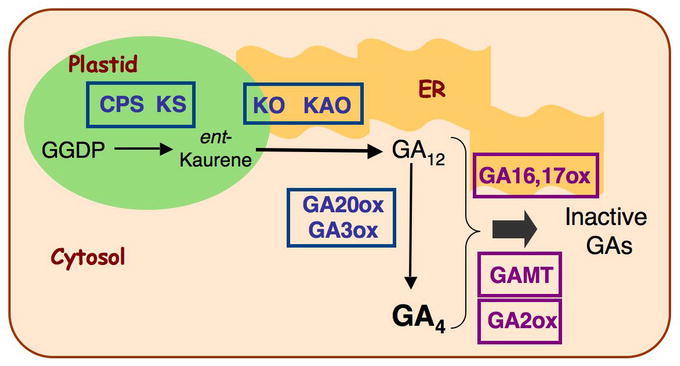
Subcellular Localization of GA Biosynthetic and Catabolic Pathways. Adapted from Yamaguchi, 2008: reprinted, with permission, from the Annual Review of Plant Biology, Vol. 59 ©2008 by Annual Reviews, www.annualreviews.org.
DEVELOPMENTAL REGULATION OF GA METABOLISM AND GA TRANSPORT
To have a complete understanding of the developmental regulation of GA metabolism, it will be necessary to conduct detailed localization studies of all genes for the entire pathway in the same species by in situ RNA hybridization, promoter-reporter fusion, and if possible, by immunostaining using specific antibodies. So far, a number of studies using in situ RNA hybridization and promoter-reporter gene fusions are beginning to reveal the sites of GA metabolism. These studies are also showing the complex mechanisms by which the amounts of bioactive GAs are modulated in different tissues to regulate plant growth and development. However, immunostaining has not been feasible because enzymes involved in GA metabolism are present at very low levels.
Seed Germination
During germination, GA can promote embryo growth and facilitate radicle protrusion through the seed coat. In Arabidopsis seed, the primary role of GA is to facilitate the breakage of the seed coat and de novo GA biosynthesis is required. Seeds of severely GA-deficient mutants, such as ga1, ga2 and ga3 single mutants and the double homozygous mutant ga3ox1 ga3ox2, fail to germinate in the dark and have only ~5% germination frequency in the light (Koornneef and van der Veen, 1980; Mitchum et al., 2006). Similarly, the triple GA receptor mutant gid1a gid1b gid1c also fails to germinate (Griffiths et al., 2006). However, when the seed coats of these mutants are damaged mechanically, the mutant embryos can germinate and grow into plants (Telfer et al., 1997; Griffiths et al., 2006). Mutations that affect abscisic acid (ABA) biosynthesis or seed coat structure in Arabidopsis and tomato rescue the seed germination defect of GA biosynthesis mutants, suggesting that GA is essential to overcome the inhibitory effects of the seed coat and ABA-related embryo dormancy on seed germination (Groot and Karssen, 1987; Debeaujon and Koornneef, 2000).
To examine the sites of GA biosynthesis in germinating seeds, expression patterns of CPS, KO and GA3ox genes have been analyzed. Using a translational gene fusion CPS promoter:CPS-GUS (CPS-GUS) in transgenic Arabidopsis, CPS promoter activity is detected in the shoot apex and provasculature in both cotyledons and embryo axis (Figure 6A; Silverstone et al., 1997b; Yamaguchi et al., 2001). In contrast, transcripts of KO, GA3ox1 and GA3ox2 are detected in the cortex and endodermis in the embryo axis by promoter-GUS analysis and/or in situ hybridization (Figures 6B–6C). These results indicate that the synthesis of bioactive GAs from ent-kaurene occurs in the cortex and endodermis. The cortical cells in embryo axis begin to expand prior to radicle protrusion. Therefore, the site of bioactive GA synthesis seems to occur in rapidly expanding cells, which are likely to be the GA-responding cells. GA3ox1 and GA3ox2 appear to have partially redundant functions because their spatial expression patterns are similar in the germinating embryo, and only the double mutant shows a germination defect (Mitchum et al., 2006). However, only GA3ox1 expression is under feedback control by the GA response pathway, suggesting that GA3ox2 may be required for the embryo to produce a higher level of bioactive GA to ensure efficient germination. Interestingly, early (CPS) and later (KO, GA3ox) GA biosynthetic genes are expressed in distinct cell types. The physical separation of steps in the GA biosynthetic pathway in germinating embryos implies that the synthesis of active GAs requires intercellular transport of a pathway intermediate. The transported intermediate is likely ent-kaurene because biochemical studies suggest that CPS and KS form a complex in catalyzing the two consecutive cyclization reactions (West et al., 1982).
Figure 6.
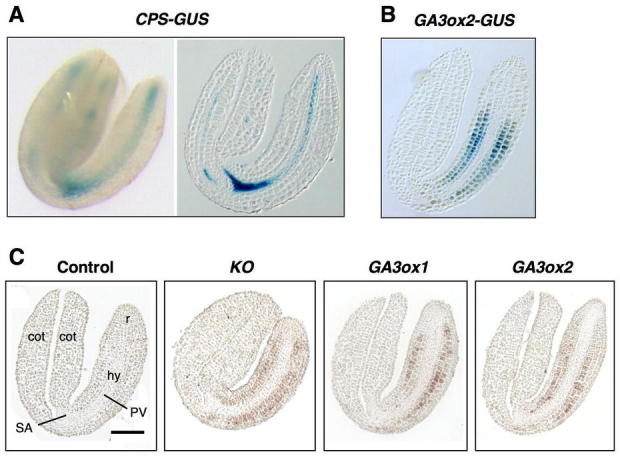
Physical Separation of Early and Later Steps in GA Biosynthesis in Germinating Embryos.
(A) X-gluc staining of the CPS-GUS line. left, whole embryo; right, a longitudinal section.
(B) X-gluc staining of the GA3ox2-GUS line (a longitudinal section of the embryo).
(C) In situ hybridization analysis of KO, GA3ox1 and GA3ox2 mRNAs. Wild-type embryo sections were incubated with the sense GA3ox1 (control), anti-sense KO or GA3ox1 or GA3ox2 probes. PV, provasculature; r, radicle; hy, hypocotyl; SA, shoot apex; cot, cotyledon. Scale bar = 100μmm.
In (A–C), all seeds were imbibed for 24 h under constant light. Dissected embryos were then sectioned or X-gluc stained. This figure was adapted from Yamaguchi et al. (2001) with permission from Blackwell Publishing.
Vegetative Phase
Following germination, bioactive GAs promote hypocotyl and stem elongation, leaf expansion and root growth. Severely GA-deficient mutants, such as ga1, ga2 and ga3 single mutants (defective in CPS, KS and KO, respectively) and the triple GA receptor mutant gid1a gid1b gid1c are extreme dwarfs with much reduced hypocotyl and root lengths and small rosette leaves (Figures 7A–7B; Koornneef and van der Veen, 1980; Silverstone et al., 1997a; Griffiths et al., 2006). GA3ox1 and GA3ox2, among the 4 GA3ox genes, are expressed during vegetative growth. The ga3ox1 single mutant is a semi-dwarf (Figure 7A), whereas the ga3ox2 mutant has no obvious phenotype, indicating that GA3ox1 plays a predominant role in vegetative growth. The ga3ox1 ga3ox2 double mutant is more dwarfed than ga3ox1, although it is not as severe as the ga1 mutant (Figures 7A and 7C; Mitchum et al., 2006). The GA4 levels in ga3ox1, ga3ox1 ga3ox2 and ga1 are 50%, 20% and <10% of WT, respectively, consistent with the plant phenotypes (Mitchum et al., 2006). The trace amount of GA4 produced in ga1 may be due to uptake of ent-kaurene synthesized by nearby plants because this hydrocarbon was shown to be released into the atmosphere and can be taken up by the ga1 mutant (Otsuka et al., 2004). In the case of the ga3ox1 ga3ox2 double mutant, the extremely elevated GA9 level (24 folds) may facilitate the conversion of GA4 by a small amount of GA3ox3 or GA3ox4 even though their transcripts are not detectable in the rosette leaves.
Figure 7.
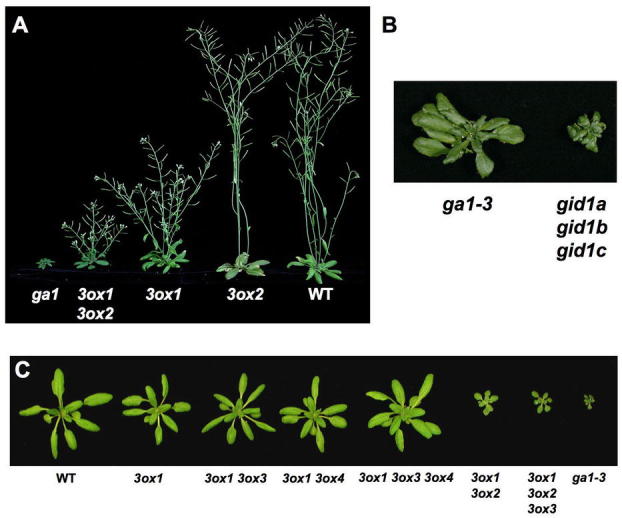
Phenotypes of GA Biosynthesis and Receptor Mutants.
(A) 40-day-old plants. Reproduced from Mitchum et al. (2006) with permission from Blackwell Publishing.
(B) The triple gid1 mutant is more severely dwarfed than ga1–3. Reprinted from Griffiths et al. (2006).
(C) 24-day-old plants. Reproduced from Hu et al. (2008). Lacking GA3ox3 and/or GA3ox4 functions in the ga3ox1 mutant background do not alter rosette and stem growth because GA3ox3 and GA3ox4 are mainly expressed in flowers and/or siliques. All mutants are homozygous for the labeled genotype(s).
qRT-PCR and/or RNA blot analyses indicated that CPS, KS and KO mRNAs in Arabidopsis are accumulated more in young seedlings compared to older vegetative tissues (Silverstone et al., 1997b; Helliwell et al., 1998; Yamaguchi et al., 1998a). Expression studies using the CPS-GUS transgenic Arabidopsis showed that CPS is expressed in shoot and root apices, and the vasculature of petioles and leaves (Figure 8). In most vegetative tissues, expression of either GA3ox1-GUS and/or GA3ox2-GUS overlaps with CPS-GUS (Figure 8). In two day-old seedlings, CPS-GUS is detected in the two-to-four-cell-layer-thick juvenile shoot apical meristem (SAM) and in the region below the SAM that is undergoing cell division, elongation and differentiation (Silverstone et al., 1997b). Interestingly, in five-day-old seedlings, CPS-GUS is only expressed in the region below the SAM, but not in the adult SAM (Figure 8A). GA20ox1-GUS and GA3ox1-GUS are expressed in the rib meristem below the SAM and leaf primordia, but not or only weakly expressed in the SAM (Figure 8B; Hay et al., 2002; Mitchum et al., 2006).
Figure 8.
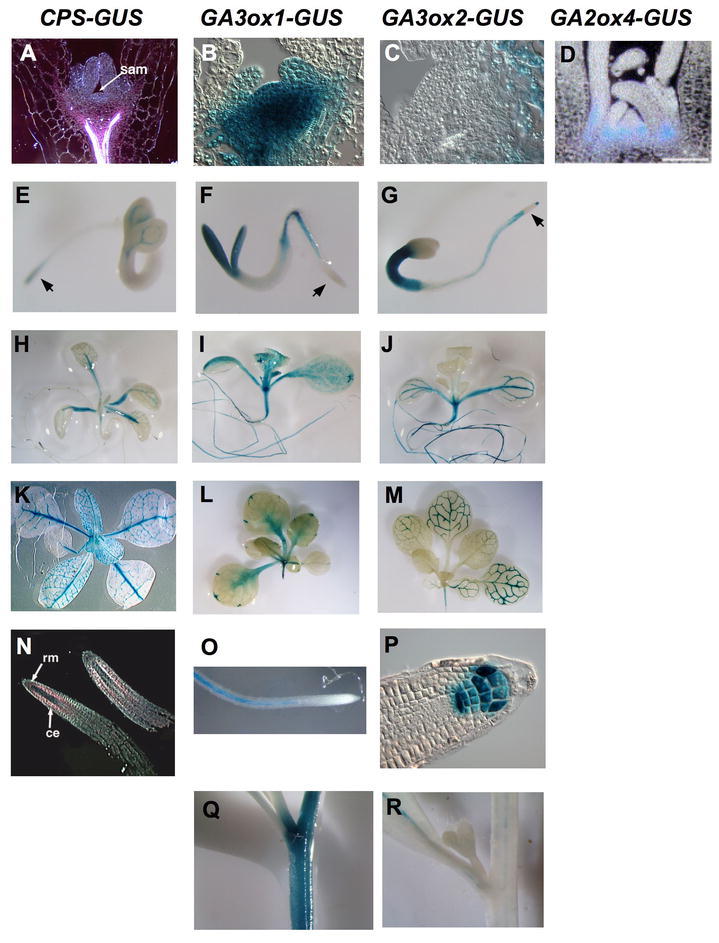
Histochemical Localization of CPS, GA3ox1, GA3ox2 and GA2ox4 Promoter
Activity in Transgenic Arabidopsis Containing Promoter-GUS Gene Fusions. Tissues were stained with X-gluc.
(A–D) shoot apices of seedlings. Longitudinal sections through apical shoot meristem.
(E–G) Three-day-old seedlings. Arrows indicate the elongation zone.
(H–J) Ten-day-old seedling.
(K)Four-week-old rosette.
(L–M) Two-week-old rosettes.
(N–P) roots: longitudinal sections in (N) and (P), whole root in (O).
(Q–R) Inflorescence stems. The X-gluc staining is pink when viewed under dark-field microscopy in (A) and (N). sam, shoot apical meristem; ce, cortex and endodermis; rm, root meristem.
CPS-GUS images reprinted from Silverstone et al. (1997b) with permission from Blackwell Publishing.
GA3ox-GUS images reprinted from Mitchum et al. (2006) with permission from Blackwell Publishing.
GA2ox4-GUS image reprinted from Jasinski et al. (2005) with permission from Elsevier.
Histochemical Localization of CPS, GA3ox1, GA3ox2 and GA2ox4 Promoter
KNOTTED-LIKE HOMEOBOX (KNOX) transcription factors appear to reduce bioactive GA levels in the SAM. In simple-leaved species (e.g., Arabidopsis and tobacco), most of the KNOX genes are expressed in the SAM, but are not active in the lateral organ primordia (reviewed in Reiser et al., 2000). In vivo and in vitro studies demonstrated that the KNOX genes modulate GA biosynthesis in the SAM by inhibiting expression of the GA20ox genes (Tanaka-Ueguchi et al., 1998; Sakamoto et al., 2001; Hay et al., 2002). The tobacco KNOX protein NTH15 binds to a 5′ upstream region and the first intron of a tobacco GA20ox gene (Sakamoto et al., 2001). On the other hand, GA production at the flank of SAM may promote organized cell proliferation and consequently induces the determination of cell fate. In addition, KNOX induces expression of GA2ox genes at the base of the SAM in rice and Arabidopsis, which may prevent the influx of active GAs from leaf primordia (Figure 8D; Jasinski et al., 2005). Therefore, expression of KNOX in the SAM leads to low GA levels in order to maintain the indeterminate state of the corpus. In complex-leaved species (e.g., tomato), KNOX regulates leaf shape by inhibiting GA20ox expression during early leaf development (Bharathan et al., 2002; Hay et al., 2002).
In roots, CPS and GA3ox genes appear to be expressed in separate locations. CPS-GUS activity is detected in the root meristem and in endodermis and cortex of the elongation zone, but not in the root cap or root vascular tissue (Figures 8E and 8N; Silverstone et al., 1997b). In contrast, both GA3ox1- and GA3ox2-GUS are expressed in the vasculature of non-elongating regions of roots (Figures 8F, 8G and 8O; Mitchum et al., 2006). GA3ox2-GUS is also detected in the elongation zone, quiescent center and columella cells (Figures 8G and 8P). These observations suggest that bioactive GA is synthesized in multiple sites in Arabidopsis roots and transport of GA intermediates is required.
Reproductive Phase
GA promotes flowering in many species. In Arabidopsis, the ga1-3 mutant fails to flower under SD, indicating that GA is required for flower initiation under the non-inductive conditions (Wilson et al., 1992). Expression of the flower meristem identity gene LEAFY (LFY) and the flowering time gene SUPRESSOR OF OVEREX-PRESSION OF CO 1 (SOC1) is induced by GA (Blazquez et al., 1998; Moon et al., 2003). Single leaf treatment assays showed that GA4 is the most active GA in promoting LFY expression upon flower induction (Eriksson et al., 2006). The amount of GA4 increases sharply in the shoot apex immediately prior to flower initiation. However, qRT-PCR analysis of transcript levels of GA metabolism genes (GA20ox, GA3ox and GA2ox) in the shoot apex did not support elevated de novo synthesis of GA4, suggesting that it is transported from other tissues.
To investigate the sites of GA biosynthesis in inflorescence stems after bolting, CPS-GUS and GA3ox-GUS lines were analyzed. GA3ox1 is the only GA3ox gene that is highly expressed in the stem (Figure 8Q). GA3ox1-GUS activity is not detectable in the inflorescence meristem (IM), but is strongly expressed in the elongation region below the IM (Figure 9A; Mitchum et al., 2006). Interestingly, CPS-GUS is highly expressed in IM and early floral primordia (Figure 9A; Silverstone et al., 1997b). The expression pattern of GA3ox1 is consistent with the above-mentioned GA transport hypothesis during floral induction. However, these studies only examined GA3ox-GUS and CPS-GUS expression after bolting, but did not analyze their expression prior to or during the time of flower initiation.
Figure 9.
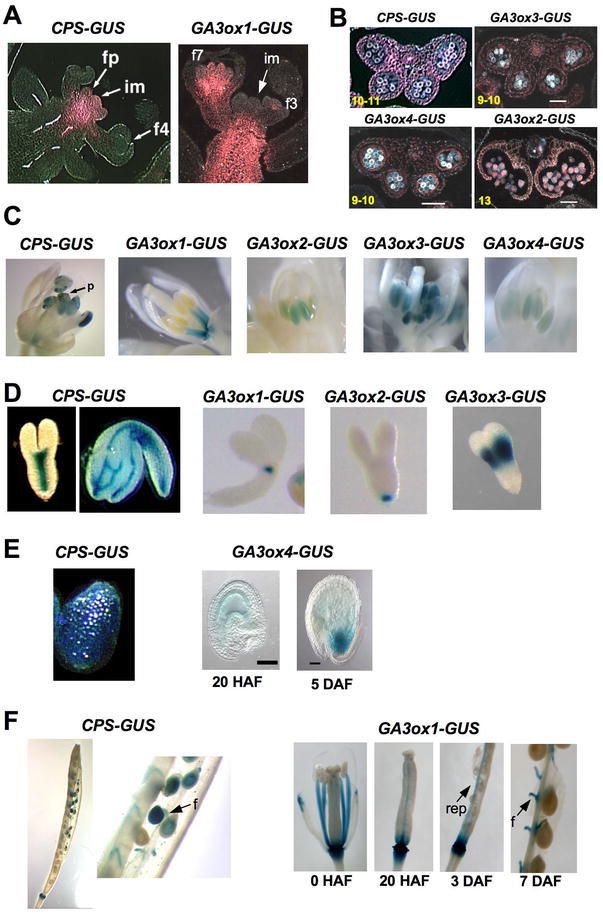
CPS-GUS and GA3ox-GUS Expression Patterns during Reproductive Growth by X-Gluc Staining.
CPS-GUS and GA3ox-GUS Expression Patterns during Reproductive Growth by X-Gluc Staining.
(A) Infloresence stems. im, inflorescence meristem; fp, floral primordia; f#, flower stage #. The X-gluc staining is pink when viewed under dark-field microscopy.
(B) Transverse sections of anthers expressing CPS-GUS and GA3ox-GUS, viewed under dark-filed microscopy. Numbers at the left corners indicate anther stages.
(C) Developing flowers. p, pollen on the stigma.
(D) Developing embryos.
(E) Endosperm staining.
(F) Siliques and developing seeds. f, funiculus; rep, replum.
(E–F) HAF, hr after flowering; DAF, day after flowering.
CPS-GUS images reprinted from Silverstone et al. (1997b) with permission from Blackwell Publishing.
GA3ox-GUS images reprinted from Mitchum et al. (2006) with permission from Blackwell Publishing and Hu et al. (2008).
During Arabidopsis flower development, GA is essential for the development of stamens and petals (Koornneef and van der Veen, 1980). Scanning electron microscopy showed that the petal and stamen growth of the ga1-3 mutant flower is arrested at floral stage 10 (Figure 10A; Cheng et al., 2004). The stamen phenotype includes reduced filament length (due to shorter epidermal cell length) and pollen defect (due to microsporogenesis arrest after meiosis at anther stage 7). Flower development in the triple mutant gid1a gid1b gid1c arrests even earlier (before flower stage 9) than ga1-3 and the pistils are very short, indicating that GA also controls pistil growth (Figure 10A; Griffiths et al., 2006).
Figure 10.
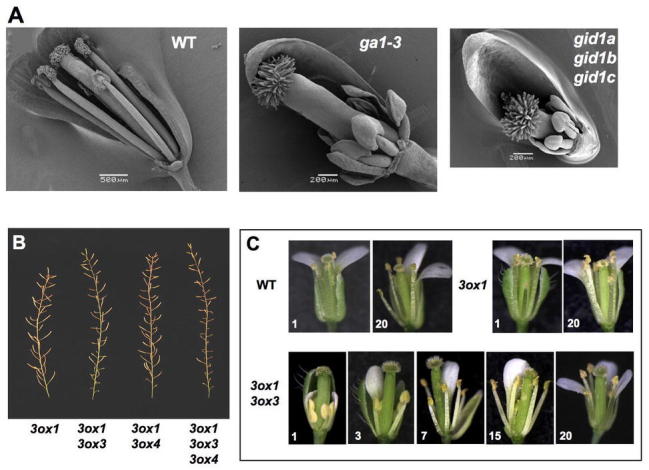
Flower and Silique Phenotypes of ga1-3, gid1a gid1b gid1b and ga3ox Mutants.
(A) Scanning electron microscopy images of WT and mutant flowers. Sepals and petals have been removed to visualize the anthers and pistil. Reproduced from Griffiths et al. (2006)
(B) Primary inflorescence stems of ga3ox mutants. Reproduced from Hu et al. (2008).
(C) Gradual improvement in the development of the ga3ox1 ga3ox3 mutant flowers on primary inflorescence. Numbers at the bottom corner of each picture indicate the positions of the flowers on the main stem (bottom to top). Reproduced from Hu et al. (2008).
To determine the potential sites of bioactive GA synthesis in developing flowers, expression patterns of CPS and all four GA3ox genes were analyzed by in situ hybridization (only for GA3ox1), qRT-PCR analysis and promoter-reporter (GUS) fusion in transgenic Arabidopsis (Silverstone et al., 1997b; Gomez-Mena et al., 2005; Mitchum et al., 2006; Hu et al., 2008). The CPS promoter is active in the floral receptacle, entire anther including pollen prior to dehiscence (Figures 9B–9C). GA3ox1 is expressed in the floral receptacle and stamen filaments once they are differentiated whereas the other GA3ox's are expressed in entire anther including pollen starting at anther stage 6 (with GA3ox3 showing the highest expression, GA3ox4 and GA3ox2 having low and extremely low expression, respectively) (Figures 9B–9C). Mutant analysis further supports that GA3ox1 and GA3ox3 are the major contributors of GA3ox function for stamen development. The mutants lacking both GA3ox1 and GA3ox3 functions (e.g., the double ga3ox1 ga3ox3 and the triple ga3ox1 ga3ox3 ga3ox4 mutants) have dramatically reduced fertility (especially in the earlier formed flowers) due to defects in stamen filament elongation and in pollen development (Figures 10B–10C; Hu et al., 2008). Therefore, de novo synthesis of bioactive GAs in stamen filaments and anthers is necessary for stamen development in Arabidopsis. GA synthesized in the stamen appears to also control petal development. None of the GA3ox genes are expressed in petals, although mutants lacking both functional GA3ox1 and GA3ox3 have severe petal defects (Hu et al., 2008). These observations suggest that active GAs (mainly GA4) synthesized in the stamens are transported to petals to promote their growth. This mechanism may be commonly present in plants since a previous study in petunia showed that emasculation of stamens caused reduced corolla growth, which could be rescued by GA application (Weiss and Halevy, 1989).
In developing carpels, CPS-GUS is expressed in the four main veins and the funiculi. After pollination, both CPS-GUS and GA3ox1-GUS are expressed in the funiculi and silique receptacles, and GA3ox1-GUS is also detected in the replums of developing siliques (Figure 9F) (Silverstone et al., 1997b; Hu et al., 2008). In some species (e.g., pea, tomato and Arabidopsis), GA is required for seed development (Swain and Singh, 2005). Ectopic expression of a pea GA2ox gene in Arabidopsis results in seed abortion and pollen tube growth defect (Singh et al., 2002), suggesting that GA is required for seed development and pollen tube growth in Arabidopsis. During seed development, CPS-GUS is expressed in developing endosperm and embryo axis (Figures 9D–9E). In the mature embryo, CPS-GUS activity is also detected in the vasculature in the cotyledons (Figure 9D). GA3ox1, GA3ox2 and GA3ox3 are expressed in distinct locations in the embryo (Figure 9D). In contrast, GA3ox4 promoter is active in the endosperm of the seed starting 20 h after flowering (but not in the embryo), and this endosperm expression becomes localized at the chalazal region in seeds at heart stage (Figure 9E). The data of qRT-PCR analysis of temporal and spatial expression of GA3ox genes in mature carpels and developing siliques are consistent with the GUS staining patterns of these genes, although the amount of GA3ox1 transcript present in the placenta sample (including replums, septums and funiculi) is much more abundant than that in the receptacle (Hu et al., 2008).
GA promotes fruit set and subsequent growth in many species (Gillaspy et al., 1993; Ozga and Reinecke, 2003). Mutants and pollination analyses demonstrate that GA3ox1 has a major role in promoting fruit (i.e., silique) elongation by acting in maternal tissues (Hu et al., 2008). In addition, endosperm-expressed GA3ox4 also promotes fruit growth, suggesting that active GAs are transported from the seed endosperm to the surrounding maternal tissues where they promote growth.
REGULATORS OF GA METABOLISM GENES
As discussed in the previous section, individual gene encoding GA metabolism enzyme displays a distinct developmental expression profile. Although the molecular mechanisms involved are largely unclear, several regulators of GA metabolism genes have been identified. Three transcription factors are found to regulate GA and/or ABA metabolism during embryo development. LEAFY COTYLEDON2 (LEC2) and FUSCA3 (FUS3) are transcription factors with a B3 DNA binding domain, and are necessary for embryogenesis. Recent reports suggest that LEC2 and FUS3 function to decrease GA levels in developing seeds (Curaba et al., 2004; Gazzarrini et al., 2004). The lec2 and fus3 mutant seeds have higher active GA levels and display ectopic expression of GA3ox2-GUS in the embryo axis. Induction of FUS3 expression during early embryogenesis leads to reduced GA3ox1 and GA20ox1 mRNA levels, and increased ABA levels. Interestingly, GA appears to inhibit FUS3 function by promoting its degradation. The MADS box transcription factor AGAMOUS-LIKE 15 (AGL15) is also expressed during embryogenesis and seed germination. Overexpression of AGL15 causes GA-deficient phenotypes (Wang et al., 2004). Chromatin-immunoprecipitation (ChIP) studies show that AGL15 binds to the promoter of GA2ox6 in vivo, suggesting that AGL15 may reduce active GA levels by inducing GA2ox6 expression. In addition, a GATA zinc finger transcription factor BME3 (for Blue Micropylar End 3) positively regulates seed germination by increasing GA biosynthesis (Liu et al., 2005). Compare to WT, seeds of the knockout bme3 mutants are less responsive to cold stratification-induced germination. GA20ox3 and GA3ox1 transcript levels are lower in the mutant seed than in WT after stratification, although GA treatment can rescue the germination defect.
For SAM maintenance, KNOX transcription factors reduce bioactive GA levels by inhibiting GA20ox1 expression in the corpus of SAM and inducing GA2ox2 and GA2ox4 expression at the base of the SAM (Hay et al., 2002; Jasinski et al., 2005). In developing flowers, the organ identity gene AGAMOUS (AG) regulates initiation of petal and stamen primordia. Microarray analysis identified GA3ox1 to be one of the AG-induced genes (Gomez-Mena et al., 2005). Gel-shift and ChIP experiments further indicate that AG directly binds to the GA3ox1 promoter. These results suggest that AG increases active GA production, which may promote differentiation of petals and stamens.
To elucidate the GA feedback mechanism, a cis-element that is required for GA3ox1 feedback response was identified by promoter deletion analysis in transgenic Arabidopsis (Matsushita et al., 2007). An AT-hook protein, AGF1 (AT-hook protein of GA feedback regulation), binds to this cis element in yeast one-hybrid and in vitro gel-shift assays. Overexpression of AGF1 results in a higher accumulation of GA3ox1 mRNA levels in the presence of a GA biosynthesis inhibitor (paclobutrazol, PAC) and a reduced downregulation by GA. These observations suggest that AGF1 promotes GA3ox1 expression in response to feedback control. However, the molecular mechanism involved is unclear, as AGF1 mRNA levels are not affected by GA, and T-DNA insertion agf1 mutant has no visible phenotype, likely due to functional redundancy of homologous genes. The nuclear proteins DELLAs, a class of GA signaling repressors (to be discussed in the next section), have also been found to induce transcription of GA3ox1 and GA20ox2 (Zentella et al., 2007).
GA RECEPTORS AND GA SIGNAL TRANSDUCTION PATHWAY
Recent genetic, molecular and biochemical approaches have identified a class of GA receptors as well as several early components of the GA signaling pathway (Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007a). Mutants those are defective in GA responses display GA-unresponsive dwarf phenotype, which is similar to GA biosynthesis-mutants, except that they cannot be rescued completely by GA treatment. Recessive dwarf mutants define positive components, whereas dominant mutations help to identify negative regulators in the GA signaling pathway. On the other hand, mutants with elevated GA responses show a slender phenotype, which resembles WT plants that have been treated with an excessive amount of GA. Elevated GA responses are due to loss-of-function (recessive) mutations in the negative regulators of GA signaling or gain-of-function (dominant) mutations in positive components of GA signaling pathway.
GA Receptors (GIBBERELLIN INSENSITIVE1, GID1)
The GA receptor GID1 was first identified in rice by the studies of a GA-insensitive dwarf mutant (GID1) (Ueguchi-Tanaka et al., 2005). Arabidopsis contains three GID1 orthologs (GID1a, GID1b and GID1c) (Table 1; Nakajima et al., 2006). In vitro binding assays showed that the GID1 proteins in rice and Arabidopsis function as soluble GA receptors (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). GID1s show sequence similarity to hormone-sensitive lipases (HSL), and contain the conserved HSL motifs (HGG and GXSXG) (Figure 11). However, a conserved residue His near the C-terminus that is essential for HSL catalytic function is absent and HSL-like enzymatic activity has not been detected. GID1-GFP is localized to both the cytoplasm and nucleus in transgenic plants (Ueguchi-Tanaka et al., 2005; Willige et al., 2007). GID1s bind to bioactive GAs with high affinity, but not to inactive GA derivatives. Loss-of-function gid1 mutations in rice result in a GA-insensitive severe dwarf phenotype, and the aleurone cells fail to respond to GA in α-amylase gene induction. These results demonstrate that GID1 is the major GA receptor in rice. Early biochemical studies in the cereal aleurone system suggest that GA signal is perceived on the external face of the plasma membrane (Hooley et al., 1991; Gilroy and Jones, 1994). It remains to be determined whether a transmembrane GA receptor is also present.
Figure 11.
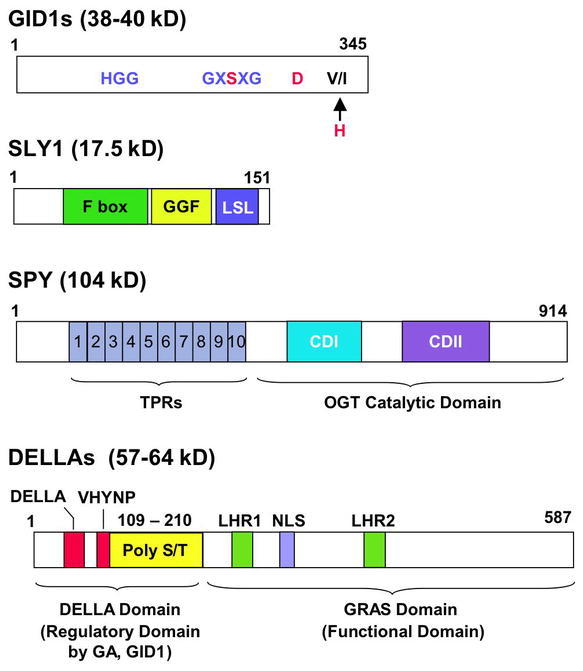
Schemes of GID1, SLY1, SPY and DELLA.
GID1s: The conserved motifs in HSL are shown in blue. GID1s also contain two of the three residues (in red) that are required for the HSL catalytic function. The “H” residue is substituted by V or I in GID1s.
SLY1: The substrate specificity is presumably provided by the C-terminal GGF and LSL motifs, which are conserved between SLY1 and GID2.
SPY: The C-terminal conserved domains (CDI and CDII) are part of a UDP-GlcNAc binding pocket and participate in the transfer of GlcNAc.
DELLAs: DELLA and VHYNP, the two highly conserved motifs among DELLAs, are required for GID1 binding and GA-induced degradation. PolyS/T, multiple Ser/Thr residues (but variable among DELLAs); LHR, Leu heptad repeat; NLS, nuclear localization signal. The C-terminal GRAS domain is the functional domain (presumably for transcriptional regulation), which also contains the SLY1 binding site. Numbers above each scheme represent the amino acid residues for each protein (GID1a and RGA are shown for GID1 and DELLA, respectively).
The triple mutant gid1a gid1b gid1c in Arabidopsis is a non-germinating extreme dwarf, whose phenotype is even more severe than the GA-deficient ga1-3 mutant (Figure 7B; Griffiths et al., 2006; Iuchi et al., 2007; Willige et al., 2007). As expected, this triple mutant is impaired in flower initiation. The gid1a-1 gid1b-1 gid1c-1 mutant flowers at 38.7 d, even later than ga1-3 (30.8 d) in LD (WT flowers at 19.9 d) (Griffiths et al., 2006). Another triple gid1 mutant with the gid1c-2 allele does not flower under similar LD conditions (Willige et al., 2007). The gid1c-1 allele contains a T-DNA insertion in the intron, whereas the T-DNA in the gid1c-2 allele is located in the second exon. The phenotypic difference could be due to differences in experimental conditions or allelic severity. As mentioned earlier, the triple gid1 mutant is also defective in flower development (including stamen, petal and pistil) (Figure 10A). The three GID1 genes are partially redundant because none of the single gid1 mutants have obvious phenotype. The double mutants gid1a gid1c and gid1a gid1b display shorter stem height and reduced fertility, respectively, although not as severe as the triple gid1 mutant (Figure 12). The mechanism by which GA activates GID1 and its signaling pathway will be discussed later.
Figure 12.

The Phenotypes of the Single, Double and Triple gid1 Mutants.
(A) Aerial portions of 37-d-old wild-type and homozygous mutant plants.
(B) and (C) Close-up views of WT Col-0 (B) and gid1a-1 gid1b-1 (C) flowers. Sepals and petals have been removed to reveal the anthers and pistil. Reproduced from Griffiths et al. (2006).
Positive Regulators of Early GA Signaling
The F-box protein SLEEPY1 (SLY1) in Arabidopsis (and its ortholog in rice GID2): The recessive sly1 and gid2 mutants are GA-unresponsive dwarfs (Steber et al., 1998; Sasaki et al., 2003). In addition, sly1 displays increased seed dormancy and reduced fertility (Steber et al., 1998; Dill et al., 2004). The gid2 mutant shows reduced GA-induced α-amylase expression and is infertile (Sasaki et al., 2003). SLY1 and GID2 encode homologous proteins containing a F-box motif in their N-termini (McGinnis et al., 2003; Sasaki et al., 2003). Many F-box proteins function as part of the SCF (SKP1, CULLIN1, F-Box)-type E3 ubiquitin ligase complexes, which recruit and target specific proteins for ubiquitination and degradation through the 26S proteasome (Gagne et al., 2002; Smalle and Vierstra, 2004). The F-box motif (~44 amino acids) contains the SKP1 binding site, whereas the C-terminal domain of the F-box protein often interacts directly with its target protein (Figure 11). SLY1 and GID2 appear to activate GA signaling by targeting GA signaling repressors, DELLA proteins (described further below), for ubiquitination and degradation (Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). SLY1-yellow fluoresence protein, when transiently expressed in onion epidermal cells, is nuclear localized, suggesting that SLY1 functions in the nucleus. The loss-of-function sly1 mutant is a semi-dwarf (Figure 13), suggesting that functional homolog(s) of SLY1 may be present in the Arabidopsis genome. An activation-tagging mutant screen showed that overexpression of SNEEZY (SNE), another F-box protein in Arabidopsis, can rescue the sly1 semi-dwarf phenotype (Strader et al., 2004). SNE transcript is most abundant in flowers, whereas SLY1 is expressed in all tissues examined. Future characterization of the loss-of-function sne mutant will verify its role in GA signaling.
Figure 13.

The Semi-Dwarf Phenotype of sly1 Is Rescued by rga and gai Null Mutations. Reproduced with modifications from Dill et al. (2004).
Negative Regulators of Early GA Signaling
SPINDLY (SPY): The loss-of-function spy mutations in Arabidopsis cause elevated GA signaling, which partially rescues all defects resulting from GA deficiency (Figure 14A; Jacobsen and Olszewski, 1993; Silverstone et al., 1997a). SPY and its orthologs in several crops share high sequence similarity to mammalian O-linked N-acetylglucosamine (O-GlcNAc) transferases (OGT) (Jacobsen et al., 1996; Filardo and Swain, 2003). The animal OGTs modify target proteins by adding a GlcNAc monosaccharide to Ser/Thr residues, which either interfere or compete with kinases for phosphorylation sites (Slawson and Hart, 2003). The N-terminal region of each OGT contains multiple copies of tetratricopeptide (34-amino acid) repeats (TPRs) that are protein-protein interaction domains, and serve as scaffolds for assembly of protein complexes. Specific blocks of TPR motifs provide specificity for interaction with different proteins (Young et al., 1998).
Figure 14.

The spy Mutation Partially Suppresses ga1 and rga-Δ17 phenotypes, but does not affect GFP-RGA nuclear localization.
(A) 35 day-old plants as labeled. spy and rga synergistically suppress the ga1-3 phenotype. Reprinted from Silverstone et al. (1997a) with permission from the Genetics Society of America.
(B) spy does not alter GFP-RGA nuclear localization. GFP fluorescence in root tips was visualized by confocal loser microscopy under an identical setting for the images. Reprinted from Silverstone et al. (2007).
(C) spy partially rescues the dwarf phenotypes of rga-Δ17. All plants in (C) are 36 days old and have been treated (+) or not treated (−) with GA3 as indicated. Reprinted from Silverstone (2007).
SPY contains 10 N-terminal TPRs as well as the C-terminal OGT catalytic domain (Figure 11; Thornton et al., 1999). The recombinant SPY protein exhibited OGT activity in vitro (Thornton et al., 1999), and like animal OGTs, SPY is localized both to the cytoplasm and nucleus (Swain et al., 2002). Target proteins of SPY have not been identified, but potential candidates include the DELLA proteins (Shimada et al., 2006; Silverstone et al., 2007).
All of the available spy alleles display elevated GA respones (Filardo and Swain, 2003). However, these mutants show additional phenotypes (such as altered phyllotaxy), suggesting that SPY also regulates other cellular pathways (Swain et al., 2001; Tseng et al., 2004; Greenboim-Wainberg et al., 2005). Characterization of the phenotypes and molecular lesions of 14 new spy alleles indicates that TPRs 6, 8 and 9, and the OGT catalytic domain are crucial for GA-regulated stem elongation, floral induction and fertility (Silverstone et al., 2007). Thus, SPY may inhibit GA signaling through specific protein-protein interactions via these 3 TPRs and its OGT catalytic activity. The other TPRs may be important for SPY to function in other pathways.
DELLA proteins: The DELLA proteins are a subfamily of the plant-specific GRAS (for GAI, RGA and SCARECROW) family of regulatory proteins (Pysh et al., 1999). DELLAs were initially identified by studies of the Arabidopsis GA-response mutants rga (for repressor of ga1-3) and gai (for GA-insensitive) (Peng et al., 1997; Silverstone et al., 1998). Functional DELLA orthologs have been found in many crops, e.g., wheat (Rht), maize (D8), rice (SLR1) and barley (SLN1) (reviewed in Thomas and Sun, 2004). Arabidopsis contains 32 GRAS family members (Bolle, 2004; Tian et al., 2004), which have a conserved C-terminal GRAS domain. Their N-termini are more divergent, and probably specify their roles in different cellular pathways. DELLA proteins were named after a conserved DELLA motif near their N-termini (Peng et al., 1997; Silverstone et al., 1998; Pysh et al., 1999), which is absent in other GRAS members. DELLA proteins appear to repress all aspects of GA responses, although their effects on GA-promoted vegetative growth are the most extensively documented in different species (Thomas and Sun, 2004). Arabidopsis contains 5 DELLA protein genes [RGA, GAI, RGA-Like1 (RGL1), RGL2 and RGL3]; each displays overlapping, but also some distinct functions in repressing GA responses. RGA and, to a lesser extent, GAI mRNAs are expressed ubiquitously in all tissues, whereas RGL1, 2 and 3 transcripts are present at high levels only in germinating seeds and/or flowers and siliques (Tyler et al., 2004).
RGA was identified by the ability of loss-of-function rga alleles to partially suppress the dwarf phenotype of the GA biosynthetic mutant ga1-3 (Figure 14A; Silverstone et al., 1997a). The gain-of-function gai-1 mutant has a GA-unresponsive dwarf phenotype (Koornneef et al., 1985), whereas the loss-of-function gai-t6 allele confers resistance to the GA biosynthesis inhibitor PAC in vegetative growth (Peng et al., 1997). Removing both RGA and GAI functions in the ga1-3 background completely rescues vegetative growth and floral initiation, indicating that these two DELLAs are the major repressors that modulate these processes (Figure 15A; Dill and Sun, 2001; King et al., 2001). However, seed germination or flower development of ga1-3 was not improved by the rga gai null mutations. In contrast, rgl2 partially rescues seed germination of ga1-3, indicating that RGL2 plays a major role in seed germination in the light (Lee et al., 2002). RGA, GAI, RGL1 and RGL3 also regulate seed germination because mutations in these genes increase the germination percentage in the rgl2 ga1-3 mutant background (Cao et al., 2005; Tyler, 2006). RGA, RGL1 and RGL2 are all involved in modulating floral development (Cheng et al., 2004; Tyler et al., 2004; Yu et al., 2004). Approximately 60% of siliques produced by the quadruple mutant ga1-3 rga rgl1 rgl2 are fertile, in contrast to complete sterile phenotype of ga1-3 (Figures 15B–15C).
Figure 15.

Phenotypes of Wild-Type and Homozygous ga1-3 and della Mutant Plants.
Each mutant line is homozygous for the allele(s) listed.
(A) All plants are 52 days old except for rga-24/gai-t6, which is 37 days old. Reprinted from Dill and Sun (2001) with permission from the Genetics Society of America.
(B) Primary inflorescences. Reproduced from Tyler et al. (2004). (C) Flowers. Reproduced from Tyler et al. (2004).
Recent studies suggest that DELLA proteins are transcriptional regulators (Zentella et al., 2007). The N-terminal DELLA domain is a regulatory domain in response to GA, whereas the C-terminal GRAS domain functions to regulate transcription [Figure 11; Sun and Gubler, 2004)]. The N-terminal DELLA domain contains two highly conserved motifs (named DELLA and VHYNP) and a variable region that is rich in Ser and Thr (Poly S/T). DELLA and VHYNP motifs are essential for regulating DELLA stability in response to the GA signal (Figure 16A; Gomi and Matsuoka, 2003; Sun and Gubler, 2004). Deletion mutations in these sequences (e.g., gai-1, rga-Δ17 and rgl1-Δ17) result in a GA-unresponsive dwarf phenotype (Figure 17; Peng et al., 1997; Dill et al., 2001; Wen and Chang, 2002; Feng et al., 2008). Similar gain-of-function della mutations in crops also resulted in reduced stature. The most notable example is the semi-dwarf wheat cultivars, crucial components of the “Green Revolution”, which all contain deletions in the DELLA domain of an Rht gene (Peng et al., 1999). Transgenic Arabidopsis plants expressing mutated gai genes carrying similar mutations as in the crops display GA-insensitive dwarf phenotypes (Willige et al. 2007). The Poly S/T region contains putative target sites of phosphorylation and/or glycosylation. The GRAS domain contains Leu heptad repeats (LHR) that may mediate protein-protein interactions, and a putative nuclear localization signal (NLS) (Figure 11). All DELLA-GFP fusion proteins examined are nuclear localized when expressed in transgenic plants (Figure 16A; Silverstone et al., 2001; Fleck and Harberd, 2002; Gubler et al., 2002; Itoh et al., 2002; Wen and Chang, 2002). DELLA proteins do not have a clearly identified DNA-binding domain, and they may associate with their target promoters by interacting with other transcription factors (Zentella et al., 2007).
Figure 16.
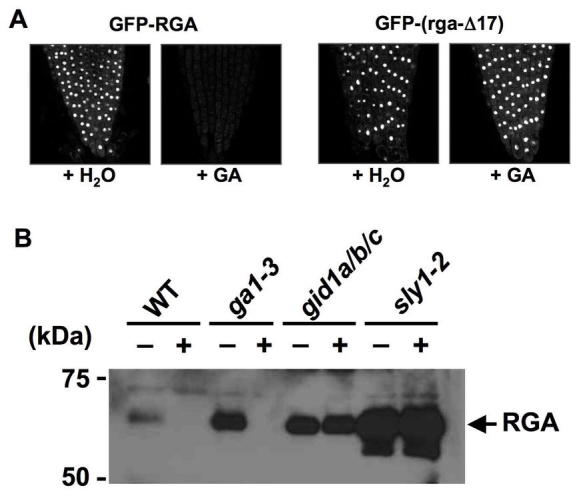
The DELLA Motif Is Essential GA-Induced Proteolysis of DELLA Proteins.
(A) GA induces proteolysis of GFP-RGA, but not GFP-(rga-Δ17) that lacks the DELLA motif. Nuclear fluorescence in root tips was visualized by confocal loser microscopy under an identical setting for all images. Reproduced with modifications from Dill et al. (2001).
(B) SLY1 and GID1 Are Required for GA-Induced Degradation of RGA. The protein blot contains proteins extracted from WT and mutant seedlings that were treated with water (−) or GA (+). The blot was probed with anti-RGA antibodies. Reproduced from Griffiths et al. (2006).
Figure 17.

Deletions of the DELLA Motif Cause GA-Insensitive Dwarf Phenotype.
(A) All plants are homozygous for the genotypes indicated, and are 36 days old. They have been treated (+) or not treated (−) with GA3 as indicated. Reproduced with modifications from Dill et al. (2001).
(B) WT and a 35S:rgl1-Δ17 plant (hemizygous T1). Reproduced from Wen and Chang (2002).
GA-Induced Proteolysis of DELLA Proteins via Ubiquitin-Proteasome Pathway
Examination of both endogenous RGA and GFP-RGA fusion protein revealed that GA induces rapid degradation of this GA signaling repressor (Figure 16; Silverstone et al., 2001). Subsequent studies in Arabidopsis and other plants indicate that degradation of DELLA proteins is a common mechanism for GA to de-repress its signaling pathway (reviewed in Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2007a). DELLA proteins accumulate to high levels in the triple gid1 mutant and the sly1 mutant (Figure 16B), supporting that both GID1 and SLY1 are required for GA-induced DELLA degradation (McGinnis et al., 2003; Griffiths et al., 2006). Consistent with this idea, rga and gai null mutations rescue the dwarf phenotypes of sly1 (Figure 13) and the triple gid1 mutant (Figure 18). In vitro pull-down, yeast two-hybrid and biomolecular fluorescence complementation (BiFC) assays demonstrate that GID1 directly binds to DELLAs in a GA dose-dependent manner (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007b; Feng et al., 2008). SLY1 also interact weakly with all five Arabidopsis DELLAs, as observed using yeast two-hybrid and in vitro pull-down assays (Dill et al., 2004; Tyler et al., 2004). In addition, a novel gain-of-function sly1-d mutation [formerly named gar2 (for gai revertant); (Wilson and Somerville, 1995)], increases GA signaling by enhancing the interaction between sly1-d and DELLAs and, therefore, reducing DELLA protein levels (Dill et al., 2004; Fu et al., 2004). Although the three dimensional structure of the GA-GID1-DELLA complex has not been solved by x-ray chrystallography, Alanine scanning mutagenesis on the rice GID1 protein suggests that GA and DELLA seem to interact with regions of GID1, which are predicted to correspond to the lid and substrate-binding pocket of HSL (Ueguchi-Tanaka et al., 2007b). In vitro binding assays further showed that the presence of DELLA stabilizes GA-GID1 interaction. Based on these observations, a model for the GA-GID1-DELLA complex formation was proposed (Figure 19). By yeast three-hybrid assays, the formation of the GA-GID1-DELLA complex was shown to enhance the interaction of DELLA with SLY1, and presumably promotes polyubiquitination by SCFSLY1. The highly conserved N-terminal DELLA and VHYNP motifs in DELLAs are necessary for the interaction with GID1 (Griffiths et al., 2006; Willige et al., 2007), whereas the GRAS domain is required for SLY1 binding (Dill et al., 2004). Thus, both the DELLA/VHYNP motifs and the GRAS domain play roles in GA-induced proteolysis. GA-GID1 binding to DELLA presumably alters the conformation of DELLA, which allows SLY1 interaction with the GRAS domain of DELLA (Figure 20). Interestingly, recent study by Ariizumi and Steber (2007) showed that the sly1 mutant seed can germinate even when RGL2 protein levels remain high, suggesting that an alternative DELLA inactivation mechanism other than protein destruction may also exist. It is possible that GA-induced GID1 binding to DELLA may be able to directly inhibit DELLA function. GA and GID1-dependent post-translational modifications (such as phosphorylation) may also inactivate DELLA (see the next section).
Figure 18.

The Severe Dwarf Phenotype of gid1a gid1b gid1c Is Rescued by rga and gai Null Alleles.
All plants are 6-week-old. Reproduced from Willige et al. (2007).
Figure 19.
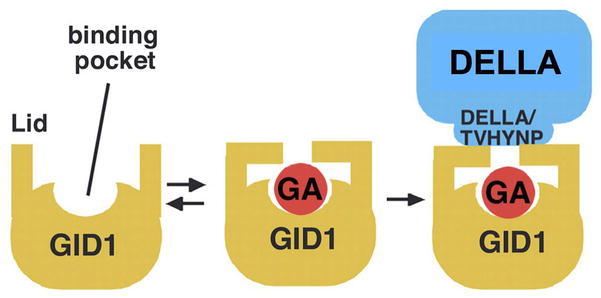
A Model for the GA-GID1-DELLA Complex.
GID1 appears to contain the substrate binding pocket and lid, similar to those present in the bacterial HSLs. Active GAs bind to the substrate binding pocket with the aid of the lid. Binding of the DELLA protein to GA-GID1 via its N-terminal DELLA and VHYNP motifs stabilizes the complex. Reproduced with modifications from Ueguchi-Tanaka et al. (2007).
Figure 20.
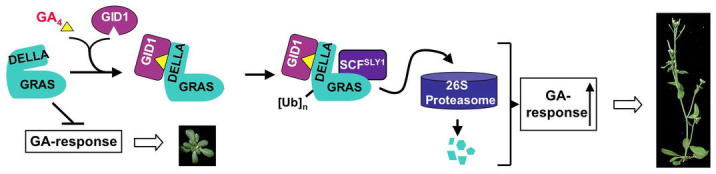
GA-GID1-DELLA Binding Promotes SLY1-DELLA Interaction and DELLA Degradation.
GA-GID1 binding to the DELLA domain may trigger conformational changes that enhance SLY1 (a subunit of the SCFSLY1 complex) binding to the GRAS domain (in the DELLA protein). Once recruited by SLY1, DELLA will be polyubiquitinated and then degraded via the ubiquitin-proteasome pathway.
Potential Roles of Phosphorylation and/or O-GlcNAcylation of DELLA
DELLA proteins are present as both phosphorylated and unphosphorylated forms in planta, although the role of this modification is less clear (Fu et al., 2004; Ueguchi-Tanaka et al., 2007a). By in vivo labeling of rice seedlings with 32PO4−, phosphorylated Ser was detected in the DELLA protein (SLR1) (Itoh et al., 2005). Deletion of DELLA, VHYNP and PolyS/T abolished the phosphorylation signal, suggesting that these regions are required for phosphorylation to occur. However, some of the phosphorylation sites may be in other regions. Phosphorylated DELLA proteins were initially suggested to promote the F-box protein recognition (Sasaki et al., 2003). However, Itoh et al. (2005) showed that phosphorylation of SLR1 in rice is independent of GA-induced degradation of this protein. In contrast, another study by Hussain et al. (2005) suggested that dephosphorylation of RGL2 (a DELLA in Arabidopsis) is required for its degradation. This was based on their observations that in tobacco BY2 cells, phosphatase inhibitor treatment blocks GA-induced RGL2 degradation, and that Ser-to-Asp or Thr-to-Glu substitutions in RGL2 stabilizes the mutant proteins. Therefore, the role of phosphorylation on DELLA needs further investigation.
As described earlier, O-GlcNAc-modification (termed O-Glc-NAcylation) of Ser and Thr residues by the enzyme OGT often interferes with phosphorylation of nearby protein kinase sites (Slawson and Hart, 2003). In animal systems, GlcNAcylation of target proteins can promote nuclear localization, protein stability or directly affect protein activity. SPY, a plant OGT, functions as a repressor of GA signaling (Filardo and Swain, 2003). The loss-of-function rga and spy-8 alleles have a synergistic effect in rescuing the dwarf phenotype of ga1-3 (Figure 14A; Silverstone et al., 1997a). Because spy-8 is not a null allele, RGA and SPY may act in the same pathway or in two parallel pathways. The following observations support that SPY may increase DELLA activity by Glc-NAcylation: (i) DELLAs contain several putative GlcNAc sites, mainly in the Poly S/T region; (ii) The paralog of SPY, SEC (SECRET AGENT), GlcNAc-modifies the Poly S/T region of RGA in a heterologous system (L. Hartweck and N.E. Olszewski, personal communication); (iii) Loss-of-function spy mutations do not alter nuclear localization of GFP-RGA fusion protein or destabilize the RGA protein, eliminating the possibility that the nuclear localization and stability of RGA depend on SPY function (Figure 14B; Silverstone et al., 2007). In addition, 2D-SDS PAGE followed by immunoblot analysis showed that reduced SPY expression by RNA interference (RNAi) in the rice gid2 mutant leads to elevated phosphorylation of DELLA without changing protein levels (Shimada et al., 2006). SPY-RNAi also partially rescues the dwarf phenotype of gid2. Therefore, phosphorylated DELLA may be the inactive (or less active) form, and SPY may activate DELLA by GlcNAcylation, which then inhibits phosphorylation of DELLA. Consistent with this model, loss-of-function spy mutations partially rescue the dwarf phenotype of the gain-of-function gai-1 and rga-Δ17 mutants (Figure 14C; Wilson and Somerville, 1995; Jacobsen et al., 1996; Silverstone et al., 2007). Future biochemical studies will be needed to verify this model.
GA-Responsive Genes and DELLA Target Genes
Although GA regulates numerous developmental processes, only a few GA-responsive genes were identified using conventional approaches (reviewed in Huttly and Phillips, 1995; Sun and Gubler, 2004). As described earlier, some of the GA biosynthetic genes GA3ox and GA20ox are down-regulated by GA, whereas GA2ox genes for deactivation of GA are up-regulated by GA. In addition, GA induces stem elongation by increasing transcription of several genes encoding proteins that are involved in cell elongation and division in rice and Arabidopsis, such as expansins (Cho and Kende, 1997), xyloglucan endotransglycosylases (Xu et al., 1996; Uozu et al., 2000) and cyclin-dependent protein kinases (Fabian et al., 2000).
GA-promoted flower initiation is mediated by the flowering time gene SOC1 and the meristem identify gene LFY. Both SOC1 and LFY are GA-induced genes, and overexpression of SOC or LFY rescues the delayed flowering defect of ga1-3 (Blazquez et al., 1998; Moon et al., 2003). GA is essential for normal petal and stamen development in Arabidopsis. Recent studies demonstrate that GA induces expression of the B and C function genes APETALLA3, PISTILLATA and AG (Yu et al., 2004). GAMYB, a R2R3 domain transcription factor, was initially identified in barley aleurone to mediate GA-induced transcription of α-amylase genes (Gubler et al., 1995; Gubler et al., 1999). Mutant studies in barley, rice and Arabidopsis indicate that GAMYB also modulates GA-regulated floral development (Murray et al., 2003; Kaneko et al., 2004; Millar and Gubler, 2005). GAMYB expression is induced by GA, although this gene is unlikely to be a direct target of DELLA because there is 1-hr lag time between DELLA protein degradation and GAMYB mRNA induction in aleurone cells. For trichome initiation, GA activates expression of GLABROUS1, another MYB gene (Perazza et al., 1998)
To elucidate the gene regulatory network in modulating GA responses, several recent microarray experiments have examined the effects of GA and/or DELLA on global transcription profiles in germinating seeds, seedlings and flowers in Arabidopsis (Ogawa et al., 2003; Cao et al., 2006; Nemhauser et al., 2006). These experiments have taken advantage of mutants with defects in the GA biosynthetic pathway and/or in DELLA genes. From these studies, a large number of GA-regulated genes were uncovered (Ogawa et al., 2003; Cao et al., 2006; Nemhauser et al., 2006), and about half are also DELLA-dependent (Cao et al., 2006). GA treatment also affects genes that are involved in synthesis, transport and signaling of other plant hormones, revealing potential crosstalk among different hormone pathways. However, this type of analysis could not distinguish early DELLA targets from those that are located further downstream in the GA response pathway. To identify early DELLA-responsive genes, another microarray study monitored global transcript profiles before and after elevated expression of a dominant DELLA mutant protein (rga-Δ17) using a dexamethasone-inducible system. This approach uncovered 14 putative DELLA direct targets in Arabidopsis seedlings (Zentella et al., 2007). ChIP-qPCR studies provided evidence for in vivo association of DELLA with promoter sequences of eight target genes, supporting the role of DELLA in transcriptional regulation (Zentella et al., 2007). Interestingly, expression of all 14 putative DELLA target genes are DELLA-induced and GA-repressed. Four DELLA targets are GA biosynthesis genes (GA3ox1 and GA20ox2) and GA receptor genes (GID1a and GID1b), suggesting that DELLA directly impacts GA homeostasis by feedback regulation (Figure 21). Other DELLA targets encode previously uncharacterized transcription factors/regulators and ubiquitin E2/E3 enzymes that are likely negative regulators of the GA response pathway. In addition, one putative DELLA target, XERICO (encoding a putative RING E3 ligase), promotes accumulation of ABA that antagonizes GA effects (Ko et al., 2006; Zentella et al., 2007). Therefore, DELLA may restrict GA-promoted processes by modulating both GA and ABA pathways. Future characterization of the putative DELLA targets will reveal their molecular functions in GA signaling.
Figure 21.
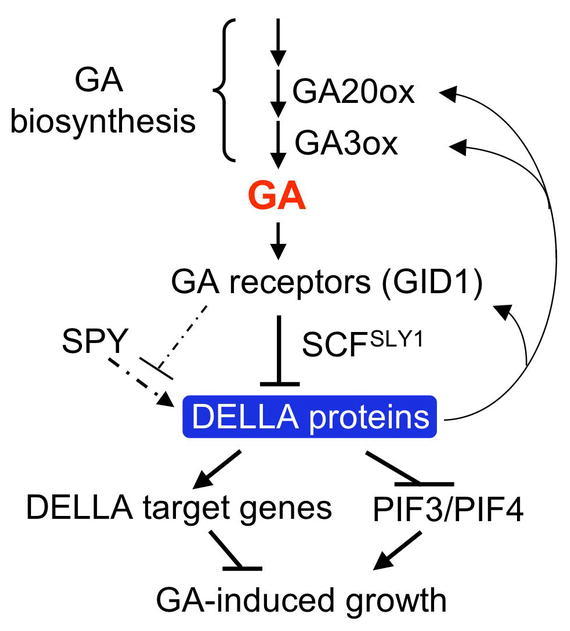
Model of GA Signaling in Arabidopsis.
The GA-GID1 complex targets DELLA proteins for ubiquitination and degradation by the 26S proteasome via the SCFSLY1 E3 ligase. SPY activates DELLA proteins by GlcNAcylation, which is inhibited by GA-induced phosphorylation. DELLA inhibits seedling growth by two mechanisms: activation of its downstream target genes (presumably encoding GA signaling repressors) and inhibition of PIF3 and PIF4 functions by direct binding to these proteins. DELLA also directly feedback induces transcript levels of GA20ox2, GA3ox1 and GID1.
ENVIRONMENTAL REGULATION OF GA METABOLISM & SIGNALING
Light and Temperature Regulation during Seed Germination
Light (mediated by phytochromes) and cold stratification are known to induce seed germination in many species. In Arabidopsis, light treatments that activate PhyA and/or PhyB lead to increased expression of GA biosynthesis genes GA3ox1 and GA3ox2 and decreased expression of GA2ox2, and elevated GA4 levels in the seeds (Yamaguchi et al., 1998b; Oh et al., 2006; Seo et al., 2006; Oh et al., 2007). In a similar way, cold-treatment of dark-imbibed after ripened Arabidopsis seeds also affect expression of GA metabolism genes (Yamauchi et al., 2004).
PIL5, a basic helix-loop-helix (bHLH) transcription factor, inhibits dark germination by affecting both GA metabolism and GA responses (Oh et al., 2004; Oh et al., 2006; Oh et al., 2007). PIL5 appears to reduce GA4 levels by inhibiting expression of GA biosynthesis genes (GA3ox1 and GA3ox2), and activating expression of GA-deactivation gene GA2ox2 in the dark (Penfield et al., 2005; Oh et al., 2006). At the same time, PIL5 promotes ABA accumulation by regulating ABA metabolism genes. However, the effect of PIL5 on GA and ABA metabolism is indirect because the results of ChIP experiments do not support direct association of PIL5 with promoters of GA or ABA metabolism genes (Oh et al., 2007). Conversely, PIL5 induces expression of two DELLA genes (RGA and GAI) by directly binding to their promoters, suggesting that PIL5 inhibition of dark germination is partly mediated by elevated RGA and GAI levels. This is consistent with the observations showing rga gai null alleles rescue germination of rgl2 ga1-3 seeds in the dark (Cao et al., 2005). Thus, light induces seed germination by promoting both GA4 accumulation and GA signaling pathways via multiple mechanisms (Yamaguchi et al., 1998b; Oh et al., 2006; Seo et al., 2006; Oh et al., 2007). Phytochrome mediates light-dependent PIL5 protein degradation, which in turn reduces DELLA (RGA and GAI) transcript levels, and increases the amount of GA4. Elevated GA4 levels then trigger degradation of DELLA proteins.
Another bHLH transcription factor SPATULA (SPT) is also a repressor of seed germination (Penfield et al., 2005). Although it is a close homolog of PIL5, SPT is photostable and mainly controls germination in response to cold stratification. Freshly harvested loss-of-function spt mutant seeds do not require a cold treatment to germinate. qRT-PCR analysis shows that SPT inhibits GA3ox1 and GA3ox2 expression in dormant seeds.
Light Regulation during Photomorphogenesis
In contrast to seed gemination, light inhibits GA4 accumulation and GA signaling to prevent hypocotyl elongation during photomorphogensis of Arabidopsis seedlings. Photomorphogensis of de-etiolated seedlings is mediated by phytochromes and cryptochromes (blue light receptors). Accumulating evidence supports that light-dependent inhibition of hypocotyl elongation is through interaction between light and GA pathways. Under GA-deficient conditions, photomorphogenesis is partially de-repressed in the dark (Alabadi et al., 2004). In addition, blue light inhibits GA20ox1 and GA3ox1 expression whereas it increases GA2ox1 expression in de-etiolated seedling (Zhao et al., 2007). These changes in GA metabolism genes correlate with transient reduction in GA4 levels. It appears that DELLA is required for light inhibition of hypocotyl growth (Achard et al., 2007a), and light promotes DELLA accumulation by decreasing the amount of GA4 in the hypocotyl. Two bHLH transcription factors PIF3 and PIF4, close homologs of PIL5 and SPT, promote hypocotyl cell elongation in dark-grown seedlings by inducing expression of their target genes (de Lucas et al., 2008; Feng et al., 2008). Like PIL5, PIF3 and PIF4 are degraded rapidly in the light. Loss-of-function of PIF3 or PIF4 confers a shorter hypocotyl phenotype, and an increased sensitivity to PAC treatment (Figure 22A). PIF3 or PIF4 overexpression lines have longer hypocotyls and are more resistant to PAC. In addition, overexpression of PIF4 partially rescues the dwarf phenotypes of the ga20ox1 and gai-1 mutants (Figure 22C). Yeast two-hybrid assays and BiFC analysis indicate that PIF3/4 interact directly with DELLAs (Figure 22B). In the dark, elevated levels of active GAs will lead to DELLA degradation via GID1, and PIF3 and PIF4 will activate their target genes to promote hypocotyl growth (Figure 22D). In the light, PIF3 and PIF4 are likely inactivated by direct interaction with DELLA proteins, which accumulate in response to light. This finding is consistent with the hypothesis that DELLAs function as transcriptional regulators by interacting with other DNA binding proteins (Zentella et al., 2007). This newly discovered mechanism together with phytochrome-mediated degradation of PIF3 and PIF4 contributes to reduce hypocotyl growth in the light. Therefore, negative interaction between PIF3/PIF4 and DELLA integrates the effects of light and GA on hypocotyl elongation.
Figure 22.
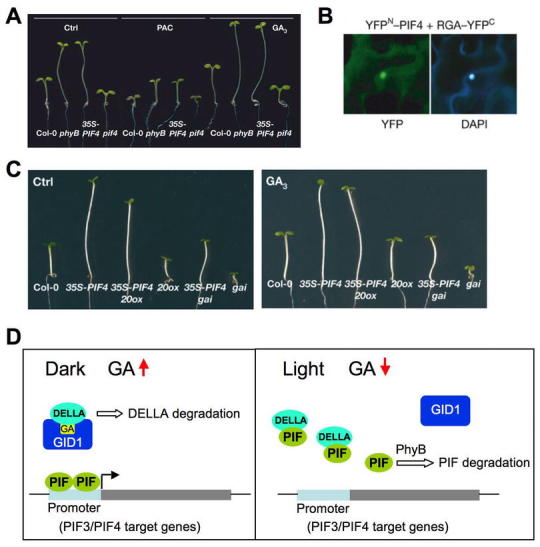
PIF3/PIF4 and DELLA Integrate Light and GA Signals in Modulating Hypocotyl Elongation.
(A) 35S:PIF4 plants are more resistant to PAC and more responsive to GA treatment, whereas pif4 is more sensitive to PAC and less responsive to GA.
(B) Direct PIF4-DELLA interaction by BiFC analysis.YFP, yellow fluorescence protein signal; DAPI, 4,6-diamidino-2-phenylindole nuclei staining.
(C) Overexpression of PIF4 partially rescues dwarf phenotypes of the ga20ox1 and gai-1 mutants. (A-C) Reprinted with permission from Macmillan Publishers Ltd: Nature, de Lucas et al (2008).
(D) A model for gene regulation through DELLA and PIF3/4 interaction in the hypocotyls of seedlings under dark vs. deetiolated (light) conditions.
Salt Stress
DWARF AND DELAYED FLOWERING1 (DDF1), an AP2 transcription factor, is related to the DREBs (dehydration responsive element-binding proteins). DDF1 overexpression lines are GA-deficient, likely by activating GA2ox7 expression (Magome et al., 2004; Yamaguchi, 2008). Interestingly, DDF1 overexpression lines as well as other dwarf mutants with reduced GA content or GA response confer salt tolerance (Magome et al., 2004; Achard et al., 2006). In addition, salt-treated WT plants have reduced amounts of bioactive GAs. DDF1 expression is induced by salt stress, further supporting its role in this stress response. The two stress hormones ethylene and ABA seem to promote salt tolerance, in part, by reducing bioactive GA levels thereby resulting in increased DELLA accumulation (Achard et al., 2006).
INTERACTIONS BETWEEN GA AND OTHER HORMONE PATHWAYS
It has been known for a long time, from physiological studies, plant hormones often have overlapping (additive or synergistic) or antagonistic roles in regulating a specific developmental process. Some of the molecular mechanisms involved in interactions between GA and other hormones are beginning to be elucidated with the available components that function in individual hormone's metabolism and response pathways (reviewed in Alvey and Harberd, 2005; Weiss and Ori, 2007).
Microarray studies showed that within a 3-hr treatment period, each of the seven plant hormones (auxin, ABA, brassinosteroid, cytokinin, ethylene, GA, jasmonic acid) appears to alter expression of other hormone metabolism genes in Arabidopsis seedlings (Nemhauser et al., 2006). However, early signal transduction genes for each hormone are largely specific. Therefore, early responses to individual hormone seem to include altered metabolism of other hormones, instead of directly affecting their signaling pathways. It is possible that additional crosstalk among different hormone signaling pathways may occur at a later stage in their signaling pathways to modulate similar developmental processes.
During seed germination, ABA is known to antagonize the promoting effect of GA. Analysis of ABA and GA contents and gene expression demonstrates that ABA and GA mutually affect each other's metabolism. The GA-deficient mutant ga1-3 seed accumulates higher amounts of ABA (Oh et al., 2007), whereas the ABA-deficient mutant seed aba2 exhibits increased GA biosynthesis by activating GA3ox1 and GA3ox2 expression (Seo et al., 2006). In seedlings, ABA may also reduce bioactive GA levels, as suggested by decreased GA20ox1 and increased GA2ox6 transcript levels after ABA treatment (Zentella et al., 2007).
Both auxin and GA promotes cell elongation in stems and roots. Recent studies suggest that shoot-apex derived auxin promotes stem and root growth, at least in part, through its effect on GA metabolism and/or signaling. In decapitated pea plants, GA3ox1 and GA2ox1 transcript levels are reduced and elevated, respectively, in elongating internodes. These changes in gene expression correlate with reduced amounts of bioactive GA1 (O'Neill and Ross, 2002). In Arabidopsis seedlings, auxin treatment leads to increased expression of GA20ox1 and GA20ox2, as well as GA2ox genes (Frigerio et al., 2006). Mutant studies further suggest that this effect of auxin depends on Aux/IAA-ARF signaling, but does not require functional DELLAs (RGA and GAI). In contrast, auxin appears to promote Arabidopsis root growth by regulating GA-induced degradation of RGA and GAI (Fu and Harberd, 2003). In decapitated seedlings, depriving shoot-derived auxin reduces the ability of GA to promote GFP-RGA disappearance in the roots.
In seedlings, interaction between ethylene and GA could be antagonistic or additive, depending on the tissue type. Ethylene inhibits root elongation, whereas GA promotes root elongation. Ethylene appears to block GA-induced root elongation by inhibiting DELLA degradation because the ctr1 mutant (with constitutive ethylene response) shows delayed GFP-RGA disappearance in GA-treated roots (Achard et al., 2003). Conversely, ethylene, auxin and GA work together to promote apical hook formation in the hypocotyl. Phenotypic analysis of GA and ethylene-related mutants (in the presence or absence of GA, PAC, ethylene precursor ACC or auxin polar transport inhibitor NPA) suggests that DELLA inhibits apical hook formation, and GA-induced deactivation of DELLA is essential for the promoting effects of ethylene and auxin on hook maintenance (Achard et al., 2003; Vriezen et al., 2004).
For SAM maintenance, low GA and high cytokinin signals are essential (Hay et al., 2002; Jasinski et al., 2005; Yanai et al., 2005). KNOX transcription factors promote SAM function by activating cytokinin biosynthesis (via upregulation of Isopentenyl transferase 7 gene expression) and inhibits GA biosynthesis (via downregulation of GA20ox gene expression) in the corpus of the SAM. Elevated cytokinin levels in the SAM then lead to increased expression of GA2ox genes at the base of the SAM, which may deactivate GAs that are transported from the leaf primordia.
For floral induction, GA promotes whereas ethylene and ABA inhibit this process. Elevated ethylene response, in ctr1, appears to delay flowering by reducing bioactive GA levels, which then cause increased DELLA accumulation (Achard et al., 2007b). ABA is likely to delay flowering via a similar mechanism. As described above, ABA treatment can lead to lower bioactive GA levels. This reduction in GA content by ABA may indirectly stabilize GFP-RGA in the wild-type GA1 (CPS) background (Achard et al., 2006; Penfield et al., 2006). This conclusion is supported by the observation that the endogenous RGA protein in the GA-deficient ga1-3 mutant is not protected by ABA treatment (Zentella et al., 2007). These observations are also consistent with the studies in barley aleurone showing that ABA interferes with the GA signaling pathway by acting downstream (instead of upstream) of DELLA (Gómez-Cadenas et al., 2001; Gubler et al., 2002). As mentioned earlier, DELLA may also induce ABA accumulation by activating XERICO expression (Zentella et al., 2007). Therefore, interactions between GA and ABA pathways occur in both their metabolic and signaling pathways to balance the antagonistic activities of these two hormones in plants (Figure 23).
Figure 23.
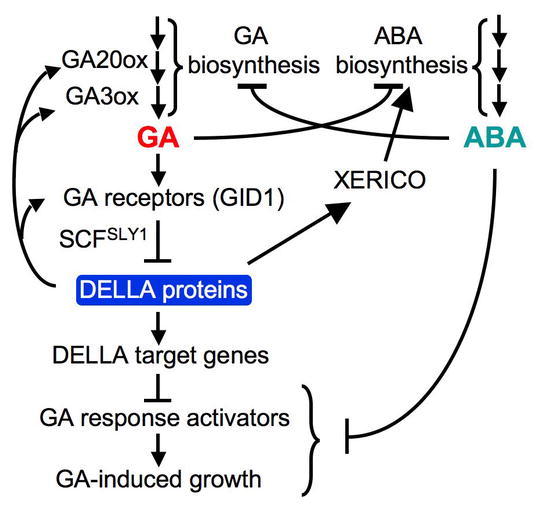
Model of GA Signaling Network and Interaction Between GA and ABA Pathways in Arabidopsis.
GA and ABA not only mutually inhibit each other's biosynthesis, but also promote each other's catabolism (not shown). Inhibition of GA-promoted processes by DELLA proteins is achieved by modulating both GA and ABA pathways. DELLA also plays a direct role in maintaining GA homeostasis by inducing genes encoding GA biosynthetic enzymes and GA receptors. ABA inhibits GA signaling downstream of DELLA, although ABA direct targets need to be elucidated. Reproduced with modifications from Zentella et al. (2007).
Acknowledgments
I would like to thank Shinjiro Yamaguchi for providing figures for GA metabolism, and Yuji Kamiya for the bakanae rice image. Our work on GA biosynthesis and GA signaling was supported by USDA National Initiative Competitive Grant 03-35304-13284 and National Science Foundation (IBN-0348814 and IOS-0641548).
Footnotes
Citation: Sun T. (2008) Gibberellin Metabolism, Perception and Signaling Pathways in Arabidopsis. The Arabidopsis Book 0:e0103. doi:10.1199/tab.0103
elocation-id: e0103
Published on: September 24, 2008
REFERENCES
- Achard P., Vriezen W. H., van der Straeten D., Harberd N. P. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell. 2003;156(1):2816–2825. doi: 10.1105/tpc.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Liao L., Jiang C., Desnos T., Bartlett J., Fu X., Harberd N. P. DELLAs contribute to plant photomorpho-genesis. Plant Physiol. 2007a;1436(1):1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Baghour M., Chapple A., Hedden P., Van Der Straeten D., Genschik P., Moritz T., Harberd N. P. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA. 2007b;1046(1):6484–6489. doi: 10.1073/pnas.0610717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N. P. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;3116(1):91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- Alabadi D., Gil J., Blazquez M. A., Garcia-Martinez J. L. Gibberellins repress photomorphogenesis in darkness. Plant Physiol. 2004;1346(1):1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvey L., Harberd N. P. DELLA proteins: integrators of multiple plant growth regulatory inputs. Physiol. Plant. 2005;1236(1):153–160. [Google Scholar]
- Ariizumi T., Steber C. M. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell. 2007;196(1):791–804. doi: 10.1105/tpc.106.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G., Goliber T. E., Moore C., Kessler S., Pham T., Sinha N. R. Homologies in leaf form inferred from KNOX1 gene expression during development. Science. 2002;2966(1):1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- Blazquez M. A., Green R., Nilsson O., Sussman M. R., Weigel D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;106(1):791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;2186(1):683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- Cao D., Hussain A., Cheng H., Peng J. Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta. 2005;2236(1):105–113. doi: 10.1007/s00425-005-0057-3. [DOI] [PubMed] [Google Scholar]
- Cao D., Cheng H., Wu W., Meng Soo H., Peng J. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 2006;1426(1):509–525. doi: 10.1104/pp.106.082289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qin L., Lee S., Fu X., Richards D. E., Cao D., Luo D., Harberd N. P., Peng J. Gibberellin regulates Ara-bidopsis floral development via suppression of DELLA protein function. Development. 2004;1316(1):1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- Chiang H-H., Hwang I., Goodman H. M. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;76(1):195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-T., Kende H. Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell. 1997;96(1):1661–1671. doi: 10.1105/tpc.9.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles J. P., Phillips A. L., Crocker S. J., García-Lepe R., Lewis M. J., Hedden P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 1999;176(1):547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., Vachon G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004;1366(1):3660–3669. doi: 10.1104/pp.104.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Daviere J. M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J. M., Lorrain S., Fankhauser C., Blazquez M. A., Titarenko E., Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;4516(1):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000;1226(1):415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Sun T-p. Synergistic de-repression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;1596(1):777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Jung H-S., Sun T-p. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA. 2001;986(1):14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S. G., Hu J., Steber C. M., Sun T-p. The Arabidopsis F-box protein SLEEPY1 targets GA signaling re-pressors for GA-induced degradation. Plant Cell. 2004;166(1):1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Bohlenius H., Moritz T., Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell. 2006;186(1):2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian T., Lorbiecke R., Umeda M., Sauter M. The cell cycle genes cycA1;1 and cdc2Os-3 are coordinately regulated by gibberellin in planta. Planta. 2000;2116(1):376–383. doi: 10.1007/s004250000295. [DOI] [PubMed] [Google Scholar]
- Feng S., Martinez C., Gusmaroli G., Wang Y., Zhou J., Wang F., Chen L., Yu L., Iglesias-Pedraz J. M., Kircher S., Schafer E., Fu X., Fan L. M., Deng X. W. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;4516(1):475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo F. F., Swain S. M. SPYing on GA signaling and plant development. J. Plant Growth Regul. 2003;226(1):163–175. [Google Scholar]
- Fleck B., Harberd N. P. Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilized by gibberellin. Plant J. 2002;326(1):935–947. doi: 10.1046/j.1365-313x.2002.01478.x. [DOI] [PubMed] [Google Scholar]
- Fleet C. M., Yamaguchi S., Hanada A., Kawaide H., David C. J., Kamiya Y., Sun T-p. Overexpression of AtCPS but not AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 2003;1326(1):830–839. doi: 10.1104/pp.103.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio M., Alabadi D., Perez-Gomez J., Garcia-Carcel L., Phillips A. L., Hedden P., Blazquez M. A. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 2006;1426(1):553–563. doi: 10.1104/pp.106.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Harberd N. P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature. 2003;4216(1):740–743. doi: 10.1038/nature01387. [DOI] [PubMed] [Google Scholar]
- Fu X., Richards D. E., Fleck B., Xie D., Burton N., Harberd N. P. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell. 2004;166(1):1406–1418. doi: 10.1105/tpc.021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J. M., Downes B. P., Shin-Han S., Durski A. M., Vierstra R. D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2002;996(1):11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and ab-scisic acid. Dev. Cell. 2004;76(1):373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Gillaspy G., Ben-David H., Gruissem W. Fruits: A Developmental Perspective. Plant Cell. 1993;56(1):1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S., Jones R. L. Perception of gibberellin and ab-scisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol. 1994;1046(1):1185–1192. doi: 10.1104/pp.104.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A., Zentella R., Walker-Simmons M., Ho T. H. D. Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell. 2001;136(1):667–679. [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mena C., de Folter S., Costa M. M., Angenent G. C., Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;1326(1):429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- Gomi K., Matsuoka M. Gibberellin signalling pathway. Curr. Opin. Plant Biol. 2003;66(1):489–493. doi: 10.1016/s1369-5266(03)00079-7. [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y., Maymon I., Borochov R., Alvarez J., Olszewski N., Ori N., Eshed Y., Weiss D. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;176(1):92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z. L., Powers S. J., Gong F., Phillips A. L., Hedden P., Sun T. P., Thomas S. G. Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis. Plant Cell. 2006;186(1):3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot S. P. C., Karssen C. M. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 1987;1716(1):525–531. doi: 10.1007/BF00392302. [DOI] [PubMed] [Google Scholar]
- Gubler F., Kalla R., Roberts J., Jacobsen J. V. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell. 1995;76(1):1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Chandler P., White R., Llewellyn D., Jacobsen J. GA signaling in barley aleurone cells: control of SLN1 and GAMYB expression. Plant Physiol. 2002;1296(1):191–200. doi: 10.1104/pp.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Raventos D., Keys M., Watts R., Mundy J., Jacobsen J. V. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;176(1):1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. The gibberellin pathway mediates KNOTTED1-type home-obox function in plants with different body plans. Curr. Biol. 2002;126(1):1557–1565. doi: 10.1016/s0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- Hedden P., Phillips A. L. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;56(1):523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Helliwell C. A., Sullivan J. A., Mould R. M., Gray J. C., Peacock J., Dennis E. S. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001a;286(1):201–208. doi: 10.1046/j.1365-313x.2001.01150.x. [DOI] [PubMed] [Google Scholar]
- Helliwell C. A., Chandler P. M., Poole A., Olive M. R., Dennis E. S., Peacock W. J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA. 2001b;986(1):2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Sheldon C. C., Olive M. R., Walker A. R., Zeevaart J. A. D., Peacock W. J., Dennis E. S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA. 1998;956(1):9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley R., Beale M. H., Smith S. J. Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta. 1991;1836(1):274–280. doi: 10.1007/BF00197799. [DOI] [PubMed] [Google Scholar]
- Hu J., Mitchum M. G., Barnaby N., Ayele B. T., Ogawa M., Nam E., Lai W-C., Hanada A., Alonso J. M., Ecker J. R., Swain S. M., Yamaguchi S., Kamiya Y., Sun T-p. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell. 2008;206(1):320–336. doi: 10.1105/tpc.107.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Raman A. S., Ream J. E., Fujiwara H., Cerny R. E., Brown S. Overexpression of 20-oxidase confers a gibberellin-overproducing phenotype in Arabidopsis. Plant Physiol. 1998;1186(1):773–781. doi: 10.1104/pp.118.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A., Cao D., Cheng H., Wen Z., Peng J. Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J. 2005;446(1):88–99. doi: 10.1111/j.1365-313X.2005.02512.x. [DOI] [PubMed] [Google Scholar]
- Huttly A. K., Phillips A. L. Gibberellin-regulated plant genes. Physiol. Plant. 1995;956(1):310–317. [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;146(1):57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Sasaki A., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Hasegawa Y., Minami E., Ashikari M., Matsuoka M. Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 2005;466(1):1392–1399. doi: 10.1093/pcp/pci152. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Suzuki H., Kim Y. C., Iuchi A., Kuromori T., Ueguchi-Tanaka M., Asami T., Yamaguchi I., Matsuoka M., Kobayashi M., Nakajima M. Multiple loss-of-function of Ara-bidopsis gibberellin receptor AtGID1s completely shuts down a gib-berellin signal. Plant J. 2007;506(1):958–966. doi: 10.1111/j.1365-313X.2007.03098.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen S. E., Olszewski N. E. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;56(1):887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S. E., Binkowski K. A., Olszewski N. E. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1996;936(1):9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005;156(1):1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Kaneko M., Inukai Y., Ueguchi-Tanaka M., Itoh H., Izawa T., Kobayashi Y., Hattori T., Miyao A., Hirochika H., Ashikari M., Matsuoka M. Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell. 2004;166(1):33–44. doi: 10.1105/tpc.017327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H., Hanada A., Kuzuyama T., Takagi M., Kamiya Y., Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gib-berellins in Arabidopsis. J. Biol. Chem. 2002;2776(1):45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- King K., Moritz T., Harberd N. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;1596(1):767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. H., Yang S. H., Han K. H. Upregulation of an Ara-bidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;476(1):343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M., van der Veen J. H. Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 1980;586(1):257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Elgersma A., Hanhart C. J., van Loenen M. E. P., van Rijn L., Zeevaart J. A. D. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 1985;656(1):33–39. [Google Scholar]
- Lee S., Cheng H., King K. E., Wang W., He Y., Hussain A., Lo J., Harberd N. P., Peng J. Gibberellin regulates Ara-bidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;166(1):646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. P., Koizuka N., Martin R. C., Nonogaki H. The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 2005;446(1):960–971. doi: 10.1111/j.1365-313X.2005.02588.x. [DOI] [PubMed] [Google Scholar]
- MacMillan J. Occurrence of gibberellins in vascular plants, fungi and bacteria. J. Plant Growth Regul. 2002;206(1):387–442. doi: 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004;376(1):720–729. doi: 10.1111/j.1365-313x.2003.01998.x. [DOI] [PubMed] [Google Scholar]
- Matsushita A., Furumoto T., Ishida S., Takahashi Y. AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 2007;1436(1):1152–1162. doi: 10.1104/pp.106.093542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K. M., Thomas S. G., Soule J. D., Strader L. C., Zale J. M., Sun T-p, Steber C. M. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin lig-ase. Plant Cell. 2003;156(1):1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. A., Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;176(1):705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum M. G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T. P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;456(1):804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Moon J., Suh S. S., Lee H., Choi K. R., Hong C. B., Paek N. C., Kim S. G., Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003;356(1):613–623. doi: 10.1046/j.1365-313x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Murray F., Kalla R., Jacobsen J., Gubler F. A role for HvGAMYB in anther development. Plant J. 2003;336(1):481–491. doi: 10.1046/j.1365-313x.2003.01641.x. [DOI] [PubMed] [Google Scholar]
- Nakajima M., Shimada A., Takashi Y., Kim Y. C., Park S. H., Ueguchi-Tanaka M., Suzuki H., Katoh E., Iuchi S., Kobayashi M., Maeda T., Matsuoka M., Yamaguchi I. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;466(1):880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser J. L., Hong F., Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;1266(1):467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- O'Neill D. P., Ross J. J. Auxin regulation of the gibberellin pathway in pea. Plant Physiol. 2002;1306(1):1974–1982. doi: 10.1104/pp.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;156(1):1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J. I., Kang C., Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;166(1):3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W. I., Choi G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;476(1):124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H-S., Sun T-p, Kamiya Y., Choi G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;196(1):1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N., Sun T-p, Gubler F. Gibberellin signaling:biosynthesis, catabolism, and response pathways. Plant Cell. 2002;146(1):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M., Kenmoku H., Ogawa M., Okada K., Mitsuhashi W., Sassa T., Kamiya Y., Toyomasu T., Yamaguchi S. Emission of ent-kaurene, a diterpenoid hydrocarbon precursor for gibberellins, into the headspace from plants. Plant Cell Physiol. 2004;456(1):1129–1138. doi: 10.1093/pcp/pch149. [DOI] [PubMed] [Google Scholar]
- Ozga J. A., Reinecke D. M. Hormonal interactions in fruit development. J. Plant Growth Regul. 2003;226(1):73–81. [Google Scholar]
- Penfield S., Gilday A. D., Halliday K. J., Graham I. A. DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 2006;166(1):2366–2370. doi: 10.1016/j.cub.2006.10.057. [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E. M., Kannangara R., Gilday A. D., Halliday K. J., Graham I. A. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 2005;156(1):1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Peng J., Harberd N. P. The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 2002;56(1):376–381. doi: 10.1016/s1369-5266(02)00279-0. [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D. E., King K. E., Cowling R. J., Murphy G. P., Harberd N. P. The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;116(1):3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Richards D. E., Hartley N. M., Murphy G. P., Devos K. M., Flintham J. E., Beales J., Fish L. J., Worland A. J., Pelica F., et al. ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;4006(1):256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Perazza D., Vachon G., Herzog M. Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 1998;1176(1):375–383. doi: 10.1104/pp.117.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A. L., Ward D. A., Uknes S., Appleford N. E. J., Lange T., Huttly A., Gaskin P., Graebe J. E., Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;1086(1):1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh L. D., Wysocka-Diller J. W., Camilleri C., Bouchez D., Benfey P. N. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;186(1):111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Reiser L., Sánchez-Baracaldo P., Hake S. Knots in the family tree: evolutionary relationships and functions of knox home-obox genes. Plant Mol. Biol. 2000;426(1):151–166. [PubMed] [Google Scholar]
- Rieu I., Ruiz-Rivero O., Fernandez-Garcia N., Griffiths J., Powers S. J., Gong F., Linhartova T., Eriksson S., Nilsson O., Thomas S. G., Phillips A. L., Hedden P. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;536(1):488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 2001;156(1):581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D-H., An G., Kitano J., Ashikari M., Matsuoka M. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;2996(1):1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- Schomberg F. M., Bizzell C. M., Lee D. J., Zeevaart J. A. D., Amasino R. A. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;156(1):151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., North H., Marion-Poll A., Sun T. P., Koshiba T., Kamiya Y., Yamaguchi S., Nambara E. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;486(1):354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Sakamoto T., Fujioka S., Takatsuto S., Yoshida S., Sazuka T., Ashikari M., Matsuoka M. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006;486(1):390–402. doi: 10.1111/j.1365-313X.2006.02875.x. [DOI] [PubMed] [Google Scholar]
- Silverstone A. L., Ciampaglio C. N., Sun T-p. The Ara-bidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;106(1):155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A. L., Mak P. Y. A., Casamitjana Martínez E., Sun T-p. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997a;1466(1):1087–1099. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A. L., Chang C-w, Krol E., Sun T-p. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 1997b;126(1):9–19. doi: 10.1046/j.1365-313x.1997.12010009.x. [DOI] [PubMed] [Google Scholar]
- Silverstone A. L., Jung H-S., Dill A., Kawaide H., Kamiya Y., Sun T-p. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;136(1):1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A. L., Tseng T-S., Swain S., Dill A., Jeong S. Y., Olszewski N. E., Sun T-p. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;1436(1):987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D. P., Jermakow A. M., Swain S. M. Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell. 2002;146(1):3133–3147. doi: 10.1105/tpc.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Hart G. W. Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr. Opin. Struct. Biol. 2003;136(1):631–636. doi: 10.1016/j.sbi.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R. D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;556(1):555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Steber C. M., Cooney S., McCourt P. Isolation of the GA-response mutant sly1 as a suppressor of ABI1–1 in Arabidopsis thaliana. Genetics. 1998;1496(1):509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Ritchie S., Soule J. D., McGinnis K. M., Steber C. M. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc. Natl. Acad. Sci. USA. 2004;1016(1):12771–12776. doi: 10.1073/pnas.0404287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;66(1):1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004;556(1):197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- Swain S. M., Singh D. P. Tall tales from sly dwarves: novel functions of gibberellins in plant development. Trends Plant Sci. 2005;106(1):123–129. doi: 10.1016/j.tplants.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Swain S. M., Tseng T-s, Olszewski N. E. Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 2001;1266(1):1174–1185. doi: 10.1104/pp.126.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S. M., Singh D. P., Helliwell C. A., Poole A. T. Plants with increased expression of ent-kaurene oxidase are resistant to chemical inhibitors of this gibberellin biosynthesis enzyme. Plant Cell Physiol. 2005;466(1):284–291. doi: 10.1093/pcp/pci027. [DOI] [PubMed] [Google Scholar]
- Swain S. M., Tseng T-s, Thornton T. M., Gopalraj M., Olszewski N. SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 2002;1296(1):605–615. doi: 10.1104/pp.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Phinney B. O., MacMillan J. Gibberellins. New York: Springer-Verlag, New York Inc; 1991. [Google Scholar]
- Tanaka-Ueguchi M., Itoh H., Oyama N., Koshioka M., Matsuoka M. Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J. 1998;156(1):391–400. doi: 10.1046/j.1365-313x.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- Telfer A., Bollman K. M., Poethig R. S. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;1246(1):645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Thomas S. G., Sun T-p. Update on gibberellin signaling. A tale of the tall and the short. Plant Physiol. 2004;1356(1):668–676. doi: 10.1104/pp.104.040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. G., Phillips A. L., Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multi-functional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA. 1999;966(1):4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T. M., Swain S. M., Olszewski N. E. Gibberellin signal transduction presents...the SPY who O-GlcNAc'd me. Trends Plant Sci. 1999;46(1):424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S., Li J., Chen M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004;546(1):519–532. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- Tseng T. S., Salome P. A., McClung C. R., Olszewski N. E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell. 2004;166(1):1550–1563. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L. Durham: Duke University; 2006. An analysis of potential negative and positive components of the gibberellin signaling pathway in Arabidopsis thaliana. Ph.D. thesis. [Google Scholar]
- Tyler L., Thomas S. G., Hu J., Dill A., Alonso J. M., Ecker J. R., Sun T-p. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;1356(1):1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 2007a;586(1):183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T. Y., Hsing Y. I., Kitano H., Yamaguchi I., Matsuoka M. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;4376(1):693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Katoh E., Ohmiya H., Asano K., Saji S., Hongyu X., Ashikari M., Kitano H., Yamaguchi I., Matsuoka M. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell. 2007b;196(1):2140–2155. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozu S., Tanaka-Ueguchi M., Kitano H., Hattori K., Matsuoka M. Characterization of XET-related genes of rice. Plant Physiol. 2000;1226(1):853–859. doi: 10.1104/pp.122.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova M., Yamaguchi S., Yang Y., McKelvey K., Hanada A., Borochov R., Yu F., Jikumaru Y., Ross J., Cortes D., Je Ma C., Noel J. P., Mander L., Shulaev V., Kamiya Y., Rodermel S., Weiss D., Pichersky E. Methylation of Gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell. 2007;196(1):32–45. doi: 10.1105/tpc.106.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen W. H., Achard P., Harberd N. P., Van Der Straeten D. Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J. 2004;376(1):505–516. doi: 10.1046/j.1365-313x.2003.01975.x. [DOI] [PubMed] [Google Scholar]
- Wang H., Caruso L. V., Downie A. B., Perry S. E. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell. 2004;166(1):1206–1219. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D., Halevy A. H. Stamens and gibberellin in the regulation of corolla pigmentation and growth in Petunia hybrida. Planta. 1989;1796(1):89–96. doi: 10.1007/BF00395775. [DOI] [PubMed] [Google Scholar]
- Weiss D., Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007;1446(1):1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C-K., Chang C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell. 2002;146(1):87–100. doi: 10.1105/tpc.010325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West C. A., Shen-Miller J., Railton I. D. Regulation of kaurene synthetase. In: Wareing P. F., editor. Plant Growth Substances 1982. 1. Vol. 6. London: Academic; 1982. pp. 81–90. [Google Scholar]
- Willige B. C., Ghosh S., Nill C., Zourelidou M., Dohmann E. M. N., Maier A., Schwechheimer C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;196(1):1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. N., Somerville C. R. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;1086(1):495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. N., Heckman J. W., Somerville C. R. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992;1006(1):403–408. doi: 10.1104/pp.100.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Campbell P., Vargheese A. V., Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J. 1996;96(1):879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;596(1):225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;416(1):251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y., Sun T-p. Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 2001;286(1):443–453. doi: 10.1046/j.1365-313x.2001.01168.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Sun T-p, Kawaide H., Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998a;1166(1):1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Smith M. W., Brown R. G. S., Kamiya Y., Sun T-p. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis Seeds. Plant Cell. 1998b;106(1):2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;166(1):367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005;156(1):1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- Young J. C., Obermann W. M., Hartl F. U. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J. Biol. Chem. 1998;2736(1):18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- Yu H., Ito T., Zhao Y., Peng J., Kumar P., Meyerowitz E. M. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA. 2004;1016(1):7827–7832. doi: 10.1073/pnas.0402377101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z. L., Park M., Thomas S. G., Endo A., Murase K., Fleet C. M., Jikumaru Y., Nambara E., Kamiya Y., Sun T. P. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;196(1):3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yu X., Foo E., Symons G. M., Lopez J., Bendehakkalu K. T., Xiang J., Weller J. L., Liu X., Reid J. B., Lin C. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007;1456(1):106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nomura T., Xu Y., Zhang Y., Peng Y., Mao B., Hanada A., Zhou H., Wang R., Li P., Zhu X., Mander L. N., Kamiya Y., Yamaguchi S., He Z. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;186(1):442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]


