INTRODUCTION
Ethylene (C2H4) is a simple gaseous hydrocarbon that has profound effects upon plant growth and development (reviewed in Mattoo and Suttle, 1991; Abeles et al., 1992). Although most popularly associated with ripening, ethylene plays a role throughout the entire life of the plant. Ethylene is a regulator of seed germination, seedling growth, leaf and petal abscission, organ senescence, stress responses, and pathogen responses.
Ethylene was one of the first plant hormones discovered (Abeles et al., 1992). In the nineteenth and early twentieth centuries, illuminating gas produced from coal was used for lighting. Leaks from pipelines carrying illuminating gas resulted in premature senescence and abscission in nearby vegetation, sometimes seriously damaging trees and greenhouse plants. Dimitry Neljubov identified ethylene as the “active” component in illuminating gas and published his results in 1901. In the 1930s, plants were demonstrated to produce ethylene themselves, thereby establishing ethylene as an endogenous regulator of plant growth and development.
By 1980, many effects of ethylene upon plant growth and development had been documented and the steps in ethylene biosynthesis elucidated (reviewed in Kende, 1993). However, little was known about how plants perceived ethylene and transduced the signal. Studies using radiolabeled ethylene demonstrated that plants had ethylene binding sites (Sisler, 1991), but the physiological relevance of these sites was debatable due in part to an inability to purify the binding component(s). Although the identification of pathway components by biochemical approaches proved recalcitrant, the ethylene signal transduction pathway proved quite tractable to genetic dissection in Arabidopsis (Bleecker et al., 1988; Guzmán and Ecker, 1990; Kieber et al., 1993; Roman et al., 1995). Many components in the primary signal transduction pathway have been identified in Arabidopsis, including ethylene receptors, pathway intermediates, and two families of transcription factors. The complete genome sequence of Arabidopsis has clarified this picture still further by indicating the size and variability of the gene families involved in ethylene biosynthesis and signal transduction.
ISOLATION OF MUTANTS IN ARABIDOPSIS
Ethylene affects many processes throughout the plant's growth and development, but most of these effects are cumbersome to use for a mutant screen. For example, although ethylene modulates the timing of leaf senescence in Arabidopsis, it is not required for the senescence syndrome to occur (Grbic and Bleecker, 1996). Thus, mutant isolation in Arabidopsis has relied almost exclusively upon one mutant screen: the triple response. Dark-grown seedlings exhibit several phenotypic responses to ethylene that are collectively termed the “triple response” (Knight et al., 1910). As shown in Figure 1, the triple response in Arabidopsis seedlings is characterized by a shortened and thickened hypocotyl, an inhibition of root elongation, and the formation of an exaggerated apical hook (Guzmán and Ecker, 1990). These features contrast sharply with the etiolated phenotype observed in dark-grown seedlings exposed to air. The readily distinguishable phenotype and the ability to screen thousands of seedlings on a Petri dish have greatly facilitated the identification of mutants that affect ethylene signaling in Arabidopsis.
Figure 1.
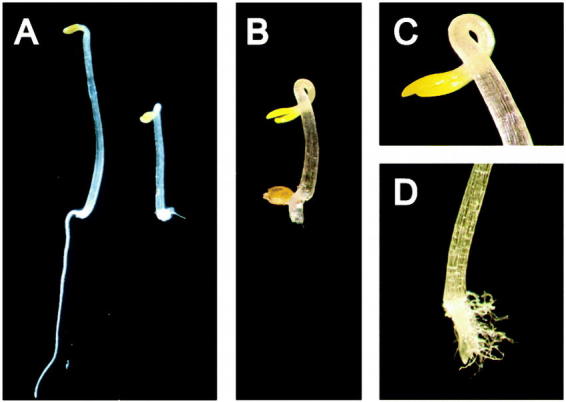
The Triple-Response to Ethylene of Dark-Grown Arabidopsis Seedlings.(A) Wild-type seedlings grown in the absence (left) or presence (right) of ethylene.(B) Wild-type seedling grown in the presence of the ethylene precursor ACC.(C) Close-up of the pronounced apical hook found with the triple response to ethylene.(D) Close-up of the shortened root found with the triple response to ethylene.
Mutations isolated using a screen for an altered triple-response to ethylene fall into two main classes: (1) mutations that render a plant insensitive to ethylene; and (2) mutations that result in a constitutive ethylene response (Figure 2, Table 1). Ethylene-insensitive mutants display a similar phenotype when grown in ethylene to what they display in air. Examples of ethylene-insensitive mutants are etr1-1, a gain-of-function mutation in an ethylene receptor (Bleecker et al., 1988; Chang et al., 1993), and ein2, a loss-of-function mutation in one element of the signal transduction pathway (Alonso et al., 1999). Constitutive ethylene-response mutants display a triple response in both air and ethylene. A constitutive ethylene response can result from ethylene over-production (eto mutants) (Guzmán and Ecker, 1990). Alternatively, mutations in the signal transduction pathway can also lead to a constitutive ethylene response as has been found with the ctr1 mutant (Kieber et al., 1993).
Figure 2.
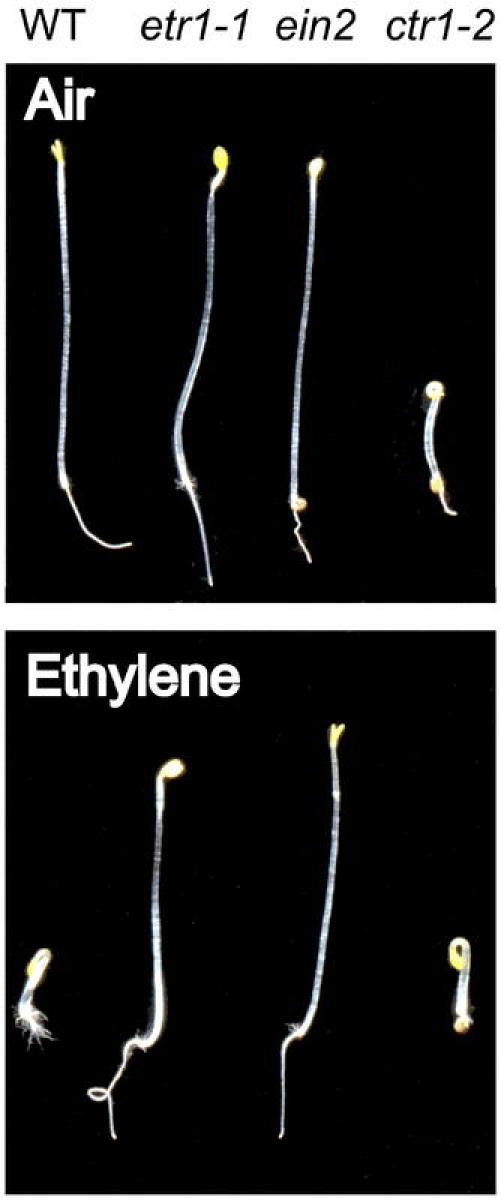
Mutants of the Ethylene Signal Transduction Pathway.The effect of ethylene upon the induction of the triple-response in dark grown seedlings is shown for wildtype, the ethylene-insensitive mutants etr1-1 and ein2, and the constitutive ethylene-response mutant ctr1-2.
The standard triple-response screen is likely saturated for the identification of viable loss-of-function mutations that affect ethylene responses. However, refinements of the screen continue to yield results. One refinement is to screen for mutations that only affect one aspect of the triple response such as hook formation (hookless; hls1) (Guzmán and Ecker, 1990; Lehman et al., 1996). Another is to screen for mutations that display an enhanced ethylene response (eer1) at a low ethylene concentration (Larsen and Chang, 2001). Another is to screen for ethylene responsiveness to a compound that interacts with the receptor but normally acts as an inhibitor of ethylene responses (response to antagonist; ran) (Hirayama et al., 1999).
ETHYLENE BIOSYNTHESIS
The ethylene biosynthetic pathway (Figure 3) was elucidated in a series of elegant studies, principally by Yang and co-workers (reviewed in: Yang and Hoffman, 1984; Kende, 1993; Zarembinski and Theologis, 1994; Bleecker and Kende, 2000). Ethylene is derived from the amino acid methionine, which in the first step is converted to S-adenyl-methionine (AdoMet) by AdoMet synthetase. AdoMet serves as an intermediate in a number of biosynthetic pathways, including the production of polyamines. ACC synthase, which converts AdoMet to 1-aminocyclopropane-1-carboxylic acid (ACC) (Adams and Yang, 1979), is the first committed and generally rate-limiting step in ethylene biosynthesis. The triple response can be induced in Arabidopsis seedlings by application of ACC (Figure 1), the applied ACC being readily converted to ethylene.
Figure 3.
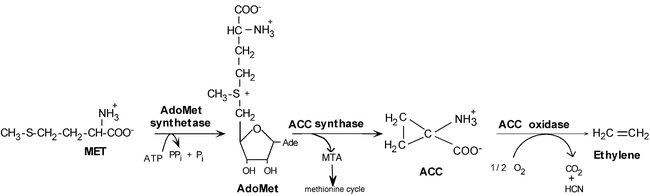
Ethylene Biosynthetic Pathway.The enzymes catalyzing each step are shown above the arrows. AdoMet: S-adenyl-methionine; Met: methionine; ACC: 1-aminocyclopropane-1-carboxylic acid; MTA: methylthioadenine.
A gene encoding ACC synthase was first cloned from zucchini by Sato and Theologis (Sato and Theologis, 1989). The Arabidopsis genome contains thirteen ACC synthase genes (ACS1-13); however the ACS3 gene is most likely a pseudogene and ACS1 encodes a non-functional ACC synthase (Liang et al., 1992; Liang et al., 1995). This is similar to the other plant species, such as tomato and rice, in which ACS is present as a multigene family. Inhibition of protein synthesis by cycloheximide treatment induces expression of several ACS genes, suggesting that these genes are under negative control by a short-lived transcriptional repressor, or that these transcripts are degraded by a short-lived nuclease (Liang et al., 1992). In other species, ACC synthase protein has been demonstrated to have a very short half-life (Zarembinski and Theologis, 1994; Bleecker and Kende, 2000), and this may be a general property of these enzymes.
The pattern of expression of the ACS2 was examined by fusions to a GUS reporter gene (Rodrigues-Pousada et al., 1993). This study suggested that ACS2 expression is high in young tissues and is switched off as the tissue matures. Also, expression of ACS2 was correlated with lateral root formation. The pattern of expression of the other ACS genes has not yet been examined carefully.
The final step of ethylene biosynthesis, the conversion of ACC to ethylene, is catalyzed by the enzyme ACC oxidase (ACO), which was previously called ethylene-forming enzyme. ACC oxidase may play an important role in regulating ethylene biosynthesis, especially during conditions of high ethylene production (Holdsworth et al., 1988; Nadeau et al., 1993; Kim and Yang, 1994; Tang et al., 1994; Barry et al., 1996; Lasserre et al., 1996). The ACC oxidase gene has been cloned from many different plant species. In Arabidopsis, ACO is present as a multigene family, but little information about these genes has been reported (Gomez-Lim et al., 1993; Raz and Ecker, 1999). An ACO gene, AtACO2, has been shown to be expressed preferentially on the outer cells of the apical hook in etiolated Arabidopsis seedlings and may play a role in regulating the differential cell elongation that is responsible for the formation of this structure (Raz and Ecker, 1999). The steady-state level of the AtACO2 transcript is elevated in response to exogenous ethylene (Kieber et al., 1993; Raz and Ecker, 1999).
Almost all plant tissues have the capacity to make ethylene, although in most cases the amount of ethylene produced is very low. In some plant species, ethylene production increases dramatically during developmental events such as germination, leaf and flower senescence and abscission and fruit ripening (Yang and Hoffman, 1984; Mattoo and Suttle, 1991; Abeles et al., 1992). In Arabidopsis, flowers and siliques produce copious levels of ethylene (Vogel et al., 1998b), though an extensive developmental profile of ethylene biosynthesis has not been done. The high level of ethylene produced by the siliques does not appear to play a major role in ripening as Arabidopsis is a non-climacteric species and ethylene-insensitive mutants do not have a major effect on silique maturation.
There is a diverse group of factors that have been described in numerous plant species that modulate the level of ethylene biosynthesis. In Arabidopsis, these inducers include auxin, cytokinin, brassinosteroids, ethylene, ozone, copper, mechanostimuli, pathogens and wounding (Botella et al., 1995; Cary et al., 1995; Liang et al., 1996; Vahala et al., 1998; Woeste et al., 1999a). As in other species, the Arabidopsis ACS genes are differentially regulated by various factors. These studies have only analyzed expression of ACS2, ACS4, ACS5 and/or ACS6, which were the first set of ACS genes identified in Arabidopsis; the remaining ACS genes have been identified solely by genomic and/or EST sequencing and have not yet been analyzed. In general, these factors act by increasing the steady-state level of ACS mRNA (Van der Straeten et al., 1990; Abel et al., 1995; Botella et al., 1995; Vahala et al., 1998). The ACS4 gene has been shown to be an auxin primary response gene and there are auxin-responsive motifs present upstream of this gene (Abel et al., 1995). ACS4 may be the primary isoform involved in mediating the auxin induction of ethylene biosynthesis. The ACS6 gene is induced by exposure to ozone, and is also upregulated by other stimuli such as wounding and ethylene (Vahala et al., 1998).
Genetic analysis of the regulation of ethylene biosynthesis in Arabidopsis has provided compelling evidence that ACC synthase can also be regulated post-transcriptionally (Vogel et al., 1998b; Woeste et al., 1999b). Cytokinin has been found to induce ethylene biosynthesis in seedlings from several plant species, including Arabidopsis (Lau and Yang, 1976; Babiker et al., 1993; Cary et al., 1995; Vogel et al., 1998a; Vogel et al., 1998b). In Arabidopsis, the elevation of ethylene biosynthesis that occurs in etiolated seedlings grown in the presence of cytokinin leads to a triple response morphology (Cary et al., 1995; Vogel et al., 1998b). This has been exploited to identify mutants that fail to increase ethylene in response to cytokinin (Vogel et al., 1998b). One gene identified in this screen was ACS5. Loss-of-function acs5 mutations severely reduce the amount of ethylene produced in response to cytokinin, suggesting that this isoform is the major target for cytokinin regulation. Northern analysis revealed that cytokinin increases ACS5 function primarily by a post-transcriptional mechanism, as cytokinin treatment had little effect on the steady-state level of ACS5 transcript.
Further evidence that ACS5 is post-transcriptionally regulated came from the analysis of the dominant eto2 mutation. This mutation results in a 20-fold increase in the level of ethylene produced by three-day-old etiolated seedlings (Kieber et al., 1993). Molecular analysis revealed that the eto2 mutation was the result of a single nucleotide insertion that disrupted the C-terminal 11 amino acids of ACS5 (Vogel et al., 1998b). As with cytokinin-induced ethylene production, the eto2 mutation did not affect ACS5 mRNA levels and thus the mutation likely acts by increasing the activity and/or stability of the ACS5 protein. Together, these results indicate that the C-terminus of ACS5 negatively regulates the function of the protein, and cytokinin may elevate ethylene biosynthesis by partially relieving this inhibition. Recent results in tomato have demonstrated that Le-ACS2 is phosphorylated on a serine residue present in the carboxy-terminus (Tatsuki and Mori, 2001); this serine residue is conserved in ACS5 and would be predicted to be disrupted by the eto2 mutation.
Two other Arabidopsis mutants have been described that elevate ethylene biosynthesis: the recessive eto1 mutation and the dominant eto3 mutation. Both eto1 and eto3 result in an increase in ethylene production, primarily in etiolated seedlings (Guzmán and Ecker, 1990; Kieber et al., 1993). Like eto2, both of these mutations are likely to affect the post-transcriptional regulation of ACS function (Woeste and Kieber, 1999). The ETO1 gene has recently been cloned (Cosgrove et al., 2000). It encodes a protein containing putative peptide binding domains and interacts with the wild-type, but not the eto2 version of ACS5 in the yeast two-hybrid system. Furthermore, ETO1 can inhibit the activity of ACS5 in vitro. ETO1 is likely to play a role in the post-translational regulation of ACS5 function. Thus, post-transcriptional control of ACC synthase may be an important mechanism regulating ethylene production in Arabidopsis.
ETHYLENE SIGNAL TRANSDUCTION
An important discovery from the analysis of ethylene signal transduction mutants was that, although mutants were isolated based on their triple-response phenotype, they affected other ethylene responses not just the triple-response (Bleecker et al., 1988). Thus, ethylene signal transduction appears to employ the same initial steps regardless of the downstream response. A genetically defined pathway for ethylene signal transduction has been determined by double-mutant analysis. For this purpose, the ctr1 mutation was essential as it initially represented the only constitutive ethylene-response mutant available in the signal transduction pathway (Kieber et al., 1993). Crosses between ctr1 and ethylene-insensitive mutants were used to determine the order of gene action. Double-mutant analysis demonstrated that a family of genes carrying dominant mutations (etr1, etr2, ers1, ers2, and ein4) functioned upstream of ctr1 (Kieber et al., 1993; Hua et al., 1995; Roman et al., 1995; Hua et al., 1998; Sakai et al., 1998), while other mutations (ein2, ein3) functioned downstream of ctr1 (Kieber et al., 1993; Roman et al., 1995). More recently the order of gene action downstream of CTR1 has been clarified by using overexpression phenotypes to aid in double-mutant analysis (e.g. if loss-of-function leads to ethylene insensitivity, one can potentially produce a gain-of-function mutant with constitutive ethylene response by overexpressing the gene) (Chao et al., 1997). The genetically defined pathway for ethylene signal transduction is shown in Figure 4.
Figure 4.
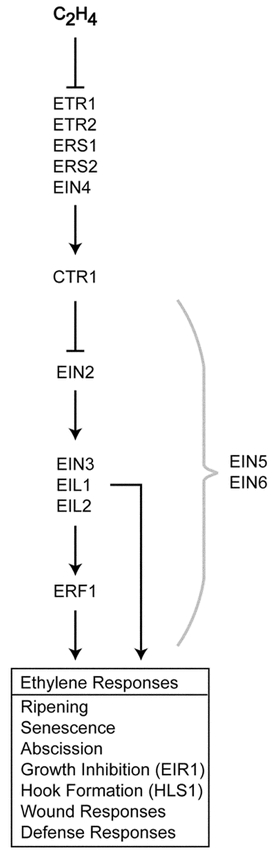
Genetically-Defined Pathway for Ethylene Signal TransductionGenes affecting the primary signal transduction pathway are shown. The order of action is based on double mutant analysis. EIN5 and EIN6 function at or downstream of CTR1 but their order of action has not been more precisely defined. EIR1 and HLS1 each affect a subset of ethylene responses.
The gene products for many of the mutations affecting ethylene signal transduction have been identified. Sequence analysis of the gene products suggests that the ethylene signal transduction pathway contains components of disparate evolutionary origin. The ethylene receptors contain features found in bacterial signal transduction systems (Chang et al., 1993; Schaller, 2000). The Raf-like kinase CTR1 may represent the first step in a Map kinase pathway, a signaling cascade found in eukaryotic but not prokaryotic systems (Kieber et al., 1993). The EIN3/EIL and ERF families of transcription factors are unique to plants (Ohme-Takagi and Shinshi, 1995; Chao et al., 1997). A model for ethylene signaling that incorporates known components into the genetically defined pathway is shown in Figure 5.
Figure 5.
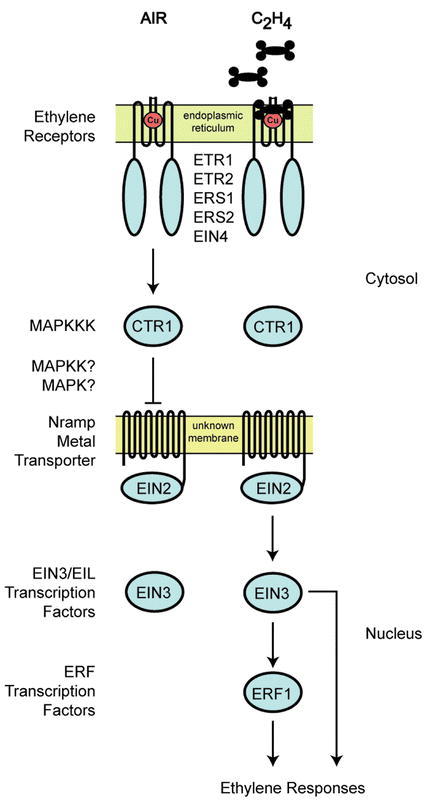
Model for Ethylene Signal Transduction.In air, ethylene receptors maintain CTR1 in an active state that serves to repress ethylene responses. In ethylene, the repression is relieved. Binding of ethylene inactivates the receptors, thereby inactivating CTR1. As a result, EIN2 is activated and a transcriptional cascade involving the EIN3/EIL and ERF transcription factors is initiated. Both families of transcription factors are involved in regulating ethylene responses. Soluble protein domains are shown as circles. The predicted transmembrane structures are shown for the ethylene receptors and EIN2. Location of pathway components is shown where known.
Components of the pathway exist in various intracellular locations. Several components contain transmembrane domains: the ethylene receptors (Schaller et al., 1995); RAN1, a protein involved in biogenesis of the ethylene receptors (Hirayama et al., 1999); and EIN2, a signaling component that functions downstream of the receptors (Alonso et al., 1999). From sequence alone it is not possible to determine to which membrane these components are localized. However, recent experimental evidence indicates that the ethylene receptor ETR1 is localized to the endoplasmic reticulum (Chen et al., 2002), a viable location for ethylene perception due to ready diffusion of ethylene in both aqueous and lipid environments (Abeles et al., 1992). Ethylene signaling also makes use of a soluble component CTR1 that is presumed to be cytosolic (Kieber et al., 1993), but may directly interact with ethylene receptors at the membrane (Clark et al., 1998). Finally, ethylene signaling makes use of nuclear-localized components such as the EIN3/EIL and ERF families of transcription factors (Ohme-Takagi and Shinshi, 1995; Chao et al., 1997). Thus, the initial steps in ethylene signal transduction form a circuit that reaches from membrane to nucleus but, unlike most hormone perception systems, the membrane involved is the endoplasmic reticulum rather than the plasma membrane.
In the following sections the mechanism of the ethylene signal transduction pathway is described in terms of the known components. This mechanistic picture is not complete and it is likely that other, as yet unidentified, components will be required to resolve some of the puzzles still remaining.
THE ETHYLENE RECEPTORS ETR1, ETR2, ERS1, ERS2, AND EIN4
As shown in Figure 6, a family of 5 proteins (ETR1, ETR2, ERS1, ERS2, and EIN4) function in ethylene perception by Arabidopsis (Chang et al., 1993; Hua et al., 1995; Hua et al., 1998; Sakai et al., 1998). The ethylene receptor ETR1 is the founding member of this family and has been characterized in the most detail. ETR1 is a membrane protein of 738 amino acid residues. ETR1 contains three aminoterminal transmembrane domains that encompass the ethylene-binding site (Schaller and Bleecker, 1995). Following the transmembrane domains is a GAF domain, a signaling motif implicated in binding of small molecules such as cGMP (Aravind and Ponting, 1997). The role of GAF domains in ethylene signaling is not known. In its carboxyterminal half, ETR1 contains regions with homology to histidine kinases and the receiver domains of response regulators (Chang et al., 1993), signaling elements originally identified as parts of bacterial two-component systems (Parkinson, 1993). The functional unit for ethylene perception is likely to be a receptor dimer (Schaller et al., 1995). In the case of ETR1, homo-dimerization is mediated in part by cysteine residues near the aminoterminus that are capable of forming disulfide bonds (Schaller et al., 1995).
Figure 6.
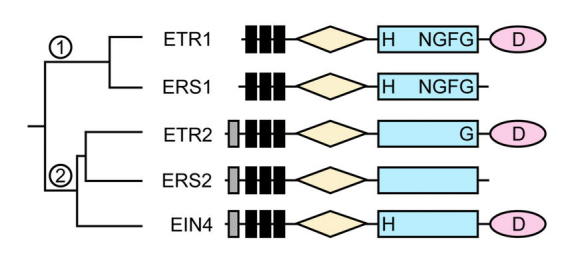
The Ethylene Receptor FamilyPrimary structures of the five-member family are indicated. Black bars represent transmembrane segments. Gray bars represent putative signal sequences. Diamonds indicate GAF domains. Rectangles indicate histidine kinase domains. Ovals indicate receiver domains. The conserved phosphorylation sites upon histidine (H) and aspartate (D) are indicated if present. Conserved motifs (NGFG) within the histidine kinase domain are indicated if present. There are two subfamilies of ethylene receptors (subfamily 1 and 2) based on sequence and phylogenetic analysis.
The other ethylene receptors have similarity at both the sequence and structural level to ETR1 (Figure 6) (Hua et al., 1995; Hua et al., 1998; Sakai et al., 1998). However, there are enough differences to support the existence of two subfamilies of ethylene receptors (Hua et al., 1998). Subfamily 1 is composed of ETR1 and ERS1. Subfamily 2 is composed of ETR2, ERS2, and EIN4. All five proteins contain three predicted transmembrane segments in the aminoterminal region, where the ethylene-binding site has been localized in ETR1. However, members of subfamily 2 contain a putative signal sequence that could target the proteins to the secretory pathway. All five proteins contain a histidine kinase domain. In subfamily 1 there is complete conservation of the residues considered essential for histidine kinase activity; on the other hand, in subfamily 2 the histidine kinase domains lack some essential residues (Figure 6). Three of the proteins (ETR1, ETR2, and EIN4) contain a receiver domain at the carboxyterminus. All five proteins contain the cysteine residues implicated in formation of the disulfide-linked dimer.
Ethylene binding has been demonstrated for both ETR1 and ERS1 (Schaller and Bleecker, 1995; Hall et al., 2000). The affinity of ETR1 for ethylene (dissociation constant, Kd, of 0.04 µL L−1 of ethylene) parallels the affect of ethylene upon the seedling growth response (Schaller and Bleecker, 1995). Moreover, inhibitors of ethylene responses in plants also inhibit ethylene binding by ETR1 and ERS1 (Schaller and Bleecker, 1995; Hall et al., 2000). These binding characteristics are consistent with what would be expected for an ethylene receptor. The ethylene binding site is novel, and by deletion analysis of ETR1 has been localized to the aminoterminal region containing the three transmembrane domains (Schaller and Bleecker, 1995; Rodriguez et al., 1999). Interestingly, the cyanobacterium Synechocystis (strain 6803) contains a protein (slr1212) with a similar ethylene-binding site, suggesting that plant ethylene receptors may have originated from an ancestral chloroplast genome (Rodriguez et al., 1999). A model for the ethylene-binding site is shown in Figure 7. In this model, there is one ethylene-binding site per receptor homo-dimer (Rodriguez et al., 1999). High-affinity binding of ethylene is mediated by a copper co-factor that is coordinated by two conserved amino acids (Cys65 and His69) (Schaller and Bleecker, 1995; Rodriguez et al., 1999). These amino acids are present in the second transmembrane domain, which would place them adjacent to each other on the same face of an alpha helix.
Figure 7.
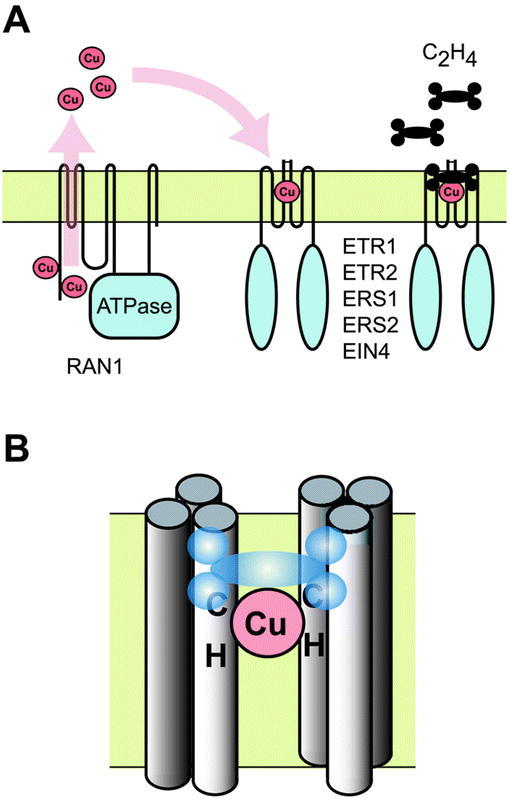
Model of Ethylene Binding Site(A) Copper loading of ethylene receptors by RAN1. RAN1 is a copper transporting ATPase. Copper is bound to RAN1 by two aminoterminal metal-binding motifs and then transported across the membrane. Copper is delivered to the ethylene receptor apoproteins. One copper ion is coordinated per receptor homo-dimer. Upon coordinating copper, the ethylene receptors are competent for ethylene binding.(B) Transmembrane structure of ethylene binding site. There is one copper binding site and consequently one ethylene-binding site per receptor homodimer. Each monomer of the receptor homodimer contains three transmembrane segments. Cys65 and His69 coordinate the copper ion within the second transmembrane domains of each monomer.
Ethylene receptors were originally identified based on mutations that resulted in a dominant ethylene-insensitive phenotype (Chang et al., 1993; Hua et al., 1995; Hua et al., 1998; Sakai et al., 1998). These mutations turn out to be missense mutations within the sensory portion of the receptors. Indeed, the first ethylene mutant isolated in Arabidopsis (etr1-1) affects an amino acid crucial for ethylene binding (Bleecker et al., 1988; Chang et al., 1993). The etr1-1 mutation results in a conversion of Cys65 to Tyr (Chang et al., 1993). As previously mentioned, Cys65 coordinates a copper co-factor required for ethylene binding (Rodriguez et al., 1999). Thus, as demonstrated biochemically, the etr1-1 mutant protein is unable to chelate copper, and as a consequence can not bind ethylene (Schaller and Bleecker, 1995; Rodriguez et al., 1999). The missense mutations etr1-3 and etr1-4 have also been found to reduce or eliminate ethylene binding (Hall et al., 1999). Interestingly, the mutation etr1-2, which results in a missense mutation in the third transmembrane domain, does not eliminate ethylene binding and thus may impede intramolecular transduction of the ethylene signal (Hall et al., 1999). Mutations that affect the ethylene-binding site need not be confined to the receptors themselves. For example, mutations in a copper-transporting ATPase (RAN1) in Arabidopsis perturb ethylene responses or induce a constitutive ethylene response (Hirayama et al., 1999; Woeste and Kieber, 2000). RAN1 is thought to deliver copper for the assembly of functional ethylene receptors (Figure 7).
Signal output by the ethylene receptors is likely to be mediated by the carboxyterminal half of the proteins. The striking feature of this region is the similarity to signaling elements found in two-component systems. In two-component systems, histidine kinases autophosphorylate on a conserved histidine residue, often in response to an environmental stimulus; this phosphate is then transferred to an aspartic acid residue within a receiver domain. Receiver domains are sometimes found joined to histidine kinases and are sometimes found in separate proteins referred to as response regulators (Parkinson, 1993; Swanson et al., 1994). The ethylene receptor ETR1 contains both histidine kinase and receiver domains, with all the conserved residues required for activity. Histidine kinase activity of ETR1 has been biochemically confirmed (Gamble et al., 1998). The receiver domain of ETR1 has been crystallized and shown to have the tertiary structure of known receiver domains (Muller-Dieckmann et al., 1999). Interestingly, the crystal structure indicates that the receiver domains of ETR1 form dimers, with dimerization mediated by the carboxyterminal end of the protein. The role of histidine kinase activity in ethylene signal transduction is unclear. ETR1 could participate in a phosphorelay that then regulates activity of the Raf-like kinase CTR1. A precedent for such a mechanism is found in the osmosensing pathway of yeast (Posas et al., 1996). However, the output domains of ETR1 and ERS1 are capable of physically interacting with CTR1 (Clark et al., 1998), raising the possibility that information could be passed from receptors to downstream signaling components by allosteric mechanisms. If not directly involved in the primary ethylene signal transduction pathway, histidine kinase activity might allow for cross-talk between ethylene perception and other two-component signaling pathways such as cytokinin signal transduction (Inoue et al., 2001).
The missense mutations within the ethylene receptors that lead to ethylene-insensitive phenotypes are gain-of-function mutations. To directly assess the role of the ethylene receptor family in ethylene perception, loss-of-function mutations have been isolated in four of the five gene members of the family (Hua and Meyerowitz, 1998). Single loss-of-function mutations have little or no effect upon ethylene signal transduction. However, in combination with the ETR1 loss-of-function mutation, the mutants show constitutive ethylene responses and this effect is most pronounced in triple and quadruple loss-of-function mutations. These results indicate that there is functional overlap among the receptor family members. These results also indicate that the receptors serve as negative regulators of the ethylene response pathway since elimination of receptors activates ethylene responses.
In hormone perception, varying the concentrations of hormone, receptor, or downstream signaling elements can modulate signal transduction. Extensive research has demonstrated that ethylene biosynthesis is regulated by environmental stresses and other plant hormones (Abeles et al., 1992; Woeste et al., 1999), and that levels of ethylene vary in different plant tissues and at different developmental stages (Abeles et al., 1992). Dynamic regulation of ethylene receptor levels would provide another means to activate or repress ethylene signal transduction. Interestingly, ethylene itself has been found to regulate transcription of the receptors ERS1, ERS2, and ETR2. Because the receptors are negative regulators of the pathway (Hua and Meyerowitz, 1998), an increase in the number of receptors could result in desensitization of the pathway. This mechanism of reducing ethylene sensitivity has been proposed to limit the spread of necrosis following pathogen infection in tomato (Ciardi et al., 2000). Ethylene-induced biosynthesis of new receptors could also play an additional role in ethylene signal transduction. Evidence indicates that the ethylene receptors are capable of binding ethylene very tightly, the apparent half-life for dissociation of ethylene from ETR1 is at least 12.5 hours (Schaller and Bleecker, 1995). However, plants respond within minutes to a decrease in ethylene levels (Abeles et al., 1992), an observation difficult to account for if all receptors are still occupied by ethylene. The production of new “empty” receptors may account for the re-sensitization of the plant to ethylene. If ethylene levels have decreased, the newly synthesized receptors will not bind ethylene and consequently, being negative regulators, will suppress ethylene responses in the plant.
CTR1, A RAF-LIKE PROTEIN KINASE
The CTR1 gene encodes a protein of 821 amino acids (Kieber et al., 1993). The carboxyl half of the protein contains a kinase domain with similarity to the Raf family of serine/threonine protein kinases that function in mitogen-activated protein kinase (MAPK) cascades in animals (Kieber et al., 1993). Based on this similarity, CTR1 may function as a mitogen-activated protein kinase kinase kinase (MAPKKK) in analogous fashion to Raf, and initiate signaling through a MAPK cascade in Arabidopsis. Consistent with this possibility, ethylene stimulates MAPK-like activity in Arabidopsis (Novikova et al., 2000; Ouaked et al., 2003). In addition, there is precedent for a two-component signal transduction system interacting with a MAP kinase cascade, as found in the osmosensing pathway of Saccharomyces cerevisiae (Posas et al., 1996). However, although MAPKs and MAPKKs exist in Arabidopsis, none have yet been demonstrated by loss-of-function mutations to play a role in the initial steps of the ethylene signal transduction pathyway.
A variety of mutations isolated in CTR1 are predicted to result in a loss of function (Kieber et al., 1993). For example, ctr1-5 is a T-DNA insertion, ctr1-3 arises from a premature stop codon positioned prior to the kinase domain, and ctr1-1 and ctr1-4 are in highly conserved kinase residues predicted to be required for activity. The ctr mutants are recessive and result in constitutive ethylene responses, indicating that CTR1 is a negative regulator of ethylene responses. Phosphorylation of substrates by CTR1 is apparently required to suppress ethylene responses.
Based on genetic analysis, the CTR1 gene functions at or downstream of the ethylene receptors (Kieber et al., 1993; Hua et al., 1995; Roman et al., 1995; Hua et al., 1998; Sakai et al., 1998). Physical evidence supports the genetic analysis as CTR1 is capable of directly interacting with the ethylene receptors ETR1 and ERS1, based on two-hybrid analysis and in vitro binding experiments (Clark et al., 1998). The region of CTR1 involved in the interaction lies within the aminoterminal half of the protein, a region that based on the Raf-kinase model could be involved in regulation of kinase activity. CTR1 was found capable of interacting with both the histidine kinase and receiver domains of ETR1, and the histidine kinase domain of ERS1 (which lacks a receiver domain). These results suggest that the site of action for CTR1 might be at the membrane and support the existence of a signaling complex involved in ethylene perception. These results also raise several possibilities for regulation of CTR1 activity: (1) CTR1 could directly respond to the phosphorylation state of the receptors; (2) CTR1 could be regulated by conformational changes in the receptors; or (3) CTR1 could be complexed with receptors but regulated through intermediary proteins.
The ctr1 mutants induce a constitutive ethylene response based on phenotype and the induction of ethylene-regulated molecular markers such as chitinase and the E1305 gene (Kieber et al., 1993). However, based on two lines of evidence, the ethylene signal transduction pathway is not completely dependent upon activity of CTR1. First, plants containing ctr1 loss-of-function mutations still display some ethylene responsiveness (Roman et al., 1995; Larsen and Chang, 2001). Second, plants containing loss-of-function mutations in four ethylene receptors display a more severe phenotype than ctr1 loss-of-function mutations (Hua and Meyerowitz, 1998). A possible explanation for these results is that other CTR1-like proteins act in the ethylene signal transduction pathway.
EIN2, AN NRAMP-LIKE PROTEIN
EIN2 acts downstream of CTR1 and encodes an integral membrane protein of 1294 amino acids (Alonso et al., 1999). EIN2 contains 12 predicted transmembrane domains in the aminoterminal third of the polypeptide, a region that exhibits significant similarity to the Nramp family of cation transporters. The carboxyterminal portion of EIN2 is a novel sequence that is predicted to be soluble and cytosolic. A role for the EIN2 carboxyterminal region in ethylene signaling has been confirmed by overexpressing just this domain in an EIN2 null background (Alonso et al., 1999). Plants expressing the EIN2 carboxyterminal region display constitutive ethylene-response phenotypes as adults and constitutively express ethylene-regulated genes. Interestingly, these results were only obtained with plants grown in the light; expression of the EIN2 carboxyterminal region was unable to induce the triple response in dark-grown seedlings. It is hypothesized that the aminoterminal region of EIN2 represents an input domain, interacting with upstream signaling factors, while the carboxyterminal region represents an output domain, interacting with downstream signaling factors.
Mutations have been isolated throughout the coding sequence of EIN2 and are predicted to result in a loss of function (Alonso et al., 1999). The ein2 mutants exhibit the strongest ethylene-insensitive phenotype of all the ethylene insensitive mutants isolated in Arabidopsis, supporting a critical role for EIN2 in ethylene signaling. In spite of this, it is still not clear how EIN2 mechanistically fits into the signal transduction pathway. There is no precedent for an Nramp-like protein operating downstream of a MAP kinase pathway. In addition, although related to the Nramp family, it is still questionable whether EIN2 functions in an analogous manner. Experiments to detect metal transporting activity in EIN2 have failed and, in contrast to other members of the Arabidopsis Nramp-like family, EIN2 is unable to complement metal-uptake deficient yeast strains (Thomine et al., 2000).
THE EIN3/EIL AND ERF FAMILIES OF TRANSCRIPTION FACTORS
Based on genetic evidence, EIN3 acts downstream of EIN2 (Chao et al., 1997). EIN3 encodes a protein of 628 amino acids that has the features of a transcription factor (Chao et al., 1997). EIN3 contains acidic, proline-rich, and glutamine-rich domains such as have been found in transcriptional activation domains, and is nuclear localized. Loss-of-function mutations in EIN3 result in an ethylene insensitive phenotype, indicating that EIN3 is a positive regulator of ethylene signal transduction. EIN3 is the founding member of a family that contains at least three additional EIN3-like proteins (EILs) (Chao et al., 1997). The finding that EILs complement a loss-of-function mutation in EIN3 that leads to ethylene insensitivity supports a role for the EILs in ethylene signal transduction. In addition, overexpression of EIN3 and EIL genes confers a constitutive ethylene response phenotype on dark-grown seedlings as well as plants grown in the light.
Direct evidence EIN3 and EILs function as transcription factors comes from the finding that they bind to a promoter element in the ERF1 gene (Solano et al., 1998). This primary ethylene response element (PERE) is a degenerate palindrome and the EIN3/EILs bind to the target sequence as homodimers. The PERE is similar to a motif that confers ethylene-regulated gene expression in other plant species. This motif is found in the promoter of the tomato E4 gene that is involved in fruit ripening (Montgomery et al., 1993) and in the promoter of the carnation GST1 gene that is involved in senescence (Itzhaki et al., 1994).
The ERF1 protein induced by the EIN3/EIL transcription factors is itself a transcription factor, indicating that ethylene signal transduction involves a transcriptional cascade (Solano et al., 1998). ERF1 belongs to a family of ethylene response element binding factor (ERF) proteins (Fujimoto et al., 2000), a family that has also been referred to in the literature as ethylene response element binding proteins (EREBPs) (Ohme-Takagi and Shinshi, 1995). ERFs bind to a GCC box found in the promoters of several pathogenesis-related genes such those encoding β-1,3-glucanase, basic chitinase, and defensin (PDF1.2) (Ohme-Takagi and Shinshi, 1995; Solano et al., 1998). Overexpression of ERF1 induced a partial triple-response phenotype in dark-grown seedlings, the seedlings lacking the exaggerated apical hook normally associated with the triple response (Solano et al., 1998). This result indicates that (1) the role of ERF1 is not confined to the pathogenesis response and (2) other factors besides ERF1 are required for initiating a full ethylene response. Analysis of the ERF family in Arabidopsis indicates that some members such as ERF1 function as transcriptional activators while other members function as repressors (Fujimoto et al., 2000). It should also be noted that not all ERFs are regulated by the EIN3/EIL transcription factors. Some ERFs contain GCC boxes in their promoters indicating that they are targets for other members of the ERF family, and suggesting the existence of a tertiary level of transcriptional regulation in ethylene signal transduction (Solano et al., 1998). A model for how this transcriptional cascade might operate is shown in Figure 8.
Figure 8.
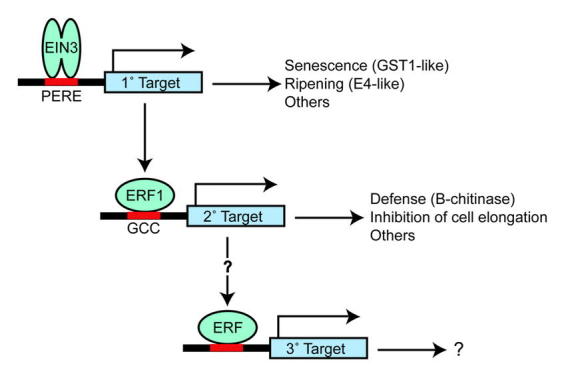
Transcriptional Regulation of Ethylene ResponsesEIN3 functions as a homodimer and binds to a primary ethylene response element (PERE) in the promoters of various targets. Targets found in various plant systems include genes involved in senescence, ripening, and the Arabidopsis transcription-factor ERF1. The transcription factor ERF1 binds to a GCC box found in the promoters of genes involved in defense, cell elongation, and other members of the ERF transcription-factor family. (Figure adapted from Solano et al., 1998)
The uncovering of this transcriptional cascade represents an important step in unraveling the means by which ethylene regulates gene expression, and gives a glimpse into the complexity that we can expect in this and other systems. Primary, secondary, and potentially tertiary targets for transcriptional regulation are now known in ethylene signal transduction. However, not all genes induced by ethylene contain PERE or GCC box promoter elements, suggesting the existence of additional classes of transcription factors. The existence of a transcriptional cascade with multi-member families of transcription factors provides many points for modulation by other regulatory pathways. Consistent with this is the finding that the expression of ERFs is regulated by other factors besides ethylene such as cold, drought, salt stress, wounding, and pathogens (Fujimoto et al., 2000). In addition, the ERF AtEBP is able to physically interact with a bZIP transcription factor involved in the plant defense response (Buttner and Singh, 1997). Microarray analysis such as that performed with SA, MJ, and ethylene should clarify this complex web of interactions among the signal transduction pathways (Schenk et al., 2000).
NEGATIVE REGULATION IN THE ETHYLENE SIGNAL TRANSDUCTION PATHWAY
Genetic analysis of ethylene signal transduction in Arabidopsis has uncovered both positive and negative regulators of the pathway (Table 1, Figure 5). Negative regulation of the pathway is evident in the initial steps of ethylene signal transduction as loss-of-function mutations in the ethylene receptors or CTR1 result in the induction of ethylene responses. In air, the ethylene receptors maintain CTR1 in an “on” state that serves to repress the ethylene responses. Binding of ethylene to the receptors turns CTR1 “off” and results in derepression of the pathway. Specifically, inactivation of CTR1 would result in the activation of EIN2, and consequently activation of the transcriptional cascade that involves the EIN3/EILs and the ERFs.
Gain-of-function mutations can be understood in the context of each signaling element's participation in the pathway as a positive or negative regulator. An ethylene-insensitive mutant of the ethylene receptor such as etr1-1 represents one such gain-of-function mutation. The etr1-1 mutant receptor cannot bind ethylene and, as a result, the receptor is unable to turn CTR1 “off” even when ethylene is present. The activated CTR1 continuously represses the ethylene pathway, resulting in the ethylene-insensitive phenotype. In contrast, overexpression of positive regulators such as the EIN2 carboxyterminus, the EIN3/EILs, and the ERFs results in phenotypes consistent with a constitutive ethylene response.
OTHER GENETICALLY DEFINED LOCI THAT AFFECT ETHYLENE SIGNALING
The mutations described in the previous sections affect key components in ethylene signal transduction. These components can be modeled as parts of a linear pathway that transduces initial perception of the ethylene signal. Other mutations have also been identified that affect ethylene signal transduction. Some of these have not been as fully characterized, and where they fit into the signal transduction pathway is not yet clear. Other mutations only affect a subset of ethylene responses, indicating that they function downstream of the primary signal transduction pathway.
ein5/ain1, ein 6, and ein 7: These mutations are ethylene-insensitive (Van der Straeten et al., 1993; Roman et al., 1995), and based on double-mutant analysis function downstream of CTR1 (Roman et al., 1995). ein5/ain1 and ein6 are recessive. ein7 is semidominant but, based on map position, could be allelic to ein5. The ethylene-insensitive phenotypes for these mutations are not as severe as observed with loss-of-function ein2 mutations.
eer1: The recessive mutant eer1 displays an enhanced ethylene response (Larsen and Chang, 2001). In the absence of ethylene, dark-grown mutant seedlings have slightly shorter and thicker hypocotyls than wildtype plants, consistent with a partial induction of the triple response. Expression of the ethylene-regulated gene basic-chitinase was also observed in the absence of ethylene. In the presence of ethylene, dark-grown eer1 seedlings display a more severe phenotype than that observed with wildtype seedlings, the eer1 hypocotyls being only about 50% the length of wildtype hypocotyls. The eer1 mutant phenotype is similar to that observed in sabre mutants (Aeschbacher et al., 1995).
aux1 and eir1: In these recessive mutations the hypocotyl is responsive to ethylene, but the roots show partial ethylene-insensitivity and display an altered gravitropic response (Roman et al., 1995). These mutants differ from each other in that aux1 plants are resistant to auxin but eir1 plants are responsive to auxin. AUX1 encodes an auxin influx carrier that functions in the transport and redistribution of auxin in the plant (Bennett et al., 1996; Marchant et al., 1999). The finding that aux1 mutants also have an altered ethylene response points to the importance of interactions between these two plant hormone signaling pathways. Ethylene insensitivity in the roots of aux1 and eir1 mutant plants may relate to the role of ethylene in regulating auxin transport (Suttle, 1988; Roman et al., 1995).
hls1: Dark-grown seedlings containing the hls1 mutation do not form a pronounced apical hook in response to ethylene (Guzmán and Ecker, 1990; Roman et al., 1995; Lehman et al., 1996). In addition, cells of the dark-grown cotyledons are enlarged. The hookless morphology can be phenocopied by treatment of seedlings with auxin or auxin-transport inhibitors (Lehman et al., 1996). Thus, like the aux1 mutant, the hls1 mutant suggests a link between auxin and ethylene signaling. HLS1 encodes a putative N-acetyltransferase and could potentially acetylate a protein or compound involved in auxin signaling or transport (Lehman et al., 1996).
INTERACTION WITH OTHER SIGNAL TRANSDUCTION PATHWAYS
At a party once, several plant scientists took it upon themselves to come up with alternative and more fitting names for the plant hormones. Peter McCourt (University of Toronto) proposed that ethylene be called “James Brown.” James Brown is known as the hardest working man in show business, and a good case can be made for ethylene being the hardest working hormone in the plant. Ethylene functions as a regulator throughout plant growth and in the responses to biotic and abiotic factors. For example, ethylene mediates many of the effects of other plant hormones, acting as a “second messenger” in these cases. Historically, the role of ethylene as a second messenger led to some confusion in defining what the primary responses were for the plant hormones (Abeles et al., 1992). For example, auxin, cytokinin, and brassinosteroid are each able to elevate ethylene biosynthesis in etiolated Arabidopsis seedlings, thereby inducing the triple-response phenotype (Cary et al., 1995; Woeste et al., 1999).
It would, however, be misleading to think of ethylene as only acting downstream of other plant hormones. In many cases ethylene is an equal partner in signaling and the physiological response is the result of a complex interaction between the different signaling pathways. This is readily apparent in the pathogen defense response in plants which is coordinated by ethylene, jasmonic acid (JA), and salicylic acid (SA). These three signaling factors are sometimes required individually and sometimes in concert for mobilizing defense responses to different pathogens (Glazebrook, 1999). For example, studies with Arabidopsis have shown that both ethylene and JA are required for induction of the defensin gene PDF1.2 in response to the avirulent fungal pathogen Alternaria brassicicola (Penninckx et al., 1998). Microarray analysis supports coordination among the signal transduction pathways (Schenk et al., 2000). SA, methyl jasmonate (MJ), and ethylene display individual effects upon gene expression, but also overlap significantly with some mRNAs regulated by multiple signaling molecules. An overlap was most apparent between ethylene and MJ, in which 50% of the genes induced by ethylene treatment were also induced by MJ treatment. Thus, it is possible that by careful regulation of signaling molecules a defense response fine-tuned to the particular pathogen can be initiated by the plant.
Interactions between the hormone signaling pathways can also be dependent upon the developmental state of the plant and the specific response evaluated, as evidenced by studies on the interaction between the abscisic acid (ABA) and ethylene signaling pathways. Mutant screens for genes involved in abscisic acid signaling uncovered ctr1 as an enhancer and ein2 as a suppressor of ABA-resistant seed germination (Beaudoin et al., 2000; Ghassemian et al., 2000), thereby providing genetic evidence for interactions between ethylene and ABA signaling. This interaction differs depending on whether seed germination or seedling root growth is being examined. Ethylene appears to promote seed germination by altering endogenous ABA levels and/or by decreasing the sensitivity of the seed to ABA; this is consistent with the ABA signal transduction pathway operating downstream of the ethylene signal transduction pathway (Beaudoin et al., 2000; Ghassemian et al., 2000). On the other hand, ABA inhibition of root growth requires an active ethylene signal transduction pathway, although apparently not ethylene itself; in this case the ABA signal transduction pathway operates upstream of the ethylene signal transduction pathway (Beaudoin et al., 2000; Ghassemian et al., 2000).
AGRONOMIC RELEVANCE OF STUDIES IN ARABIDOPSIS
This review began with the almost offhand remark that ethylene was most popularly associated with ripening such as is observed with tomatoes, bananas, and other climacteric fruit. Of course, ethylene-induced ripening is a developmental program that cannot be explicitly studied in Arabidopsis, which is a non-climacteric species. However, the finding that a group of genetically-defined components in Arabidopsis regulate multiple ethylene responses suggests a model for ethylene signaling in which there is a single primary response pathway that then branches into multiple secondary response pathways. In such a case, ripening would be predicted to make use of the same components of the primary response pathway found in Arabidopsis, but would then branch into a secondary pathway not found in Arabidopsis. Consistent with this prediction was the discovery that a tomato fruit-ripening mutant (never-ripe) arose due to a mutation in an ethylene receptor similar to ERS1 of Arabidopsis (Wilkinson et al., 1995).
Conservation of the primary response pathway for ethylene signal transduction has been borne out in studies on many plants. A search of GenBank indicates that ethylene receptors analogous to those of Arabidopsis have been identified in dicots such as tomato (accession number AF043084), melon (AB052228), mango (AF227742), banana (AF113748), peach (AF124527), grape AF243474), apple (AF032448), broccoli (AF032448), cucumber (AB026498), tobacco (AF022727), geranium (AF141929), carnation (AB035806), and rose (AF127221), as well as in the monocot rice (AF013979). The mutant ethylene receptor etr1-1 of Arabidopsis can confer dominant ethylene-insensitivity when expressed in such heterologous systems as tomato and petunia (Wilkinson et al., 1997). As shown in Figure 9, transgenic expression of etr1-1 in petunia delays the natural senescence of corollas so that more flowers are open and in color compared to the non-transgenic control. This demonstrates that the mechanism of ethylene perception is conserved among these different plant species. It also demonstrates the utility of the etr1-1 mutant as a tool for modulating ethylene responses. Continued dissection of the ethylene signal transduction pathway in Arabidopsis will undoubtedly lead to new discoveries with agronomic impact.
Figure 9.

Transgenic Expression of the Arabidopsis etr1-1 Mutant Gene in PetuniaOn the left is a transgenic petunia line carrying the etr1-1 gene expressed using a CaMV35S promoter (line 44568 from Wilkinson et al., 1997). On the right is a control wild-type petunia line. Due to delayed corolla senescence, the transgenic line 44568 has 7 flowers open compared to the 2 open in the control line. (Photograph courtesy of David Clark, University of Florida)
Acknowledgments
We thank Bernie Glick and Yi-Feng Chen for critical reading of the manuscript, and Xue-Chu Zhao, Xiang Qu, and David Clark for assistance with figures. Research in the authors' laboratories has been supported by grants from the National Science Foundation and the U. S. Department of Agriculture.
Footnotes
Citation: Schaller G.E., and Kieber J.J. (2002) Ethylene. The Arabidopsis Book 1:e0071. doi:10.1199/tab.0071
elocation-id: e0071
Published on: March 27, 2002
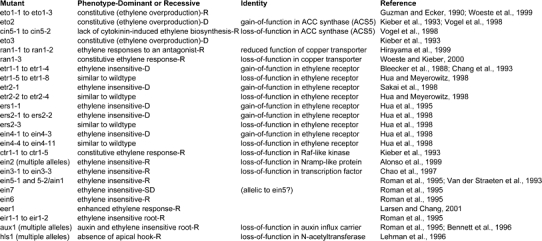
Table 1.
Mutations that affect ethylene signaling.
REFERENCES
- Abel S., Nguyen M. D., Chow W., Theologis A. ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J. Biol. Chem. 1995;2701(1):19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- Abeles F. B., Morgan P. W., Saltveit M. E., Jr Ethylene in Plant Biology, 2nd Edition. 1992. (San Diego: Academic Press)
- Adams D. O., Yang S. F. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA. 1979;761(1):170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeschbacher R. A., Hauser M-T., Feldman K. A., Benfey P. N. The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 1995;91(1):330–340. doi: 10.1101/gad.9.3.330. [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Hirayama T., Roman G., Nourizadeh S., Ecker J. R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;2841(1):2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Aravind L., Ponting C. P. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 1997;221(1):458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- Babiker A. G. T., Butler L. G., Ejeta G., Woodson W. R. Enhancement of ethylene biosynthesis and germination by cytokinins and 1-aminocyclopropane-1-carboxylic acid in Striga astiatica seeds. Physiol. Plant. 1993;891(1):21–26. [Google Scholar]
- Barry C. S., Blume B., Bouzayen M., Cooper W., Hamilton A. J., Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;91(1):525–35. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- Beaudoin N., Serizet C., Gosti F., Giraudat J. Interactions between the abscisic acid and ethylene signaling cascades. Plant Cell. 2000;121(1):1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. J., Marchant A., Green H. G., May S. T., Ward S. P., Millner P. A., Walker A. R., Schulz B., Feldmann K. A. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;2731(1):948–50. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bleecker A. B., Estelle M. A., Somerville C., Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;2411(1):1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Bleecker A. B., Kende H. Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell. Dev. Biol. 2000;161(1):1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Botella J. R., Arteca R. N., Frangos J. A. A mechanical strain-induced 1-aminocyclopropane-1-carboxylic acid synthase gene. Proc. Natl. Acad. Sci. USA. 1995;921(1):1595–8. doi: 10.1073/pnas.92.5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M., Singh K. B. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA. 1997;941(1):5961–6. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary A. J., Liu W., Howell S. H. Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 1995;1071(1):1075–82. doi: 10.1104/pp.107.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M. Arabidopsis ethylene response gene ETR1: Similarity of product to two-component regulators. Science. 1993;2621(1):539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J. R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;891(1):1133–44. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen Y-F., Randlett M. D., Findell J. L., Schaller G. E. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 2002;2771(1):19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Ciardi J. A., Tieman D. M., Lund S. T., Jones J. B., Stall R. E., Klee H. J. Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 2000;1231(1):81–92. doi: 10.1104/pp.123.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. L., Larsen P. B., Wang X., Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS1 ethylene receptors. Proc. Natl. Acad. Sci. USA. 1998;951(1):5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J., Gilroy S., Kao T., Ma H., Schultz J. C. Plant Signaling 2000. Cross talk among geneticists, physiologists, and ecologists. Plant Physiol. 2000;1241(1):499–506. doi: 10.1104/pp.124.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S. Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;121(1):393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble R. L., Coonfield M. L., Schaller G. E. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;951(1):7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;121(1):1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis. Curr. Opin. Plant Biol. 1999;21(1):280–6. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- Gomez-Lim M. A., Valdes-Lopez V., Cruz-Hernandez A., Saucedo-Arias L. J. Isolation and characterization of a gene involved in ethylene biosynthesis from Arabidopsis thaliana. Gene. 1993;1341(1):217–21. doi: 10.1016/0378-1119(93)90096-l. [DOI] [PubMed] [Google Scholar]
- Grbic V., Bleecker A. B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1996;81(1):595–602. [Google Scholar]
- Guzm´n P., Ecker J. R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;21(1):513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. E., Chen Q. G., Findell J. L., Schaller G. E., Bleecker A. B. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 1999;1211(1):291–299. doi: 10.1104/pp.121.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. E., Findell J. L., Schaller G. E., Sisler E. C., Bleecker A. B. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000;1231(1):1449–58. doi: 10.1104/pp.123.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Kieber J. J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J. M., Dailey W. P., Dancis A., Ecker J. R. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell. 1999;971(1):383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- Holdsworth M. J., Schuch W., Grierson D. Organization and expression of a wound/ripening-related small multi-gene family from tomato. Plant Mol. Biol. 1988;111(1):81–88. doi: 10.1007/BF00015661. [DOI] [PubMed] [Google Scholar]
- Hua J., Chang C., Sun Q., Meyerowitz E. M. Ethylene sensitivity conferred by Arabidopsis ERS gene. Science. 1995;2691(1):1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J., Meyerowitz E. M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;941(1):261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J., Sakai H., Nourizadeh S., Chen Q. G., Bleecker A. B., Ecker J. R., Meyerowitz E. M. EIN4 and ERS2 are members of the putative ethylene receptor family in Arabidopsis. Plant Cell. 1998;101(1):1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;4091(1):1060–3. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Itzhaki H., Maxson J. M., Woodson W. R. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA. 1994;911(1):8925–9. doi: 10.1073/pnas.91.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;441(1):283–307. [Google Scholar]
- Kieber J. J., Rothenberg M., Roman G., Feldman K. A., Ecker J. R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;721(1):427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim W. T., Yang S. F. Structure and expression of cDNAs encoding 1-aminocyclopropane-1-carboxylate oxidase homologs isolated from excised mung bean hypocotyls. Planta. 1994;1941(1):223–9. [PubMed] [Google Scholar]
- Knight L. I., Rose R. C., Crocker W. Effects of various gases and vapors upon etiolated seedlings of the sweet pea. Science. 1910;311(1):635–636. [Google Scholar]
- Larsen P. B., Chang C. The Arabidopsis eer1 Mutant Has Enhanced Ethylene Responses in the Hypocotyl and Stem. Plant Physiol. 2001;1251(1):1061–73. doi: 10.1104/pp.125.2.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre E., Bouquin T., Hernandez J. A., Bull J., Pech J. C., Balague C. Structure and expression of three genes encoding ACC oxidase homologs from melon (Cucumis melo L.). Mol. Gen. Genet. 1996;2511(1):81–90. doi: 10.1007/BF02174348. [DOI] [PubMed] [Google Scholar]
- Lau O. L., Yang S. F. Stimulation of ethylene production in the mung bean hypocotyls by cupric ion, calcium ion, and kinetin. Plant Physiol. 1976;571(1):88–92. doi: 10.1104/pp.57.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A., Black R., Ecker J. R. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;851(1):183–94. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Liang X., Abel S., Keller J. A., Shen N. F., Theologis A. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1992;891(1):11046–50. doi: 10.1073/pnas.89.22.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Oono Y., Shen N. F., Kohler C., Li K., Scolnik P. A., Theologis A. Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene. 1995;1671(1):17–24. doi: 10.1016/0378-1119(95)00694-x. [DOI] [PubMed] [Google Scholar]
- Liang X., Shen N. F., Theologis A. Li(+)-regulated 1-aminocyclopropane-1-carboxylate synthase gene expression in Arabidopsis thaliana. Plant J. 1996;101(1):1027–36. doi: 10.1046/j.1365-313x.1996.10061027.x. [DOI] [PubMed] [Google Scholar]
- Marchant A., Kargul J., May S. T., Muller P., Delbarre A., Perrot-Rechenmann C., Bennett M. J. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;181(1):2066–73. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo A. K., Suttle J. C. The Plant Hormone Ethylene. 1991. (Boca Raton: CRC Press, Inc.)
- Montgomery J., Goldman S., Deikman J., Margossian L., Fischer R. L. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc. Natl. Acad. Sci. USA. 1993;901(1):5939–43. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Dieckmann H. J., Grantz A. A., Kim S. H. The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure Fold. Des. 1999;71(1):1547–56. doi: 10.1016/s0969-2126(00)88345-8. [DOI] [PubMed] [Google Scholar]
- Nadeau J. A., Zhang X. S., Nair H., O'Neill S. D. Temporal and spatial regulation of 1-aminocyclopropane-1-carboxylate oxidase in the pollination-induced senescence of orchid flowers. Plant Physiol. 1993;1031(1):31–9. doi: 10.1104/pp.103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova G. V., Moshkov I. E., Smith A. R., Hall M. A. The effect of ethylene on MAPKinase-like activity in Arabidopsis thaliana. FEBS Lett. 2000;4741(1):29–32. doi: 10.1016/s0014-5793(00)01565-9. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M., Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;71(1):173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouaked F., Rozhon W., Lecourieux D., Hirt H. AMAPK pathway mediates ethylene signaling in plants. EMBOJ. 2003;221(1):1282–1288. doi: 10.1093/emboj/cdg131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993;731(1):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Penninckx I. A., Thomma B. P., Buchala A., Metraux J. P., Broekaert W. F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;101(1):2103–13. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Wurgler-Murphy S. M., Maeda T., Witten E. A., Thai T. C., Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two component” osmosensor. Cell. 1996;861(1):865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Raz V., Ecker J. R. Regulation of differential growth in the apical hook of Arabidopsis. Development. 1999;1261(1):3661–8. doi: 10.1242/dev.126.16.3661. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Pousada R. A., De Rycke R., Dedonder A., van Caeneghem W., Engler G., van Monagu M., Van der Straeten D. The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell. 1993;51(1):897–911. doi: 10.1105/tpc.5.8.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F. I., Esch J. J., Hall A. E., Binder B. M., Schaller G. E., Bleecker A. B. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;2831(1):996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Roman G., Lubarsky B., Kieber J. J., Rothenberg M., Ecker J. R. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics. 1995;1391(1):1393–409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Hua J., Chen Q. G., Chang C., Medrano L. J., Bleecker A. B., Meyerowitz E. M. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;951(1):5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Theologis A. Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc. Natl. Acad. Sci. USA. 1989;861(1):6621–5. doi: 10.1073/pnas.86.17.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller G. E. Histidine kinases and the role of two-component systems in plants. Adv. Bot. Res. 2000;321(1):109–148. [Google Scholar]
- Schaller G. E., Bleecker A. B. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;2701(1):1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schaller G. E., Ladd A. N., Lanahan M. B., Spanbauer J. M., Bleecker A. B. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 1995;2701(1):12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Schenk P. M., Kazan K., Wilson I., Anderson J. P., Richmond T., Somerville S. C., Manners J. M. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA. 2000;971(1):11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisler E. C. Ethylene-binding components in plants. 1991;1(1):81–99. In The Plant Hormone Ethylene, A. K. Mattoo and J. C. Suttle, eds. (Boca Raton: CRC Press), pp. [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J. R. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;121(1):3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle J. C. Effect of ethylene treatment onn polar IAA transport, net IAA uptake and specific binding of N-1-naphthylphthalamic acid in tissues and microsomes isolated from etiolated pea epicotyls. Plant Physiol. 1988;881(1):795–799. doi: 10.1104/pp.88.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R. V., Alex L. A., Simon M. I. Histidine and aspartate phosphorylation: two-component systems and the limits of homology. Trends Biochem. 1994;191(1):485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- Tang X., Gomes A. M. T. R., Bhatia A., Woodson W. R. Pistil-specific and ethylene-regulated expression of 1-aminocyclopropane-1-carboxylate oxidase gene in petunia flowers. Plant Cell. 1994;61(1):1227–1239. doi: 10.1105/tpc.6.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M., Mori H. Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J. Biol. Chem. 2001;241(1):24. doi: 10.1074/jbc.M101543200. [DOI] [PubMed] [Google Scholar]
- Thomine S., Wang R., Ward J. M., Crawford N. M., Schroeder J. I. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA. 2000;971(1):4991–6. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala J., Schalgnhaufer C. D., Pell E. J. Induction of ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol. Plant. 1998;1031(1):45–50. [Google Scholar]
- Van der Straeten D., Djudzman A., van Caenegeghem W., Smalle J., van Montagu M. Genetic and physiological anlaysis of a new locus in Arabidopsis that confers resistance to 1-aminocyclopropane-1-carboxylic acid and ethylene and specifically affects the ethylene signal transduction pathway. Plant Physiol. 1993;1021(1):401–408. doi: 10.1104/pp.102.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Straeten D., Van Wiemeersch L., Goodman H. M., Van Montagu M. Cloning and sequence of two different cDNAs encoding 1-aminocyclopropane-1-carboxylate synthase in tomato. Proc. Natl. Acad. Sci. USA. 1990;871(1):4859–63. doi: 10.1073/pnas.87.12.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. P., Schuerman P., Woeste K., Brandstatter I., Kieber J. J. Isolation and characterization of Arabidopsis mutants defective in the induction of ethylene biosynthesis by cytokinin. Genetics. 1998a;1491(1):417–27. doi: 10.1093/genetics/149.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. P., Woeste K. E., Theologis A., Kieber J. J. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA. 1998b;951(1):4766–4771. doi: 10.1073/pnas.95.8.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson J. Q., Lanahan M. B., Clark D. G., Bleecker A. B., Chang C., Meyerowitz E. M., Klee H. J. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nature Biotech. 1997;151(1):444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- Wilkinson J. Q., Lanahan M. B., Yen H-C., Giovannoni J. J., Klee H. J. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;2701(1):1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Woeste K., Kieber J. J. Characterization of Arabidopsis ethylene-overproducing mutants. 1999;1(1):37–43. In Biology and Biotechnology of the Plant Hormone Ethylene II, A. K. Kanellis, C. Chang, H. Klee, A. B. Bleecker, J. C. Pech and D. Grierson, eds. (Dordecht: Kluwer Academic Publishers), pp. [Google Scholar]
- Woeste K. E., Vogel J. P., Kieber J. J. Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol. Plant. 1999a;1051(1):478–484. [Google Scholar]
- Woeste K. E., Ye C., Kieber J. J. Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 1999b;1191(1):521–30. doi: 10.1104/pp.119.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste K. E., Kieber J. J. A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell. 2000;121(1):443–55. doi: 10.1105/tpc.12.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. F., Hoffman N. E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984;351(1):155–189. [Google Scholar]
- Zarembinski T. I., Theologis A. Ethylene biosynthesis and action: a case of conservation. Plant Mol. Biol. 1994;261(1):1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]


