Introduction
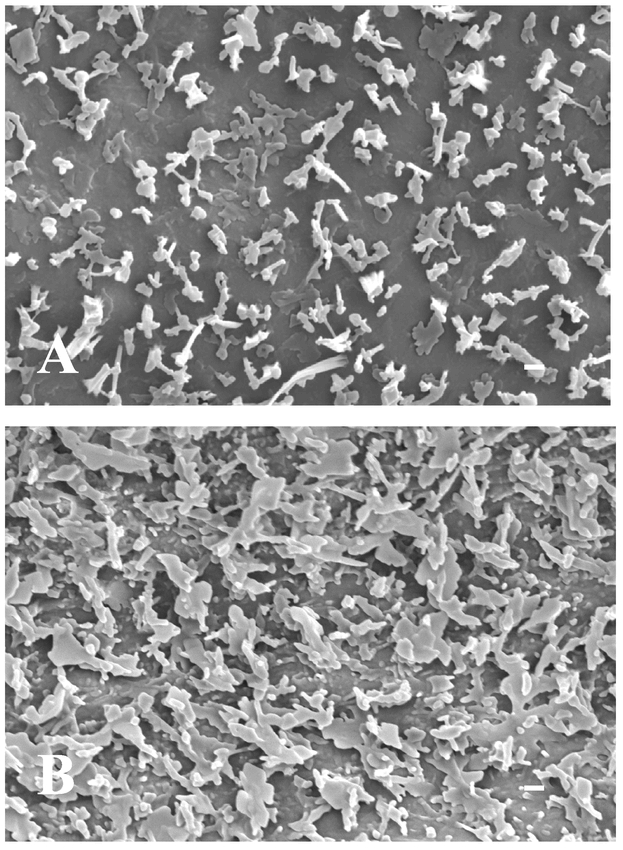
As in most terrestrial plants, the cuticle on Arabidopsis thaliana (L.) Heynh. forms a continuous lipid membrane over the apical epidermal cell walls of essentially all aerial plant organs. Epicuticular waxes form the outermost layer over this membrane and are visible on Arabidopsis inflorescence stem and silique surfaces as a bluish-white colored coating called glaucousness or waxy bloom. Intracuticular waxes are intermeshed within the cuticle membrane and not visible to the naked eye. Close examination of epicuticular waxes on Arabidopsis stems and siliques using scanning electron microscopy (SEM) at around 3000× magnification best reveals their diverse crystalline structures (Figure 1A). The stem and silique epicuticular wax morphology is composed primarily of columnar-shaped crystals (of ∼1.0 µm diameter), although rods, tubes, vertical plates, dendritic-, and umbrella-like structures are also typically present. The non-glaucous rosette and cauline leaf surfaces of Arabidopsis lack wax crystals detectable at the level of SEM (Figure 1B). Interestingly, other organs possess epicuticular wax crystals, including those of the pistil (Bowman, 1993). Descriptions of wax crystalline morphology on other flower parts, seeds, seedlings, roots, and other organs of Arabidopsis have not been reported.
Figure 1.
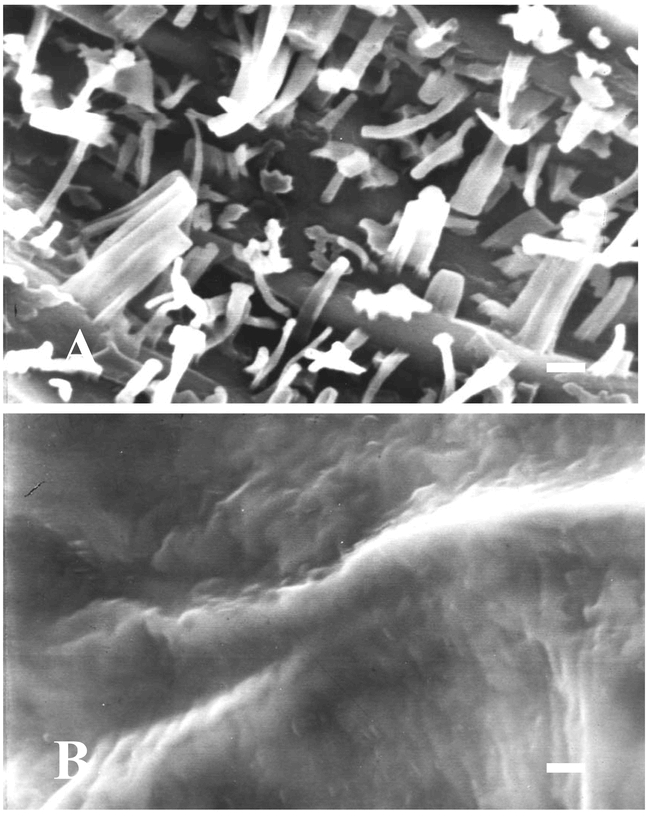
Surface morphology of air-dried stem and leaf surfaces of wild-type Arabidopsis thaliana ecotype Wassilewskija produced using scanning electron microscopy. A. Flowering stem. B. Abaxial blade surface between midrib and margin. Bar equals 1 µm.
Whereas the term epicuticular wax is used to describe wax crystals above of the cuticle, cuticular wax is being used here to describe those long chain lipids extracted by submersion of tissues in solvents like hexane and chloroform since this extraction procedure likely removes both epicuticular and intracuticular waxes. The chemical composition of cuticular waxes on Arabidopsis leaves and stems are typical of those on many dicotyledonous plants, being composed primarily of saturated free fatty acids, aldehydes, alkanes, primary alcohols, secondary alcohols, ketones, and wax esters (Figure 2). Within these component classes, homologues occur as aliphatic chains of between 16 and 33 carbons, except the wax esters, which are composed of even more carbons. The dominant wax class on Arabidopsis leaves and stems is the alkanes, although primary alcohols comprise a significant wax fraction on these surfaces. Stems and siliques possess relatively high amounts of ketones and secondary alcohols, whereas rosette and cauline leaves possess these constituents in trace or undetectable amounts. Arabidopsis lacks the wax hydroxy-ß-diketones, ß-diketones, and alkan-2-ol esters found on certain monocots (Bianchi and Bianchi, 1990), the estolides only reported in gymnosperms, and other minor constituents that occur idiosyncratically in plants that have been examined (Walton 1990). Inflorescence stems of wild-type Arabidopsis can produce over ten fold more total wax per area than leaves, and stem wax chain length distribution is shorter than leaves, with the C29 alkane homologue dominating stem waxes but the C31 alkane dominating leaf waxes (Jenks et al., 1995). Rosette leaves possess lower relative amounts of primary alcohols than cauline leaves, whereas rosette and cauline leaves, and siliques, have lower relative amounts of the C30 aldehydes than the stems (Todd et al., 1999). Interestingly, pollen grains are also coated with waxes, these being dominated as for stems by the C29 alkanes, secondary alcohols, and ketones, and the C30 aldehydes (Preuss et al., 1993; Fiebig et al., 2000). Wax composition on other Arabidopsis organs have not been reported.
Figure 2.
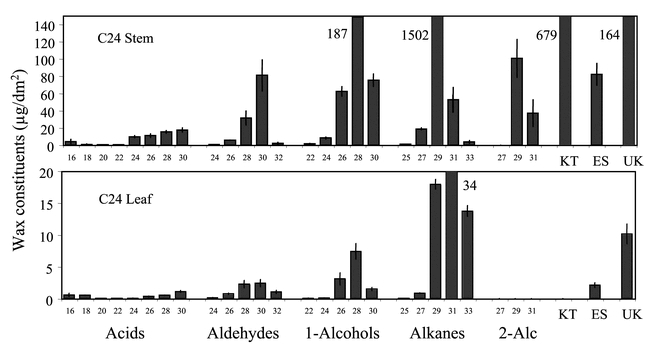
Cuticular wax composition of stems and leaves of Arabidopsis thaliana ecotype C24. Values represent cuticular wax load in µg/dm2 of stem and leaf blade area ± s.d. Chemical classes and chain lengths are labeled on the horizontal axis. Where cuticular wax constituent amount was off the scale, a number designating actual value is next to the bar. 2-Alc = secondary alcohol, KT = ketones, ES = esters, and UK = unknowns. Wax ester chain length distributions for Arabidopsis thaliana have not been determined.
Little is known about metabolic and regulatory processes associated with synthesis of cuticular waxes by Arabidopsis. Similarities between wax constituents on Arabidopsis and many other plant species however suggest that the basic mechanisms for wax production are highly conserved within the plant kingdom. The Arabidopsis close-relative Brassica oleraceae L. has been used to describe many of these reactions (see Kolattukudy, 1996). According to the developing model (Figure 3), early wax precursors are short acyl chains activated by a soluble plastidic Acyl Carrier Protein (ACP). Acyl chains are elongated by a single plastidic Fatty Acid Synthetase (FAS) complex which condenses acetal groups from malonyl-ACP onto a growing chain (Ohlrogge and Jaworski, 1997). Once C16 and C18 acyl-ACPs are synthesized in the plastids, acyl-ACP thioesterases cleave the ACP and release free C16 and C18 acids (palmitic and stearic acids) into the cytoplasm, where they are activated by Acyl-CoenzymeA (CoA) Synthetase via condensation with CoA. Of those C16 and C18 acyl-CoAs destined for conversion to waxes, most enter a membrane-associated pathway where they are modified by what may be a series of cytoplasmic enzyme complexes called elongases. Unlike short chain synthetases of the plastid, elongases use malonyl-CoA as the two-carbon donor, instead of malonyl-ACP (Agrawal et al., 1984; Bessoule et al., 1989). Studies using chemical inhibitors, used to target single elongation steps, suggested that, unlike short acyl-chain elongation, very long wax acyl-CoAs may be elongated by numerous chain-length-specific acyl-CoA elongase complexes (Mikkelsen, 1978; Agrawal et al., 1984; Wettstein-Knowles, 1985; 1995). Mutations in Arabidopsis and other plant species provide additional evidence for multiple elongases, as individual gene disruptions have been shown to affect single elongation steps primarily, either C24, C26, C28, or C30 acyl-chain elongation (Bianchi et al., 1979; Macey and Barber, 1970a; 1970b; Avato et al., 1982; Wettstein-Knowles, 1982; Jenks et al., 1995; 2000; Rashotte et al., 2001). Whether multiple elongase complexes are present however has not been confirmed. Once the very long acyl-CoA chains are synthesized, they are converted to cuticular waxes after either 1) deactivation by acyl-CoA thioesterases to release free acids (Liu and Post-Beittenmiller, 1995), 2) conversion to aliphatic esters by condensation of the acyl moiety with a primary alcohol by a putative acyl-CoA:fatty alcohol transacylase (Kolattukudy, 1967), or 3) entry into one of two reductive pathways that either convert acyl-CoAs to primary alcohols, or convert acyl-CoAs to aldehydes (Vioque and Kolattukudy, 1997). Clearly, the activity and regulation of very-long-chain acyl-CoA elongation reactions defines a central control point in plant wax biosynthesis since all cuticular waxes are derived from these reactions. Aldehydes generated by acyl-CoA reduction are likely converted, in large part, to alkanes by a putative decarbonylase (Cheesbrough and Kolattukudy, 1984). Much of the alkanes are then converted to secondary alcohols by a putative alkane hydroxylase, and then to ketones by a putative secondary alcohol oxidase (Kolattukudy et al., 1973). Much work is still needed to fully elucidate how the very-long-chain fatty acyl-CoAs are elongated in Arabidopsis, and how their acyl moieties are then shunted through the various networks of the wax biosynthetic pathway.
Figure 3.
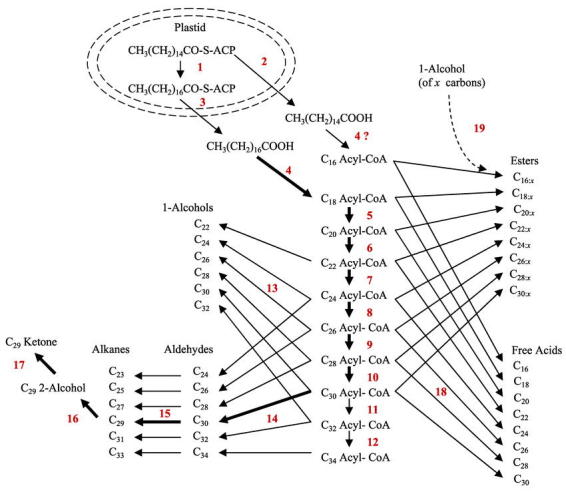
Conversion of major constituents in the stem cuticular wax biosynthetic pathway. [1] 3-ketoacyl-ACP synthetase II (KASII), [2] palmitoyl-ACP thioesterase, [3] stearoyl-ACP thioesterase, [4] acyl-CoA synthetase, [5–12] each a unique chain-length-specific acyl-CoA elongase complex (presumed similar to FAS having four major subunits), [13] primary alcohol generating acyl-CoA reductase, [14] aldehyde generating acyl-CoA reductase, [15] aldehyde decarbonylase, [16] alkane hydrolase, [17] secondary alcohol oxidase, [18] acyl-CoA thioesterase, [19] fatty acyl-CoA:fatty alcohol transacylase (Kolattukudy, 1996).
It is presently unclear how cuticular waxes of Arabidopsis get to the surface from locations of initial synthesis in epidermal plastids. Likely, Arabidopsis waxes are secreted according to conserved mechanisms as described in comparative studies using plant, yeast, and mammalian cells (Moore et al., 1991; Barinaga, 1993). Wax precursors formed in epidermal plastids appear to be transported into the cytoplasm where various microsomal elongases have been localized (Agrawal et al., 1984; Bessoule et al., 1989). Once the precursors are transferred out of the cytoplasm through the apical plasmalemma via exocytosis (Jenks et al., 1994b), they must then traverse the cell wall and cuticle layers where decarbonylase activity has been localized (Cheesbrough and Kolattukudy, 1984). The pathway most likely involves endoplasmic reticulum, transport vesicles, substrate ligands, vesicle receptors, and many other secretory factors (Barinaga, 1993; Jenks et al., 1994b). For example, Jenks et al., (1994b) showed a dramatic increase in endoplasmic reticulum and vesicle density below wax secretory sites in the Sorghum bicolor (L.) Moench leaf epidermis during wax induction by light. Other plant studies demonstrated that lipid transfer proteins (LTPs) were the most abundant proteins within Brassica cuticular waxes, providing evidence that LTPs may be involved in wax transport to the surface (Pyee et al. 1994). Arabidopsis mutants altered in both cell wall structure and cuticular waxes were recently reported, and a premise was set forth that mutants bearing these phenotypes may possess mutations that alter common processes in secretion of epidermal cell wall and cuticular lipid constituents (Jenks et al. 1996a). Notwithstanding, Arabidopsis cuticular wax secretory mutants, if discovered, would provide a model system for studying gene involvement in basic secretory processes used by plants.
Genetics of Arabidopsis Cuticular Wax
Studies of cuticular wax composition on inflorescence stems of 40 Arabidopsis ecotypes recently provided an indication of cuticular wax variability within the Arabidopsis thaliana species (Rashotte et al., 1997). Total wax amount on stems varied by almost 2-fold among ecotypes, however the proportional amounts of individual wax constituents for all but one ecotype varied little. Only the CT-1 ecotype showed a significant change in chain length distribution, having higher 22 and 24 carbon primary alcohols relative all other ecotypes. Differences in total wax amount within populations of plant species like Oryza sativa L. (rice), Musa spp. (banana and plantain), and Leymus angustus (Trin.) Pilger Dewey (Altai wildrye) showed clear polygenic inheritance (Haque et al. 1992; Ortiz et al., 1995; Jefferson, 1994). These species also responded to genetic selection for increased total wax quantity. Whether variations in wax amounts and primary alcohol chain-length distributions among Arabidopsis ecotypes are heritable has not been determined.
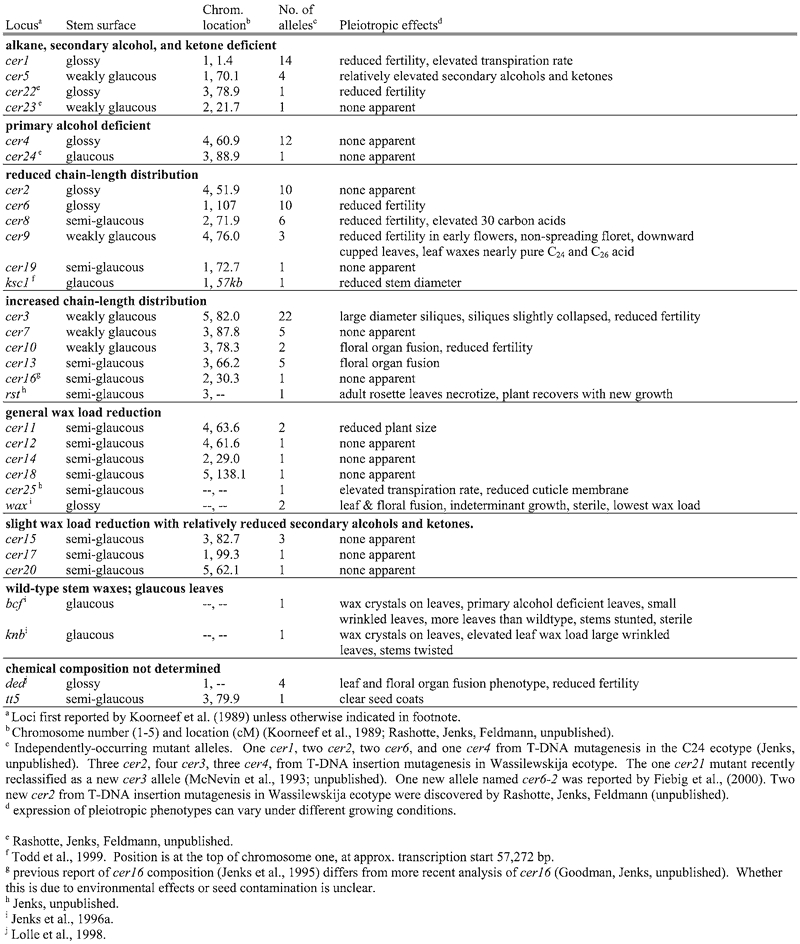
Various mutagenesis approaches have been used to produce Arabidopsis mutants with reduced visible depositions of epicuticular wax on inflorescence stems. While wild-type Arabidopsis has glaucous stems, these mutant stem surfaces range from semi-glaucous to very glossy (Figure 4). Dellaert et al., (1979) reported the first cuticular wax mutants in Arabidopsis and named them eceriferum (cer), which in latin is “without wax” (a term applied to fine stone sculptures lacking “wax” fillers). The first allelism studies were published by Koorneef et al. (1989), in which 21 new recessive cer loci were described. Since then, mutagenesis approaches have been used to identify 120 cuticular wax mutants representing a total of 31 recessive wax mutant loci (Table 1); 24 cer, transparent testa (tt5), wax, deadhead (ded), knobhead (knb), bicentifolia (bcf), 3-ketoacyl-CoA synthase1 (kcf1), and resurrection (rst). To date, dominant wax gene mutations have not been reported in Arabidopsis, even though naturally-occurring dominant wax alleles producing glossy leaf traits in Triticum turgidum L. var. durum (durum wheat) (Clarke et al., 1994), Musa spp. (Ortiz et al., 1995), and Brassica (Eigenbrode, unpublished data) have been described.
Figure 4.
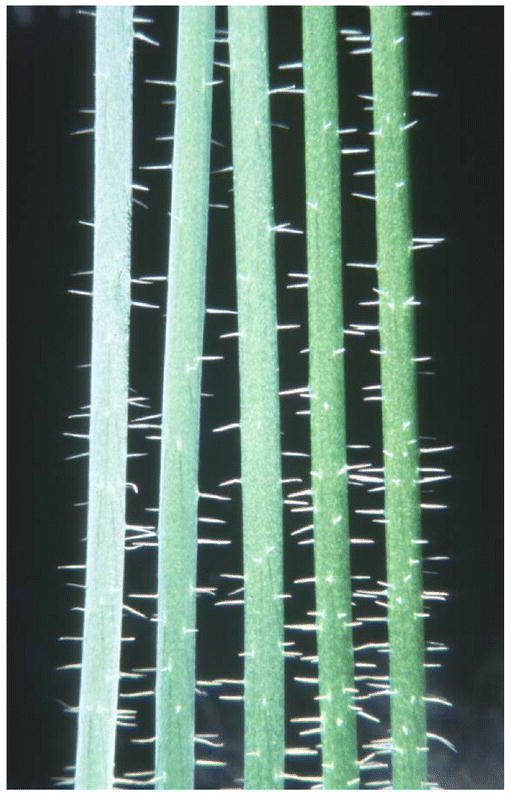
Photograph of the stems of Arabidopsis thaliana Wassilewskija (left) and isogenic cer3, cer4, cer1, and cer2 (from left to right) from McNevin et al. (1993).
Most Arabidopsis cer mutants have visual morphology similar to wildtype, except for their glossy stems. However, some mutants show distinct pleiotropic phenotypes (Table 1). A common pleiotropic effect of wax mutations is reduced fertility that can be restored by growth in high humidity environments, as occurs in cer1, cer2, cer3, cer6, cer8, cer9, cer10, cer22, cer25, ded, and wax1 (Koorneef et al., 1989). Studies using cer2 and cer6 revealed that this sterility is male gamete specific and that the mutant pollen coats lack certain waxes (Preuss et al., 1993). The altered lipids on mutant pollen may disrupted normal stigma-pollen communication since mutant pollen induced non-compatibility reactions (callose synthesis) in normally compatible stigmas, wild-type and mutant pollen germinated in an artificial medium similarly, and wild-type and mutant pollen placed on the same stigma induced release of stigma water leading to hydration and “near” normal development of the mutant pollen. The glossy-stemmed mutants wax (Jenks et al., 1996a) and ded (Lolle et al., 1998), and the semi-glaucous cer10 and cer13, have organs that show postgenital fusion early in development. Fusion by cer10 and cer13 is less severe than that which occurs in wax and ded, wherein cer10 and cer13 fusion occurs primarily in the flowers. Three genes have been cloned whose mutations result in postgenital fusions, Arabidopsis' FIDDLEHEAD (FDH) (Yephremov et al. 1999; Pruitt et al., 2000) and LACERATA (LCR) (Wellesen et al., 2001), and Zea mays' CRINKLY4 (CR4) (Becraft et al., 2001). Based on sequence analysis alone, two of these genes are clearly associated with metabolism of lipids like those found in the plant cuticle. FDH codes for a FAE-like gene involved in aliphatic lipid elongation, whereas LCR codes for a P450 monooxygenase that catalyzes w-hydroxylation of fatty acids perhaps involved in cutin monomer synthesis. The CR4 gene encodes a receptor-like kinase, an enzyme likely involved with developmental regulation (Jin et al., 2000; Becraft et al., 2001). More detailed studies are needed to determine whether cuticle lipids on the fdh, lcr, and cr4 mutants have been altered. Interestingly, expression of the cutinase gene from Fusarium solani f sp pisi in Arabidopsis lead to disruption in cuticle membrane ultrastructure and postgenital fusion (Seiber et al., 2000). Based on these reports, it appears that waxes or other cuticle membrane lipids may play a critical role as suppressors of postgenital fusion early in organ development. Another set of unique mutants, knb and bcf, have barely detectable reductions in their stem glaucousness, but increased leaf surface glaucousness due to presence of plate-like wax crystals (Jenks et al., 1996a). Knb and bcf also have a wrinkled leaf and stem morphology, and bcf has significantly more leaves than wildtype. The rst mutant (Jenks, unpublished) has a semi-glaucous stem and reduced wax load, and expresses a unique developmental program wherein adult but not juvenile phase leaves are round (lacking polar growth) and express visually large amounts of anthocyanins (purple) just before complete early senescence. One to two weeks later however, rst generates a new inflorescence stem, hence the name resurrection, apparently from lateral nodes of the rosette. This phenotype is leaky as a minor proportion of the rst mutants can produce more normal leaves and inflorescences in certain environments. Importantly, rst is highly susceptible to Erisphe orontii (powdery mildew) whereas its isogenic C24 ecotype parent is immune, indicating potential alteration of a constitutive mildew susceptibility factor at the surface. How single genes control the unique arrays of pleiotropic phenotypes in these wax mutants will be an important focus of future studies.
Most existing Arabidopsis wax mutant loci occur with only one or few alleles (Table 1), suggesting that many more wax loci could be discovered in additional mutagenesis studies. Koorneef et al (1989) used cer mutation frequencies to predict that ∼65 eceriferum (cer) mutants (visible stem wax mutants) could be found in Arabidopsis. More rigorous screening methods to select for alterations in chemical composition, microscopic crystal structure, or other physiology-based responses of both leaves and stems might greatly increase the number of wax mutants. Additional work to rescue the completely sterile cer mutants from mutagenized populations (an approach apparently not yet utilized) would likewise increase available wax loci. Lundqvist and Lundqvist (1988) identified over 79 cer loci in Hordeum vulgare (barley). Post-Beittenmiller (1996) suggested that well over 30 enzymatic steps are involved in just the basic wax metabolic pathway. If proteins involved in secretion, lipid transfer, organ and developmental regulation, or synthesis of minor wax compounds are also considered, then the number of wax gene loci in Arabidopsis may be well in the hundreds.
Whereas stem wax mutants (cer) have been isolated (Koorneef et al., 1989), most of these cer mutants have less significant alterations in their leaf waxes, or no differences at all (Jenks et al., 1995). Leaves are the primary photosynthetic organs, comprise the primary biomass of most agronomic and horticultural crops, and are often severely damaged by pests and other environmental stresses. Development of new research approaches to identify leaf wax genes involved in stress tolerance could be valuable for crop improvement. Studies of Arabidopsis representing worldwide distribution revealed no glaucous leaf types in over 80 ecotypes (Jenks, unpublished), and only little variation for stem wax composition in 40 ecotypes (Rashotte et al., 1997). Visual screening of mutagenized populations of Arabidopsis to find mutants having increased leaf glaucousness or glossiness due to alterations in cuticular waxes have had limited success (Jenks et al., 1996; Jenks, unpublished). Recent work has highlighted the potential of using new genomics-based approaches to identify and clone valuable new genes from close relatives of Arabidopsis thaliana (Zhu, 2001), particularly for the isolation of genes conferring traits not apparent in Arabidopsis thaliana, like leaf glaucousness. Unlike Arabidopsis thaliana, Thellungiella halophila (C.A. Meyer) O.E. Schultz and Thellungiella parvula (Schrenk) Al-Shehbaz & O'Kane (both previously included within the genus Arabidopsis, Al-Shehbaz et al., 1988; Rollins, 1993) produce a highly glaucous leaf coating (Al-Shehbaz and O'Kane, 1995; Teusink et al., 2002). Studies using SEM revealed that both Thellungiella species possess dense wax crystals over their leaves, while Arabidopsis leaves have none (Figure 5). Gas chromatography/mass spectrometry (GC/MS) was used to reveal that these Thellungiella species produce over 12-fold more total wax on their leaves than Arabidopsis thaliana. Like Arabidopsis stems, the leaves of both Thellungiella species had a large relative proportion of secondary alcohols, ketones, and esters which were trace or undetectable on Arabidopsis leaves (Teusink et al., 2002). Mutagenesis of Thellungiella would likely generate many glossy-leaved mutants easily identified among the glaucous wildtypes in mutagenized populations, and thereby provide a means to identify leaf wax genes. Moreover, since wax mutants having alterations in leaf, stem, or both organs would likely arise from a Thellungiella mutagenized population, genes involved in differential regulation of leaf and stem pathways would also likely be identified. Of the wax genes cloned, only CER2 (Negruk et al., 1996a; Xia, 1996), CER3 (Hannoufa et al., 1996), and GL15 (Moose and Sisco, 1996) may be regulatory genes.
Figure 5.
Surface morphology of air-dried adaxial leaf surfaces of A. Thellungiella halophila and B. T. parvula produced using scanning electron microscopy. Bar equals 1 µm.
Certain Thellungiella species have extremely high drought tolerance suggesting that wax genes associated with drought response might likewise be selected for. Potentially, the more drought tolerant Thellungiella species might survive mutations causing more severe alteration in its cuticular layers than Arabidopsis. If so, mutagenesis could be used to generate many additional, and more physiologically significant, wax mutants in populations of Thellungiella than might be created in Arabidopsis. Finally, it should be noted that in addition to Thellungiella halophila and Thellungiella parvula, many other species within the Brassicaceae have been found in native habitats that suggest adaptation to extreme environments, with many of these having waxy leaf coatings (Rollins, 1993). The usefulness of this biological material for molecular genetic dissection of wax genetics and physiology has not been evaluated.
Wax Crystalline Morphology of Wax Mutants
SEM has been used to examine wax crystal structures on nearly all of the known wax mutants in Arabidopsis thaliana (Figure 6). Generally, wax mutations reduce the density of wax crystals on mutant stems compared to wild-type stems, and also alter wax crystal shape and size. For example, cer4 has a very high density of vertically oriented (relative to the horizontal cuticle surface) plate-like waxes (Figure 6D), cer19 that has a very high proportion of highly branched dendritic structures (Figure 6L), and cer2 (Figure 6C) and cer6 (Figure 6F) have very tiny or completely lack wax crystals. What factors determine how these secreted waxes polymerize on the surface to form their unique crystalline shapes is quite uncertain. Likely, secretion site morphology (Jenks et al., 1994b), chemical composition (Rashotte et al., 1998), growth phase or organ development, and environment (Wettstein-Knowles, 1974) all play some role. Studies show how wax crystal formation involves complex molecular interactions which are themselves greatly effected by rates of secretion and environmental conditions (Adamson, 1976; Jeffree et al., 1976). Rashotte and Feldmann (1998) compared waxes on 22 Arabidopsis wax mutants and two wild-type ecotypes to reveal some broad correlations between wax composition and structure. For example, the shorter carbon chain waxes correlated best with dendritic structures, while long chain constituents correlated best with umbrella-like wax structures. However, as these authors and others (Jetter and Riederer, 1995) have concluded, even minor variations in specific wax constituents can dramatically alter crystal shape. The diversity of wax crystalline morphologies among Arabidopsis wax mutants could nevertheless make Arabidopsis an excellent tool for elucidating the physico-chemical basis for wax crystallization patterns.
Figure 6.
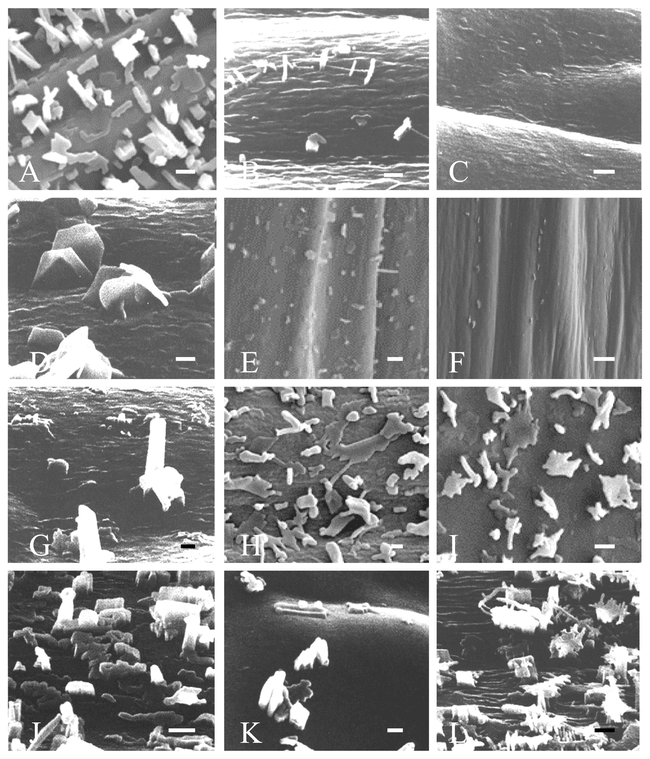
Scanning electron micrographs showing the microscopic surface morphology of air-dried stem surfaces of wild-type Arabidopsis thaliana ecotype Landsberg erecta and 11 representative wax mutants. A. Ler. B. cer1. C. cer2. D. cer4. E. cer5. F. cer6. G. cer7. H. cer8. I. cer9. J. cer15. K. cer16. L. cer19. Bar equals 1 µm.
Wax Chemical Composition of Wax Mutants
Detailed description of wax chemical composition on wax mutants has proved useful in predicting gene function, even before the respective gene is cloned, sequenced, and studied further (Jenks et al. 1995; 1996a; 1996b). Wax gene function can be elucidated by examining the respective gene mutation's effect on the size of substrate pools utilized by the wax metabolic pathway. The effect of mutation on the relative pool sizes can be used to predict where flux in the network has been suppressed or enhanced. For example, the cer4 mutant is nearly lacking all primary alcohols, and has a slight increase in alkanes and aldehydes with no other significant wax changes (Jenks et al., 1995). Thus, lack of the CER4 product may inhibit acyl-CoA reductase activity (see step 13 on Figure 3) described by Vioque and Kolattukudy, (1997). Larger aldehyde and alkane pool sizes on cer4 are easily explained by the shunting of wax precursors away from the primary alcohol- to the alkane-synthesizing branch of the pathway. The cer2 and cer6 mutants have higher relative amounts of shorter chain length constituents that indicate suppression of C26 acyl-CoA elongation specifically (see step 9 on Figure 3). While cer6 blocks both leaf and stem elongation, cer2 only effects stem waxes (having wild-type leaf wax) indicating that CER2 may be associated with a stem-specific regulator of the C26 elongase (Jenks et al., 1995). The cer19 mutant is similar to cer6 except that the C28 elongation step (see step 10 on Figure 3) appears blocked. Thus, cer19 may suppress activity of the C28 elongase. By comparison, cer1 has very little alkane but elevated aldehydes. Initially, it was proposed that CER1 knocked out step 15 in Figure 3 above, the step catalyzed by an aldehyde decarbonylase. Closer examination of the wax profile however revealed that alcohols were also reduced on cer1. Based on the current pathway model, the cer1 mutant would be expected to have elevated primary alcohols instead due to increase shunting into the primary-alcohol synthesizing pathway. Thus, the function of the CER1 gene product is difficult to predict based on compositional analysis alone.
Wax mutations in Arabidopsis have been described whose effects on wax composition indicate that their functions are linked with many steps in the wax biosynthetic pathway (Figure 3), including steps earlier than step 4. Table 1 provides an overview of the mutants and their primary effects on wax composition. The multiple pleiotropic effects of many wax mutants (see Table 1) indicate that wax genes may be involved in, not just wax synthesis, but many other aspects of overall plant development and cellular functions like secretion.
Molecular Biology of Cuticular Waxes
Thus far, nine genes involved in wax deposition have been isolated from Arabidopsis and Zea. Three of these appear to encode regulatory loci (CER2, CER3, GL15) while four encode enzymes (CER60, CER6, KCS1, GL8), and two may be involved in the transport of wax compounds (CER1, GL1). The first wax biosynthesis genes cloned in Arabidopsis and Zea, CER2 and GL2 respectively, encoded orthologs of a novel protein (Tacke et al. 1995; Negruk et al., 1996a; 1996b; Xia et al., 1996). Alignment of the predicted amino acid sequences of these genes revealed numerous conserved motifs (Negruk et al., 1996a). Cell fractionation and immunoblot analysis using polyclonal antibodies raised against a peptide whose sequence was based on the predicted amino acid sequence of CER2 suggest that this novel gene product is soluble and nuclear localized (Xia et al., 1997). Considering that the predicted amino acid sequence of CER2 does not have a known nuclear localization sequence, Xia et al. (1997) suggested that it may enter the nucleus with the aid of another protein. Reporter gene fusions of the CER2 promoter region with the ß-glucoronidase gene (GUS) suggest that CER2 expression is highest in epidermal layers of a many organs. As shown by Figures 7a and 7b, expression in leaves appears confined to the guard cells, trichomes, and petioles. Application of 1 µM 6-benzylaminopurine (BAP) induces ectopic expression of CER2-GUS in all cell types of the leaves that emerge following this treatment (Figures 7a and 7b). Consistent with this observation, the amount of CER2 protein also increased after BAP treatment. In contrast, other stimuli including, light, drought stress, salt stress, wounding, salicylic acid, heat or cold shock, and abscisic acid failed to induce either the reporter gene or synthesis of the CER2 protein. Thus, whereas CER2 is potentially linked to growth regulation via cytokinin response, CER2 may not play a role in environmental stress response.
Figure 7.
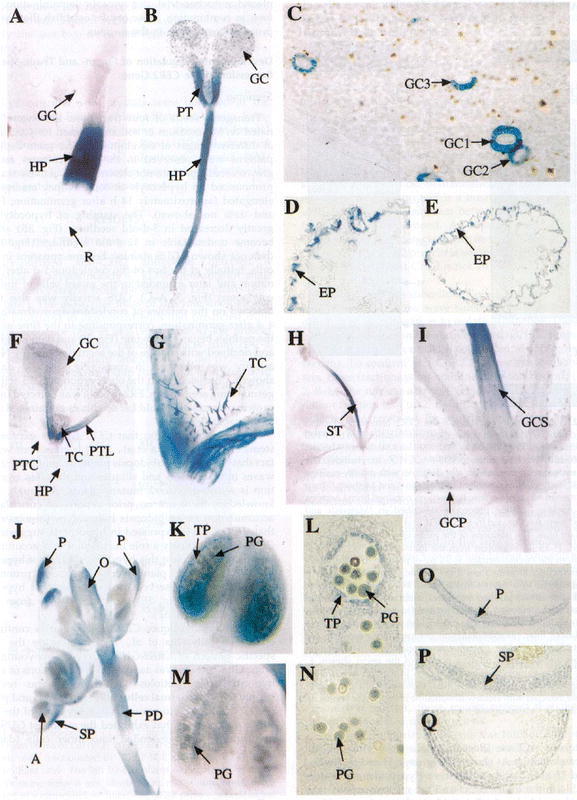
Staining pattern in transgenic plants containing a CER2-GUS fusion. In Panel A, one-day-old seedlings were germinated on MS medium and stained for GUS activity in the hypocotyl (HP) and guard cells (GC of the leaf). Panel B shows a five day-old seedling with GUS activity in HP, GC and petiole (PT). Panel C is a close-up of the cotyledon from a 5 day-old seedling showing the guard cell staining in the GC of both the adaxial (GC1, GC3) and abaxial (GC2) surfaces of the leaf. Panel D is a cross-section of the hypocotyl of a 5 day-old seedling showing GUS activity mostly in the epidermal layer (EP). Panel E is a cross section of a cotyledonary petiole from a 5 day-old seedling that also shows activity mostly in the EP. Panel F is a 12 day-old plant grown in soil showing GUS activity in the petiole of leaves (PTL), developing trichomes (TC), and GC. Note the lack of activity in the petioles of the cotyledon (PTC) and HP. Panel G is a close-up of the plant in F showing activity in the TC. Panel H is a 22 day-old plant showing strong GUS activity in the stem. Panel I is a close-up view of the plant shown in H that shows strong expression in the guard cells of the petiole (GCP) and stem (GCS). Panel J shows GUS activity in the petals (P), sepals (SP), anthers (A) ovaries (O) and pedicel (PD) of the developing inflorescence. Panel K is a stage 12 anther showing activity in the tapetal layer (TP) and pollen grain (PG). Panel L is a cross-section of a stage 12 anther showing activity in the TP and PG. Panels M and N are a stage 13 anther and cross section showing activity in the PG only. Panels O, P and Q are cross sections of the petal, sepal and ovary showing GUS activity in the EP of petals, across the section in sepals and in the EP of the ovary. Panel R shows activity in the HP of 2 day-old etiolated seedlings but no activity in the root (R). Panel S shows that only cells of the outside of the apical hook (AH) have GUS activity and that the cotyledon (CL) did not stain. Panel T shows staining in a 2 day-old seedling placed in MS medium supplemented with 0.5 mM sodium salicylate for 12 hours. Panel U, shows a 7 day-old seedling transferred to 4 mM BAP in which GUS activity can be seen in all cells of the leaves (TL) grown after BAP induction. Panel V, shows a 7 day-old seedling grown in the absence of BAP shows activity in the petioles and guard cells alone. Panel W is a cross-section of a leaf from a plant grown under the same conditions as the plant in panel U. Note: the staining in all layers differs from that in panel C. Panel X shows a lesser induction of GUS by BAP in a 15 day-old plant grown on BAP-supplemented medium when compared to panel U. Panel Y shows staining in a 10 day-old plant grown for 5 days in MS medium and transferred to MS supplemented with 4 mM BAP for 5 days. In the newly emerging leaves both the trichomes and epidermal cells stain. Taken from Xia et al. (1997).
The CER3 gene encodes a protein of 795 amino acids and is transcribed in roughly equal amounts in stems, leaves, roots, flowers, and apical meristems (Hannoufa et al., 1996). The predicted amino acid sequence of CER3 contains two domains found in alpha-type E3 ubiquitin-protein ligase from a range of organisms (Kwon et al., 1998). The first is a ring-H2 finger with a Cys/His domain implicated in the specificity of protein-protein interaction. The second, located at the COOH-terminal region is an Asp/Glu acid domain. CER3 is smaller than other E3 ubiquitin-protein ligases and is lacking certain domains normally found in these proteins, so if it is part of the ubiquitin pathway, it must act as part of a multi-component system. A heterodimeric NEDD8 family, E1 ubiquitin activating enzyme has been reported in Arabidopsis (Del Pozo et al., 1998). Moreover, the 45 kDa PRT1 gene of Arabidopsis, a protein known to participate in the N-end rule of protein degradation, is believed to encode an E3 ubiquitin-protein ligase smaller than those seen in other organisms. The unusual temporal and spacial expression pattern of CER3 suggests that it may regulate multiple aspects of plant development.
GLOSSY15 (GL15) regulates the transition from juvenile to adult shoot development, and affects a variety of leaf epidermal cell traits including epicuticular waxes, leaf hairs, and cell wall chemistry (Moose and Sisco, 1996). The GL15 gene was isolated using plant DNA flanking the site of insertion of a defective Suppressor-mutator (dSpm) element insertion as a transposon-tag. Transcripts from GL15 are found in the epidermal layer of juvenile leaves and the level of these mRNAs is regulated by upstream factors such as CORNGRASS1. The GL15 gene encodes a putative transcription factor with significant sequence similarity to the Arabidopsis regulatory genes APETALA2 and AINTEGUMENTA. Small but significant differences in wax composition exist among juvenile and adult growth phase rosette leaves, and cauline leaves, of Arabidopsis (Jenks, unpublished). Whether any of the existing Arabidopsis wax mutants have leaf wax alterations that occur in a leaf developmentally specific manner has not been investigated.
Millar et al. (1999) identified 15 Arabidopsis Expressed Sequence Tags (ESTs) (Newman et al. 1994; Cooke et al. 1996) with significant sequence similarity to Fatty Acid Elongation1 (FAE1), a gene involved in the synthesis of very long chain length fatty acids in seeds (James et al. 1995). The predicted amino acid sequence of these ESTs shared homology with those of condensing enzymes (Chalcone Synthase, Stilbene Synthases, and ß-ketoacyl-Acyl Carrier Protein Synthase III). The N-terminal hydrophobic regions of these genes appeared to be transmembrane domains. One of the genes coding FAE1-like proteins, CER6 (syn. CUT1), mapped to a location at 107 cM on chromosome one (Millar et al., 1999; Fiebig et al., 2000). A highly homologous gene, designated CER60, has exons that share 88% identity and 94% similarity with those of CER6, and occurs in a general region around 38 cM on chromosome one (Fiebig et al., 2000). Ten-fold more ESTs correspond to CER6 compared to CER60, suggesting that CER6 is expressed at higher levels than CER60. No mutant exists with alteration in the CER60 gene. Thirty-six out of 46 transgenic plants containing an antisense construct of CER6 had stems with a shiny bright green appearance, indicating reduction in the epicuticular wax layer (Millar et al., 1999). SEM failed to detect wax crystals on CER6 antisense plants. In addition to reduction in stem wax amount, 32 of these transformants had seedless short siliques whereas four were partially sterile, producing <200 seeds each. When the sterile transgenic plants were placed in a highly humid environment, 14 of them were able to set seed. This type of conditional male sterility has been previously associated with cer mutants in which the absence of waxes in the trypine layer of the pollen grain disrupts pollen–pistil interactions (Preuss et al. 1993). Similar as cer mutants, cross-pollination of the sterile antisense CER6 transgenics with wild-type pollen resulted in normal seed set. DNA sequencing of the cer6-1 mutant allele indicates that it has a three-base deletion eliminating the glutamate at the 319th codon. The cer6-2 is an A-to-C transversion that changes a histidine to a proline at codon 148. The cer6-1 deletion lies in one of the conserved regions identified by Millar et al. (1999). The H148P mutation in cer6-2 is not located in such a region, however, the new proline residue may alter the structure of the protein. Cer6-2 has the more pronounced phenotype. Fiebig et al. (2000) isolated six intragenic suppressors of the cer6-2 allele, cer6-2R, that partially restored pollen coat lipids but did not rescue the stem wax deposition defect. This demonstrates that low amounts of very long chain lipids are sufficient for pollen hydration and germination. Although these suppressors were isolated as independent mutants, DNA sequencing revealed the same reversion event in all the lines: i.e., a C-to-T transition that generated a ser at position 148. Like cer6, the kcs1 mutant has an alteration in a unique FAE1-like gene near the top of chromosome one that causes reduction in wax chain lengths (Todd et al., 1999). In contrast however, the kcs1 mutant did not have reduction in visual stem glaucousness or wax quantity relative to wildtype as did cer6, suggesting potential redundancy in elongation activity. The KCS1 gene is expressed in many tissues including stems, siliques, leaves, flowers, cotyledons, and roots. Whether KCS1 plays a role in metabolism of other non-wax lipids in roots and other tissues is yet unclear. Interestingly, the fiddlehead (fdh) mutant showing postgential fusion was caused by mutation in another FAE1 gene on chromosome two that is highly similar to CER6 and KCS1 (Yephremov et al., 1999; Pruitt et al., 2000). Unlike cer6 and kcs1, the major cuticular wax constituents on fdh stems appear unchanged from wildtype. Whether FDH plays a role in metabolism of other lipids, perhaps those playing a role in cell-cell communication, has not been established.
The GL8 gene encodes a 1.4-kb transcript present in wild-type seedling leaves and, in lesser amounts, in other organs and at other developmental stages (Xu et al., 1997). The amino acid sequence deduced from a cDNA of this gene exhibits highly significant sequence similarity to a group of enzymes from plants, eubacteria, and mammals that catalyzes the reduction of ketones. This finding and the chemical composition of the wax on the surface of the gl8 mutant suggests that the GL8 protein probably functions as a 3-ketoacyl-CoA reductase during fatty acid elongation in the cuticular wax biosynthetic pathway.
The CER1 gene, which encodes a novel protein involved in the deposition of long chain alkanes on the plant surface, was cloned after gene tagging with the heterologous maize transposable element system Enhancer-inhibitor, also known as Suppressor-mutator. Transcripts from this gene are present in stems, flowers, and fruits. A CER1-like gene, located next to CER1, is expressed in flowers but not in stems or fruits. The CER1 protein belongs to a group of integral membrane enzymes that process highly hydrophobic molecules. The presence of HX3H HX2HH and HX2HH motifs (where X stands for any amino acid) prompted Aarts et al. (1995) to speculate that CER1 may encode a decarbonylase enzyme (Aarts et al. 1995). The Zea ortholog of CER1 (GLOSSY1) does not have the third histidine rich domain. The rice (Oryza sativa L.) and Senecio odora [Forssk] Defl orthologs of CER1 are incomplete. Secondary structure predictions suggest that these proteins are integral membrane proteins with either 7 (CER1) or 5 (GL1) transmembrane helices on the NH2-terminal end of the protein and a water-soluble, globular cluster, predicted to reside on the cytoplasmic side at the COOH-terminal. This arrangement is similar to that found in a large family of integral membrane receptors involved in G-protein interactions via their COOH-terminal domains. A small subset of these receptors, called the ß-chemokine receptors, share homology to the plant CER1 orthologs. Rather than microsomal membranes however, Cheesbrough and Kolattukudy (1984) localized the decarbonylation activity to a cell wall/cuticle fraction. Together, these observations suggest that CER1 may be involved in transport (or secretion) of wax components rather than decarbonylation.
Functional genomic approaches may help identify additional genes directly or indirectly involved in the deposition of waxes on the plant surface. Recently, Irian and Lemieux (2002) undertook a gene expression survey of transcripts in stems of the Arabidopsis cer3 mutant and compared those to transcripts from wild-type stems. As CER3 is expressed in sub-epidermal layers of the stem, its product may be involved in controlling development of the epidermis. A total of 40,174 SAGE tags were collected from the stems of Arabidopsis Wassilewskija and BRL1 (a null allele of cer3) plants. These represented, potentially, more than 8,000 genes. Analysis of the tags with two or more representative members revealed: (i) 67% matched a known or predicted gene, (ii) 1.8% did not have a perfect 14-mer match, and potentially represented inter-ecotype polymorphism, (iii) 31.2% matched genomic sequences. The majority (93%) of the latter were within a distance of 500 bp of the 3´ of the predicted ORFs while the remaining 7% represented 11 novel or NORF (non-annotated open reading frames) genes for which predicted ORFs of 30 to 90 amino acids were identified by the authors. Four genes encoding lipid transfer proteins were among the most differentially expressed genes. The LTP3, LTP4, and LTP5 genes were more highly expressed in BRL1. These are only three of the more than 15 lipid transfer proteins identified in Arabidopsis (Arondel et al., 2000). In situ hybridization has revealed that, LTP3 is not restricted to the epidermis but is also expressed in subepidermal layers of the stem, immediately below the flowers (Clark and Bohnert, 1999). The fourth putative lipid transfer protein displaying differential transcript accumulation encodes the Arabidopsis homolog of the Zea GL1, a gene reported to be involved in wax metabolism in seedlings (Hansen et al., 1997). Transcripts from this gene could not be detected in BRL1. Sixteen differentially expressed genes belong to a wide variety of pathways ranging from several calcium binding proteins to transcription regulatory factors. The biological role of these gene products should be further investigated by reverse genetics (Krysan et al., 1999). The differences in the transcript levels of a large number of genes of the light-harvesting complex suggest a possible role for waxes in photoprotection. The two genes potentially related to photosynthesis, BCS1P and FERROCHELATASE-I, appeared strongly induced in BRL1 stems. AtMRP2 transcripts were moderately more abundant in BRL1 stems. This gene is involved in the transport of tetrapyrroles (derived from the catabolism of chlorophyll) to the vacuoles for degradation (Lu et al., 1998). Finally, three genes belonging to the photorespiratory (C2) cycle were more abundant in BRL1. The complex interactions that control how diverse genes and their associated pathways affect, or are affected by, wax metabolism needs further elucidation.
Ecology of Cuticular Waxes
Arabidopsis as a Model System to Study the Ecology of Cuticular Waxes
Plant cuticular waxes are important ecologically due to their location at the interface between the environment and the aerial soma of all terrestrial plants. Besides their role in preventing plant desiccation in water limiting environments (Chatterton et al., 1975; Schönherr, 1976; Riederer and Markstädter, 1996; Riederer and Schreiber, 2001), cuticular waxes may protect plants from supra-optimal levels of ultraviolet radiation (Reicosky and Hanover, 1978; Mulroy, 1979, mechanical damage (Eglinton and Hamilton 1967), freezing temperatures (Thomas and Barber 1974), phytopathogens (Jenks et al. 1994a; Carver et al. 1996), and herbivorous insects (Eigenbrode and Espelie 1995; Eigenbrode 1996). Diverse functions for plant waxes are suggested by their large intra- and inter-specific variation in crystalline morphology and chemistry (Jenks and Ashworth, 1999; Barthlott et al., 1998; Jeffree, 1996). Isogenic mutants of Brassica and Sorghum having altered cuticular waxes have been useful plant models for revealing the ecological functions of waxes (Blum, 1975; Eigenbrode et al., 1995, 1998; Jenks et al., 1994a). Similar ecological studies using the isogenic wax mutants of Arabidopsis however have received relatively little research focus. This chapter section will rely largely on unpublished studies, with which we shall attempt to show the potential of Arabidopsis wax mutants for working out the mechanisms of plant surface wax ecology.
Cuticle Waterproofing
Although it is practically beyond dispute that plant surface waxes are essential for waterproofing the cuticle, how cuticular waxes function in providing this water barrier is yet unclear. For example, little is known about the role that wax amount, composition, crystallization pattern, physico-chemical interactions between waxes and the cutin polymer of the cuticle membrane, or other wax properties play in plant water use. Previous studies have not shown clear associations between the rate of transpiration and the total amount of cuticular wax. In some studies, greater wax amount was associated with lower transpiration rates (O'Toole et al., 1979; Clarke and Richards, 1988; Johnson et al., 1983; Jordan et al., 1984; Premachandra et al., 1992, 1994; Lownds et al., 1993). Other studies however showed that greater wax amount had no effect on, or even a slight positive correlation with, transpiration rate (Bengston et al. 1978; Jefferson et al. 1989; Hadley and Smith, 1990; Araus et al., 1991; Riederer and Schreiber, 2001). Based on these studies, cuticular control over water loss appears to be more complicated than simple diffusion through a homogenous lipoidal cuticle layer. Nonetheless, cer-type mutations in Brassica (Denna, 1970; Eigenbrode, unpublished), Arabidopsis (Todd et al., 2000; Jenks, unpublished), and other plants (Jenks and Ashworth, 1999; Eigenbrode, unpublished) often exhibit increased susceptibility to limited soil or atmospheric water availability. For example, Todd et al. (2000) reported that a wax mutant kcs1 in Arabidopsis had reduced wax load, especially C26 and C30 alcohols, and was less tolerant of low humidity stress at a young age. Excised stem and whole plant in-pot transpiration studies revealed that some cer mutants lost water more rapidly than wildtype (Goodwin and Jenks, unpublished). Interestingly, there was no correlation between total wax or wax class/constituent quantity and the rate of water loss. In fact, some cer mutants with the highest water loss rates had wax amounts that were significantly higher than other cer lines having some of the lowest water loss rates. Recent studies in the Jenks lab reveal that the chemical composition or ultrastructure of the cuticle membrane itself, rather than wax only, was altered in certain of these high water loss cer mutants. Schönherr (1976a) showed that water flow through the cuticle might be directed through preferred sub-microscopic polar pathways. Potentially, more or larger polar pathways in a cuticle could lead to greater amounts (and rates) of water loss. The function of these polar pathways in water transport appears to be influenced by the amount and composition of intracuticular waxes. For example, removal of soluble waxes from cuticular membranes greatly increased membrane permeability (Grncarevic and Radler, 1967; Schönherr 1976a, 1976b; Knoche et al. 2000). Baur et al. (1999) provided evidence that waxes serve as cuticle matrix fillers that increase the path length or tortuosity of the diffusion pathways through which solutes travel in the cuticle framework created by the cutin polyester. Studies of water movement through artificial membranes showed that wax hydrocarbons, alcohols, and aldehydes effectively restricted water movement whereas acids restricted permeability only slightly and wax triterpenoid conjugates had no effect (Grncarevic and Radler, 1967). Moreover, alkanes, alcohols, ketones, and esters were shown to repel water interactions with the cuticle more than other wax constituents (Holloway 1969; Peter et al. 1987). Other studies suggest that both polar and non-polar pathways for solute movement are formed within the cuticle membrane by the organization of cuticle components having polar and non-polar functional groups (Schönherr 1976a; Hess and Foy 2000). Thus, water and other hydrophilic substances appear to pass through different cuticular pathways than lipophilic substances. There appears then to exist a complex association between intracuticular waxes and cutin meshwork that defines the water transport function of the cuticle membrane. Most previous work on cuticle barrier function has been done using isolated cuticles in vitro. Further studies are needed to investigate what role cuticle lipid structure and composition has on transpiration and subsequent plant drought tolerance in situ. The wax mutants of Arabidopsis should provide an excellent model system to investigate these questions.
Induction of Cuticular Waxes
Although studies using mutant isolines have not always displayed a clear correlation between wax quantity/composition and transpiration rate, additional circumstantial evidence for a role of plant wax in controlling transpiration rate is nonetheless provided by their inducibility in response to water stress. Leaves, bracts, and bolls of Gossypium hirsutum L. (cotton) produced up to 41% more total wax per area and longer-chain-length alkanes in drought stressed field plots than plants in irrigated plots (Bondada et al., 1996). Likewise, leaves of Rosa cultivars (rose) that developed during drought produced up to 14% more total wax per area and longer-chain-length alkanes (Williams et al., 2000; Jenks et al., 2001). Whether the relatively improved performance of these plants after removal of the drought stress was due to subsequent reduction in epidermal conductance to water vapor needs further examination. Similar drought treatments resulted in 25% more wax on Abutilon theophrasti Medic. (velvetleaf) (Levene and Owen, 1995), 23% more on Agropyron desertorum (Fisch.) Schult. (crested wheat grass) and 33% more on Medicago sativa L. (alfalfa) (Jefferson et al., 1989), and likewise various Triticum spp. (Johnson et al., 1983) and Sorghum (Premachandra et al., 1992) had more wax after exposure to drought stress. Low atmospheric humidity likewise appears to induce wax synthesis (Baker, 1974; Maier and Post-Beittenmiller, 1998). We recently showed, as for other plants species, that water stress results in increased wax quantity per stem surface area on Arabidopsis (Eigenbrode et al., unpublished). In our experiment, plants at the bolting stage were provided with either adequate water (15ml 50ml soil−1 plant−1) or reduced water (5ml 50ml soil−1 plant−1) daily for one week. All plants were then provided control levels of water (15ml 50ml soil−1 plant−1) for an additional week, and then harvested for analysis of cuticular waxes. Total wax amount increased 1.25-fold on the stems of water-stressed plants (Figure 8). This induction was strongest (1.6-fold) for long-chain fatty acids, intermediate for reduction products (primary alcohols) and least for decarbonylation products (alkanes, ketones, and secondary alcohols). Within each class, chain lengths were differentially induced. Potentially, the changes in cuticular waxes induced by water limiting environments reveal an ecological role for specific waxes to reduce water loss during drought. Wax inducibility, if genetic, would also reveal the presence of drought responsive elements in wax genes. New Arabidopsis mutants altered in the normal drought induction of wax could provide a means to dissect genetic mechanism regulating wax drought inducibility. Effective screening methods to isolate wax induction mutants are still needed.
Figure 8.
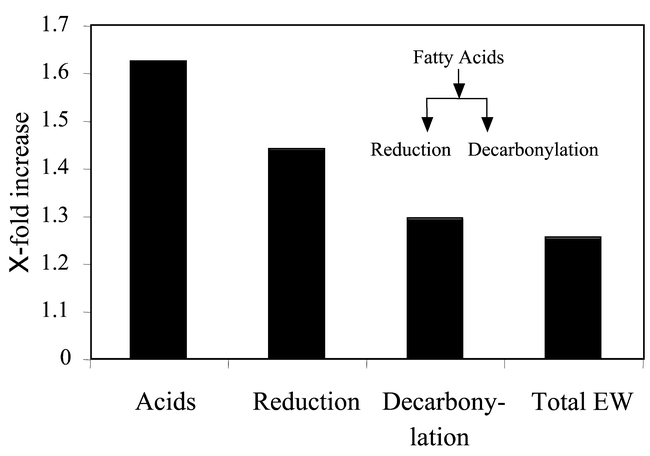
Water-stress treatment effects on inflorescence stem epicuticular waxes of Arabidopsis thaliana ecotype Landsberg erecta.
Interactions with Insects
Plant epicuticular wax morphology can affect insects (Eigenbrode, 1996; Eigenbrode and Espelie, 1995). The prominent waxy bloom on leaves of Brassica and Pisum sativum L. (pea) reduces attachment by leaf feeding beetles and provides protection against herbivory (Stork, 1980; Bodnaryk, 1992). The cer mutations in Brassica and Pisum reduced waxes and render plants vulnerable to damage by beetles (Stoner, 1990; White and Eigenbrode, 2000). Flea beetles (Chrysomelidae: Alticini) are important herbivores of Arabidopsis grown in the field (Mauricio et al., 1997, R. Mauricio, pers. comm.). Whether cer mutations of Arabidopsis affect flea beetle susceptibility has not been tested. In the field, waxy bloom, on at least two plant species, affects predatory insects, apparently leading to wax-mediated differences in the densities of their prey (Eigenbrode, 1996; Eigenbrode et al., 1998; 2000). This complex multitrophic effect of plant waxes on insect herbivores has not been tested using Arabidopsis cer mutants.
Plant wax chemical composition can influence host acceptance for feeding and oviposition by insect herbivores (reviewed in Eigenbrode and Espelie, 1995; with recent examples in Cervantes et al., in press; and Morris et al., 2000). Arabidopsis cer mutations, which alter wax composition, could help elucidate the role of specific wax components that influence insect behavior. Oviposition by the diamondback moth, Plutella xylostella L. appears to be influenced by the chemical composition of Arabidopsis waxes. Moths lay more eggs on cer4 plants than on cer2 plants in the same background (Eigenbrode unpublished, Figure 9). Waxes from cer4 plants extracted and deposited on aluminum foil models also receive more eggs than models treated with wild-type waxes. Compositional differences between cer4 and cer2 waxes, possibly their greatly divergent primary alcohol compositions, may influence the moth's oviposition behavior. The cabbage aphid, Brevicoryne brassicae L., shows evidence of an antixenotic reaction to waxes from the cer3 Arabidopsis mutant. The aphids probe less and walk more on the stems of cer3 plants (Figure 10) (Rashotte, 1999). This response is associated with reduced fecundity of the aphids on cer3, suggesting that reduced acceptance of the host leads to decreased performance. Waxes of cer3 plants extracted and deposited on wild-type plants also caused increased walking and apparent decreased acceptance of the plants by cabbage aphids. Cer3 waxes have notably increased C30 primary alcohol (triacontanol) amounts as compared with the other Arabidopsis types tested. Application of physiological concentrations of pure triacontanol to wild-type plants caused increased walking by the aphids. Thus, the characterized cer3 mutation helped to identify a chemical repellent for the cabbage aphid present in surface waxes of Arabidopsis. A set of diverse Arabidopsis ecotypes varying slightly in surface wax characteristics, but not in triacontanol concentration, did not differ in acceptability for the cabbage aphid (Rashotte et al., 1997, Rashotte, 1999).
Figure 9.
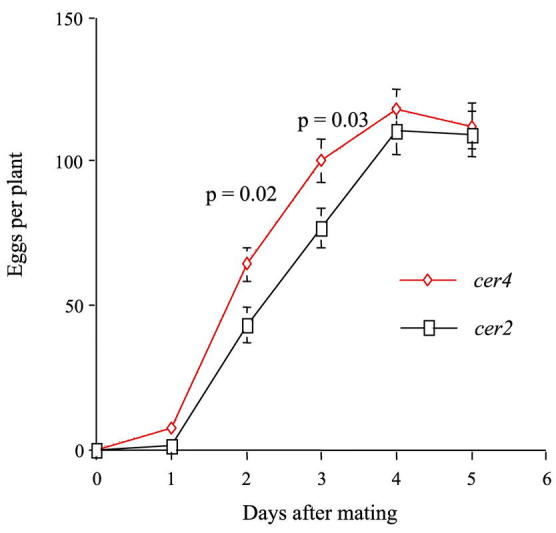
Oviposition by diamondback moth (Plutella xylostella) on cer2 and cer4 mutants of Arabidopsis thaliana ecotype Wassilewskija.
Figure 10.
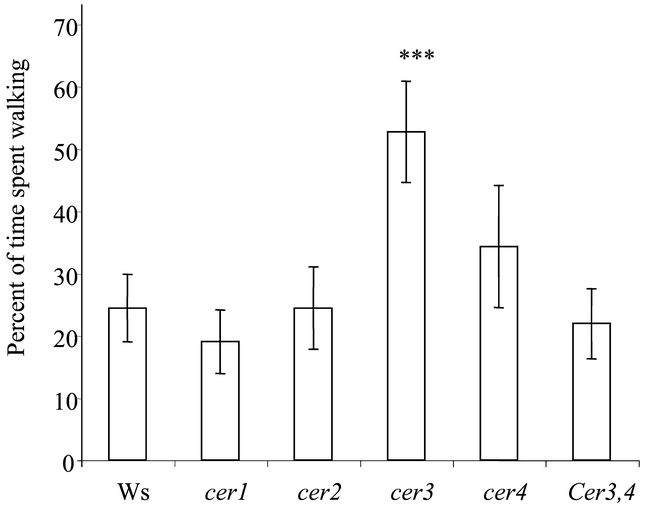
Walking time by cabbage aphid (Brevicoryne brassica) on wild-type Arabidopsis ecotype Wassilewskija and five cer mutants (Rashotte, 1999), including a new double wax loci mutant cer3,4.
Other ecological roles of cuticular waxes
The roles of Arabidopsis cuticular waxes in providing protection from UV and cold temperatures, and in mediating interactions with pathogens have not been examined. As the cer mutants and others like them are better characterized, they should become increasingly useful for testing hypotheses about the diverse ecological roles of cuticular waxes in Arabidopsis, and provide a model system for revealing similar plant-environment interactions in other plants systems.
Footnotes
Citation: Jenks M.A., Eigenbrode S.D., and Lemieux B. (2002) Cuticular Waxes of Arabidopsis. The Arabidopsis Book 1:e0016. doi:10.1199/tab.0016
elocation-id: e0016
Published on: August 12, 2002
Figure 7.
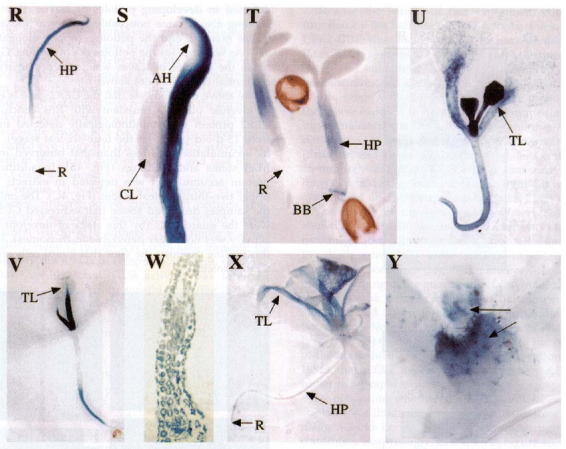
Continued.
Table 1.
Arabidopsis thaliana cuticular wax mutants grouped by distinguishing alteration in chemical composition of cuticular waxes on inflorescence stems.
References Cited
- Aarts M. G., Keijzer C. J., Stiekema W. J., Pereira A. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell. 1995;71(1):2115–2127. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson A. W. Physical chemistry of surfaces. 3rd edition. 1990. John Wiley and Sons, New York.
- Agrawal V. P., Lessire R., Stumpf P. K. Biosynthesis of very-long-chain fatty acids in microsomes of epidermal cells of Allium porrum L. Archives of Biochemistry and Biophysics. 1984;2301(1):580–589. doi: 10.1016/0003-9861(84)90438-7. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz I. A., O'Kane S. L. Placement of Arabidopsis parvula in Thellungiella (Brassicaceae). NOVON. 1995;51(1):309–310. [Google Scholar]
- Al-Shehbaz I. A. The genera of Sisymbrieae (Cruciferae; Brassicaceae) in the Southeastern United States. J. Arnold Arboretum. 1988;691(1):213–237. [Google Scholar]
- Araus J. L., Febrero A., Vendrell P. Epidermal conductance in different parts of durum wheat grown under mediterranean conditions: the role of epicuticular waxes and stomata. Plant Cell Environ. 1991;141(1):545–558. [Google Scholar]
- Arondel V., Vergnolle C., Cantrel C., Kader J. C. Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci. 2000;1571(1):1–12. doi: 10.1016/s0168-9452(00)00232-6. [DOI] [PubMed] [Google Scholar]
- Avato P., Mikkelsen J. D., Wettstein-Knowles Pvon. Synthesis of epicuticular primary alcohols and intracellular fatty acids by tissue slices from cer-j59 barley leaves. Carlsberg Res. Commun. 1982;471(1):377–390. [Google Scholar]
- Baker E. A. The influence of environment on leaf wax development in Brassica oleracea var. gemmifera. New Phytol. 1974;731(1):955–966. [Google Scholar]
- Barinaga M. Secrets of secretion revealed. Science. 1993;2601(1):487–489. doi: 10.1126/science.8475382. [DOI] [PubMed] [Google Scholar]
- Barthlott W., Neinhuis C., Cutler D., Ditsch F., Muesel I., Theisen I., Wilhelm H. Classification and terminology of plant epicuticular waxes. Bot. J. Linn. Soc. 1998;1261(1):237–260. [Google Scholar]
- Baur P., Marzouk H., Schönherr J. Estimation of path lengths for diffusion of organic compounds through leaf surfaces. Plant Cell Environ. 1999;221(1):291–299. [Google Scholar]
- Becraft P. W., Kang S. H., Suh S. G. The maize CRINKLY4 receptor kinase controls a cell-autonomous differentiation response. Plant Physiol. 2001;1271(1):486–496. [PMC free article] [PubMed] [Google Scholar]
- Bengtson S., Larsson S., Liljenberg C. Effects of water stress on cuticular transpiration rate and amount and composition of epicuticular wax in seedlings of six oat varieties. Physiol. Plant. 1978;441(1):319–324. [Google Scholar]
- Bessoule J. J., Lessire R., Cassagne C. Partial purification of the acyl-CoA elongase of Allium porrum leaves. Arch. Biochem. Biophys. 1989;2681(1):475. doi: 10.1016/0003-9861(89)90315-9. [DOI] [PubMed] [Google Scholar]
- Bianchi G., Avato P., Salamini F. Glossy mutants of maize. IX. Chemistry of glossy4, glossy8, glossy15 and glossy18 surface waxes. Heredity. 1979;421(1):391–395. [Google Scholar]
- Bianchi A., Bianchi G. Surface lipid composition of C3 and C4 plants. Biochem. System. Ecol. 1990;181(1):533–537. [Google Scholar]
- Blum A. Effect of bm genes on epicuticular wax and the water relations of Sorghum bicolor. Isr. J. Bot. 1975;241(1):50. [Google Scholar]
- Bodnaryk R. P. Distinctive leaf feeding patterns on oilseed rapes and related Brassicaceae by flea beetles, Phyllotreta cruiciferae (Goeze) (Coleoptera: Chrysomelidae). Can. J. Plant Sci. 1992;721(1):575–581. [Google Scholar]
- Bondada B. R. Effect of water stress on the epicuticular wax composition and ultrastucture of cotton (Gossypium hirsutum L.) leaf, bract and boll. Environ. Exp. Bot. 1996;361(1):61–69. [Google Scholar]
- Bowman J. Arabidopsis. An Atlas of Morphology and Development. 1993;1(1):450. Springer-Verlag, NY. Pp. [Google Scholar]
- Carver T. L. W., Ingerson S. M., Thomas B. J. Influences of host surface features on the development of Erysiphe graminis and Erysiphe pisi. 1996;1(1):255–266. In: Kertiens G (ed). Plant Cuticles, An Integrated Functional Approach. BIOS, Oxford, UK, pp. [Google Scholar]
- Cervantes D., Eigenbrode S. D., Ding H., Bosque-Perez N. Oviposition preference of Hessian fly, Mayetiola destructor, on winter wheats varying in surface waxes. J. Chem. Ecol. 2002. (in print) [DOI] [PubMed]
- Chatterton N. J., Hanna W. W., Powell J. B., Lee D. R. Photosynthesis and transpiration in bloom and bloomless sorghum. Can. J. Plant Sci. 1975;551(1):641–643. [Google Scholar]
- Cheesbrough T. M., Kolattukudy P. E. Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum. Proc. Nat. Acad. Sci. 1984;811(1):6613–6617. doi: 10.1073/pnas.81.21.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. M., Bohnert H. J. Cell-specific expression of genes of the lipid transfer protein family from Arabidopsis thaliana. Plant Cell Physiol. 1999;401(1):69–76. doi: 10.1093/oxfordjournals.pcp.a029476. [DOI] [PubMed] [Google Scholar]
- Clark J. M., McCaig T. M., DePauw R. M. Inheritance of glaucousness and epicuticular wax in Durum wheat. Crop Sci. 1994;341(1):327–330. [Google Scholar]
- Clark J. M., Richards R. A. The effects of glaucousness, epicuticular wax, leaf age, plant height, and growth environment on water loss rates of excised wheat leaves. Can. J. Plant Sci. 1988;681(1):975–982. [Google Scholar]
- Clark J. A., Levit J. The basis of drought resistance in the soybean plant. Physiol. Plant. 1956;91(1):598–606. [Google Scholar]
- Cooke R. Further progress toward a catalogue of all Arabidopsis genes: Analysis of a set of 5000 non-redundant ESTs. Plant J. 1996;91(1):101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- Dellaert L. M. W. Eceriferum mutants in Arabidopsis thaliana (L.) Heynh. II. Phenotypic and genetic analysis. Arabid. Inf. Serv. 1979;161(1):10–26. [Google Scholar]
- Del Pozo J. C., Timpte C., Tan S., Callis J., Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;2801(1):1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Denna D. W. Transpiration and the waxy bloom in Brassica oleracea L. Aust. J. Biol. Sci. 1970;231(1):27–31. [Google Scholar]
- Eglinton G., Hamilton R. J. Leaf epicuticular waxes. Science. 1967;1561(1):1322–1335. doi: 10.1126/science.156.3780.1322. [DOI] [PubMed] [Google Scholar]
- Eigenbrode S. D., Rayor L., Chow J., Latty P. Effects of wax bloom variation in Brassica oleracea on foraging by a vespid wasp. Entomol. Exp. Appl. 2000;971(1):161–166. [Google Scholar]
- Eigenbrode S. D., Pillai S. K. Neonate Plutella xylostella L. responses to surface wax components of a resistant cabbage (Brassica oleracea L.). J. Chem. Ecol. 1998;241(1):1611–1627. [Google Scholar]
- Eigenbrode S. D. Plant surface waxes and insect behaviour. 1996;1(1):201–222. In: Kerstiens G (ed). Plant cuticles: an intergrated functional approach. Bios Press, Oxford, pp. [Google Scholar]
- Eigenbrode S. D., Espelie K. E. Effects of plant epicuticular lipids on insect herbivores. Ann. Rev. Entomol. 1995;401(1):171–194. [Google Scholar]
- Fiebig A., Mayfield J. A., Miley N. L., Chau S., Fischer R. L., Preuss D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell. 2000;121(1):2001–2008. doi: 10.1105/tpc.12.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grncarevic M., Radler F. The effect of wax components on cuticular transpiration - model experiments. Planta. 1967;751(1):23–27. doi: 10.1007/BF00380835. [DOI] [PubMed] [Google Scholar]
- Hadley J. L., Smith W. K. Influence of leaf surface wax and leaf area to water content ratio on cuticular transpiration in western conifers, U.S.A. Can. J. For. Res. 1990;201(1):1306–1311. [Google Scholar]
- Hannoufa A., Negruk V., Eisner G., Lemieux B. The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. Plant J. 1996;101(1):459–467. doi: 10.1046/j.1365-313x.1996.10030459.x. [DOI] [PubMed] [Google Scholar]
- Hansen J. D., Pyee J., Xia Y., Wen T. J., Robertson D. S., Kolattukudy P. E., Nikolau B. J., Schnable P. S. The GLOSSY1 locus of maize and an epidermis-specific cDNA from Kleinia odora define a class of receptor-like proteins required for the normal accumulation of cuticular waxes. Plant Physiol. 1997;1131(1):1091–1100. doi: 10.1104/pp.113.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque M. M., Mackill D. J., Ingram K. T. Inheritance of leaf epicuticular wax content in rice. Crop Sci. 1992;321(1):865–868. [Google Scholar]
- Hess F. D., Foy C. L. Interaction of surfactants with plant cuticles. Weed Technol. 2000;141(1):807–813. [Google Scholar]
- Holloway P. J. Chemistry of leaf waxes in relation to wetting. J. Sci. Fd. Agri. 1969;201(1):124–128. [Google Scholar]
- Irian S., Lemieux B. Comparative expression analysis of stems from Wassilewskija and the eceriferum3 mutant BRL1. Genome Biology. 2002. (in prep)
- James D. W., Jr, Lim E., Keller J., Plooy I., Ralston E., Dooner H. K. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell. 1995;71(1):309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson P. G. Genetic variation for epicuticular wax production in Altai wildrye populations that differ in glaucousness. Crop Sci. 1994;341(1):367–371. [Google Scholar]
- Jefferson P. G., Johnson D. A., Rumbaugh M. D., Asay K. H. Water stress and genotypic effects on epicuticular wax production of alfalfa and crested wheatgrass in relation to yield and excised leaf water loss rate. Can. J. Plant Sci. 1989;691(1):481–490. [Google Scholar]
- Jeffree C. E. Structure and ontogeny of plant cuticles. 1996;1(1):33–82. In: Kerstiens G (ed). Plant Cuticles, An Integrated Functional Approach. BIOS Scientific Pubishers, Oxford, pp. [Google Scholar]
- Jeffree C. E., Baker E. A., Holloway P. J. Origins of the fine structure of plant epicuticular waxes. 1976;1(1):118–158. In: Dickinson, C.H., and Preece, T.F. (eds). Microbiology of Aerial Plant Surfaces. Academic Press, London, pp. [Google Scholar]
- Jenks M. A., Anderson L., Teusink R., Williams M. H. Leaf cuticular waxes of potted rose cultivars as effected by plant development, drought and paclobutrazol treatments. Physiol. Plant. 2001;1121(1):61–69. doi: 10.1034/j.1399-3054.2001.1120109.x. [DOI] [PubMed] [Google Scholar]
- Jenks M. A., Rich P. J., Rhodes D., Ashworth E. A., Axtell J. D., Ding C. K. Chemical composition of leaf sheath cuticular waxes on bloomless and sparse-bloom mutants of Sorghum bicolor (L.) Moench. Phytochemistry. 2000;541(1):577–584. doi: 10.1016/s0031-9422(00)00153-9. [DOI] [PubMed] [Google Scholar]
- Jenks M. A., Ashworth E. N. Plant epicuticular waxes: function, production, and genetics. 1999;1(1):1–68. In: Janick J (ed). Horticultural Reviews, Vol. 23. John Wiley and Sons, Inc., New York, pp. [Google Scholar]
- Jenks M. A., Tuttle H. A., Rashotte A. M., Feldmann K. A. Mutants in Arabidopsis altered in epicuticular waxes and leaf morphology. Plant Physiol. 1996a;1101(1):377–385. doi: 10.1104/pp.110.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M. A., Tuttle H. A., Feldmann K. A. Changes in epicuticular waxes on wildtype and eceriferum mutants in Arabidopsis during development. Phytochemistry. 1996b;421(1):29–34. [Google Scholar]
- Jenks M. A., Tuttle H. A., Eigenbrode S. D., Feldmann K. A. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 1995;1081(1):369–377. doi: 10.1104/pp.108.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M. A., Joly R. J., Peters P. J., Rich P. J., Axtell J. D., Ashworth E. A. Chemically-induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol. 1994a;1051(1):1239–1245. doi: 10.1104/pp.105.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks M. A., Rich P. J., Ashworth E. N. Involvement of cork cells in the secretion of filaments on Sorghum bicolor (L.) Moench. Int. J. Plant Sci. 1994b;1551(1):506–518. [Google Scholar]
- Jenks M. A., Rich P. J., Peters P. J., Axtell J. D., Ashworth E. N. Epicuticular wax morphology of bloomless (bm) mutants in Sorghum bicolor. Int. J. Plant Sci. 1992;1531(1):311–319. [Google Scholar]
- Jetter R., Schäffer S., Riederer M. Leaf cuticular waxes are arranged in chemically and mechanically distinct layers: evidence from Prunus laurocerasus L. Plant, Cell Environ. 2000;231(1):619–628. [Google Scholar]
- Jetter R., Riederer M. In vitro reconstitution of epicuticular wax crystals: formation of tubular aggregates by long-chain secondary alkanediols. Bot. Acta. 1995;1081(1):111–120. [Google Scholar]
- Jin P., Guo T., Becraft P. W. The maize CR4 receptor-kinase mediates a growth factor-like differentiation response. Genesis. 2000;271(1):104–116. doi: 10.1002/1526-968x(200007)27:3<104::aid-gene30>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Johnson D. A., Richards R. A., Turner N. C. Yield, water relations, gas exchange and surface reflectances of near-isogenic wheat lines differing in glaucousness. Crop Sci. 1983;231(1):318–325. [Google Scholar]
- Jordan W. R., Shouse P. J., Blum A., Miller F. R., Monk R. C. Environmental physiology of sorghum. II. Epicuticular wax load and cuticular transpiration. Crop Sci. 1984;241(1):1168–1173. [Google Scholar]
- Knoche M., Peschel S., Hinz M., Bukovac M. J. Studies on water transport through the sweet cherry fruit surface: characterizing conductance of the cuticular membrane using pericarp segments. Plant. 2000;2121(1):127–135. doi: 10.1007/s004250000404. [DOI] [PubMed] [Google Scholar]
- Kolattukudy P. E. Biosynthetic pathways of cutin and waxes, their sensitivity to environmental stresses. 1996;1(1):83–108. In: Plant Cuticles, An Integrated Functional Approach, Kersteins G (ed), BIOS Scientific Publishers Ltd., Oxford. pp. [Google Scholar]
- Kolattukudy P. E. Mechanisms of synthesis of waxy esters in broccoli (Brassica oleracea). Biochemistry. 1967;61(1):2705–2717. doi: 10.1021/bi00861a010. [DOI] [PubMed] [Google Scholar]
- Kolattukudy P. E., Buckner J. S., Liu T. Y. J. Biosynthesis of secondary alcohols and ketones from alkanes. Arch. Biochem. Biophys. 1973;1561(1):613–620. doi: 10.1016/0003-9861(73)90312-3. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., Thiel F. A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis thaliana. J. Hered. 1989;801(1):118–122. [Google Scholar]
- Krysan P. J., Young J. C., Sussman M. R. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;111(1):2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. T., Reiss Y., Fried V. A., Hershko A., Yoon J. K., Gonda D. K., Sangan P., Copeland N. G., Jenkins N. A., Varshavsky A. The mouse and human genes encoding the recognition component of the N-end rule pathway. PNAS. 1998;951(1):7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene B. C., Owen M. D. K. Effect of moisture stress and leaf age on bentazon absorption in common cocklebur (Xanthium strumarium) and velvetleaf (Abutilon theophrasti). Weed Sci. 1995;431(1):7–12. [Google Scholar]
- Liu D., Post-Beittenmiller D. Discovery of an epidermal stearoyl-acyl carrier protein thioesterase: its potential role in wax biosynthesis. J. Biol. Chem. 1995;2701(1):16962–16969. doi: 10.1074/jbc.270.28.16962. [DOI] [PubMed] [Google Scholar]
- Lolle S. J., Hsu W., Pruitt R. E. Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics. 1998;1491(1):607–619. doi: 10.1093/genetics/149.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lownds N. K., Banaras M., Bosland P. W. Relationships between postharvest water loss and physical properties of pepper fruit (Capsicum annuum L.). HortScience. 1993;281(1):1182–1184. [Google Scholar]
- Lu Y. P., Li Z. S., Drozdowicz Y. M., Hortensteiner S., Martinoia E., Rea P. A. AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with AtMRP1. Plant cell. 1998;101(1):267–282. doi: 10.1105/tpc.10.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist U., Lundqvist A. Mutagen specificity in barley for 1580 eceriferum mutants localized to 79 loci. Hereditis. 1988;1081(1):1–12. [Google Scholar]
- Macey M. K., Barber H. N. Chemical genetics of wax formation on leaves of Pisum sativum. Phytochemistry. 1970a;91(1):5–12. [Google Scholar]
- Macey M. J. K., Barber H. N. Chemical genetics of wax formation on leaves of Brassica oleracea. Phytochemistry. 1970b;91(1):13–23. [Google Scholar]
- Maier C. G. A., Post-Beittenmiller D. Epicuticular wax on leek in vitro developmental stages and seedlings under varied growth conditions. Plant Sci. 1998;1341(1):53–67. [Google Scholar]
- Mauricio R. M., Rausher M. D., Burdick D. S. Variation in the defense strategies of plants: are resistance and tolerance mutually exclusive? Ecology. 1997;781(1):1301–1311. [Google Scholar]
- McNevin J. P., Woodward W., Hannoufa A., Feldmann K. A., Lemieux B. Isolation and characterization of eceriferum (cer) mutants induced by T-DNA insertions in Arabidopsis thaliana. Genome. 1993;361(1):610–618. doi: 10.1139/g93-082. [DOI] [PubMed] [Google Scholar]
- Mikkelson J. D., Wettstein-Knowles Pvon. Biosynthesis of b-diketones and hydrocarbons in barley spike epicuticular wax. Arch. Biochem. Biophys. 1978;12881(1):178–181. doi: 10.1016/0003-9861(78)90370-3. [DOI] [PubMed] [Google Scholar]
- Millar A. A., Clemens S., Zachgo S., Giblin E. M., Taylor D. C., Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;111(1):825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P. J., Swords K. M., Lynch L. A., Staehelin A. Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus of plants. J. Cell. Biol. 1991;1121(1):589–602. doi: 10.1083/jcb.112.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose S. P., Sisco P. H. GLOSSY15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 1996;101(1):3018–3027. doi: 10.1101/gad.10.23.3018. [DOI] [PubMed] [Google Scholar]
- Morris B. D., Foster S. P., Harris M. O. Identification of 1-octacosanal and 6-methoxy-2-benzoxazolinone from wheat as ovipositional stimulants for Hessian fly, Mayetiola destructor. J. Chem. Ecol. 2000;261(1):859–867. [Google Scholar]
- Mulroy T. W. Spectral properties of heavily glaucous and non-glaucous leaves of a succulent rosette-plant. Oecologia. 1979;381(1):349–357. doi: 10.1007/BF00345193. [DOI] [PubMed] [Google Scholar]
- Negruk V., Yang P., Subramanian M., McNevin J. P., Lemieux B. Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J. 1996a;91(1):137–145. doi: 10.1046/j.1365-313x.1996.09020137.x. [DOI] [PubMed] [Google Scholar]
- Negruk V., Eisner G., Lemieux B. Addition-deletion mutations in transgenic Arabidopsis thaliana generated by the seed co-cultivation method. Genome. 1996b;391(1):1117–1122. doi: 10.1139/g96-140. [DOI] [PubMed] [Google Scholar]
- Newman T., De Bruijn F. J., Green P., Keegstra K., Kende H., McIntosh L., Ohlrogge J., Raikhel N., Somerville S., Thomashow M., Retzel E., Somerville C. Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;1061(1):1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J. B., Jaworski J. G. Regulation of fatty acid synthesis. Ann. Rev. Plant Physiol. Mol. Biol. 1997;481(1):109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- O'Reilly C., Arnott J. T., Owens J. N. Effects of photoperiod and moisture availability on shoot growth, seedling morphology, and cuticle and epicuticular wax features of container-grown western hemlock seedlings. Can. J. For. Res. 1989;191(1):122–131. [Google Scholar]
- Ortiz R., Vuylsteke D., Ogburia N. M. Inheritance of pseudostem waxiness in banana and plantain (Musa spp.). J. Hered. 1995;861(1):297–299. [Google Scholar]
- O'Toole J. C., Cruz R. T., Seiber J. N. Epicuticular wax and cuticular resistance in rice. Physiol. Plant. 1979;471(1):239–244. [Google Scholar]
- Peter J., Stevens G., Baker E. A. Factors affecting the foliar absorption and redistribution of pesticides. 1. Properties of the leaf surfaces and their interactions with spray droplets. Pestic. Sci. 1987;191(1):265–281. [Google Scholar]
- Post-Beittenmiller D. Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;471(1):405–430. doi: 10.1146/annurev.arplant.47.1.405. [DOI] [PubMed] [Google Scholar]
- Premachandra G. S., Hahn D. T., Axtell J. D., Joly R. J. Epicuticular wax load and water-use efficiency in bloomless and sparse-bloom mutants of Sorghum bicolor L. Environ. Exp. Bot. 1994;341(1):293–301. [Google Scholar]
- Premachandra G. S., Saneoka H., Kanaya M., Ogata S. Cell membrane stability and leaf surface wax content as affected by increasing water deficits in maize. J. Exp. Botany. 1991;421(1):167–171. [Google Scholar]
- Preuss D., Lemieux B., Yen G., Davis R. W. A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Develop. 1993;71(1):974–985. doi: 10.1101/gad.7.6.974. [DOI] [PubMed] [Google Scholar]
- Pruitt R. E., Vielle-Calzada J. P., Ploense S. E., Grossniklaus U., Lolle S. J. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. PNAS. 2000;971(1):1311–1316. doi: 10.1073/pnas.97.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyee J., Yu H., Kolattukudy P. E. Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biochem. Biophys. 1994;3111(1):460–468. doi: 10.1006/abbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- Rashotte A. M., Jenks M. A., Feldmann K. A. Cuticular waxes on eceriferum mutants of Arabidopsis thaliana. Phytochemistry. 2001;571(1):115–123. doi: 10.1016/s0031-9422(00)00513-6. [DOI] [PubMed] [Google Scholar]
- Rashotte A. M. Epicuticular wax in Arabidopsis thaliana: a study of the genetics, chemistry, structure, and interactions with insects. 1999. Ph.D. Dissertation, Plant Science. University of Arizona, Tucson.
- Rashotte A. M., Feldmann K. A. Correlations between epicuticular wax structures and chemical composition in Arabidopsis thaliana. Int. J. Plant Sci. 1998;1591(1):773–779. [Google Scholar]
- Rashotte A. M., Jenks M. A., Nguyen T. D., Feldmann K. A. Epicuticular wax variation in ecotypes of Arabidopsis thaliana. Phytochemistry. 1997;451(1):251–255. doi: 10.1016/s0031-9422(96)00792-3. [DOI] [PubMed] [Google Scholar]
- Reicosky D. A., Hanover J. W. Physiological effects of surface waxes. 1. Light reflectance for glaucous and non-glaucous Picea pungens. Plant Physiol. 1978;621(1):101–104. doi: 10.1104/pp.62.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M., Schreiber L. Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp. Bot. 2001;521(1):2023–2032. doi: 10.1093/jexbot/52.363.2023. [DOI] [PubMed] [Google Scholar]
- Riederer M., Markstadter C. Cuticular waxes: a critical assessment of current knowledge. 1996;1(1):189–200. In: Plant Cuticles, An Integrated Functional Approach, Kersteins G (ed), BIOS Scientific Publishers Ltd., Oxford. pp. [Google Scholar]
- Rollins R. C. The Cruciferae of Continental North America. 1993. Stanford University Press, California.
- Schönherr J. Water permeability of isolated cuticular membranes: The effect of cuticular waxes on diffusion of water. Planta. 1976a;1311(1):159–164. doi: 10.1007/BF00389989. [DOI] [PubMed] [Google Scholar]
- Schönherr J. Water permeability of isolated cuticular membranes: The effect of pH and cations on diffusion, hydrodynamic permeability, and size of polar pores in the cutin matrix. Planta. 1976b;1281(1):113–126. doi: 10.1007/BF00390312. [DOI] [PubMed] [Google Scholar]
- Sieber P., Schorderet M., Ryser U., Buchala A., Kolatukudy P., Metraux J. P., Nawrath C. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell. 2000;121(1):721–737. doi: 10.1105/tpc.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner K. A. Glossy leaf wax and host-plant resistance to insects in Brassica oleracea L. under natural infestation. Environ. Entomol. 1990;191(1):730–739. [Google Scholar]
- Stork N. E. Role of waxblooms in preventing attachment to brassicas by the mustard beetle, Phaedon cochleariae. Entomol. Exp. Appl. 1980;281(1):100–107. [Google Scholar]
- Tacke E., Korfhage C., Michel D., Maddaloni M., Motto M., Lanzini S., Salamini F., Doring H. P. Transposon tagging of the maize GLOSSY2 locus with the transposable element En/Spm. Plant J. 1995;81(1):907–917. doi: 10.1046/j.1365-313x.1995.8060907.x. [DOI] [PubMed] [Google Scholar]
- Teusink R. S., Rahman M., Bressan R. A., Jenks M. A. Cuticular waxes on Arabidopsis thaliana close-relatives Thellungiella halophila and Thellungiella parvula. Int. J. Plant Sci. 2001. (in print)
- Thomas D. A., Barber H. N. Studies on leaf characteristics of a cline of Eucalyptus urnigera from Mount Wellington, Tasmania. I. Water repellency and the freezing of leaves. Aust. J. Bot. 1974;221(1):501–512. [Google Scholar]
- Todd J., PostBeittenmiller D., Jaworski J. G. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;171(1):119–126. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- Vioque J., Kolattukudy P. E. Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch. Biochem. Biophys. 1997;3401(1):64–72. doi: 10.1006/abbi.1997.9932. [DOI] [PubMed] [Google Scholar]
- Walton T. J. Waxes, cutin, and suberin. 1990;41(1):105–158. In: Methods in Plant Biochemistry. Academic Press Limited, New York. [Google Scholar]
- Wellesen K., Durst F., Pinot F., Benveniste I., Nettesheim K., Wisman E., Steiner-Lange S., Saedler H., Yephremov A. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid w-hydroxylation in development. PNAS. 2001;981(1):9694–9699. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein-Knowles Pvon. Biosynthesis and genetics of waxes. 1995;1(1):91–129. In R.J. Hamilton, ed, Waxes: Chemistry, Molecular Biology and Functions, The Oily Press LTD, Dundee, Scotland, pp. [Google Scholar]
- Wettstein-Knowles Pvon. Effects of inhibitors on synthesis of esterified alkan-2-ols in barley spike epicuticular wax. Carlsberg Res. Commun. 1985;501(1):239–262. [Google Scholar]
- Wettstein-Knowles Pvon. Elongases and epicuticular wax biosynthesis. Physiol. Veg. 1982;201(1):797–809. [Google Scholar]
- Wettstein-Knowles Pvon. Ultrastructure and origin epicuticular wax tubes. J. Ultrastruct. Res. 1974;461(1):483–498. doi: 10.1016/s0022-5320(74)90069-0. [DOI] [PubMed] [Google Scholar]
- White C., Eigenbrode S. D. Effects of surface wax variation in Pisum sativum on herbivorous and entomophagous insects in the field. Environ. Entomol. 2000;291(1):773–780. [Google Scholar]
- Williams M. H., Rosenqvist E., Buchhave M. The effect of reducing production water availability on the post-production quality of potted miniature roses (Rosa x hybrida). Postharv. Biol. Technol. 2000;181(1):143–150. [Google Scholar]
- Xia Y., Nikolau B. J., Schnable P. S. Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol. 1997;1151(1):925–937. doi: 10.1104/pp.115.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Nikolau B. J., Schnable P. S. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell. 1996;81(1):1291–1304. doi: 10.1105/tpc.8.8.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Dietrich C. R., Delledonne M., Xia Y., Wen T. J., Robertson D. S., Nikolau B. J., Schnable P. S. Sequence analysis of the cloned GLOSSY8 gene of maize suggests that it may code for a beta-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 1997;1151(1):501–510. doi: 10.1104/pp.115.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A., Wisman E., Huijser P., Huijser C., Wellesen K., Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;111(1):2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. Plant salt tolerance. Trends in Plant Science. 2001;61(1):66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]


