Abstract
During embryogenesis a single cell gives rise to a functional multicellular organism. In higher plants, as in many other multicellular systems, essential architectural features, such as body axes and major tissue layers are established early in embryogenesis and serve as a positional framework for subsequent pattern elaboration. In Arabidopsis, the apicalbasal axis and the radial pattern of tissues wrapped around it are already recognizable in young embryos of only about a hundred cells in size. This early axial pattern seems to provide a coordinate system for the embryonic initiation of shoot and root. Findings from genetic studies in Arabidopsis are revealing molecular mechanisms underlying the initial establishment of the axial core pattern and its subsequent elaboration into functional shoots and roots. The genetic programs operating in the early embryo organize functional cell patterns rapidly and reproducibly from minimal cell numbers. Understanding their molecular details could therefore greatly expand our ability to generate plant body patterns de novo, with important implications for plant breeding and biotechnology.
1. INTRODUCTION
The generation of a functional organism from a single cell requires the spatially coordinated acquisition of numerous cell identities. Molecular genetic studies of embryo development in animals have not only elucidated many of the underlying processes, but have also greatly advanced the general understanding of eukaryotic cell signaling and gene regulation. Because of the fundamental differences between animal and plant cells, the study of plant embryo pattern formation may have a similar dual impact. In addition to improving our understanding of the organized growth of plant cells and consequently our ability to manipulate these patterns, research in this field has and will continue to reveal entirely novel modes of eukaryotic cell communication.
In animals, the importance of understanding pattern formation in the embryo is self—evident, as the mature embryo usually constitutes a miniature version of the adult organism. In plants, by contrast, embryogenesis generates only a less complex core structure, the seedling (Figure 1), while virtually the entire adult plant morphology is generated by the activities of the apical meristems. This raises the question of why, in plants, should emphasis be placed on pattern formation in the embryo. As we will discuss in this review, the seedling is not merely an unstructured support device for autonomously patterning apical meristems. Rather, it can be regarded as the initial unit of a reiterative patterning process, in which existing structures serve as positional references for those that follow (Figure. 2). Moreover, the seedling pattern is itself generated with reference to an early axial embryonic pattern, which is already visible in young embryos comprising about hundred cells. Uniquely therefore, the study of early embryo pattern formation may elucidate mechanisms through which plant cells are able to generate growth axes and functional body patterns in the absence of pre-structuring multicellular templates.
Figure 1.
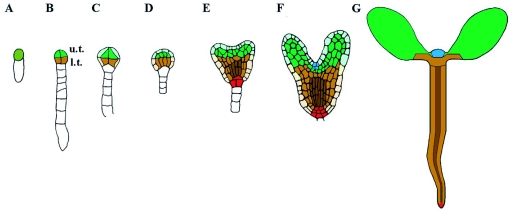
Embryonic origin of seedling structures.
The reproducibility of Arabidopsis embryo development enables tracing the origin of seedling organs and tissues to progenitor cells in the early embryo. Colors identify corresponding regions in embryo and seedling. A detailed description of embryonic stages is given in Figure 3. Radial subdivision; grayscale: vascular tissues, dark; ground tissue, plain color; epidermis, lightly shaded (missing in G). Apical-basal subdivision; upper and lower tier descendants are colored in green and brown, respectively. The most distal part of the root meristem (red) originates from the uppermost suspensor cell (hypophyseal cell. Section 4.2).
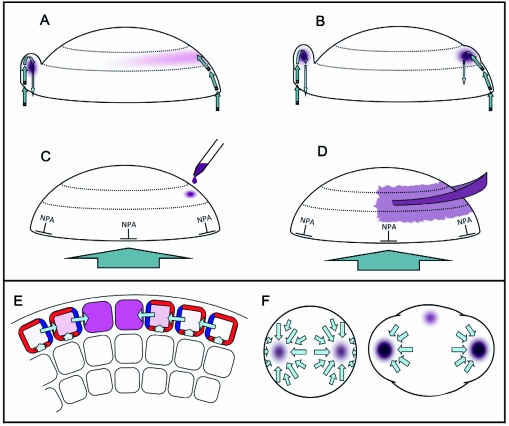
Figure 2.
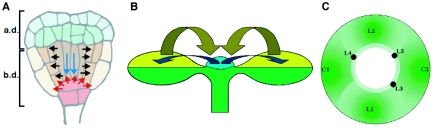
Positional references provided by the early embryo pattern.
Arrows indicate presumed pattern transmission mechanisms, which are discussed in sections 4 and 5.
(A) Triangular-stage embryo with central vascular cylinder (narrow cells in the center). Black arrows indicate signaling from the vascular cylinder to induce radial patterning in the overlying ground tissue (Section 5.3.); blue arrows indicate the likely dependence of hypophyseal cell fate acquisition on apical signals (Section 4.2.2.). Signals from hypophyseal derivatives (red arrows) confer stem cell identity in the root meristem (a.d. and b.d. = the apical and basal domains respectively. Section 4).
(B) Signals from the shoot meristem promote adaxial-abaxial polarity in leaves, while conversely, adaxial cell fate in leaf primordia promotes shoot meristem development, yellow arrows (Section 4.3.1).
(C) Positioning of lateral shoot organs. Primordia are restricted to the peripheral zone of the meristem. Initiated with an arbitrarily positioned cotyledon primordium (C1), graded lateral inhibition (indicated by green gradients) could restrict C2 to a position opposite C1 (Section 4.3.2). As subsequent leaf primordia (L1–L5) are produced, less synchronous differential inhibition would lead to a gradual transition of phyllotactic angles from 180° to 137°.
De novo generation of body patterns from individual cells other than the zygote is a well-known and widely exploited property of plant cells. Gardeners as well as entire industries exploit the amazing regenerative capacities of plant cells, but the process remains stochastic and its molecular basis elusive. Only a few cells in an aggregate participate in the initiation of a multicellular pattern and species-specific differences in regeneration properties are still poorly understood. By contrast, the genetic program initiated in zygotic embryos creates a highly reproducible axial pattern from a single cell often within ten cell divisions. In Arabidopsis, embryos of the late globular stage (Figure 3E) are overtly structured in the apical-basal and radial dimensions and are in the process of developing two properly spaced cotyledons. In this review, we will focus on the establishment of this axial pattern in the early embryo and on mechanisms, through which it acts as a foundation for shoot and root development. Other aspects of embryogenesis and overlapping activities of the apical meristems will be addressed in chapters on ovule, seed and apical meristem development, respectively. Further aspects of angiosperm embryo development have also been discussed in a number of thoughtful reviews (Raghavan, 2000; Jurgens, 2001; Laux et al., 2004; Willemsen and Scheres, 2004; Weijers and Jurgens, 2005; Jenik et al., 2007; Chandler et al., 2008; Nawy et al., 2008) as has somatic embryogenesis (Raghavan, 2006; Rose and Nolan, 2006).
Figure 3. Stages of Arabidopsis embryogenesis.
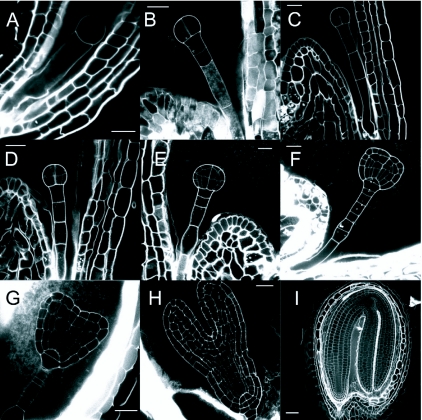
(A) Early embryo, with a single cell in the embryo proper.
(B) Early embryo with 2 cells in the embryo proper.
(C) Octant stage; four of eight cells in two tiers are visible. Cells of the upper and lower tier (u.t. and l.t.) of the octant will give rise to specific parts of the seedling (see Figure 1). Together with descendants of the uppermost suspensor cell (hypophyseal cell) the eight ‘octant’ cells will form all the structures of the seedling.
(D) Dermatogen stage. A tangential division of each of the eight ‘octant’ cells produces inner cells and epidermis (protoderm) cells.
(E) Early globular stage; divisions of the inner cells immediately after the dermatogen stage are oriented in the apical-basal dimension, endowing the embryo with a morphologically recognizable axis.
(F) Triangular stage; now a polarized pattern of major elements is recognizable (see text): u.t. cells have generated two symmetrically positioned cotyledon primordia and l.t. cells a radially patterned cylinder (comprising epidermis, ground tissue and vascular tissue). Additional divisions distinguish the ‘hypophyseal cell’ from other suspensor cells. Its descendants will ultimately form the quiescent center of the primary root meristem and the columella initials.
(G) Heart stage; cotyleldon outgrowth. Subsequently, cells between the outgrowing cotyledons initiate the primary shoot meristem.
(H) Mid-torpedo stage; enlargement of cotyledons and hypocotyl and further elaboration of the radial pattern.
(I) Bent cotyledon stage embryo with elaborated radial pattern in different organs. In the cotyledons a single adaxial subepidermal layer of elongated cells (palisade mesophyll) can be distinguished from underlying mesophyll cells. The radial pattern of the hypocotyl is comprised of a single cell layer of epidermal cells, two cortex layers, one endodermis and one pericycle layer enclosing the vascular cylinder.
Bar is 5 µm in A, 10 µm in B, G and H, 15 µm in C and E, 20 µm in D and F, 50 µm in I.
Images kindly provided by J. Runions, are also available at: https://www.brookes.ac.uk/lifesci/runions/HTMLpages/Embryo%20development.html!
2. STAGES OF ARABIDOPSIS EMBRYO DEVELOPMENT
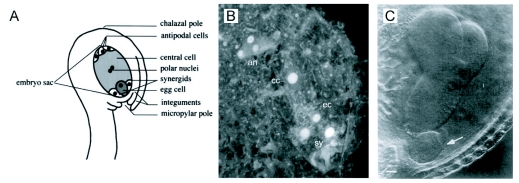
The embryo develops from a fertilized egg cell, positioned within the embryo sac, which is itself embedded in the protective maternal tissue of the ovule (Figure 4A; reviewed in Yadegari and Drews, 2004; Kagi and Gross-Hardt, 2007). All three structures are elongated and polarized along the long axis. The polar organization of the Arabidopsis ovule comprises a small nucellus, harboring the embryo sac, and the chalaza, from which two integuments of unequal size grow out to enclose most of the embryo sac, leaving only a small opening at the opposite end, the micropyle. Within the embryo sac the egg cell and the synergids are located at the micropylar end, while the antipodal cells occupy the opposite position (chalazal end). To complete fertilization, the pollen tube enters the ovule through the micropyle and delivers two haploid nuclei, one of which fuses with the nucleus of the egg cell, while the other combines with the central cell. This double fertilization event initiates the development of two intimately interconnected multicellular structures, the embryo and the endosperm, which are derived from the zygote and the fertilized central cell, respectively. The complex development of the endosperm is described in several informative reviews (Brown et al., 1999; Berger, 2003; Olsen, 2004).
Figure 4.
Maternal polarity and embryo development.
(A) The egg cell develops at the micropylar end of the embryo sac and its basal pole points towards the outside. From Mordhorst et al. (1997)
(B) Confocal image of an Arabidopsis thaliana mature embryo sac, showing the central cell nucleus (cc) after fusion of the polar nuclei, the location of the antipodal cells (an) at the chalazial pole, the egg cell nucleus (ec), and the synergids (sy) at the mycropylar pole. From Capron et al. (2003)
(C) The twin mutants develop two embryos of opposite polarity. Arrow points at the basal end of a second embryo developing from a suspensor cell. From Vernon and Meinke (1994).
In Arabidopsis embryogenesis, the pattern of cell division follows the Capsella variation of the Onagrad type, commencing with an approximately 3-fold elongation of the zygote and followed by an asymmetric cell division, yielding a smaller apical and a larger basal cell (Mansfield and Briarty, 1991; Figures 1A, 3A). These two cells differ profoundly in their internal composition and in their subsequent division patterns. The apical cell contains dense cytoplasm and is the site of very active protein synthesis, whereas the basal cell and its descendants are highly vacuolated. The apical cell undergoes two vertical and one horizontal divisions to produce the octant, comprising eight cells. The octant is divided between an upper and a lower tier (u.t. and l.t. in the following) which will later give rise to the apical and much of the basal portion of the seedling respectively (Figure 1B and 3C). The sequence of these early divisions of the apical cell descendants is highly reproducible. By contrast, the basal cell divides horizontally producing a filamentous structure, the suspensor. Most of the mature embryo is thus derived from the apical cell. However, parts of the root apex originate from the basal cell, as the uppermost suspensor cell (the hypophysis) becomes incorporated in the formation of the embryonic root meristem (Figure 1A–F and Section 4.2.2.).
At the transition from the octant stage embryo, a single round of tangential divisions separates an outer layer of eight epidermal precursor (or protoderm) cells from eight inner cells (Figure 1C and 3D). Protoderm and inner cells soon become histologically distinguishable and due to predominantly anticlinal divisions in the outer layer, the protoderm remains essentially separated from inner cells throughout development. While the external shape of the embryo remains globular for a while, cell divisions of inner cells already reveal axis formation and regional differentiation. Initially, all inner cells adopt a common orientation of cell division, in which newly formed cell walls are aligned along the apical basal axis (Figure 1D and 3E). Therefore, the inner cells remain organized in two tiers, but amplify the number of cells in each tier. Further, the common orientation of division endows the still globular embryo with an anatomically recognizable apical-basal axis.At this stage, there is little indication of differential cell behavior along this axis (u.t. versus l.t.). Thus, the embryo proper at this stage can be considered as ‘axialized’ but not yet asymmetrically ‘polarized’ in the apical-basal dimension. However, immediately succeeding rounds of oriented cell division generate narrow cells specifically in the center of the l.t.. Thereby, in addition to radially subdividing the emerging cylindrical cell pattern into central vascular and surrounding ground tissue, these divisions also reflect apical-basal polarity through differential cell behavior of u.t. and l.t. cells. Further divisions of l.t. descendants are strictly oriented either parallel or perpendicular to the apical-basal axis, generating continuous cell files in increasing numbers of concentric cell layers (Figure 3F to 3I). Cell divisions of u.t. descendants are less strictly oriented and these cells also remain largely isodiametric prior to the initiation of the cotyledons (Figure 3F). At late globular stage, when the number of cells has increased to more than a hundred, the embryo gradually assumes a triangular shape due to localized growth at two opposite positions in the apical region (Figure 1E, 3F). The early heart stage embryo (also referred to as ‘triangular’ stage) comprises approximately 200 cells and the primordia of most major seedling organs, cotyledons, hypocotyl and primary root, as well as the basic tissue types, provascular, protoderm and cortex are anatomically discernible.
The development of the suspensor is somewhat variable, but the two terminal cells adopt invariant fates. The uppermost cell, the hypophysis, undergoes a sequence of reproducible divisions giving rise to part of the primary root meristem, comprising the quiescent center and the central (columella) root cap initials (Scheres et al., 1994). The most basal suspensor cell enlarges dramatically and has abundant contact with surrounding maternal tissues, likely facilitating the supply of nutrients to the embryo. It is interesting to note that in a number of species no plasmodesmata connecting the embryo to maternal tissue have been observed, in sharp contrast to the extensive symplastic connections initially present within the embryo (Kim and Zambryski, 2005). Therefore, it is unlikely that high molecular weight molecules of maternal origin can be transferred to the embryo.
Further refinement of the embryonic pattern occurs during succeeding developmental stages, in which the embryo adopts sequentially “heart,” “torpedo” then “bent cotyledon” shapes (Figure 3G, H and I). In post heart-stage embryos the shoot meristem becomes discernible as three distinct cell layers that will subsequently attain a tunica-corpus organization (Barton and Poethig, 1993). In torpedo and bent-cotyledon stage embryos, provascular tissues also become recognizable within cotyledon primordia and the cellular organization of hypocotyl and root is completed (Figure 3I). Although cells in most tissues will complete differentiation after germination, the complexity of the tissue pattern in the bentcotyledon stage embryo basically equals that of the seedling.
3. CONCLUSIONS FROM VARIABLE DEVELOPMENT
3.1. Is there maternal control?
Maternal influences on embryonic development in Arabidopsis have been reported (Ray, 1998), but apparently these are not involved in specifying basic architectural features of the embryo, such as the establishment of apical-basal polarity. There is obvious polarity alignment within the reproductive apparatus (Figure 4A, B), and it therefore seems reasonable to suggest that the polarity of the ovule impinges on the polar orientation of the embryo sac, the egg cell and thereby the zygotic embryo. However, apical-basal pattern formation in the embryo is not dependent upon this external influence and can be uncoupled from it. This can be demonstrated not only in somatic embryos, which display properly ordered apical-basal patterns in the absence of any fixed spatial relationship to maternal structures, but also in zygotic embryos, which can develop normally in abnormal orientation relative to maternal structures. In twin mutants for example (Schwartz et al., 1994; Zhang and Somerville, 1997), multiple embryos from the same zygote can develop inverted apical-basal patterns (Vernon and Meinke, 1994), excluding the possibility that the polarity of the zygote stringently predisposes the polarity of the embryo (Figure 4C).
3.2 Lineage or position?
The reproducible sequence of cell divisions in the Arabidopsis embryo effectively results in the specification of cell fates along predictable cell lineages. For example, characteristic early divisions suggest a hierarchy of partitioning events, in which early established lineages contribute only to particular structures in the mature embryo (Section 2; Figure 1). Does this predictability reflect lineage-imposed cell fate restriction? Obviously, lineage-imposed fate restriction cannot be recognized in an invariant system, since here its consequences cannot be distinguished from those of reproducible positional specification. However, in Arabidopsis mutants where the regular sequence of cell divisions is dramatically disturbed and major organs and tissues remainproperly positioned (Torres-Ruiz and Jurgens, 1994). These, and many similar observations suggest that the specification of the basic embryonic pattern is largely, if not entirely, based on positional cues, which are remarkably indifferent to cell boundaries, cell numbers and overall dimensions. The interpretation is consistent with the highly variable development observed in embryos of certain angiosperm species (Johri, 1984) and of Arabidopsis embryos developing in culture (Luo and Koop, 1997). However, it does not exclude a role for lineage-dependent cell fate specification in local patterning processes at later stages.
3.3. A second level of pattern control?
If the sequence of cell divisions has no bearing on the resulting pattern, why then is embryonic cell division so invariant? What evolutionary pressure could have generated a second, tighter level of control specifying each individual division so precisely? Perhaps the most attractive explanation is that the stereotyped pattern of divisions in unperturbed Arabidopsis development constitutes the product of an optimization process to produce functional cell patterns from a minimum number of cells. While it seems possible to generate a seedling pattern in various ways, it is hard to imagine how this can be accomplished with fewer cells. In the Arabidopsis seedling, individual tissue layers are often only one cell wide, which obviously constrains the variability of a functional patterning system. Consistent with this interpretation, the generation of the seedling pattern through variable cell divisions is typically associated with larger cell numbers. Therefore, in small embryos there could be a second level of control, probably involving numerous short-range interactions, which together specify the course of embryo development in great detail.
4. GENERATING THE APICAL-BASAL PATTERN
The seedling pattern is often viewed as the superimposition of an apical-basal and a radial pattern. In this review, we will follow this scheme as a suitable formal categorization, but it should be emphasized that the available evidence does not support a strict separation of patterning cues along these axes. Rather, it appears that individual genes can be involved in patterning events along both axes (Sections 4.2.1 and 5.2) and that patterning in a single dimension may involve the piecemeal action of independent signaling processes (Section 5.1 and 5.2).
The main elements to be positioned along the apical-basal axis are the shoot apical meristem (SAM), cotyledons, hypocotyl, radicle and the root apical meristem (RAM). For the most part, SAM and parts of the cotyledons originate from the u.t., hypocotyl and radicle (including most RAM initials) from the l.t., and the quiescent center and columella from the hypophyseal cell (Scheres et al., 1994; Figure 1). As will be discussed below, the primary shoot and root meristems are generated as integral parts of signaling processes defining an apical and a basal embryo domain, respectively. In the following sections, we will begin by discussing the establishment of apical-basal polarity and the embryo axis and then examine the signaling processes within the basal and the apical domains themselves.
4.1 Apical-basal polarity
The decisions that separate the apical and basal domains of the embryo establish a reference framework for subsequent patterning events. These crucial fate decisions rely upon domain-specifying gene expression and coordination between domains by cell-to-cell communication. Recent research has made considerable progress in unraveling the pathways specifying the domains along the apical-basal axis and the communication events that coordinate development.
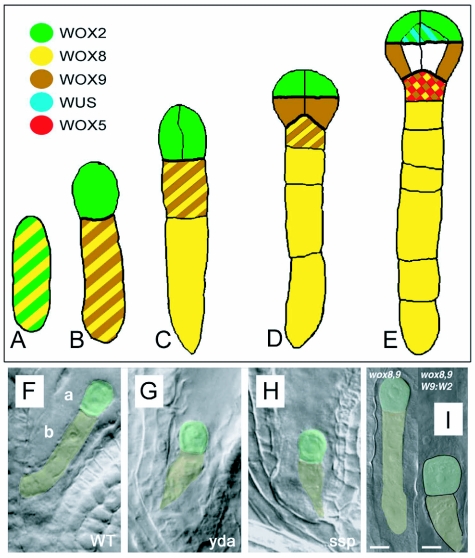
Early cell fate decisions that separate differing domains along the apical-basal axis are reflected in the differential expression of members of the WUSCHEL (WUS) family of homeodomain transcription factors ((Haecker et al., 2004); Figure 5A–E). After the initial asymmetric division of the zygote the apical cell is marked by WUSCHEL RELATED HOMEOBOX2 (WOX2) expression and the basal cell by expression of WOX8 and WOX9. During subsequent divisions, WOX2 comes to mark the u.t., WOX9 the outer cells of the l.t. and the hypophysis, and WOX8 the hypophysis and suspensor (Haecker et al., 2004). WOX8 and WOX9 are closely related homologues that affect patterning and cell division in both the apical and basal cell lineages (Haecker et al., 2004; Wu et al., 2007; Breuninger et al., 2008). Functions of WOX8 and WOX9 in the apical domain appear to be at least partly mediated by downstream activation of WOX2 expression, which is absent in the embryo of wox8 wox9 double mutants (Breuninger et al., 2008). WOX2 expression alone appears sufficient to confer important characteristics of the apical cell lineage (Breuninger et al., 2008).
Figure 5.
Apical-basal WOX expression domains.
(A-E) Expression domains of WUSCHEL-RELATED HOMEOBOX (WOX) genes during the early stages of embryogenesis and development from the single-celled zygote. Images A–E are redrawn after Haecker et al. (2004) and Nawy et al. (2008).
(A) The zygote expressing WOX2 (green) and WOX8 (yellow), which subsequently mark the apical and basal daughter cells of the first division (B).
(B) WOX9 is upregulated in the basal cell at this time.
(C) After division of the basal cell, WOX9 is expressed only in the more apical cell, while WOX8 is expressed in both daughter cells.
(D) At the octant stage, the upper and lower tiers of embryo proper are marked by WOX2 and WOX9 expression, respectively. Both WOX8 and 9 are expressed in the hypophysis.
(E) At the dermatogen stage WOX9 expression is downregulated in the embryo proper, except for the outer cells of the lower tier. Simultaneously, WUSCHEL is turned on in the inner cells of the upper tier and WOX5 within the hypophysis.
(F–I) The fundamental fate decision that leads to the separation of apical and basal cells with differing characteristics is dependent on the YODA/MPK (YDA/MPK) pathway and may also be regulated by WOX gene expression.
(F) Apical (a) and basal (b) cell proportions in wildtype.
(G, H) Loss of YDA or SHORT SUSPENSOR (SSP) function appears to allow apical cell fate to be expressed in the basal cell and its descendants. In these mutants elongation of the zygote is suppressed and subsequent division and development of the suspensor from the basal cell is affected. Images F–H reproduced from Bayer et al. (2009) with permission from the American Association for the Advancement of Science.
(I) In wox8 wox9 double mutants, WOX2 and other apical cell lineage features are not expressed. Expression of WOX2 seems to be instrumental in apicalfate acquisition, because targeted mis-expression of WOX2 confers apical-cell features. Scale bars in I=10µm. Image reproduced from Breuninger et al. (2008) with permission from Elsevier Limited.
Misexpression of WOX2 in the wox8 wox9 background reduces the asymmetry of the initial division of the zygote and promotes aspects of the apical lineage in the basal cell (Figure 5). This phenotype is mirrored in loss-of-function mutations affecting the mitogen-activated protein kinase kinase (MPKK) YODA (YDA) wherein the zygote fails to elongate properly and divides to produce an abnormally small basal cell (Figure 5; Lukowitz et al., 2004). Subsequent divisions of the basal daughter cells are disorganized and prevent the formation of a distinct suspensor. Two MPKs, MPK3 and MPK6 have been shown to function downstream of YDA in epidermal cell specification and are also redundantly required to coordinate apical and basal cell fates in embryo development (Wang et al., 2007). A third gene SHORT SUSPENSOR (SSP) is believed to act upstream of the YDA/MPK pathway (Wang et al., 2007; Bayer et al., 2009). Recent evidence suggests that SSP transcripts transferred from the male parent during fertilization are important in activating the YDA expression in the zygote (Bayer et al., 2009). Interestingly, mutant analysis suggests that WOX-dependent regulation and the MPK/YDA genes do not operate on a single linear pathway (Breuninger et al., 2008).
The plant hormone auxin is also known to play a vital role in apical-basal patterning and the establishment of the embryo axis. Auxin is transported in a directional manner by membrane spanning proteins that mediate the influx and efflux of this signaling molecule into and out of cells (reviewed in Fleming, 2006; Kramer and Bennett, 2006; Leyser, 2006; Teale et al., 2006; Boutte et al., 2007; Zazimalova et al., 2007; Kleine-Vehn and Friml, 2008; Robert and Friml, 2009) and has long been implicated in basic patterning processes in plants, such as axis formation (Figure 6). The localization of members of the PINFORMED (PIN) family of auxin efflux facilitators within cells reflects the direction of auxin transport in contrast to other classes of proteins implicated in the auxin transport (Galweiler et al., 1998; Petrasek et al., 2006; Wisniewska et al., 2006) (reviewed in Weijers and Jurgens, 2005; Friml et al., 2006; Benjamins and Scheres, 2008; Chandler et al., 2008; Nawy et al., 2008; Benkova et al., 2009). In the absence of more direct evidence for the position and polarity of major auxin transport routes, such polar avenues decorated by PIN protein expression have been used to establish plausible models linking dynamic patterns of auxin transport to patterning processes in all parts of the plant, beginning with the formation of the main body axis in the early globular embryo (Friml et al., 2003).
Figure 6.
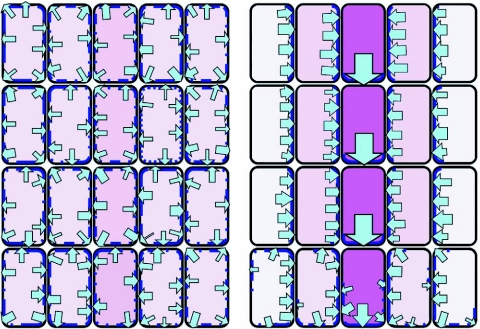
Integration of cell polarity through auxin transport.
A highly schematic view. Rectangles represent cells and arrows of different strength represent the Intensity of auxin flow. For simplicity it is assumed that Intensity and direction of auxin flow is solely controlled through the quantity and distribution of auxin efflux carriers (dark blue) in the plasma membrane. Routes of preferred auxin transport have been associated with sites of vascular differentiation (dark purple).
The central proposition is that auxin flow and cell polarization are connected in a positive feedback loop, Illustrated here by restricting auxin efflux to the basal side of each cell as an expression of cell polarization. Thereby, cells in a given region, Including cells newly formed by division, would Integrate polarity. The feedback system could further Include the stabilization of auxin sources or sinks. Note that the same cellular feed-back mechanism would progressively enhance Initial differences in auxin conductivity leading to the specification of different cell types in the radial dimension. Drawn after Sachs (1991).
The perception/response to localized auxin accumulation can be visualized with the synthetic auxin response promoter element DR5 (Ulmasov et al., 1997). During the globular stage of embryonic development, PIN-mediated auxin flux appears to flow in a basal to apical direction through the suspensor and into the proembryo (Friml et al., 2003) (Figure 7, adapted from Jenik et al., 2007). The vital role played by the PIN proteins and polar auxin transport in early embryogenesis is only revealed when enough of these partially redundantly acting genes are knocked out, resulting in embryos with impaired polarities (Friml et al., 2003). Further evidence supporting the critical role played by directional auxin transport in establishing apical-basal polarity is provided by work with the EMBRYO DEFECTIVE30/GNOM (EMB30/GN) mutant. Loss of EMB30/GN function may produce ball-shaped seedlings with fully differentiated, but randomly oriented, vascular cells at the center, indicative of a selective loss of apical-basal polarity (Mayer et al., 1993), a phenotype that can be mimicked by application of high concentrations of auxin transport inhibitors (Hadfi et al., 1998; Friml et al., 2003). EMB30/GN encodes an Adenosyl ribosylation factor Guanine nucleotide Exchange Factor (ARF GEF), which functions as an endosomal regulator of vesicle budding. This function appears critical for auxin transport and cell polarity through a role in the positioning of multiple auxin transport membrane proteins (Steinmann et al., 1999). Consistent with this Interpretation, EMB30/GN mutations were shown to interfere with the coordinated polar localization of PIN1 proteins (Geldner et al., 2003) (Figure 8B) (for a detailed review, see Kleine-Vehn and Friml, 2008). The role of endocytic recycling in establishing polar localization of PIN proteins has been further elaborated by manipulation of the Arabidopsis Rab5 GTPase pathway (Dhonukshe et al., 2008). The critical role of the Rab5 GTPase pathway in endocytosis has been well characterized in mammalian systems and two homologues of Rab5 in Arabidopsis, ARA7 and RHA1, have been shown to localize to the endosome (Ueda et al., 2004). Although single ara7 or rha1 mutants show no obvious phenotypic defects and the double ara7 rha1 mutant is gametophytic lethal, an inducible dominant negative version of ARA7 has been used to demonstrate a requirement for Rab5-mediated endocytosis in the polarized distribution of PIN1 and PIN2 proteins (Dhonukshe et al., 2008).
Figure 7.
Auxin flux and auxin perception maxima during embryogenesis.
In the early stages of embryogenesis, localization of the PIN7 auxin efflux facilitator (cyan) to the apical membranes of basal cell and subsequently suspensor cells appears to drive auxin flux (arrows) upwards. A weak DR-5 marked auxin perception maximum (light purple) suggestive of auxin accumulation is seen in the apical parts of the developing embryo up to and including the dermatogen stage. During the globular stage PIN1 (dark blue) localization in the basal membranes of the inner cells of the embryo is associated with a switch in the apparent direction of auxin flux to apical — basal. At this time PIN7 is now seen concentrated in the basal membranes of the hypophysis and suspensor cells. A stronger auxin perception maximum (purple) also appears in the hypophysis and apical cells of the suspensor, which becomes restricted to the daughter cells of the hypophysis. PIN1 distribution in the cells of the L1 layer is associated with the generation of auxin perception maxima at the positions of incipient cotyledon initiation. Figure redrawn from Jenik et al. (2007) with permission from Annual Reviews.
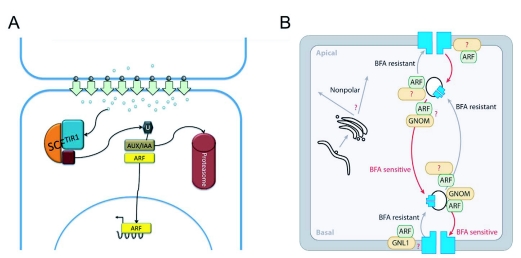
Figure 8.
Auxin Signaling and GNOM-dependent PIN1 localization.
(A) Auxin accumulates in certain cells through coordinated transport mediated by the PIN proteins and this accumulation leads to the activation of the ARFs. At low auxin concentration, ARFs are maintained in a complex with the Aux/IAA proteins that act as transcriptional repressors. At high auxin concentrations, auxin bound to a TIR1 related F-Box protein within a SCF complex stabilizes the interaction between the complex and its target, the Aux/IAA protein. The SCF complex catalyzes the ubiquitination (U) of the Aux/IAA target and marks it for degradation by the proteasome, releasing ARF activity. This, in turn, may affect PIN genes expression and auxin transport properties of the respective cells, leading to potentially complex mutual influences between auxin distribution and transport patterns.
(B) Polar targeting of PIN1: The original transport of PIN1 from the ER/Golgi (on the right side of the figure) is non-polar. However, an ARF GEF-dependent transcytosis mechanism then targets the PIN1 protein to the apical or basal side of the cell. The ARF GEF GNOM is involved in the basal targeting of PIN1. Image reproduced from Kleine-Vehn and Friml (2008) with permission from Annual Reviews.
PIN1 is represented in blue, ARF in green and ARF GEF in yellow.
In conclusion, the WOX cascade, YDA/MPK pathway and directional auxin transport all appear to play critical roles in establishing apical-basal polarity in early embryonic development. However, much remains to be elucidated regarding the potential interdependences or Interactions between these processes. There is some evidence that the WOX cascade may impact upon subsequent processes and the directional flux of auxin, since WOX8 and 9 are required for normal PIN1 expression and formation of auxin perception maxima in the early globular stage (Breuninger et al., 2008).
4.2. Patterning the basal domain
The basal domain comprises the hypocotyl, the radicle and the primary root meristem (Figure 1 and 2A). Essential steps in basal patterning are the formation of a radially structured cylinder in the late globular-stage embryo and the subsequent establishment of a complex stem cell system at the basal end of this cylinder. We will refer to these two processes as the formation of the embryo axis and of the primary root meristem, respectively. Despite signaling overlap, they will be discussed in separate sections.
4.2.1. Formation of the embryo axis
Most of the body of the seedling takes the form of a cylinder comprised of concentrically arrayed tissue layers. The hypocotyl and radicle are sections along this cylinder, with distinguishable anatomical and physiological properties. The Initiation of the embryo axis occurs through strictly oriented divisions in the early globular embryo. Divisions oriented along the apical-basal axis predominate among l.t. cells and generate a cylinder of parallel cell files. This cylinder becomes radially structured by further divisions in the center, which generate narrow procambial cells of the embryonic stele (Figure 3E).
Auxin signaling and directional auxin transport are essential for normal patterning in the basal domain. The hormone elicits context-dependent effects on gene transcription that are mediated by members of the AUXIN RESPONSE FACTOR (ARF) family of genes (for a more comprehensive treatment, see Guilfoyle and Hagen, 2007; Benjamins and Scheres, 2008; Mockaitis and Estelle, 2008). Briefly, in the absence of an auxin signal, ARF proteins are bound and inactivated by transcriptional repressors of the Auxin/Indole Acetic Acid (Aux/IAA) gene family. Auxin signaling triggers the release of ARF activity by rapid degradation of Aux/IAA proteins. The auxin signal is initially sensed by receptors of the auxin-signaling F-box (AFB) family, such as TRANSPORT INHIBITOR RESPONSE1 (TIR1) (Dharmasiri et al., 2005). These F-box proteins are components of the SKP1-CULLIN-F-Box (SCF) E3 ubiquitin ligase complex which recognizes the domain II motif of Aux/IAAs proteins and target them for degradation. Auxin greatly increases the affinity of the F-box component for the target Aux/IAA protein and thus promotes its degradation (Figure 8A; Dharmasiri et al., 2005; Kepinski and Leyser, 2005).
Loss of function in one specific ARF gene, MONOPTEROS (MP/ARF5), leads to severe defects in formation of the embryo axis. The vascular systems in these mutants are severely reduced, the cotyledons fused, and hypocotyl and root replaced by a short undifferentiated basal peg (Berleth and Jurgens, 1993). Similar phenotypes are observed in mutations suppressing degradation of the closely related Aux/IAA proteins BODENLOS (BDL/IAA12) and IAA13 (Weijers et al., 2005b). MP and BDL proteins have been shown to interact in vivo (Hamann et al., 2002; Weijers et al., 2006) and it seems likely that both BDL and IAA13 inhibit MP activity. Mutant analysis has also revealed that a second closely related ARF, NON-PHOTOTROPHIC HYPOCOTYL4 (NPH4/ARF7) appears to act redundantly with MP in embryonic patterning. Loss of NPH4 function dramatically enhances the mp phenotype, abolishing organized structures along the apical-basal axis (Hardtke et al., 2004). Initially wider expression of MP, BDL and IAA13 becomes restricted to the central cylinder (provascular cells) of the midglobular stage embryo. This pattern of expression is mirrored by the PIN FORMED1 (PIN1) auxin efflux facilitator, which marks the formation of the central procambial axis in the globular embryo (Steinmann et al., 1999; Friml et al., 2003). The cellular localization of PIN efflux carriers in this region appears to drive the apical to basal auxin flux (Figure 7). This flux is critical for subsequent patterning along the embryo axis (Friml et al., 2003), as is timely expression of MP and BDL in these cells (Weijers et al., 2006).
The single mutants dornroschen (drn) and dornoschen-like (drnl) display a variety of cotyledon defects such as single or fused cotyledons, some aberrant divisions in the embryo and in rare instances a phenotype similar to mp (Chandler et al., 2007). Mutant dm embryos are also disturbed in auxin responsiveness (Chandler et al., 2007). In a double mutant combining a strong dm allele with a weak drnl, the penetrance of all three phenotypes is greatly increased, whereas a homozygous double mutant combining a strong drn allele and a strong drnl allele shows a pin-like phenotype (Chandler et al., 2007). DRN and DRNL encode AP2-like transcription factors and have been shown to interact with PHAVOLUTA (PHV) and BES interacting Myc-like protein 1 (BIM1) (Chandler et al., 2007; 2009), two other transcription factors involved in embryo patterning. DRN itself has also been shown to be a direct target of MP in the cotyledons (Cole et al., 2009). Although a lot of data has been gathered on the DRN genes, their exact function in the establishment of the embryo axis and formation of the cotyledons remains elusive.
While mp and bdl mutants lack the basal pole, a mutation in the TOPLESS (TPL) gene results in the loss of the apical pole, which is instead replaced with another basal pole (Long et al., 2002). Moreover, the semi-dominant tpl1 mutation can suppress many aspects of the bdl phenotype (Figure 9, from (Szemenyei et al., 2008). TPL has been shown to interact physically with the BDL/IAA12 protein via an ETHYLENE-RESPONSE FACTOR (ERF) —associated amphilic repression (EAR) motif (Szemenyei et al., 2008) and may thereby modulate the transcriptional activity of MP/ARF5. Since root initiation and the expression of master regulators of root fate (Section 4.2.2) are positively controlled by auxin signaling, inhibition of elements of auxin-induced gene expression by TPL and 3 TPL-related proteins may act to repress root fates in shoot tissues.
Figure 9.
A mutation in TOPLESS (tpl-1) rescues aspects of the bodenlos/iaa12 (bdl) mutant phenotype.
A, E, I: Wildtype (WT). B, F, J: The tpl-1 mutant. C, G, K: bdl. D, H, L: tpl1 bdl double mutant.
(A) Wildtype seedling with normal root (pale blue) and vascular development in the cotyledons.
(B) The temperature sensitive tpl-1 mutant, showing the weaker apical defects at permissive temperatures on the left. On the right, at non-permissive temperatures, a complete homeotic transformation of the shoot into a root takes place.
(C) The bdl mutant showing the replacement of the primary root with an undifferentiated basal peg, and defects in cotyledon vasculature.
(D) Introduction of tpl-1 into the bdl background permits improved development of the root. In the apical domain, it ameliorates some of the vascular defects present in the single bdl mutant. Heart stage embryos of WT (E, I), tpl-1 (F, J), bdl (G, K) and tpl-1 bdl (H, L). Images A–D drawn after Osmont and Hardtke (2008).
(E–H) The lens-shaped cell and derivatives that normally gives rise to the quiescent center of the RAM are outlined.
(I–L) Associated auxin perception maxima marked by DR5rev::GFP. Introducing tpl-1 to the bdl background restores the correct formation of the lensshaped cell (compare H with G) and reinstates DR5rev::GFP expression in this region (compare L with K). Images E–L reproduced from Szemenyei et al. (2008) with permission from the American Association for the Advancement of Science.
Overall therefore, auxin signal transduction and regulated auxin transport are required for oriented cell divisions and vascular differentiation in the basal domain, and these patterning events constitute prerequisites for subsequent radial differentiation (Section 5.2.) and root meristem formation (Section 4.2.2)
4.2.2. Formation of the primary root meristem
The RAM cell pattern consists of concentrically arrayed stem cells (initials in the following) that extend the radial pattern in the growing root, the four cells of the quiescent center (QC), which divide only infrequently, and most distally, the initials of the central root cap (columella) (Figure 10A). This highly organized cell pattern is not only generated in the embryo, but also post-embryonically in the formation of lateral roots. As these are initiated from the pericycle at considerable distance from the meristem of the higher order root, the cell pattern seems to be generated de novo in each lateral root primordium, in response to shoot derived signals (Casimiro et al., 2001; Bhalerao et al., 2002).
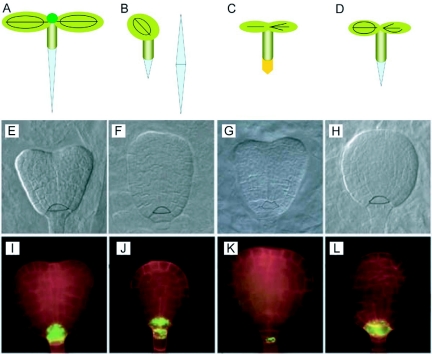
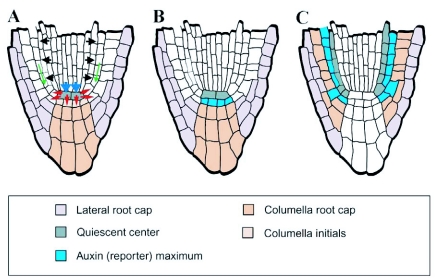
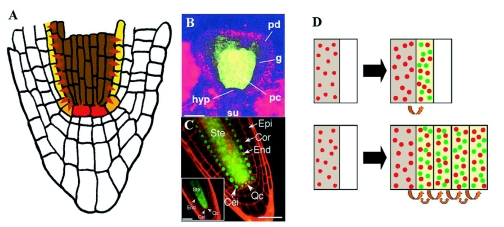
Figure 10.
Cell fate specification in the root meristem.
(A) Organization of cell types in the root meristem. Centrally located QC cells (grey) are flanked by initials of various tissues: initials extending tissue layers in the growing root and, laterally and basally, initials replenishing cells in the lateral (violet) and central root cap (orange). Blue arrows indicate that the acquisition of QC cell fate seems to be dependent on signals from the shoot (compare Figure 2A); red arrows the dependence of stem cell fate on signals from the QC. Black arrows represent endodermis inducing signals from the stele (Figure 18) and green arrows the stabilization of tissue identity within each layer.
(B) An auxin-response reporter gene detects a maximum (blue) at the position of the columella initial cells.
(C) When the auxin response maximum is displaced (e.g. because of auxin transport inhibition), the positions of all three cell types in relation to the stele and auxin response maximum are maintained, suggesting an important role for auxin distribution in root meristem patterning. From Scheres (2000).
Patterning the RAM involves the establishment and maintenance of a stable stem cell pool at the root tip, specification of proper tissue identity of initials in their respective positions and, in order to get the system started, some mechanistic link to a localized signal from the shoot (Figure 10A). It has long been suspected that cell fate in the RAM is largely controlled by positional cues and single-cell ablation has been employed to trace the origin of some of these cues (summarized in Scheres and Heidstra, 1999). One, originating from the QC, seems to confer stem cell identity to cells surrounding the QC (Figure 10A). Ablation of individual QC cells is associated with the loss of stem cell identity in neighboring cells, which instead acquire characteristics of their differentiated daughter cells (van den Berg et al., 1997). Short-range signaling from the QC would obviously be a very suitable mechanism to ensure that a stem cell pool of constant size is maintained at the root tip.
Another type of positional signal seems to help integrate the tissue identity of all cells in a given layer (Figure 10A). Cell ablation experiments revealed that cortex tissue continuity is required to maintain the identity of the cortex-endodermal initials, suggesting that signals from more mature cells are transmitted within a tissue layer to reinforce tissue identity of less mature cells (van den Berg et al., 1995). Many Arabidopsis tissues comprise only single cell layers, but their functions often critically depend on tissue continuity. Therefore, integrating signals passed along individual tissue layers (in combination with highly flexible radial patterning mechanisms, Section 5.3.) could help stabilize an otherwise fragile radial pattern.
If the QC has a central role in conferring stem cell identity on neighboring cells, how is the QC itself positioned? Localized accumulation of auxin is thought to act as a positional signal in proximal-distal patterning in the RAM, specifying the positions of the QC and columella initials relative to the vascular cylinder (reviewed in Jiang and Feldman, 2005). This view is supported by work studying the expression and localization of PIN proteins, which has demonstrated their involvement in meristem patterning and patterning in the basal domain of the embryo (Friml et al., 2003; Blilou et al., 2005; Weijers et al., 2005a). Auxin supplied from the proembryo and polarly transported in an apical basal direction results in the formation of an auxin response maxima in the nearby suspensor cells, the uppermost of which assumes a hypophyseal fate. The lens-shaped daughter cell of the hypophysis subsequently gives rise to the QC. The importance of PIN-mediated auxin flux and local auxin accumulation in promoting ‘hypophyseal’ identity is supported by analysis of mp and bdl embryos where the uppermost suspensor cell fails to adopt this fate. Remarkably, expression of the auxin signaling genes MP and BDL occurs in the basal cells of the embryo proper, right above the hypophyseal cell, from where it triggers the response in the adjacent hypophyseal cell (Weijers et al., 2006). In line with the above described molecular function of TPL as a co-repressor of ARF5/MP, loss of TPL function restores correct development of the lens shaped cell that gives rise to the QC and the DR5 auxin perception maxima associated with it in a bdl background (Szemenyei et al., 2008).
Another intriguing network of genes involved in the establishment of the RAM is the PLETHORA (PLT) gene family. PLTs are believed to play a key role in the acquisition of the QC fate. PLT1 and 2 were first identified as regulators of root stem cell fate (Aida et al., 2004) and have been found to cooperate with two other APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor family members, PLT3 and BABY BOOM (BBM) to initiate the embryonic root (Galinha et al., 2007). Multiple mutant combinations may not produce root or hypocotyl, in a similar fashion to embryos with defective auxin perception. Conversely, conditional overexpression of PLT2, can cause the homeotic transformation of cells in the apical domain to express root markers (Galinha et al., 2007). The proper expression of PLT1 and PLT2 rely on the establishment of an auxin response maximum through the activity of the PIN auxin efflux facilitators (Blilou et al., 2005), as well as functional ARF-dependent downstream signaling (Aida et al., 2004). Overall, the PLTs genes seem to work as integrators of positional information provided by the auxin accumulation in the organization of the root meristem. The PLTs genes are expressed throughout the l.t. of the globular embryo and then in the pro-vascular cylinder of the heart embryo where they perform their function (Figure 11).The GRAS-domain genes SHORT-ROOT (SHR) and SCARECROW (SCR) also play a critical role in the specification of the QC fate. SHR is expressed in a domain Embryogenesis: Pattern Formation from a Single Cell largely overlapping the PLTs in the early globular embryo (Figure 11) while the SCR gene is expressed in the hypohysis (Figure 11). In the late globular embryo, the SHR gene is transcribed in the pro-vascular cells but its protein is found in the lens-shaped cell, together with SCR (Figure 11). The presence of SHR and SCR in the lens-shaped in crucial in specifying the QC fate (Aida et al., 2004). Therefore, the establishment of the QC and then the RAM in the embryo depends on several signaling networks, both auxin-dependent (PLTs) and auxin independent (SHR/SCR). Another identified factor in the QC establishment and maintenance is WOX5. Absence of WOX5 leads to defects in the mature RAM, apparently originating from improper early specification of the QC in the embryo (Sarkar et al., 2007). Moreover, WOX5 expression depends on both SHR and SCR as well as on the MP-BDL pathway, but not on the PLTs (Sarkar et al., 2007). Therefore, it seems WOX5 also functions as an integrator of the auxin-dependent and auxin independent signaling leading to the establishment and maintenance of QC fate in the embryo.
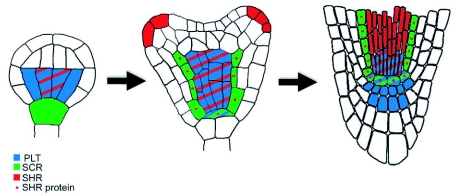
Figure 11.
Specification of the QC initial.
In the globular embryo, the PLT genes are widely expressed in the l.t., while SHR mRNA is present in its the central cells. SCR is expressed in the hypophysis. In the heart embryo the PLT genes are expressed throughout the future vascular cylinder, as well as in the lens-shaped progenitor cell of the QC. SHR mRNA is expressed in the stele, but the SHR protein is found in ground tissue surrounding the stele and the lens shaped cell, where it promotes SCR expression. SCR and the PLT genes promote QC fate of the lens-shaped cell. Redrawn after Stahl and Simon (2005).
Obviously, auxin is not the only plant hormone with critical roles in embryo patterning. Auxin and cytokinin signaling have long been hypothesized to play antagonistic roles, which may be reflected in the observation of genetic suppression of mp and high-cytokinin altered meristem program 1 (amp1) mutations (Vidaurre et al., 2007). Thus, loss of AMP1 makes MP dependent auxin-signal transduction less stringently required in the formation of the primary root as well as other instances. Interestingly, visualization of cytokinin signaling output with a synthetic response element (Two-Component Sensor output, TCS) coupled to a GFP reporter (Muller and Sheen, 2008) is detected in the hypophyseal progenitor cell at the 16 cell stage of embryogenesis. Subsequently, expression of TCS is suppressed in basal daughter cell and maintained in the lens-shaped cell that will give rise to the QC. Auxin signaling visualized by the DR5 reporter shows an inverse expression pattern. Auxin appears to antagonize cytokinin signaling by transcriptional activation of the Arabidopsis thaliana RESPONSE REGULATOR7 (ARR7) and ARR15 genes, which act as feedback repressors of cytokinin signaling. Loss of both ARR7 (by inducible RNAi) and ARR15 (loss-of-function mutant) or ectopic cytokinin signaling activation in the basal cell during early embryogenesis results in a defective RAM.
In summary, the largely cylindrical architecture of the basal domain seems to be initiated in response to directional signals from the apical domain, and apical signals also seem responsible for inducing QC identity. A properly localized QC may in turn anchor the position of initials elongating the cell files of the growing root.
4.3. Patterning the apical domain
The formation of the apical pattern comprises; (I) generation of cotyledons and SAM (4.3.1), (N) proper spacing of lateral shoot organs (4.3.2) and (Ni) adaxial-abaxial patterning within each lateral organ primordium (4.3.3). Although the two latter processes do not fall under apical-basal patterning in a strict sense, the gene activities organizing all three aspects of the apical pattern are integrated to such a degree as to preclude independent discussion.
4.3.1. Generating the cotyledons and the shoot apical meristem
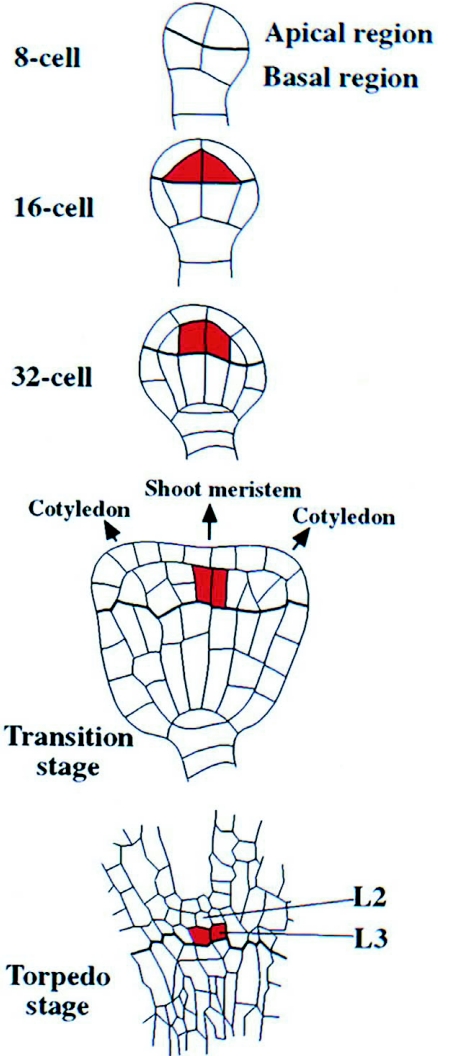
Formation of a permanent shoot apical meristem within the apical domain requires localized signals to confer stem cell identity to a small population of cells between the cotyledon primordia (Clark, 2001; Tucker and Laux, 2007; Bowman and Floyd, 2008). Continuous meristematic activity further requires stable zonation within the SAM, with stem cell identity restricted to cells within a small central zone, while cells displaced to the periphery undergo differentiation. The delicate balance between the pools of stem cells and differentiating cells in the permanent SAM is maintained through negative feedback interactions between antagonistic activities.
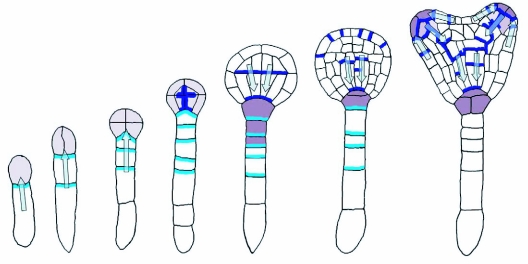
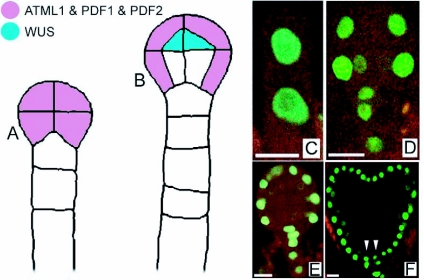
As a positive regulator of SAM development, the homeobox transcription factor WUS is expressed early during embryo development at the 16 cells stage in the upper tier of the embryo and becomes restricted to the innermost upper tier cells (Figure 12; Mayer et al., 1998). In wus mutants, the stem cell pool is reduced, but not absent and the formation of the SAM is delayed, although SAMs will eventually arise in abnormal positions after germination (Laux et al., 1996). WUS expression is negatively regulated by the stem cells through the CLAVATA (CLV) pathway (Schoof et al., 2000) (details reviewed in Jun et al., 2008). This group of three genes was identified by loss-of-function mutants with oversized meristems and constitutes the ligand and receptor parts of a distinct signaling pathway. CLV1 is a receptor kinase (Clark et al., 1997) forming a heterodimer with CLV2, a receptor kinase-like protein (Jeong et al., 1999). There is evidence that CLV3 binds to the CLV1/CLV2 complex (Trotochaud et al., 1999) and is the ligand of CLV1 (Ogawa et al., 2008).
Figure 12.
Embryonic expression of WUS.
WUS transcripts are first detected in 16-cell dermatogen-stage embryos (red). Initially expressed in all subepidermal cells of the apical domain, WUS transcripts become gradually restricted to more central positions and deeper layers at the base of the shoot meristem. No functions have been assigned to WUS expression prior to the heart-stage and the molecular basis of WUS regulation is unclear. Precise regulation of WUS expression is critical to the formation of the apical pattern, since ectopic expression of WUS seems to confer stem cell identity at inappropriate sites (Gallois et al., 2004). Modified from Mayer and Jurgens (1998).
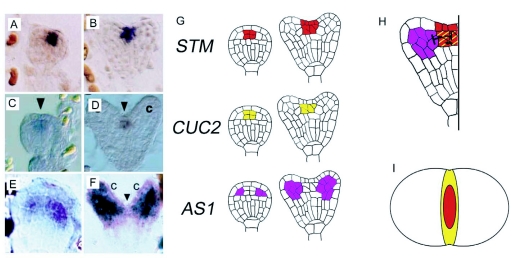
Another key network in establishing the SAM involves the CUP-SHAPED COTYLEDONS (CUC), SHOOTMERISTEMLESS (STM) and the ASYMETRIC LEAVES (AS) genes. The CUC genes encode transcription factors homologous to the petunia gene NO APICAL MERISTEM (NAM) (Aida et al., 1997) and function to form a boundary between the two cotyledons in the central domain. This function seems to be performed with the help of the TCP (TBP1-CYC-PCF1/2) proteins. The TCP and CUC genes appear to have mutually exclusive domains, with the TCP genes occupying the lateral parts and the CUCs the central part. This pattern seems necessary for the correct formation of the SAM and the cotyledons (Palatnik et al., 2003; Koyama et al., 2007). The expression of CUC genes is progressively restricted to the central domain in the early heart embryo, beginning from an initial domain comprising the whole upper tier of the 16 cell stage (Figure 13C, D, G). While single cuc1, cuc2 and cuc3 mutants do not show marked defects, double mutants display a fused collar around the apex and no SAM (Aida et al., 1997; Vroemen et al., 2003). Proper expression of CUC1 appears dependent on auxin, as the CUC1 domain expands in pin1 mutant. In a double mutant pin1 pinoid (pid), the effect is even more pronounced. The serine/threonine protein kinase PID, in turn, has been shown to control the polar localization of PIN1 (Friml et al., 2004). In a triple mutant pin1 pid cuc1, the collar shaped expansion is formed again (Furutani et al., 2004). Therefore, it appears that a role of auxin is to restrict expression of CUC1 to the central domain. Expression of CUC1, as well as of CUC2 but not CUC3 is also controlled by miRNA164 (Laufs et al., 2004; Mallory et al., 2004).
Figure 13.
Regulatory interactions in SAM development.
(A–F) In situ hybridizations of STM (A, B), CUC2 (C, D) and AS1 (E, F), in globular (A, C, E) and heart (B, D, F) stage embryos. Arrowheads in C and D point to the protoderm, where CUC2 is absent. The arrowhead in F indicates the SAM initials. Images A, B reproduced with permission from Macmillan Publishers Ltd: Nature, Long et al. (1996); C, D reproduced/adapted from Aida et al. (1999) with permission from The Company of Biologists; E, F reproduced with permission from Macmillan Publishers Ltd: Nature, Byrne et al. (2000).
(G) Schematic representation of the expression domain shown in A–F.
(H) Model of STM/CUC/AS interactions. The expression domains of STM and CUC2 (and the other CUC genes) are largely overlapping at the globular stage. Activity of CUCs promotes STM expression (A, C). Conversely, STM downregulates the CUCs. (B, D). STM activity promotes the establishment and maintenance of the SAM. This is in part accomplished by another function of STM: to inhibit expression of the genes AS1 and AS2 (here represented by AS1) in the SAM. The counter-acting activities of CUCs and STM lead to the formation of a torus of CUC expression around a STM domain centered on the SAM in the bent cotyledon stage, depicted in I.
(I) Schematic transverse section through the apex of a bent cotyledon embryo showing the inner STM expression domain surrounded by the CUC2 domain.
The CUC genes promote the expression of STM in the central domain (Figure 13A, B, G) (Aida et al., 1999). In a negative feedback loop, STM downregulates the expression of the CUC genes, leading to the formation of a ring of CUC expression around the emerging SAM (Figure 13H). This creates a boundary between the SAM and the future cotyledons (Aida et al., 1999; Aida et al., 2002). Loss-of-function mutations in STM result in a disturbance of CUC2 expression in late embryogenesis (Aida et al., 1999) and produce seedlings with slightly fused cotyledons that fail to produce a SAM (Barton and Poethig, 1993). Post-embryonically, STM is expressed in the SAM center and turned off in cells as they are recruited towards the formation of lateral organ primordia. The (AS) genes seem to act antagonistically to STM, as loss-of-function mutations in the AS1 and AS2 genes restore meristem formation in an stm background (Byrne et al., 2000; Byrne et al., 2002). AS1 is expressed in the primordia of lateral organs and cotyledons (Figure 13E, F, G). Thus, STM function seems to pro-WUS mote SAM formation primarily by counteracting AS1 and AS2 activity, which helps to maintain undifferentiated cell fate in the meristem center. Since AS1 is already expressed in the cotyledons, this counteracting function could explain why STM is required for the embryonic initiation of the SAM (Figure 13H).
There are several other genes whose expression is important for proper SAM and cotyledons formation. GURKE (GRK) (TorresRuiz et al., 1996) and PASTICCINO (PAS) (Faure et al., 1998; Vittorioso et al., 1998) are two examples. The phenotypes of these mutants are relatively pleiotropic and their gene products cannot easily be placed in a signaling context. GRK is an acetyl-CoA carboxylase, suggesting metabolites derived from malonyl-CoA could be important for patterning in apical domain (Kajiwara et al., 2004). PAS is a FK506-binding protein. Evidence suggests that PAS interacts with a NAC-like transcription factor FAN to target it to the nucleus and where FAN acts to repress cytokinin-dependent cell division.
4.3.2. Positioning of lateral organs
The cotyledon primordia are initiated at opposed positions as part of the transition from the radially symmetrical late-globular stage of embryogenesis to the bilaterally symmetrical heart-stage. Initiation and outgrowth of these primordia appear to be driven by epidermal convergence of auxin, similar to the initiation of all lateral shoot organs after germination. Analysis of these analogous postembryonic processes form the basis for our understanding of auxin-mediated positioning of lateral shoot organs.
Several lines of evidence support a central role for auxin in shoot lateral organ initiation. First, mutations in auxin signal transduction genes MP, BDL, AUXIN RESISTANT6/CULLIN1 (AXR6/CUL1) (Hardtke and Berleth, 1998; Hamann et al., 1999; Hobbie et al., 2000; Hellmann et al., 2003) result in cotyledon fusions. Moreover, loss of both MP/ARF5 and NPH4/ARF7 completely prevents the formation of cotyledon primordia (Hardtke et al., 2004). Second, lateral organ formation does not only require proper auxin perception but also auxin transport. Chemical inhibition of auxin transport and mutations in the auxin efflux facilitator gene PIN1 (Galweiler et al., 1998) result in lateral organ fusions or the complete absence of lateral organs in a dose dependent manner (Liu et al., 1993; Benkova et al., 2003; Friml et al., 2003). Additionally, a gain-of-function mutation in IAA18, which causes aberrant cotyledon placement, appears to disrupt PIN1 distribution in the apical domain of the embryo, and transgenic studies suggest the protein inhibits the activity of MP/ARF5 (Ploense et al., 2009). Finally, local auxin application has been shown to induce shoot organ formation at the site of application (Sachs, 1993; Reinhardt et al., 2000). The response is restricted to the peripheral zones of meristems, but within this zone it is the location and quantity of applied auxin that defines the position and width of the emerging lateral organ (Reinhardt et al., 2000). Normal organ formation requires a highly regulated, local auxin signal. This signal appears to be dependent upon a precise balance between local auxin supply and auxin removal, achieved via transport or metabolic processes. If the accumulation of auxin at a particular site to promote lateral organ initiation occurs by convergence of auxin, it could also be expected that draining auxin from surrounding cells to the site of initiation will inhibit primordium formation in the vicinity of such a convergence point (Figure 14). In this way, the formation of local auxin maxima and associated depletion of auxin from their surroundings could fit well with established models of phyllotaxis, which are based on the lateral inhibition of new primordia by existing ones (lateral inhibition) (Schoute, 1913; Turing, 1952; Meinhardt, 1982). Recent progress in integrating auxin transport phenomena with computational modeling of phyllotaxis can be found in reviews by Kuhlemeier (2007) and Heisler and Jonsson (2007).
Figure 14.
Auxin and lateral organ initiation.
(A–D) Model for lateral organ initiation by auxin.
(A) Convergent auxin transport within the L1 layer predicts positions of lateral organ initiation. Basipetal auxin transport routes appear to develop within and beneath these new organs. As this removes auxin from the L1 areas near the emerging primordia, the process will most likely reiterated furthest away from the existing primordia.
(B) The localization of PIN efflux proteins in the L1 layer concentrates auxin at sites of subsequent lateral organ initiation. PIN proteins also mediate basipetal transport from the site of initiation.
(C) Auxin transport inhibitors (such as NPA) prevent organ initiation. This block can be overcome and lateral organ initiation can be achieved by localized application of auxin.
(D) If excessive auxin is added to NPA treated meristems, threshold levels of the hormone are achieved over a wide area of the peripheral zone and enlarged organs are produced. A–D drawn from model by Reinhardt et al. (2003).
(E) Localization of PIN auxin efflux facilitators (dark blue) in the L1 layer is consistent with the converging auxin transport and accumulation at sites of incipient lateral organ initiation. Auxin influx associated proteins (red) are thought to help scavenge intracellular auxin and retain it in the cells of the L1 layer. Drawn from Reinhardt et al. (2003) and Reinhardt (2005).
(F) Model for positioning of cotyledons. In the late-globular stage embryo cotyledons appear to be initiated in rapid succession (Woodrick et al., 2000). Auxin accumulation at the first site of initiation ensures that only in the opposed position can sufficient auxin be accumulated to initiate the second cotyledon. Post-germination the first true leaf is initiated at one of two positions between the two cotyledon primordia, where sufficient auxin can accumulate to initiate a new lateral organ.
This interpretation, originally derived from auxin application experiments is well supported by auxin transport visualization studies in embryos (Benkova et al., 2003; Michniewicz et al., 2007). Prior to cotyledon outgrowth, the PIN1 protein is preferentially localized to lateral membranes of cells in the epidermal (L1) layer in a pattern consistent with the transport of auxin toward the incipient primordia (Figure 7 and Figure 14E; Benkova et al., 2003; Friml et al., 2003; Heisler et al., 2005). In lateral shoot organ initiation, these epidermal events are accompanied by the sub-epidermal formation of central auxin transport routes (Figure 7; Benkova et al., 2003; Reinhardt et al., 2003; Scarpella et al., 2006). Further evidence for a critical role for auxin localization by PIN1 is provided by analysis of the PID1 gene, which appears to be required for correct redistribution of PIN1 in the apex at the transition from globular to heart stage. Evidence suggests that the PID1 AGC kinase mediates the membrane localization of PIN proteins by phosphorylating the central hydrophilic loop (Michniewicz et al., 2007). This process is antagonized by PP2A phosphatase, which is required for normal embryonic and seedling development (Michniewicz et al., 2007). While pid1 mutations affect lateral organ initiation only in the adult plant (Christensen et al., 2000), simultaneous elimination of PID1 along with three other members of the gene family (PID2, WAG1 & WAG2) also abolishes cotyledon initiation, demonstrating the general role of this gene family in lateral organ initiation (Cheng et al., 2008).
In post-embryonic growth, correct positioning of lateral organ initiation by auxin transport mediated processes has also been shown to depend on a family of auxin influx proteins (AUXIN PERMEASE1 (AUX1) LIKE AUX1, 2 3 (LAX1, 2, 3)), (Bainbridge et al., 2008). Generally, the influx carriers appear to buffer PIN-mediated patterning (Bainbridge et al., 2008). Although single mutants in auxin influx carrier genes have not been reported to affect lateral organ positioning, the quadruple aux1 lax1 lax2 lax3 mutant was found to have irregular divergence angles and clusters of primordia (Bainbridge et al., 2008). In line with the morphological phenotype, auxin perception maxima and coordinated PIN localization are reduced in the quadruple mutant background (Bainbridge et al., 2008). Further, experiments with auxin application to the pin1 aux1 double mutant produces larger primordia than pin1 (Reinhardt et al., 2003) and auxin influx inhibitors have been found to cause wider primordia (Stieger et al., 2002).
Mutations affecting auxin synthesis have also been found to impact on lateral shoot organ positioning. There are eleven members of the flavin monooxygenase family YUCCA-LIKE (YUC genes) in Arabidopsis, which catalyze a rate limiting step in auxin biosynthesis (Zhao et al., 2001). A yuc1 yuc4 double mutant background was found to enhance the pin1 defective lateral organ initiation phenotype (Cheng et al., 2007). Subsequent genetic screens for enhancers of yuc1 yuc4 and pid1 mutants led to the cloning of the NAKED PINS IN YUC MUTANTS1/ENHANCER OF PINOID (NPY1/ENP) gene (Cheng et al., 2007). The NPY1 gene is expressed mainly in the apical region of embryos and encodes a BTB-NPH3-like protein. Double mutants for npy1 pid1 produce no cotyledons (Treml et al., 2005) and yuc1 yuc4 npy1 triple mutants produce pin-like inflorescences (Cheng et al., 2007).
In summary, impressive progress has been made in recent years in elaborating the role of auxin transport and metabolism in patterning the initiation of lateral shoot organs during embryonic and post-embryonic development. Additionally, interactions between established pathways regulating of meristem function or lateral organ formation with auxin transport and signaling continue to emerge (Section 4.2.1)
4.3.3. Adaxial-abaxial patterning
The development of laminar lateral organs involves an additional patterning step, the specification of adaxial and abaxial cell identities (Figure 2B). Histological and gene expression studies in vegetative leaves suggest that leaf anlagen are initially uniform, but become polarized in the adaxial-abaxial dimension by the time that the primordium becomes morphologically distinct (reviewed in Bowman, 2000). Surgical and genetic data indicate the existence of extrinsic signals from the meristem center as well as of intrinsic signaling within the primordium. An increasing number of regulatory genes is being implicated in dorso-ventral patterning and their roles have been recently reviewed (Bowman and Floyd, 2008).
Although cotyledons are polarized and adaxial-abaxial patterning is therefore an integral part of embryonic patterning, available experimental data are mainly derived from post-embryonic studies and therefore more appropriately covered in the chapter on shoot development. However, there are two observations which are important for the discussion of apical-basal patterning in the embryo. First, it has long been recognized that dorso-ventral patterning uses apical-basal polarity as a reference, evidenced by the fact that lateral organs fail to differentiate adaxial-abaxial polarity if insulated from apical signals through vertical incisions in the meristem (Sussex, 1954). Secondly, adaxial-abaxial patterning seems to feed back on the regulation of apical-basal patterning (Figure 2B). Mutations causing an abaxialization of the lateral primordium negatively affect SAM development, while adaxialization of the primordium is associated with promoting SAM development (McConnell and Barton, 1998; Siegfried et al., 1999). Therefore, adaxial-abaxial polarity seems to be initiated relative to apical-basal coordinates, but once established, stabilizes apical-basal patterning in the shoot apex.
As noted in 4.3.1, several genetic networks are involved in the establishment of a central/adaxial side and a lateral/abaxial fates in the embryo apex: the TCP expression domain defines the lateral side and is bordered by the CUC expression domain encircling the SAM (Aida et al., 1999; Aida et al., 2002; Palatnik et al., 2003; Koyama et al., 2007). Furthermore, the class III HD-ZIP, along with the KANADI (KAN) and YABBY (YAB) genes play a critical role in the process.
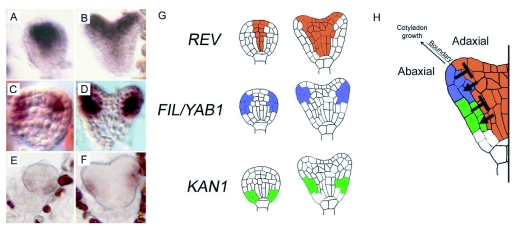
Expression of the class III HD-ZIP marks the central domain of the embryo from the globular stage and onward (Figure 15A, B, G). Although single mutants have no abnormal phenotype, double and triple mutants cannot properly generate a SAM (McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005). The classIII HD-ZIP expression domain is restricted by microRNA, namely the miR165/6 family (Williams et al., 2005). The KANADI genes, KAN1, 2, and 3 (but not KAN4) are initially expressed throughout the early globular stage embryo, but their domain of expression becomes restricted to the basal peripheral region. By the heart stage this domain comes to mark the basal abaxial portion of the cotyledon primordia (Fig 15E, F, G) (Eshed et al., 2004; McAbee et al., 2006). A triple mutant kan1 kan2 kan4 displays an enlarged SAM as well as a loss of adaxial-abaxial polarity of the cotyledons. Furthermore, ectopic expression of auxin reporters hints at the KANADI genes role as modulator of auxin response through control of auxin flux (Izhaki and Bowman, 2007).
Figure 15.
Establishment of central/peripheral and adaxial/abaxial polarity.
(A–F) In situ hybridization of REV (A, B), FIL (C, D) and KAN1 (E, F), in globular (A, C, E) and heart (B, D, F) stage embryos. Images A, B reproduced from Emery et al. (2003) with permission from Elsevier Limited; C, D reproduced/adapted from Siegfried et al. (1999) with permission from The Company of Biologists; E, F reproduced with permission from Macmillan Publishers Ltd: Nature, Kerstetter et al. (2001).
(G) Schematic representation of the expression domain shown in A–F. The class III HD-ZIP genes, here represented by REVOLUTA, are expressed in the central domain at the globular and heart stage, while the YABBY genes, represented by FILAMENTOUS FLOWER, and the KANADI genes, represented by KANANDI1, mark the peripheral domain. The activity of the class III HD-ZIPs promotes a central/adaxial fate in their domain of expression, which is also crucial for initiation of the SAM, while the KAN/YAB genes promote a peripheral/abaxial fate simultaneously limiting the expansion of the SAM. The balance of these antagonistic activities is critical for aspects of radial patterning in the embryo axis as well as for the appropriate adaxial/abaxial polarity in lateral organs, including cotyledons.
Finally, the YAB genes seem to specify abaxial fate in cotyledon primordia in a highly redundant manner. Loss-of-function mutants in the YABBY genes FILAMENTOUS FLOWER (FIL) or YAB3 have no abnormal phenotype and even the double mutant fil yab3 is only mildly distorted. However, the antagonistic, gain-of-function genotypes caused by ectopic expression of FIL or YAB3 in which the SAM stops functioning support an important role for the YAB genes in promoting abaxial fate (Siegfried et al., 1999). A current model describing this network has the class III HD-ZIP genes defining the central region of the embryo, as well as the adaxial side of the cotyledons, while the KAN and YAB genes define the periphery of the embryo and the abaxial side of the cotyledon primordia as shown in figure 15H (Kerstetter et al., 2001; McConnell et al., 2001).
In summary, a complex network of interdependent processes regulates patterning in the apical domain. Early partitioning events in the apical domain effectively set up systems of balanced controls that ensure the stable maintenance of a small central stem cell pool and the recruitment of cells exiting this pool into lateral organs. The phyllotactic positioning of these lateral organs in the periphery of the apex seems to be based on self-regulatory mechanisms involving auxin convergence in the epidermis in conjunction with narrowed subepidermal auxin transport routes that will turn into vascular tissues (Figure 6; Section 5.2). Finally, there seems to be a mutually promoting influences exchanged between cells at the adaxial side of lateral organs and the shoot meristem integrating leaf polarity into the apical-basal context.
5. THE RADIAL PATTERN
Radial patterning diversifies cell identities in the radial dimension. The first radial patterning event at the dermatogen-stage separates a surface layer and inner cells of a sphere, while subsequent radial patterning occurs within the radial symmetry of a cylinder (Figure 16). Therefore, it is possible that the separation of the epidermis and subsequent radial patterning are mechanistically unrelated.
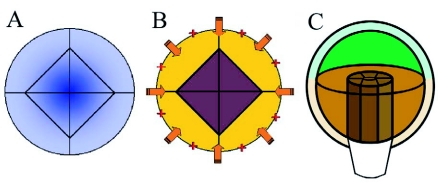
Figure 16.
Hypothetical mechanisms to separate epidermis and inner cell fate.
(A) A stable morphogen gradient (blue) could specify radial cell fates. This mechanism would allow for the concentration dependent specification of several cell identities, but only two fates, outer (epidermal) and inner cells, are specified at this stage.
(B) Alternatively, epidermal cell fate (yellow) could depend on signals from outside. External signals could originate from the surrounding milieu (orange arrows) or could be stored in the outer cell wall (red crosses) inherited from the zygote. Cells excluded from the external signal would switch to an inner cell ground state (purple).
(C) Subsequent radial patterning occurs along the axis of an elongating cylinder and may therefore be specified by a separate mechanism. This process is initiated with the formation of procambial tissue in the basal domain (brown, versus apical domain green).
5.1. Separating epidermal and inner cells
The mechanism responsible for differential cell fate acquisition by outer and inner cells at the dermatogen stage is not clear, and no mutation has been identified that completely abolishes this first step in radial patterning. Several models can be envisaged. For example, a concentration gradient of an unknown signal molecule could function as a morphogen (Wolpert, 1989; Gurdon and Bourillot, 2001) to specify cell fates in the radial dimension. Even production of the signal substance throughout the globular embryo could result in a central concentration maximum and cells could acquire distinct cell identities as a function of their distance from the center (Figure 16A). Alternatively, for example, an external signal from the endosperm could specify epidermal fate (Figure 16B). A corollary of such an ‘outside-in’ signaling mechanism is that the acquisition of inner cell fate could be dependent on proper tangential cell divisions to insulate inner cells from the external signal. This would also be true of a variation on an ‘outside-in’ model, in which epidermal fate information is deposited within the cell wall of the zygote. In this model the epidermal cells remain in contact with the outer cell wall, whereas the inner cells become separated with the first tangential division.
Since the molecular signals are unknown, none of the above models can be systematically explored. However, there is some circumstantial evidence for ‘outside-in’ signaling. Certain marker genes, such as the homeobox gene Arabidopsis thaliana MERISTEM LAYER1 (ATML1), are expressed in the apical daughter cell of the zygote, but are turned off as soon as divisions separate cells from the embryo surface (Lu et al., 1996; Sessions et al., 1999). Moreover, mutations in at least two genes, KNOLLE (KN) and KEULE (KEU), are associated with incomplete cell wall formation during cytokinesis (Lukowitz et al., 1996) and lead to the expression of epidermal markers in sub-epidermal tissues (Vroemen et al., 1996). Overall then, although the mechanism separating epidermal and inner cell identity remains elusive, its dependence on cytoplasmic discontinuity points towards some type of outward-in signaling.
During the initial patterning of the protoderm (Figure 17), from the 16 cell stage onwards, the expression of several genes are restricted to the central domain of the embryo, such as WUS (Mayer et al., 1998) and ZWILLE/PINHEAD (ZLL/PNH) (Moussian et al., 1998; Lynn et al., 1999) while others become preferentially expressed in the protoderm (Figure 17), such as PROTODERMAL FACTOR 1 and 2 (PDF1 and 2) (Abe et al., 1999; Abe et al., 2003) and AtML1 (Lu et al., 1996). PDF1/2 and AtML1 are expressed in the outer layer of the embryo (Figure 17) and subsequently in the outer (L1) layer of the shoot apical meristem. In the atml1 pdf2 double mutant, the protoderm can form but it fails to differentiate into a proper epidermis (Abe et al., 2003). Although the exact function of these genes remains unclear, PDF1 seems to be a cell wall protein (Abe et al., 1999) and AtML1 and PDF2 are GLABRA2-type homeodomain genes (Lu et al., 1996; Abe et al., 2003).
Figure 17.
Protodermal gene expression during embryogenesis.
(A) At the octant stage AtML 1, PDF1 and PDF2 are expressed in all the apical cells of the embryo proper.
(B) At the dermatogen stage, expression of these genes is restricted to the outer cells and the expression of other genes such as WUSCHEL and the class III HD-ZIP genes are specifically upregulated within the inner cells.
(C–F) Images of fluorescent reporter constructs for the expression of AtML1. C: AtML1 reporter expression in both the apical and basal cell after division of the zygote. Images C–F reproduced/adapted from Takada and Jürgens (2007) with permission from The Company of Biologists.
(D) At the dermatogen stage the reporter has been downregulated in the inner cells.
(E, F) In the late globular and heart stages respectively, the AtML1 reporter marks the outer protodermal layer.
Two receptor-like kinase (RLK) genes, ARABIDOPSIS CRINKLY4 (ACR4) and ABNORMAL LEAF SHAPE2 (ALE2) (Tanaka et al., 2002), have also been implicated in the development of surface cells. The molecular basis of the biological function of RLKs is likely to be mediated by dimerization between these proteins and it has been postulated that subtle regulation of receptor activity of ACR4 and related proteins may be achieved by the formation of different heterodimers (Stokes and Gururaj Rao, 2008). Mutations in ACR4 and ALE2 have a synergistic effect on protoderm differentiation and the ale2 acr4 double mutant fails to maintain expression of both PDF2 and AtML1 (Tanaka et al., 2002). Further, ACR4 and ALE2 are able to phosphorylate one another in vitro suggesting they may operate in a common pathway. Work investigating the role of ACR4 in organogenesis and pattern formation in the RAM and lateral root primordia has also shown that the gene product functions to restrict formative asymmetric cell divisions (De Smet et al., 2008). In the context of protodermal specification these aforementioned asymmetric divisions appear to be important in separating, specifying and maintaining the inner and outer cell layers from the 16 cell stage of embryogenesis onwards.
The expression of several epidermal genes, including ACR4, is dependent on AtML1 and PDF2, which may bind the L1 cisacting element in the promoters of these genes. The promoters of AtML1 and PDF2 themselves also contain L1 elements (Abe et al., 2003), which may be required for positive feedback reinforcement of expression (Abe et al., 2001; Takada and Jürgens, 2007). A candidate for helping establish the domains of AtML1 and PDF2 at the 16 cell stage is the integral membrane protein DEFECTIVE KERNEL1 (AtDEK1) (Lid et al., 2005). Loss of AtDEK1 eliminates AtML1 and ACR4 expression and prevents the formation of a defined protoderm. However, the mutant also displays irregular patterns of cell division and disorganized embryonic development, culminating in developmental arrest in the globular stage. This severe phenotype is difficult to interpret, but is consistent with the proposed crucial role for specific patterns and planes of cell division in the differentiation of the protoderm.
Less clearly defined in their role in radial patterning, the genes RECEPTOR-LIKE PROTEIN KINASE1 (RPK1) and TOADSTOOL2 (TOAD2) have been found to be redundantly necessary for this process: a double mutant rpk1 toad2 shows excessive cell division, aberrant protodermal cell morphologies and subsequently, embryonic arrest (Nodine et al., 2007). RPK1 has been previously reported as involved in ABA signaling (Hong et al., 1997; Osakabe et al., 2005) and TOAD2 is its closest homolog (Shiu and Bleecker, 2003). Although the exact role and function of the RLK1 and TOAD2 pairs remains elusive, other Leucine-Rich Repeat RLK, such as CLV1 (Section 4.3.1), have been shown to be involved in patterning.
5.2. Separating vascular and ground tissue
Immediately after the dermatogen-stage, divisions of inner cells become strictly oriented along the apical-basal axis, thereby establishing the external and internal cell layers of a cylinder (Figures 4E and 16C). Proper patterning of the vasculature relies on functional auxin signaling, as illustrated by arf5/mp and iaa12/bdl mutants. The most universal feature of these mutants in all organs is incomplete or belatedly formed vasculature (Przemeck et al., 1996; Hamann et al., 1999), which is a prerequisite for the formation of a basal auxin response and the initiation of an embryonic root (Section 4.2.2). As a second input, the vasculature is also dependent on correct specification of adaxial/abaxial patterning, because in the cylindrical organs of the plant axis, those cues specify the central-peripheral dimension. Consequently, mutations in class III HD-ZIP genes cause defects in the vascular patterning in the shoot (Emery et al., 2003) and ectopic expression of KAN1 or KAN2 prevent the formation of vasculature in the cotyledons and hypocotyl (Eshed et al., 2001).
Cytokinins have been implicated in cell division and cell differentiation within the vascular cylinder. The wooden leg (wol) mutation in the Arabidopsis Histidine Kinase4/CYTOKININ RESISTANT1/WOODEN LEG (AHK4/CRE1/WOL) gene results in reduced cell divisions in the root meristem and a dramatic reduction, or absence of, phloem tissues in the root and hypocotyl (Scheres et al., 1994). The AHK4/CRE1/WOL gene encodes a membrane associated two-component histidine kinase (Mahonen et al., 2000) and has been identified as a cytokinin receptor by conferring cytokinin responsiveness to yeast cells, when inserted into an appropriate yeast signal transduction pathway (Inoue et al., 2001). Two closely related proteins, AHK2 and AHK3 have also been identified as cytokinin receptors. Although genetic redundancy appears to prevent many single loss-of function mutations in AHK2, AHK3 or AHK4/CRE1/WOL manifesting an obvious phenotype, the triple ahk2 ahk3 ahk4 mutant displays abnormalities similar to those of the original wol allele (Nishimura et al., 2004). However, a triple ahk2 ahk3 ahk4 mutant displays abnormalities similar of those observed in the original wol alleles (Nishimura et al., 2004; Kuroha et al., 2006; Riefler et al., 2006). Therefore, control of cell division through cytokinin, appears to be essential for proper cell numbers and phloem formation in the vascular cylinder.
5.3. Radial patterning in the ground tissue
At around torpedo stage, the inner of two ground tissue layers is split by an asymmetric, periclinal division and the resulting two layers acquire distinguishable identities as endodermis and cortical parenchyma. In the root, separation of endodermal and cortical cell fates is ongoing, since both tissues are derived from a common initial in the RAM. Two genes mentioned in section 4.2.2, SHR and SCR, both of which encode GRAS domain transcription factors, were found to be required for both the critical asymmetric cell division and for the acquisition of separate cell identities by its products. Their interplay effectively uses the stele as inward positional reference for sub-structuring of the ground tissue and the intriguingly complex cellular mechanisms have been subject of some recent reviews (Ten Hove and Heidstra, 2008). In short, SHR acts upstream of SCR, because its activity is required for the expression of endodermal marker genes, including SCR (Helariutta et al., 2000). When switched on by SHR, SCR seems to promote the periclinal division of the ground tissue, associated with the suppression of cortical markers in the inner layer. This suggests that SHR acts at the top of a hierarchy specifying differential cell identities in the ground tissue. If this is so, how is SHR activity polarized to induce endodermal fate only in the inner ground tissue layer? Interestingly, SHR mRNA is expressed in the provascular cells in the heart embryo and in the stele of the mature root, rather than in the ground tissue (Figure 18; Helariutta et al., 2000). However, the SHR protein moves one cell layer away from its point of production, to the endodermis. This cell-to-cell movement is under the control of the SCR protein, which interact directly with SHR and sequesters it in the nuclei of the endodermal cells (Fig 18C, D) (Cui et al., 2007). Misexpression of SHR in the SCR domain results in extra layers of endodermis, probably by reiterating the emission of a positional signal, which in normal development is communicated only from the vascular cylinder to just one cell layer above. Expression of SHR under control of the promoter of its downstream target SCR is sufficient to convert this layer into a signal source and to reinitiate the entire ground tissue patterning process. Therefore, the SHR product seems to be the key signal in a short-range induction mechanism and in moving just one layer triggers endodermal differentiation from a cortical ground state adjacent to its vascular origin. The C2H2 zinc-finger protein JACKDAW (JKD) has been identified as partner and target of SHR and SCR (Cui et al., 2007; Welch et al., 2007). JKD expression is initiated during the globular stage in a SHR/SCR independent way, but its continued expression depends on the presence of SHR and to a lesser extent SCR. The function of JKD in the RAM seems to be to sequester SHR along with SCR in the nucleus of the QC cells. Furthermore, JKD is necessary to maintain the expression of SCR (Welch et al., 2007).
Figure 18.
Radial patterning in the ground tissue.
(A) The SHR gene is expressed in the vascular cylinder (brown), but its activity is required for radial patterning in the adjacent cortex-endodermal initials (orange) and in the endodermis (yellow). SHR protein moves (arrows) to cells surrounding the vascular cylinder, where it induces the expression of SCR and, with the exception of the QC (red), other endodermis specific genes. SCR acts downstream of SHR and is required for the periclinal divisions of ground tissue initials which yield the two layers of ground tissue and possibly, for maintaining endodermis identity in the inner layer. Modified after Helariutta et al. (2000).
(B) SHR expression in the embryo. A green fluorescent protein reporter gene under control of the SHR promoter is expressed exclusively in the central procambium of a triangular stage embryo. Expression remains associated with vascular tissues throughout development. From Helariutta et al. (2000). Abbreviations: g, ground tissue; hyp, hypophysis; pc, procambium; pd, protoderm; su, suspensor. Bar = 25 µm.
(C) SHR::GFP translational fusion protein is present in the stele, but selectively accumulates in the nuclei of endodermal cells (compare transcriptional fusion in insert). From Nakajima et al. (2001). Abbreviations: Cei, cortex/endodermis initial. Cor, cortex. End, endodermis. Epi, epidermis. Ste, stele. Qc, quiescent center. Bar = 50 µm.
(D) Ectopic expression of SHR driven by the SCR promoter results extra layers expressing endodermal markers. SHR protein (red) can move one layer beyond its site of production. In normal development (upper panel), it is produced in the vascular tissues (brown) and triggers SCR expression (green) and differentiation events in the overlying ground tissue. SCR is required for periclinal cell divisions subdividing the ground tissue and for proper differentiation of the endodermis (yellow). Expression of SHR in the in the SCR domain leads to a reiteration of the layer-amplification process and eventually to supernumerary endodermal layers.
6. TISSUE SPECIFIC TRANSCRIPT PROFILES
Recent research has confirmed that the functions of many important players controlling embryonic development are obscured in forward genetics approaches by widespread functional overlap. Partial redundancy is widespread in Arabidopsis. Indeed, a recent paper describing the isolation of a long-sought (co-)perceptor of ABA, PYRABACTIN RESISTANCE 1 (PYR1) (Park et al., 2009) shows why, without using a chemical approach to inhibit multiple family members (the PIRI-LIKEs (PYL), in this case) or creating higher-order mutants of multiple (PYL) family members, the classical forward genetics approach could not have worked. Consequently, the impressive accumulation of genomic data resources and diverse reverse-genetics tools generated in Arabidopsis in little over a decade have proven invaluable research assets certainly in the PYR1 example above, expression data for the various family members was in part used to guide in the creation of the necessary higher-order mutants to be able to observe an ABA-insensitive phenotype. The utility of these resources have been further enhanced by the ongoing development of bioinformatics and computational approaches to organize, process and analyze the enormous quantities of genetic and molecular data now available. Innovative experimental methods have also been developed and applied to identify new targets in the regulation of plant development through the determination and analysis of tissue- or cell-type specific gene expression patterns. These cell-type-specific samples may be generated notably by using a fluorescence-activate cell sorter (FACS) to separate GFP-positive cells of protoplasts derived from plants expressing GFP in those specific cell types, or by laser-capture microdissection.
Cell-type-specific expression profiling, as established using the FACS method by Birnbaum et al. (2003) for 5 root cell types at 3 developmental stages, is a powerful method for understanding plant biology at the level of individual cells. Gifford et al. (2008) subsequently used the technique to elucidate a transcriptional circuit within the root pericycle involving a small RNA, miR167, and its negatively regulated target, ARF8, included in the Arabidopsis Small RNA Project (ASRP), that mediates developmental plasticity. In short, the expression level of ARF8 was seen to increase in response to nitrogen, and that this was directly due to a nitrogen-stimulated decrease in miR167 production in root pericycle cells. The result is a high ratio of initiated lateral roots to emerged lateral roots under high nitrogen conditions; in nitrogen-depleted conditions, these initiated lateral roots can then emerge and explore the surrounding soil environment for nutrients. A similar study has also been undertaken for cell-type-specific responses of root cells to salt (Dinneny et al., 2008). Some instances exist where the signal of upregulation of a given gene in one tissue could be cancelled out by the downregulation of the same gene in another tissue if one were to examine just the RNA from the whole root in response to salt stress, as opposed to the response in specific cell types. Such is the case for At1g80830, encoding a manganese ion transporter, which is upregulated in the epidermis but down-regulated in the cortex in response to salt. Two quasi-cell-type-specific gene expression data sets for embryo development have been generated by the Lindsey laboratory using laser capture microdissection, followed by RNA amplification and hybridization to the Affymetrix ATH1 GeneChip (Casson et al., 2005; Spencer et al., 2007). These data sets encompass apical and basal regions of globular, heart and torpedo stage embryos, and are queryable for more than 22,000 genes on the ATH1 GeneChip using the eFP Browser of the Bio-Array Resource (Toufighi et al., 2005; Winter et al., 2007) or other online tools — except Genevestigator (Zimmermann et al., 2004), which removed these data sets because the amplification procedure necessarily introduced biases towards the 3′ probe sets, rendering expression signals from these amplified samples and other non-amplified samples in their database not entirely comparable. As long as one is aware of this fact, the data are still very useful, even if the resolution is more tissue- as opposed to cell-type-specific.
In conclusion, these studies illustrate the prospect and current limitations for obtaining transcript profiles of embryonic tissues. Given the high degree of functional redundancy between members of gene families in Arabidopsis, the interpretation of genetic approaches as discussed in this chapter is highly dependent on knowledge of transcript profiles, which can be expected to be detailed further in the near future.
7. CONCLULSIONS AND PROSPECTS
The still incomplete list of mechanisms coordinating cell behavior in the embryo is already substantial and includes differential cell fate acquisition after asymmetric cell division, micro-RNA regulation, long-range signaling through low-molecular weight substances, short-range signaling via transcription factor transfer, unknown tissue-identity maintenance signals and possibly signaling through extra-embryonic and/or cell wall associated determinants. The cell pattern established by these signals constitutes a foundation for further, equally complex interactions establishing and maintaining the organization of the apical meristem and regulating late-embryonic programs. Together therefore, the tight overlap of developmental programs operating in the embryo, has made the genetic analysis of this developmental phase a rich source of insight into plant cell signaling in general.
Knowledge acquired regarding the molecular mechanisms regulating early development in the Arabidopsis embryo has already proved valuable in guiding research and comparative investigations in a number of species. Impressive progress in recent years, lead by research in Arabidopsis, suggests that more comprehensive understanding and manipulation of embryonic patterning in important angiosperm species is close at hand.
Acknowledgements
We would like to thank John Runions for Fig. 3 and the following publishers for allowing the use of published images as indicated in the figure legends: Nature Publishing Group, The Company of Biologists ltd., Cell Press, The American Association for the Advancement of Science, and Elsevier Publishing. We would like to apologize for not being able to review and cite all relevant literature because of space constraints. Our research has been supported by grants from the Natural Science and Engineering Research Council of Canada to T.B.
Footnotes
Citation: Capron A., Chatfield S., and Provart N., and Berleth T. (2009) Embryogenesis: Pattern Formation from a Single Cell. The Arabidopsis Book 7:e0126. doi:10.1199/tab.0126
elocation-id: e0126
First published on November 12, 2009
This chapter is an updated version of a chapter originally published on September 30, 2002 doi:http://dx.doi.org/10.1199/tab.0051
References
- Abe M., Takahashi T., Komeda Y. Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant and Cell Physiology. 1999;407(1):571–580. doi: 10.1093/oxfordjournals.pcp.a029579. [DOI] [PubMed] [Google Scholar]
- Abe M., Takahashi T., Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;267(1):487–494. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- Abe M., Katsumata H., Komeda Y., T. Takahashi. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;1307(1):635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- ]Aida M., Ishida T., Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;1267(1):1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;97(1):841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development. 2002;1297(1):3965–3974. doi: 10.1242/dev.129.17.3965. [DOI] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;1197(1):109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Bainbridge K., Guyomarc'h S., Bayer E., Swarup R., Bennett M., Mandel T., Kuhlemeier C. Auxin influx carriers stabilize phyllotactic patterning. Genes & Development. 2008;227(1):810–823. doi: 10.1101/gad.462608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.K., Poethig R.S. Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in wild-type and in the shoot meristemless mutant. Development. 1993;1197(1):823–831. [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. Paternal control of embryonic patterning in Arabidopsis thaliana. Science. 2009;3237(1):1485–1488. doi: 10.1126/science.1167784. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. Auxin: the looping star in plant development. Annu Rev Plant Biol. 2008;597(1):443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Benkova E., Ivanchenko M.G., Friml J., Shishkova S., Dubrovsky J.G. A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trends Plant Sci. 2009;147(1):189–193. doi: 10.1016/j.tplants.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;1157(1):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Berger F. Endosperm: the crossroad of seed development. Curr Opin Plant Biol. 2003;67(1):42–50. doi: 10.1016/s1369526602000043. [DOI] [PubMed] [Google Scholar]
- Berleth T., Jurgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;1187(1):575–587. [Google Scholar]
- Bhalerao R.P., Eklof J., Ljung K., Marchant A., Bennett M., Sandberg G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 2002;297(1):325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. A gene expression map of the Arabidopsis root. Science. 2003;3027(1):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;4337(1):39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Boutte Y., Ikeda Y., Grebe M. Mechanisms of auxin-dependent cell and tissue polarity. Curr Opin Plant Biol. 2007;107(1):616–623. doi: 10.1016/j.pbi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Bowman J.L. Axial patterning in leaves and other lateral organs. Curr Opin Genet Dev. 2000;107(1):399–404. doi: 10.1016/s0959-437x(00)00103-9. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Floyd S.K. Patterning and polarity in seed plant shoots. Annu Rev Plant Biol. 2008;597(1):67–88. doi: 10.1146/annurev.arplant.57.032905.105356. [DOI] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Developmental Cell. 2008;147(1):867–876. doi: 10.1016/j.devcel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Brown R.C., Lemmon B.E., Nguyen H., Olsen O.A. Development of endosperm in Arabidopsis thaliana. Sexual Plant Reproduction. 1999;127(1):32–34. [Google Scholar]
- Byrne M.E., Simorowski J., Martienssen R.A. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;1297(1):1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. Asymmetric leaves 1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;4087(1):967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Capron A., Serralbo O., Fulop K., Frugier F., Parmentier Y., Dong A., Lecureuil A., Guerche P., Kondorosi E., Scheres B., Genschik P. The Arabidopsis anaphase-promoting complex or cyclosome: molecular and genetic characterization of the APC2 subunit. Plant Cell. 2003;157(1):2370–2382. doi: 10.1105/tpc.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inze D., Sandberg G., Casero P.J., Bennett M. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell. 2001;137(1):843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S., Spencer M., Walker K., Lindsey K. Laser capture microdissecation for the analysis of gene expression during embrypgenesis of Arabidopsis. Plant J. 2005;427(1):111–123. doi: 10.1111/j.1365-313X.2005.02355.x. [DOI] [PubMed] [Google Scholar]
- Chandler J., Nardmann J., Werr W. Plant development revolves around axes. Trends Plant Sci. 2008;137(1):78–84. doi: 10.1016/j.tplants.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Werr W. BIM1. a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol Biol. 2009;697(1):57–68. doi: 10.1007/s11103-008-9405-6. [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Grewe B., Werr W. The AP2 transcription factors DORNROSCHEN and DORNROSCHENLIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development. 2007;1347(1):1653–1662. doi: 10.1242/dev.001016. [DOI] [PubMed] [Google Scholar]
- Cheng Y.F., Qin G.J., Dai X.H., Zhao Y.D. NPY1 a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2007;1047(1):18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.F., Qin G.J., Dai X.H., Zhao Y.D. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2008;1057(1):21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;1007(1):469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Clark S.E. Cell signalling at the shoot meristem. Nature Reviews Molecular Cell Biology. 2001;27(1):276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;897(1):575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Cole M., Chandler J., Weijers D., Jacobs B., Comelli P., Werr W. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;1367(1):1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. An evolutionary conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;3167(1):421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- De Smet D., Jochmans K., Neyns B. Development of thrombotic thrombocytopenic purpura after a single dose of gemcitabine. Ann Hematol. 2008;877(1):495–496. doi: 10.1007/s00277-007-0429-9. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;4357(1):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Tanaka H., Goh T., Ebine K., Mahonen A.P., Prasad K., Blilou I., Geldner N., Xu J., Uemura T., Chory J., T. Ueda, Nakano A., Scheres B., Friml J. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature. 2008;4567(1):962–966. doi: 10.1038/nature07409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. Cell Identity Mediates the Response of Arabidopsis Roots to Abiotic Stress. Science. 2008;3207(1):942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial patterning of Arabidopsis shoots by class IMHD-ZIP and KANADI genes. Current Biology. 2003;137(1):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum S.F., Perea J.V., Bowman J.L. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;117(1):1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;1317(1):2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- Faure J.D., Vittorioso P., Santoni V., Fraisier V., Prinsen E., Barlier I., Van Onckelen H., Caboche M., Bellini C. The PASTIC-CINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development. 1998;1257(1):909–918. doi: 10.1242/dev.125.5.909. [DOI] [PubMed] [Google Scholar]
- Fleming A.J. Plant signalling: the inexorable rise of auxin. Trends Cell Biol. 2006;167(1):397–402. doi: 10.1016/j.tcb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jurgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;4267(1):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P.B., Ljung K., Sandberg G., Hooykaas P.J., Palme K., Offringa R. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;3067(1):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Friml J., Benfey P., Benkova E., Bennett M., Berleth T., Geldner N., Grebe M., Heisler M., Hejatko J., Jurgens G., Laux T., Lindsey K., Lukowitz W., Luschnig C., Offringa R., Scheres B., Swamp R., Torres-Ruiz R., Weijers D., Zazimalova E. Apical-basal polarity: why plant cells don't stand on their heads. Trends Plant Science. 2006;117(1):12–14. doi: 10.1016/j.tplants.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Furutani M., Vernoux T., Traas J., Kato T., Tasaka M., Aida M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development. 2004;1317(1):5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;4497(1):1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gallois J.L., Nora F.R., Mizukami Y, Sablowski R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004;187(1):375–380. doi: 10.1101/gad.291204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galweiler L., Guan C., Muller A., Wisman E., Mendgen K., Yephremov A., Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;2827(1):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jurgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;1127(1):219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences. 2008;1057(1):803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;107(1):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B., Bourillot P.Y. Morphogen gradient interpretation. Nature. 2001;4137(1):797–803. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- Hadfi K., Speth V., Neuhaus G. Auxin-induced developmental patterns in Brassica juncea embryos. Development. 1998;1257(1):879–887. doi: 10.1242/dev.125.5.879. [DOI] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;1317(1):657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jurgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;1267(1):1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Hamann T., Benkova E., Baurle I., Kientz M., Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes & Development. 2002;167(1):1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBOJ. 1998;177(1):1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Ckurshumova W., Vidaurre D.P., Singh S.A., Stamatiou G., Tiwari S.B., Hagen G., Guilfoyle T.J., Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;1317(1):1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Jonsson H. Modelling meristem development in plants. Curr Opin Plant Biol. 2007;107(1):92–97. doi: 10.1016/j.pbi.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;157(1):1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;1017(1):555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Hellmann H., Hobble L., Chapman A., Dharmasiri S., Dharmasiri N., del Pozo C., Reinhardt D., Estelle M. Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 2003;227(1):3314–3325. doi: 10.1093/emboj/cdg335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L., McGovern M., Hurwitz L.R., Pierro A., Liu N.Y., Bandyopadhyay A., Estelle M. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development. 2000;1277(1):23–32. doi: 10.1242/dev.127.1.23. [DOI] [PubMed] [Google Scholar]
- Hong S.W., Jon J.H., Kwak J.M., Nam H.G. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 1997;1137(1):1203–1212. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;4097(1):1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Izhaki A., Bowman J.L. KANADI and class IIIHD-zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell. 2007;197(1):495–508. doi: 10.1105/tpc.106.047472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik P.D., Gillmor C.S., Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol. 2007;237(1):207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;117(1):1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Feldman L.J. Regulation of root apical meristem development. Annu Rev Cell Dev Biol. 2005;217(1):485–509. doi: 10.1146/annurev.cellbio.21.122303.114753. [DOI] [PubMed] [Google Scholar]
- Johri B.M. Embryology of Angiosperms. 1984. (Berlin: Springer).
- Jun J.H., Fiume E., Fletcher J.C. The CLE family of plant polypeptide signaling molecules. Cell Mol Life Sci. 2008;657(1):743–755. doi: 10.1007/s00018-007-7411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J. 2001;207(1):3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagi C., Gross-Hardt R. How females become complex: cell differentiation in the gametophyte. Curr Opin Plant Biol. 2007;107(1):633–638. doi: 10.1016/j.pbi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Kajiwara T., Furutani M., Hibara K., Tasaka M. The GURKE gene encoding an acetyl-CoA carboxylase is required for partitioning the embryo apex into three subregions in Arabidopsis. Plant and Cell Physiology. 2004;457(1):1122–1128. doi: 10.1093/pcp/pch148. [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;4357(1):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;4117(1):706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- Kim I., Zambryski P.C. Cell-to-cell communication via plasmodesmata during Arabidopsis embryogenesis. Curr Opin Plant Biol. 2005;87(1):593–599. doi: 10.1016/j.pbi.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol. 2008;247(1):447–473. doi: 10.1146/annurev.cellbio.24.110707.175254. [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;197(1):473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M., Bennett M.J. Auxin transport: a field in flux. Trends Plant Sci. 2006;117(1):382–386. doi: 10.1016/j.tplants.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kuhlemeier C. Phyllotaxis. Trends Plant Sci. 2007;127(1):143–150. doi: 10.1016/j.tplants.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Kuroha T., Ueguchi C., Sakakibara H., Satoh S. Cytokinin receptors are required for normal development of auxin-transporting vascular tissues in the hypocotyl but not in adventitious roots. Plant Cell Physiol. 2006;477(1):234–243. doi: 10.1093/pcp/pci240. [DOI] [PubMed] [Google Scholar]
- Laufs P., Peaucelle A., Morin H., Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;1317(1):4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- Laux T., Wurschum T., Breuninger H. Genetic regulation of embryonic pattern formation. Plant Cell. 2004;167(1):S190–202. doi: 10.1105/tpc.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T., Mayer K.F.X., Berger J., Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;1227(1):87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Leyser O. Dynamic integration of auxin transport and signalling. Curr Biol. 2006;167(1):R424–433. doi: 10.1016/j.cub.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Lid S.E., Olsen L., Nestestog R., Aukerman M., Brown R.C., Lemmon B., Mucha M., Opsahl-Sorteberg H.G., Olsen O.A. Mutation in the Arabidopisis thaliana DEK1 calpain gene perturbs endosperm and embryo development while over-expression affects organ development globally. Planta. 2005;2217(1):339–351. doi: 10.1007/s00425-004-1448-6. [DOI] [PubMed] [Google Scholar]
- Liu C., Xu Z., Chua N.H. Auxin Polar Transport Is Essential for the Establishment of Bilateral Symmetry during Early Plant Embryogenesis. Plant Cell. 1993;57(1):621–630. doi: 10.1105/tpc.5.6.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;3797(1):66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Long J.A., Woody S., Poethig S., Meyerowitz E.M., Barton M.K. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development. 2002;1297(1):2797–2806. doi: 10.1242/dev.129.12.2797. [DOI] [PubMed] [Google Scholar]
- Lu P., Porat R., Nadeau J.A., O'Neill S.D. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell. 1996;87(1):2155–2168. doi: 10.1105/tpc.8.12.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Mayer U., Jurgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;847(1):61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell. 2004;1167(1):109–119. doi: 10.1016/s0092-8674(03)01067-5. [DOI] [PubMed] [Google Scholar]
- Luo Y., Koop H.U. Somatic embryogenesis in cultured immature zygotic embryos and leaf protoplasts ofArabidopsis thaliana ecotypes. Planta. 1997;2027(1):387–396. doi: 10.1007/s004250050141. [DOI] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., Barton M.K. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;1267(1):469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Mahonen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;147(1):2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Current Biology. 2004;147(1):1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. Early embryogenesis in Arabidopsis thaliana: I. The developing embryo. Can J Bot. 1991;697(1):461–476. [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jurgens G., Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;957(1):805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Mayer U., Jurgens G. Pattern formation in plant embryogenesis: a reassessment. Semin Cell Dev Biol. 1998;97(1):187–193. doi: 10.1006/scdb.1997.0210. [DOI] [PubMed] [Google Scholar]
- Mayer U., Buttner G., Jurgens G. Apical-basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development. 1993;1177(1):149–162. [Google Scholar]
- McAbee J.M., Hill T.A., Skinner D.J., Izhaki A., Hauser B.A., Meister R.J., Reddy G.V., Meyerowitz E.M., Bowman J.L., Gasser C.S. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant Journal. 2006;467(1):522–531. doi: 10.1111/j.1365-313X.2006.02717.x. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;1257(1):2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;4117(1):709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Models of biological pattern formation. 1982. (London: Academic Press).
- Michniewicz M., Zago M.K., Abas L., Weijers D., Schweighofer A., Meskiene I., Heisler M.G., Ohno C., Zhang J., Huang F., Schwab R., Weigel D., Meyerowitz E.M., Luschnig C., Offringa R., Friml J. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;1307(1):1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol. 2008;247(1):55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Mordhorst A.P., Toonen M.A.J., deVries S.C. Plant embryogenesis. Critical Reviews in Plant Sciences. 1997;167(1):535–576. [Google Scholar]
- Moussian B., Schoof H., Haecker A., Jurgens G., Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. Embo Journal. 1998;177(1):1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B., Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;4537(1):1094–U1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;4137(1):307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Nawy T., Lukowitz W., Bayer M. Talk global, act local - Patterning the Arabidopsis embryo. Current Opinion in Plant Biology. 2008;117(1):28–33. doi: 10.1016/j.pbi.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;167(1):1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M.D., Yadegari R., Tax F.E. RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev Cell. 2007;127(1):943–956. doi: 10.1016/j.devcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;3197(1):294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- Olsen O.A. Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell. 2004;167(1):S214–227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Maruyama K., Seki M., Satou M., Shinozaki K., Yamaguchi-Shinozaki K. Leucine-rich repeat receptor-like kinasel is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005;177(1):1105–1119. doi: 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont K.S., Hardtke C.S. The topless plant developmental phenotype explained! Genome Biol. 2008;97(1):219. doi: 10.1186/gb-2008-9-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. Control of leaf morphogenesis by microR-NAs. Nature. 2003;4257(1):257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., Alfred S.E., Bonetta D., Finkelstein R., Provart N.J., Desveaux D., Rodriguez P.L., McCourt P., Zhu J.K., Schroeder J.I., Volkman B.F., Cutler S.R. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;3247(1):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J., Mravec J., Bouchard R., Blakeslee J.J., Abas M., Seifertova D., Wisniewska J., Tadele Z., Kubes M., Covanova M., Dhonukshe P., Skupa P., Benkova E., Perry L., Krecek P., Lee O.R., Fink G.R., Geisler M., Murphy A.S., Luschnlg C., Zazimalova E., Friml J. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;3127(1):914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Ploense S.E., Wu M.F., Nagpal P., Reed J.W. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development. 2009;1367(1):1509–1517. doi: 10.1242/dev.025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;177(1):61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck G.K., Mattsson J., Hardtke C.S., Sung Z.R., Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;2007(1):229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- Raghavan V. Pattern formation in angiosperm embryos. Botanica. 2000;507(1):33–47. [Google Scholar]
- Raghavan V. Can carrot and Arabidopsis serve as model systems to study the molecular biology of somatic embryogenesis? Current Science. 2006;907(1):1336–1343. [Google Scholar]
- Ray A. New paradigms in plant embryogenesis: maternal control comes in different flavors. Trends in Plant Science. 1998;37(1):325–327. [Google Scholar]
- Reinhardt D. Regulation of phyllotaxis. Int J Dev Biol. 2005;497(1):539–546. doi: 10.1387/ijdb.041922dr. [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;127(1):507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;4267(1):255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmulling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;187(1):40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Friml J. Auxin and other signals on the move in plants. Nat Chem Biol. 2009;57(1):325–332. doi: 10.1038/nchembio.170. [DOI] [PubMed] [Google Scholar]
- Rose R.J., Nolan K.E. Genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula. In-Vitro Cellular and Developmental Biology. Plant. 2006;427(1):73–81. [Google Scholar]
- Sachs T. Cell polarity and tissue patterning in plants. Development (Suppl. ) 1991;17(1):83–93. [Google Scholar]
- Sachs T. The Role of Auxin in the Polar Organization of Apical Meristems. Australian Journal of Plant Physiology. 1993;207(1):541–553. [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;4467(1):811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;207(1):1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. Non-linear signaling for pattern formation? Curr Opin Plant Biol. 2000;37(1):412–417. doi: 10.1016/s1369-5266(00)00105-9. [DOI] [PubMed] [Google Scholar]
- Scheres B., Heidstra R. Digging out roots: Pattern formation, cell division, and morphogenesis in plants. In Current Topics in Developmental Biology. 1999;457(1):207–247. doi: 10.1016/s0070-2153(08)60317-8. [DOI] [PubMed] [Google Scholar]
- Scheres B., Wolkenfelt H., Willemsen V., Terlouw M., Lawson E., Dean C., Weisbeek P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;1207(1):2475–2487. [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jurgens G., Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;1007(1):635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schoute J. Beitrage zur Blattstellungslehre. Rec. Trav. Bot. Neerl. 1913;107(1):153–325. [Google Scholar]
- Schwartz B.W., Yeung E.C., Meinke D.W. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development. 1994;1207(1):3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant Journal. 1999;207(1):259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;1327(1):530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;1267(1):4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Spencer M.W.B., Casson S.A., Lindsey K. Transcriptional Profiling of the Arabidopsis Embryo. Plant Physiol. 2007;1437(1):924–940. doi: 10.1104/pp.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R. Plant stem cell niches. Int J Dev Biol. 2005;497(1):479–489. doi: 10.1387/ijdb.041929ys. [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C.L., Paris S., Galweiler L., Palme K., Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;2867(1):316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Stieger P.A., Reinhardt D., Kuhlemeier C. The auxin influx carrier is essential for correct leaf positioning. Plant Journal. 2002;327(1):509–517. doi: 10.1046/j.1365-313x.2002.01448.x. [DOI] [PubMed] [Google Scholar]
- Stokes K.D., Gururaj Rao A. Dimerization properties of the transmembrane domains of Arabidopsis CRINKLY4 receptor-like kinase and homologs. Arch Biochem Biophys. 2008;4777(1):219–226. doi: 10.1016/j.abb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Sussex I.M. Experiments on the Cause of Dorsoventrality in Leaves. Nature. 1954;1747(1):351–352. [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;3197(1):1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Takada S., Jurgens G. Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development. 2007;1347(1):1141–1150. doi: 10.1242/dev.02803. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Watanabe M., Watanabe D., Tanaka T., Machida C., Machida Y. ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant and Cell Physiology. 2002;437(1):419–428. doi: 10.1093/pcp/pcf052. [DOI] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;77(1):847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Ten Hove C.A., Heidstra R. Who begets whom? Plant cell fate determination by asymmetric cell division. Curr Opin Plant Biol. 2008;117(1):34–41. doi: 10.1016/j.pbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz R.A., Jurgens G. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development. 1994;1207(1):2967–2978. doi: 10.1242/dev.120.10.2967. [DOI] [PubMed] [Google Scholar]
- TorresRuiz R.A., Lohner A., Jurgens G. The GURKE gene is required for normal organization of the apical region in the Arabidopsis embryo. Plant Journal. 1996;107(1):1005–1016. doi: 10.1046/j.1365-313x.1996.10061005.x. [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady M., Austin R., Ly E., Provart N. The Botany Array Resource: e-Northerns, Expression Angling, and Promoter Analyses. Plant J. 2005;437(1):153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Treml B.S., Winderl S., Radykewicz R., Herz M., Schweizer G., Hutzier P., Glawischnig E., Ruiz R.A. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development. 2005;1327(1):4063–4074. doi: 10.1242/dev.01969. [DOI] [PubMed] [Google Scholar]
- Trotochaud A.E., Hao T., Wu G., Yang Z., Clark S.E. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell. 1999;117(1):393–406. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.R., Laux T. Connecting the paths in plant stem cell regulation. Trends in Cell Biology. 2007;177(1):403–410. doi: 10.1016/j.tcb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Turing A.M. The Chemical Basis of Morphogenesis. Philos Trans R Soc Lond B. 1952;2377(1):37–72. [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. Functional differentiation of endosomes in Arabidopsis cells. Plant J. 2004;407(1):783–789. doi: 10.1111/j.1365-313X.2004.02249.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;97(1):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hage W., Weisbeek P., Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;3787(1):62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hendriks G., Weisbeek P., Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;3907(1):287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- Vernon D.M., Meinke D.W. Embryogenic transformation of the suspensor in twin, a polyembryonic mutant of Arabidopsis. Dev Biol. 1994;1657(1):566–573. doi: 10.1006/dbio.1994.1276. [DOI] [PubMed] [Google Scholar]
- Vidaurre D.P., Ploense S., Krogan N.T., Berleth T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development. 2007;1347(1):2561–2567. doi: 10.1242/dev.006759. [DOI] [PubMed] [Google Scholar]
- Vittorioso P., Cowling R., Faure J.D., Caboche M., Bellini C. Mutation in the Arabidopsis PASTICCIN01 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Molecular and Cellular Biology. 1998;187(1):3034–3043. doi: 10.1128/mcb.18.5.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen C.W., Mordhorst A.P., Albrecht C., Kwaaitaal M.A., de Vries S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;157(1):1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen C.W., Langeveld S., Mayer U., Ripper G., Jurgens G., Van Kammen A., De Vries S.C. Pattern Formation in the Arabidopsis Embryo Revealed by Position-Specific Lipid Transfer Protein Gene Expression. Plant Cell. 1996;87(1):783–791. doi: 10.1105/tpc.8.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;197(1):63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Jurgens G. Auxin and embryo axis formation: the ends in sight? Curr Opin Plant Biol. 2005;87(1):32–37. doi: 10.1016/j.pbi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Weijers D., Schlereth A., Ehrismann J.S., Schwank G., Kientz M., Jurgens G. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Developmental Cell. 2006;107(1):265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Weijers D., Sauer M., Meurette O., Friml J., Ljung K., Sandberg G., Hooykaas P., Offringa R. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell. 2005a;177(1):2517–2526. doi: 10.1105/tpc.105.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Benkova E., Jager K.E., Schlereth A., Hamann T., Kientz M., Wilmoth J.C., Reed J.W., Jurgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005b;247(1):1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;217(1):2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V., Scheres B. Mechanisms of pattern formation in plant embryogenesis. Annu Rev Genet. 2004;387(1):587–614. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- Williams L., Grigg S.P., Xie M., Christensen S., Fletcher J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005;1327(1):3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An ‘Electronic Fluorescent Pictograph’ Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS One. 2007;27(1):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertova D., Brewer P.B., Ruzicka K., Blilou I., Rouquie D., Benkova E., Scheres B., Friml J. Polar PIN localization directs auxin flow in plants. Science. 2006;3127(1):883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Positional information revisited. Development. 1989;1077(1):3–12. doi: 10.1242/dev.107.Supplement.3. [DOI] [PubMed] [Google Scholar]
- Woodrick R., Martin P.R., Birman I., Pickett F.B. The Arabidopsis embryonic shoot fate map. Development. 2000;1277(1):813–820. doi: 10.1242/dev.127.4.813. [DOI] [PubMed] [Google Scholar]
- Wu X., Chory J., Weigel D. Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Developmental Biology. 2007;3097(1):306–316. doi: 10.1016/j.ydbio.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R., Drews G.N. Female gametophyte development. Plant Cell. 2004;167(1):S133–141. doi: 10.1105/tpc.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazimalova E., Krecek P., Skupa P., Hoyerova K., Petrasek J. Polar transport of the plant hormone auxin - the role of PINFORMED (PIN) proteins. Cell Mol Life Sci. 2007;647(1):1621–1637. doi: 10.1007/s00018-007-6566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z., Somerville C.R. Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Proc Natl Acad Sci USA. 1997;947(1):7349–7355. doi: 10.1073/pnas.94.14.7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.D., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;2917(1):306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffman M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;1367(1):2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]