INTRODUCTION
Variegation mutants have been defined as “any plant that develops patches of different colors in its vegetative parts” (Kirk and Tilney-Bassett, 1978). Some of the most common variegations have green and white (or yellow) sectors in normally-green tissues and organs of the plant. Whereas cells in the green sectors typically contain normal-appearing chloroplasts, cells in the white (or yellow) sectors contain plastids that are deficient in chlorophyll and/or carotenoid pigments. These plastids appear to be blocked at various steps of chloroplast biogenesis because they frequently lack organized internal membrane structures and/or contain only rudimentary lamellae. Despite their widespread occurrence, relatively few variegations have been characterized at the molecular level.
Variegation mutants have played a prominent role in the history of genetics (reviewed by Granick, 1955; Kirk and Tilney-Bassett, 1978). As a notable example, the finding that transmission of the variegation trait does not always obey Mendel's laws led to the discovery of non-Mendelian inheritance in the early 1900's. Whereas variegations have long been associated with differences in plastid form and function, it was not possible to gain insight into the molecular basis of this phenomenon until the 1960's and early 1970's, when compelling molecular evidence was presented that mitochondria and chloroplasts contain their own DNA and protein synthesis systems, and that organellar proteins are the products of genes in the chloroplast and mitochondrion, as well as the nucleus (reviewed by Bogorad, 1981).
Nuclear-organelle interactions
Nuclear DNA-encoded proteins that are destined for chloroplasts and mitochondria are translated as precursors on 80S ribosomes in the cytosol and transported into the organelle post-translationally, whereas proteins encoded in the mitochondrial and plastid genomes are translated on prokaryotic-like 70S ribosomes in the organelle itself (reviewed by Goldschmidt-Clermont, 1998; Leon et al., 1998). Mitochondria and plastids are derived from prokaryotic endosymbionts, and during the process of symbiogenesis most of the genes of the symbiont were lost or transferred to the host genome (Gray, 1992; Doolittle, 1998; Cavalier-Smith, 2000). For instance, current-day plastid genomes in higher plants code for less than 100 of the estimated 1900 to 2500 proteins in a typical chloroplast (Abdallah et al., 2000; Martin and Herrmann, 1998). Plastid and mitochondrial genomes are polyploid, and in higher plant cells they are dispersed among many plastids and mitochondria. For example, a typical mesophyll cell contains from 1,000 to 10,000 identical plastid DNAs distributed among 100 or more chloroplasts (Bendich, 1987).
It is thought that the dispersal of genes for plastid and mitochondrial proteins between two different compartments was the central driving force that led to the evolution of mechanisms that integrate nuclear and organellar gene expression (reviewed by Bogorad, 1981). A contributing factor might also have been the vast disparity in copy number between genes in the nuclear and organellar compartments. Much of the regulatory traffic between the nucleus and the organelle is anterograde, i.e., from the nucleus to the organelle, in the form of nuclear gene products that control the transcription and translation of mitochondrial and plastid genes (reviewed by Goldschmidt-Clermont, 1998; Leon et al., 1998). Yet, much of this traffic is also retrograde, i.e., from the organelle to the nucleus (reviewed by Oelmüller, 1989; Taylor, 1989; Mayfield, 1990; Susek and Chory, 1992; Gray et al., 1995; Hess et al., 1997; Rodermel, 2001).
Perhaps the best understood examples of retrograde trafficking in plants involve the transcriptional regulation of nuclear genes for photosynthetic proteins by plastid-to-nucleus signaling mechanisms initiated by a variety of “plastid signals” (reviewed by Rodermel, 2001). The plastid signals identified to date are intermediates or by-products of photosynthetic metabolism. Yet, poorly understood retrograde signals also control the expression of nuclear genes for a number of non-plastid proteins (reviewed by Oelmüller, 1989; Barak et al., 2001), as well as the transcription of mitochondrial genes, cell differentiation and leaf morphogenesis (reviewed by Hess et al., 1997; Hedtke et al., 1999; Rodermel, 2001). Consequently, plastid-to-nucleus signaling plays a central role in coordinating gene expression in the nucleus, plastid and mitochondrion, and in integrating pathways of cellular metabolism and development. Presumably, there is crosstalk between retrograde signaling and other signal transduction pathways (e.g., light, hormones, sugars, developmental factors) that also play a role in coordinating nuclear and organelle gene expression (e.g., Neff et al., 2000; Oswald et al., 2001).
Mechanisms of variegation
Variegations can arise by many different mechanisms (reviewed by Kirk and Tilney-Bassett, 1978; Tilney-Bassett, 1975). Some variegations are induced by external agents and are not heritable. For instance, chlorotic leaf sectors can be generated by preferential shading, pathogen attack, and nutritional deficiencies. Heritable variegations, on the other hand, mostly arise from mutations in nuclear, plastid and/or mitochondrial genes. Many of these are green-white variegations in which plastids in the white sectors fail to accumulate (or aberrantly accumulate) photosynthetic pigments. All the steps of chlorophyll and carotenoid biosynthesis occur in the plastid by nuclear gene products that are imported into the organelle post-translationally (reviewed by Bartley and Scolnik, 1995; vonWettstein, 1995). Consequently, heritable variegations arise from mutations in any compartment that affect pigment synthesis or accumulation in the plastid, either directly or indirectly.
Many variegations are caused by mutations in nuclear genes that generate defective plastids in some, but not all, cells of the plant. These variegations are Mendelian-inherited. In many cases, the defective plastids (and the cells that contain them) replicate normally, or nearly so, and sort out to produce clones of cells containing morphologically normal chloroplasts (green sectors) or abnormal plastids (white or yellow sectors) (reviewed by Kirk and Tilney-Bassett, 1978; Tilney-Bassett, 1984, 1989; Hagemann, 1986). The abnormal plastids are often, but not always, “permanently-defective”. Permanently-defective plastids are inherited in a non-Mendelian fashion, i.e. maternally (in the majority of angiosperms) or biparentally (Tilney-Bassett, 1975; Connett, 1987). In cases of maternal inheritance, the probability of transmission of a permanently-defective organelle is related to the extent of variegation of the mother plant.
In principle, permanently-defective plastids can arise in many different ways. One common way is by the generation of mutations in the plastid genome by defective nuclear gene products, as in some “chloroplast mutator” lines (reviewed by Tilney-Bassett, 1975; Kirk and Tilney-Bassett, 1978; Börner and Sears, 1986; Hagemann, 1986; Chang et al., 1996). Because plastid genomes are multicopy, it is thought that chloroplast mutator and other “plastome mutants” (the general term for mutants that contain chloroplast DNA lesions) are variegated because mutant and normal plastid chromosomes, following replication, sort out to form clones of plastids and cells containing either all-normal plastid DNAs (green sectors) or all-mutant plastid DNAs (white sectors) (Kirk and Tilney-Bassett, 1978; Tilney-Bassett, 1975; Birky, 1983). Plastids containing a single type of plastid DNA are termed “homoplasmic”, whereas ones with different types are termed “heteroplasmic”.
Another way that permanently-defective plastids can arise is by the loss of plastid 70S ribosomes, as in the iojap mutant of maize and the albostrians mutant of barley (Walbot and Coe, 1979; Börner and Sears, 1986; Coe et al., 1988; Han et al., 1992; Hess et al., 1992, 1993, 1994a, 1994b; Zubko and Day, 1998; Hedtke et al., 1999). The plastids in the white sectors of these mutants are permanently-defective because plastid ribosome biogenesis requires the ability to translate 70S ribosomal proteins encoded in the plastid genome. Hence, once a plastid has lost its ribosomes, it can't regain them even if returned to a wild type nuclear background. The primary lesions in iojap and albostrians are not well-understood, but IOJAP has been cloned and appears to code for a component of the 50S subunit of the plastid ribosome (Han et al., 1992; Han and Martienssen, 1995).
In addition to variegations that are caused by the induction of permanently-defective plastids by mutations in nuclear genes, variegations can also be caused by mutations in nuclear genes that generate permanently-defective mitochondria. Notable examples include the nonchromosomal stripe (NCS) mutants of maize and chloroplast mutator of Arabidopsis (Rédei, 1973; Martínez-Zapater et al., 1992; Sakamoto et al., 1996; Leon et al., 1998). As discussed in greater detail later, these mutants are variegated because the abnormal mitochondria secondarily affect the phenotype of the plastids in the cell. Hence, these sorts of variegations are Mendelian-inherited, but the defective mitochondria are maternally (or biparentally) inherited.
There are several other prominent mechanisms by which nuclear genes produce variegations. One is chimerism, in which different histological regions of a plant meristem, and consequently the tissues that derive from them, have different genotypes (reviewed by Kirk and Tilney-Bassett, 1978; Tilney-Bassett, 1986). This results in variegation if the genotypic differences affect pigment accumulation in the plastid. A second mechanism involves transposable element activity. In this mechanism, transposon insertion interrupts a nuclear gene required for normal chloroplast biogenesis (white sectors), while element excision reconstitutes wild type gene expression (green sectors) (reviewed by Federoff, 1989). In Arabidopsis, variegations of this type have been reported in transposon-tagging experiments using heterologous elements, such as the autonomous maize En-1 element, or the maize Dissociation (DS) transposable element / Activator (Ac) transposase system (Wisman et al., 1998; Klimyuk et al., 1995).
A final type of nuclear gene-induced variegation that will be mentioned is homology-dependent gene silencing, viz., transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS) (reviewed by Waterhouse et al., 2001). It is thought that double stranded RNAs (dsRNAs) play a role in regulating both TGS and PTGS: in TGS they regulate the methylation of promoters of target genes, whereas in PTGS they control target RNA degradation. Although the mechanisms are obscure, variegations are thought to arise when a nuclear gene required for normal chloroplast biogenesis is silenced in some cells but not others (Waterhouse et al., 2001).
Related to gene silencing is the phenomenon of variable gene expression by antisense transgenes. A well-known example of this is the production of variegated flowers in transgenic petunia that contain an antisense chalcone synthase gene (e.g., van der Krol et al., 1988; 1990). Antisense RNAs are thought to regulate sense gene expression by annealing to complementary sequences of the sense RNA, resulting in dsRNAs that affect sense RNA stability, transcription or translation (reviewed by Terryn and Rouzé, 2000). An inhibition at any of these levels of gene expression can result in reduced production of the target protein. It has been proposed that sense: antisense RNA duplexes cause variegation by serving as dsRNA signals to control TGS and PTGS.
In addition to nuclear gene-induction mechanisms, variegations can be caused by mutations in plastid or mitochondrial genomes that arise without the participation of defective nuclear gene products. For example, some plastome mutations arise spontaneously or following treatment with various chemical mutagens (reviewed by Tilney-Bassett, 1975; Börner and Sears, 1986). Plastome mutants can also be produced by chloroplast transformation (e.g., Kanevski and Maliga, 1994; Rochaix, 1997). Such “transplastomic” lines can be engineered by the integration of an antibiotic cassette into the plastid genome by homologous recombination via flanking plastid DNA sequences in the vector; the transformed plastid DNAs subsequently sort out to form homoplasmic lines. It might be anticipated that mutations in many plastid genes would generate permanently-defective plastids, because plastid genomes primarily code for core components of the photosynthetic apparatus and for proteins required for plastid gene expression (reviewed by Goldschmidt-Clermont, 1998).
It should be emphasized that in cases where variegations are caused by plastome mutations, transposable element activity, or chimerism, the mechanism of variegation is relatively straightforward and can be explained by the fact the green and white sectors have different genotypes. Variegations caused by gene silencing can be explained in a similar manner, i.e, a given gene is silenced in some cells but not others. These mechanisms stand in contrast to cases of nuclear gene-induced variegations in which the green and white cells have the same nuclear (mutant) genotype, but the mutant phenotype has limited penetrance and is seemingly expressed only in a subset of the cells (i.e., cells in the white sectors). These latter variegations will be the focus of this review.
Arabidopsis variegations
Mutagenesis experiments in A. thaliana typically generate a low frequency of color mutants (e.g., McKelvie, 1963, Reiter et al., 1994; Wisman et al., 1998). For instance, Reiter et al. (1994) identified nearly 300 color mutants in a collection of ∼8000 T-DNA tagged Arabidopsis (3.75% frequency). These mutants had a uniform pigmentation and no variegations were reported. Of 78 color mutants uncovered in a collection of 10,950 T-DNA and En-tagged lines (0.75% frequency), four were variegations (0.04% frequency) (Wisman et al., 1998; D. Maiwald, unpublished observations). In contrast to the relatively low percentage of variegations that arise in tagging experiments, McKelvie (1963) found that approximately 10% of the color mutants generated by EMS and X-ray mutagenesis of A. thaliana were variegated.
Röbbelen (1968) reported that the immutans variegation mutant (discussed below) arises with a frequency of approximately 2 × 10−5 in the M2 progeny from X-ray treated A. thaliana seeds and with a frequency of 1.4 × 10−3 in the M2 progeny from EMS-treated seeds. immutans did not arise spontaneously in any of Röbbelen's experiments. The frequency of EMS-generated immutans in these experiments is higher than the average per locus mutation frequency estimated for EMS mutagenesis in Arabidopsis (0.5 × 10−3 to 0.5 × 10−4) (Koorneef et al., 1982; Haughn et al., 1988; Vizir et al., 1996). Consistent with this finding, Rédei and Röbbelen claimed (as of 1975) that there were more alleles of immutans than any other Arabidopsis gene (greater than 100) (Röbbelen, 1968; Rédei, 1975). In summary, the Arabidopsis mutagenesis experiments, while not exhaustive, are illustrative of the general principle that variegations arise at a low, but variable frequency in Arabidopsis, and that this frequency is influenced by the type of mutagen used.
There are roughly 200 variegation mutants in the Arabidopsis stock centers at Ohio State and Notttingham. Many of these mutants are from the Rédei and Röbbelen collections. However, most of these lines (including putative “immutans” lines) have been characterized only superficially. For instance, allelism tests have been conducted with relatively few of the mutants, so it is unclear whether the various lines represent alleles of known genes or novel variegation loci. Complicating matters, we have found that some differently-labeled lines are, in fact, identical (discussed later, see Table 3). Therefore, the diversity of variegations represented in the stock centers is not clear, but merits further study.
Table 3.
var2 alleles
In the remainder of this review I wish to summarize what is known about specific Arabidopsis variegation mutants. I will restrict my attention to nuclear gene-induced variegations in which the mutant genotype is uniform in the plant. While I will focus on those that have been characterized at the molecular level, several others that have been characterized to a certain extent will also be discussed. These mutants are listed in Table 1. Several questions are of particular relevance in surveying each mutant. One would first like to know the identity of the gene product defined by each mutant locus, how it functions in plastid biogenesis, and why plastids are defective in the mutant. A second question concerns the mechanism of variegation. The fact that so few variegation mutants of this type have been documented raises the possibility that they have some underlying mechanism in common. Why are these mutants variegated rather than albino or uniformly-pigmented?
Table 1.
Arabidopsis nuclear gene-induced variegation mutants
The first mutant that will be discussed is immutans. It is among the oldest Arabidopsis mutants to be described and is represented by over 25 papers in the literature, spanning nearly half a century of work. immutans will be discussed in much greater detail than the other variegation mutants. One reason for this is that the methods used to study immutans serve as a paradigm of the sorts of experiments that have been conducted to characterize other variegation mutants. The discussion of immutans will also provide a contextual framework within which to understand the other mutants.
immutans
Phenotype of im
The immutans (im) mutant of Arabidopsis thaliana was first described and partially characterized nearly 40 years ago by Rédei, Röbbelen and co-workers (Chung and Rédei, 1974; Chung et al., 1974; Rédei, 1963, 1967a, 1967b, 1967c, 1975; Rédei et al., 1974; Röbbelen, 1968) (Figure 1). These studies showed that sectoring in im is caused by a nuclear recessive gene, and that white sector formation is enhanced when plants are grown in elevated temperatures and light intensities or when illuminated with red (versus blue) light (Rédei, 1963; Röbbelen, 1968).
Figure 1.

The immutans variegation mutant of Arabidopsis.The green sectors contain normal-appearing chloroplasts, whereas the white sectors are heteroplastidic and contain abnormal plastids that lack pigments and lamellar structures, as well as rare normal chloroplasts (Wetzel et al., 1994). The white sectors accumulate the carotenoid precursor phytoene (Wetzel et al., 1994), and white sector formation is promoted by increased light intensity and temperature (Rédei, 1963; Röbbelen, 1968).
Progeny from the selfing of im/im plants recapitulate the variegation of the parent, regardless of whether they are derived from all-green or all-white inflorescences (Rédei, 1967a). This indicates that the mutant seeds have a uniform genetic constitution, and that transposable element activity is not responsible for the im variegation. If transposition were the cause, progeny from green inflorescences would be predominantly wild type, while progeny from white branches would be white and/or variegated, depending on the frequency of the excision event. The selfing experiments also suggest that white plastids are capable of being converted into chloroplasts and vice versa, i.e., that the plastid phenotype is reversible. Consistent with this idea, abnormal plastids are not maternally inherited in im, suggesting that the plastid defect is “cured” before or during reproduction (Wetzel et al., 1994). This presumably occurs during the plastid dedifferentiation and redifferentiation events that take place during the reproductive process (e.g., during the formation of reproductive cells or during early embryo development), and which ultimately give rise to proplastids in the meristem tissues; proplastids are undifferentiated plastids that are the source of most plastid types in the mature plant (Tilney-Bassett, 1989). The phenotypic reversibility of plastids in im, and the apparent inability of the mutant to convert permanently from an all-green (“wild type-like”) to a variegated (“mutant”) phenotype led Rédei (1975) to name the mutant immutans (for “immutable”).
Recent studies have revealed that the white tissues of im are heteroplastidic and contain cells with abnormal plastids, as well as rare, normal-appearing chloroplasts (Wetzel et al., 1994) (Figure 2). The abnormal plastids are vacuolated and lack organized lamellar structures (Figure 3). Heteroplastidic (“mixed”) cells are uncommon in plants and usually arise from the incomplete sorting out of different plastid types during development (Tilney-Bassett, 1975). Complete sorting out generates homoplastidic cells with plastids of one type or the other. The finding of heteroplastidic cells indicates that plastids respond differently to factors from the nucleus-cytosol in im cells, and hence that im is “plastid autonomous” (Wetzel et al., 1994). An early idea was that some plastids receive threshold amounts of the IM protein (green chloroplasts), while others do not, thus blocking their development (white plastids) (Wetzel et al., 1994). As discussed below, this interpretation has been called into question because some im alleles are apparently null (Wu et al., 1999).
Figure 2.
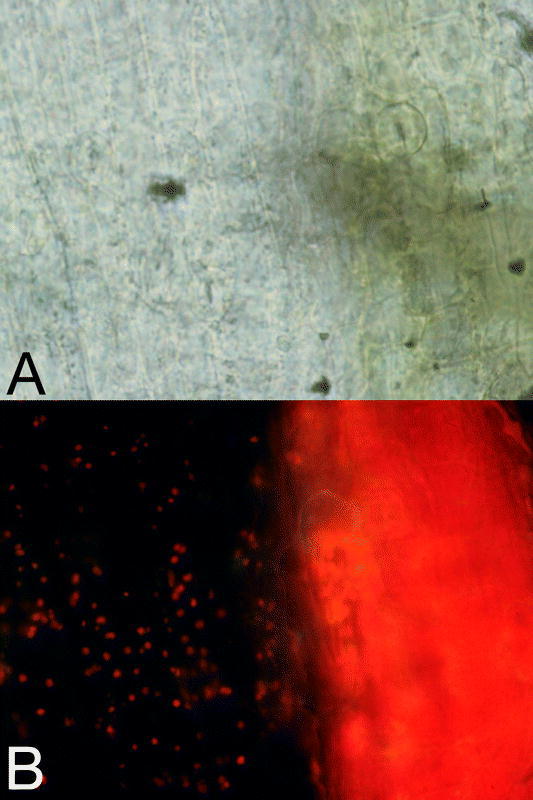
Cells in immutans white sectors are heteroplastidic.Light (A) and corresponding chlorophyll autofluorescence images (B) of variegated immutans tissue, showing a green-white interface. Chloroplasts appear red in the fluorescence photographs. Magnification is 80X.
Figure 3.
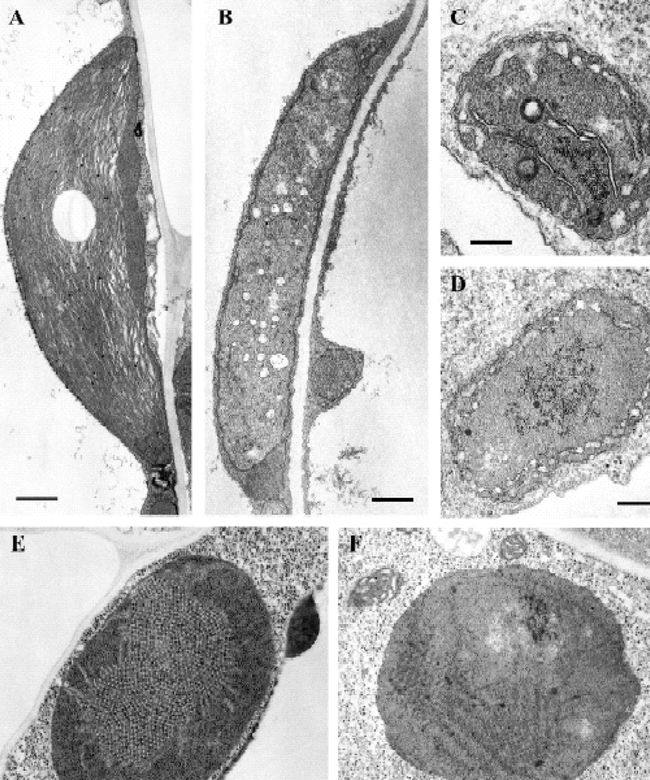
Plastid ultrastructure in immutans.Wild-type and im seedlings were grown on MS plates for 7 days under normal light conditions (A, B, C, D) or in darkness (E, F).(A) chloroplast from a wild type cotyledon, similar to chloroplasts from leaves (Wetzel et al., 1994) (Bar = 500 nm).(B) plastid from a white sector of an im cotyledon, similar to plastids from white leaf sectors of im (Wetzel et al., 1994) (Bar = 500 nm).(C) amyloplast from a wild-type root (Bar = 200 nm).(D) amyloplast from an im root (Bar = 200 nm).(E) etioplast from a wild-type cotyledon (Bar = 200 nm).(F) etioplast from an im cotyledon (Bar = 200 nm).(This figure is from Aluru et al., 2001).
Biochemical analyses have demonstrated that im white sectors accumulate phytoene, a colorless C40 carotenoid intermediate (Wetzel et al., 1994). This suggests that the mutant is impaired in the activity of phytoene desaturase (PDS), the plastid enzyme that converts phytoene to zeta-carotene (Bartley et al., 1991). Yet, IMMUTANS is not the PDS structural gene (Wetzel et al., 1994), nor does IM affect PDS expression at the level of mRNA or protein accumulation (Wetzel and Rodermel, 1998). The finding that carotenoid biosynthesis is blocked in the white tissues of im suggests that these tissues are susceptible to chlorophyll-mediated photooxidation (Oelmüller, 1989). In support of this idea is the observation that white sector formation in im can be modulated by illumination intensity. Also consistent is the finding that RNAs from various nuclear photosynthetic genes are preferentially reduced in abundance in the white im tissues (Wetzel et al., 1994). This is similar to the situation in other carotenoid-deficient, photooxidized tissues (the “plastid signal” hypothesis, discussed above) (reviewed by Oelmüller, 1989; Taylor, 1989; Mayfield, 1990; Susek and Chory, 1992; Gray et al., 1995; Hess et al., 1997; Rodermel, 2001). By contrast, the levels of RNAs from photosynthetic genes are normal in im green tissues (Wetzel et al., 1994).
In early studies, Rédei (1967a, 1975) found that the activity of a cytoplasmic acid RNase is enhanced in im, and he suggested that the primary lesion in the mutant resides in RNA metabolism. Wetzel et al. (1994) confirmed these results but found, in addition, a similar enhancement of acid RNase activity in photooxidized Arabidopsis leaf tissues produced by treatment with norflurazon, an inhibitor of PDS. It was concluded that increased cytoplasmic acid RNase activities are most probably a secondary effect of photooxidation rather than the primary lesion in the mutant. This was demonstrated conclusively by the isolation of the IM gene.
Cloning and identification of IM
We cloned the IM gene by map-based methods and found that it is a plastid member of the alternative oxidase (AOX) class of inner mitochondrial membrane proteins (Wu et al., 1999); a transposon-tagged allele of IM has also been isolated (Carol et al., 1999). AOX functions as a terminal oxidase in the alternative (cyanide-resistant) pathway of mitochondrial respiration, where it generates water from ubiquinol (reviewed by Siedow and Umbach, 1995; Vanlerberghe and McIntosh, 1997). AOX is found in all higher plants and in some algae, fungi and protists. The fact that IM bears similarity to AOX suggests that IM is a redox component of a phytoene desaturation pathway involving PDS, plastoquinol and oxygen as a final electron acceptor (Beyer et al., 1989; Mayer et al., 1990, 1992; Schulz et al., 1993; Nievelstein et al., 1995; Norris et al., 1995). Consistent with this interpretation, IM has quinol:oxygen oxidoreductase activity when expressed in E. coli (Josse et al., 2000).
IM and GH are orthologous genes
We and others (Wetzel et al., 1994; Bartley and Scolnik, 1995) reported that there are phenotypic similarities between im and the well-known ghost (gh) variegation mutant of tomato (Rick et al., 1959) (Figure 4). Like im, variegation arises in gh due to the action of a nuclear recessive gene (Rick et al., 1959); the white gh sectors accumulate phytoene (Rick et al., 1959; Mackinney et al., 1956; Scolnik et al., 1987); and white sector formation in gh is promoted by elevated light intensities (Rick et al., 1959; Scolnik et al., 1987).

Figure 4.The gh variegation mutant of tomato.
The im and gh mutants appear to arise from mutations in orthologous genes (Josse et al., 2000; R. Bae, C. Wetzel, and S. Rodermel, unpublished observations). In our experiments, we isolated a cDNA from tomato with 67% amino acid identity to IM (Genbank accession number AF302931), and found that this cDNA is from a single copy tomato gene. Mapping of the cDNA (the IM homolog) using a collection of F2 plants from an interspecific cross between Lycopersicon esculantum (L.) Mill. and L. pennelli (Correll) D'Arcy revealed that it maps to the gh locus. Figure 5 shows that transcripts from the IM homolog contain a T nucleotide insertion mutation in the gh background. It would be anticipated this would generate a premature stop codon 40 nucleotides downstream from the site of insertion. Taken together, these data suggest that GH and IM are orthologous proteins.
Figure 5.
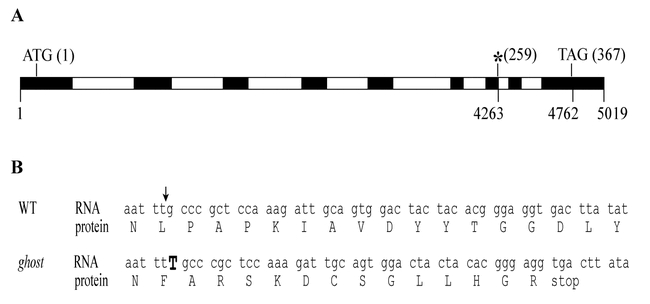
gh mRNA has an insertion mutation.(A) Schematic of the GH genomic sequence (deposited in Genbank as Accession number AF302932). Exons are shown as filled boxes, introns as open boxes. The numbering below the gene refers to base pairs (bp) in the genomic sequence (5.01 kbp), commencing with the first base of the GH cDNA. The numbering above the gene diagram refers to the codon position in the translated sequence. *, site of the insertion mutation in gh.(B) The translated sequence of the IM homolog has a T insertion in gh/gh plants, resulting in a premature stop codon.
Proposed structural model of IM, GH and AOX proteins
Phylogenetic analyses revealed that IM is a distantly related member of the AOX class of inner mitochondrial membrane proteins (Wu et al. 1999) (Figure 6). The substrates of AOX are ubiquinol and dioxygen, and iron is essential for activity (Siedow and Umbach, 1995; Vanlerberghe and McIntosh, 1997). Structural models of AOX are based on the “RNR R2” class of di-iron carboxylate proteins (named after the R2 subunit of ribonucleotide reductase) (Siedow et al., 1995; Moore et al., 1995; Andersson and Nordlund, 1999). The active sites of RNR R2-type proteins consist of a binuclear iron center coordinated by two histidines and four carboxylate residues. An early model of AOX, proposed by Siedow and colleagues (Moore et al., 1995; Siedow et al., 1995), suggested that the protein contains two transmembrane domains, with the N- and C-termini exposed to the matrix side of the membrane (Figure 7A). Of three “EXXH” motifs in the C-terminal portion of the protein (B1, B2 and B3), B2 and B3 were proposed to form part of the di-iron center because only these two would reside on the same side of the membrane. It has proven difficult to test the Siedow model, and it enjoys little unequivocal experimental support (Andersson and Nordlund, 1999).
Figure 6.
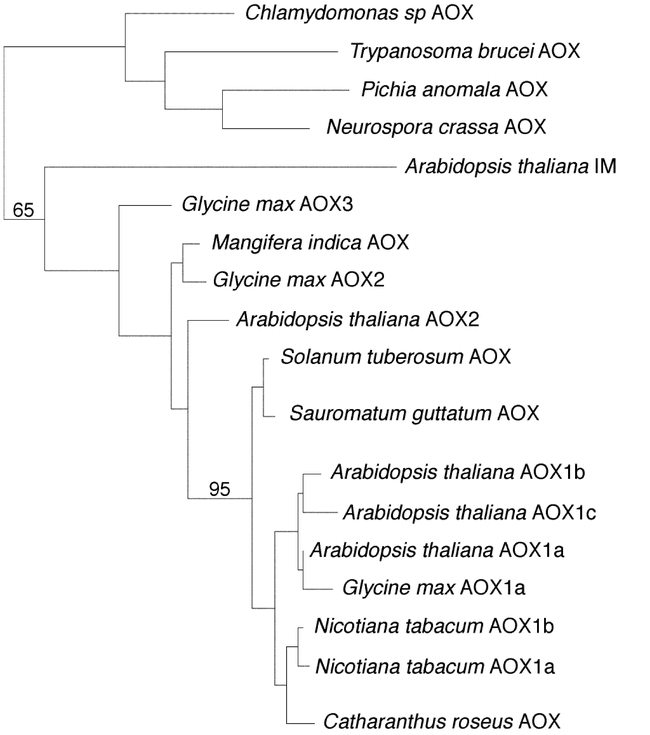
Phylogenetic relationships of AOX and IMMUTANS amino acid sequences.Relationships were determined by the neighbor-joining distance algorithm (described in Wu et al., 1999). Bootstrap values (100 replicates) are indicated above the branch nodes. The sources of the various AOX sequences are listed in Wu et al. (1999). For tree construction, amino acid sequences were limited to the conserved hydrophobic domains and the first two iron binding motifs (see Figure 7 sequence alignments). IMMUTANS and all plant AOX sequences are well-separated from the AOX genes of algae, fungi, and protists (in 65% of bootstrap replicates). This suggests that IMMUTANS shares a more recent evolutionary history with plant AOX genes. Most plant AOX sequences form a single, well-supported clade (95% bootstrap support) (the nine sequences at the bottom of the figure). Genes within this branch share approximately 75–95% amino acid identity. Although more distantly-related AOX genes are found in plants (namely, Arabidopsis AOX2, Glycine AOX2 and AOX3, and Mangifera AOX), the relationships of these genes to one other and to the other AOX genes is less clear. Even though IMMUTANS shares many conserved structural motifs with plant AOX genes, it is distantly related to all of them (as indicated by its long branch length). This suggests that IM is a novel type of AOX. (The figure is from Wu et al., 1999)
Figure 7.
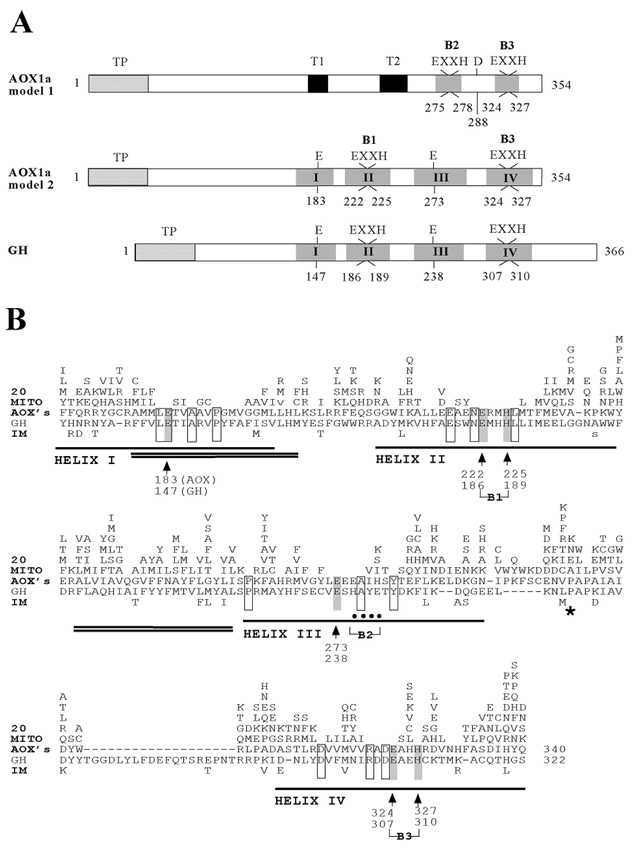
Structural and functional domains of AOX, GH and IM.(A) The predicted structure of AOX according to the Siedow model (top) (Siedow et al., 1995; Moore et al., 1995) and to the Andersson and Nordlund (1999) model (middle). The structure of GH (bottom) is modeled after the Andersson and Nordlund model. Shaded boxes (I-IV) are alpha helical regions; T1 and T2 are putative transmembrane segments; “EXXH” (B1, B2 and B3), “D” and “E” are putative Fe-binding carboxylate ligands; TP are putative transit peptides. Size of the protein is from initiating ATG (codon 1) to the termination codon.(B) Comparison of AOX, GH and IM derived amino acid sequences. Non-identical residues are shown. The sequences of GH and IM are compared downstream from codon 135 in the GH sequence. The sequences were compared with 20 AOX sequences from GenBank (Arabidopsis thaliana AOX1a, Arabidopsis thaliana AOX1b, Arabidopsis thaliana AOX1c, Arabidopsis thaliana AOX2, Glycine max AOX1, Glycine max AOX2, Glycine max AOX3, Nicotiana tabacum AOX1, Nicotiana tabacum AOX2, Oryza sativa AOX1a, Oryza sativa AOX1b, Sauromatum guttatum AOX1, Catharanthus roseus AOX, Mangifera indica AOX1, Zea mays AOX, Chlamydomonas reinhardtii AOX1, Neurospora crassa AOX, Hansenula anomala AOX, Trypanosoma brucei brucei AOX, Chlamydomonas sp AOX). Open and shaded boxes, identical amino acids among all three sequences; the six perfectly conserved Fe-ligands are indicated by the shaded boxes with the arrows underneath. Alpha helices are single-underlined; hydrophobic regions are double-underlined. *, site of the insertion mutation in gh. —, gaps in the alignment.
Taking advantage of a larger number of AOX sequences than were available when the Siedow model was proposed, Andersson and Nordlund (1999) proposed a revised structural model of AOX. They hypothesized that the hydrophobic regions of AOX are not transmembrane segments but rather, as with other RNR R2 proteins, that AOX is an interfacial membrane protein with an active site contained within a four helix bundle, with helices 1 and 3 (and helices 2 and 4) oriented anti-parallel to one another. In this model, the active site consists of a di-iron center coordinated by the B1 and B3 “EXXH” motifs on the paired second and fourth helices (Figure 7A), while the other two carboxylates are contributed by the paired first and third helices. It was proposed that these carboxylate residues are E183 on helix 1 and E274 on helix 3, based on the spacing between helices 1 and 2 (usually 30 amino acids) and between helices 3 and 4 (also, usually 30 amino acids), as found in other RNR R2 type proteins.
Because we found that IM and GH are only distantly related to AOX (Wu et al., 1999), we reasoned that phylogenetic comparisons of these sequences might offer an opportunity to test the validity of the Andersson and Nordlund model, i.e., AOX sequences that are evolutionarily conserved in GH and IM are likely important for structure and function. In Figure 7B, the sequences of 20 AOX proteins were compared with those of IM and GH in the C-terminal two-thirds of the protein. This is the most conserved region of the three proteins. Consistent with the Andersson and Nordlund model, GH (and IM) are predicted to contain four helices. Importantly, the B1 and B3 “EXXH” sites are precisely conserved between AOX, GH, and IM; the B2 site is not conserved. This suggests that these sequences provide four of the six expected Fe-ligands. The only conserved carboxylates in helices 1 and 3 are E147 and E238, respectively, suggesting that these residues serve as the other two Fe-ligands.
The strict conservation of the B1 and B3 sequences (on helices 2 and 4, respectively) indicates that if these sequences provide four of the six Fe-ligands, they must reside on the same side of the membrane to form a binuclear iron center. Considered together with the precise conservation of the E147 and E238 sequences (on helices 1 and 3, respectively), our data thus lend striking support to the Andersson and Nordlund hypothesis: the four helices in AOX are oriented anti-parallel to one another, as in other RNR R2 proteins, and AOX is an interfacial membrane protein, also as in other RNR R2 proteins. We propose that IM and GH are similar in structure to AOX, as proposed by Andersson and Nordlund (Figure 8). As with AOX, we hypothesize that the hydrophobic portions of GH and IM insert only partially through the lipid bilayer, providing a hydrophobic surface for protein/protein interactions within the bilayer, e.g., AOX is a dimer (Siedow et al., 1995). A similar structural model for IM has been proposed by Berthold et al. (2000).
Figure 8.
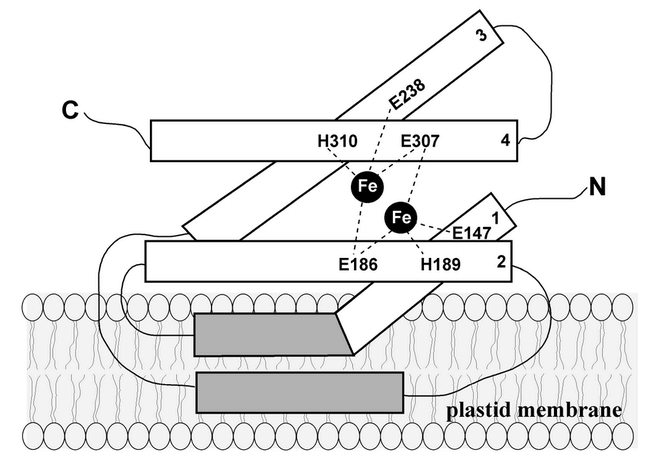
Structural model of GH.GH is proposed to be an interfacial membrane protein with a di-iron center coordinated by two EXXH motifs on helices 2 and 4 (oriented anti-parallel to one another), and two carboxylates on helices 1 and 3 (also oriented anti-parallel to one another).
Function of IM
As discussed above, phylogenetic analyses provide compelling evolutionary evidence for the validity of the Andersson and Nordlund model (1999) of the structure of AOX. Not only are the active site helices conserved between AOX, GH, IM, and RNR R2 type di-iron proteins, but evolutionary filtering allowed us to identify precisely the six carboxylates that likely act as coordinating Fe-ligands. The conservation of active site residues between GH, IM and AOX argues strongly that GH is a quinol oxidase, and suggests that these proteins have similar reaction mechanisms. This confirms biochemical analyses showing that IM has quinol oxidase activity when expressed in E.coli (Josse et al., 2000). The question arises, how does IM function in the plastid (in which biochemical pathways?) and why do lesions in IM give rise to variegated plants? Patterns of IM expression have been examined as a first approach to address these questions.
IM expression
Light shift experiments by Röbbelen (1968) and Wetzel et al. (1994) showed that the IM gene product is first active during a discrete phase of cotyledon development. This phase is coincident with seed coat breakage, and during this phase the phenotype of the cotyledons is irreversibly determined by the light intensity perceived by the growing seedling.
A powerful way to gain insight into IM function is to examine the phenotype of im plants. Three null im alleles have been reported (Wu et al., 1999), and thus the function of IM can be assessed in plants that completely lack IM activity. As discussed above, studies on im have focused on leaf variegation; in leaves, IM affects the functioning of the carotenoid biosynthetic pathway and appears to play a role in chloroplast biogenesis. Recent studies have shown that other organs of Arabidopsis (both green and non-green) are impaired in im, suggesting that IM is required for normal plant growth and development (Aluru et al., 2001). This impairment is due, in part, to a blockage of plastid differentiation in diverse cell types, including cells from cotyledons, roots and etiolated seedlings (Figure 3).
In support of the notion that IM plays a role in many types of plastids, RNA gel blot experiments (Figure 9) and analyses of transgenic plants with IM promoter:GUS reporter gene fusions have revealed that IM expression is ubiquitous in Arabidopsis tissues and organs (Aluru et al., 2001). Expression levels are generally high in tissues that accumulate carotenoids, such as leaves and cotyledons (Figure 9), consistent with the idea that IM is a redox component involved in carotenogenesis. However, IM is also expressed at appreciable levels in some tissues, such as roots, that accumulate only trace carotenoid amounts. This raises the possibility that IM is a general electron sink in plastid membranes. In agreement with this hypothesis, recent evidence in Chlamydomonas suggests that IM serves as a terminal oxidase in chlororespiration (Cournac et. al., 2000).
Figure 9.
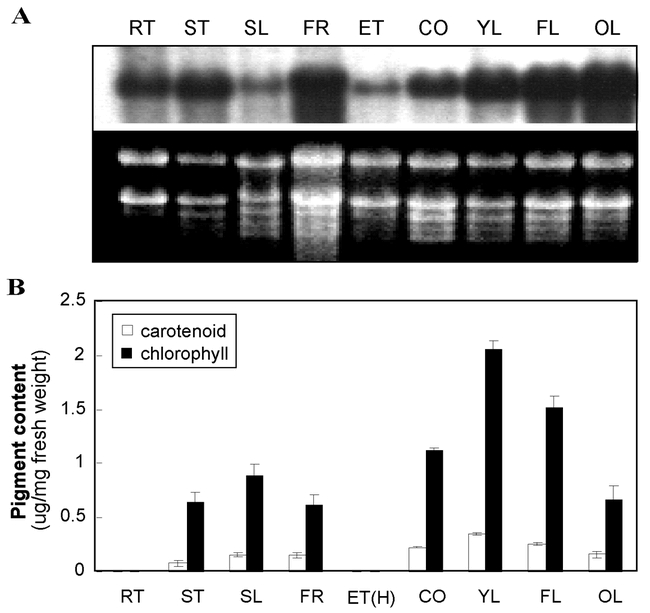
Expression analysis of IM mRNA and pigment levels.(A) Equal amounts of total cell RNA were electrophoresed through formaldehyde gels and blotted onto a membrane filter (Aluru et al., 2001). The RNA gel was stained with ethidium bromide to show rRNA (loading control). The blot was probed with a radiolabeled IM cDNA (Wu et al., 1999).(B) Total carotenoids and chlorophylls were extracted from Arabidopsis (Aluru et al., 2001). Values are an average of three separate experiments ± SD. The samples in A and B are from 4–5 wk old plants grown under normal light conditions (100 μmol·m−2·s−1); exceptions are samples from dark-grown seedlings (ET). RT, root; ST, stem; SL, green silique; FR, flowers (petals + green sepals); ET, 7-day-old etiolated seedling (cotyledon + hypocotyl); ET(C), cotyledons from 7-day old etiolated seedlings; CO, 7-day-old cotyledon; YL, young leaf (5 mm length); FL, just fully-expanded leaf (40 mm length); OL, senescing, late-fully expanded leaf. (This figure is from Aluru et al., 2001).
Leaf morphogenesis and plastid signals
Mesophyll cell morphogenesis is affected in both the green and white leaf sectors of im (Figure 10). The white sectors have a normal leaf thickness but the palisade cells fail to expand normally. As mentioned above, a considerable body of evidence supports the notion that the transcription of some nuclear genes, especially those for photosynthetic proteins, is controlled by the developmental and/or metabolic state of the plastid (the “plastid signal” hypothesis) (reviewed by Oelmüller, 1989; Taylor, 1989; Mayfield, 1990; Susek and Chory, 1992; Gray et al., 1995; Hess et al., 1997; Rodermel, 2001). Consistent with this hypothesis, we have reported that plastids in the white sectors of im have reduced rates of Lhcb transcription and decreased Lhcb mRNA levels (Meehan et al., 1996).
Figure 10.
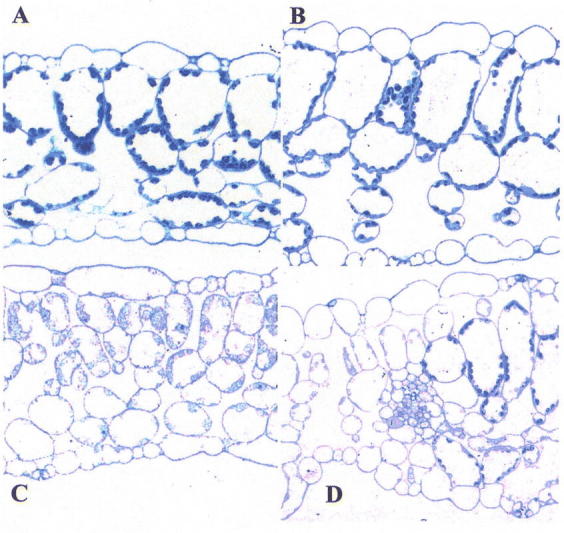
Anatomy of leaves of immutans and wild type plants.Light microscopy was conducted on fully-expanded leaves from wild-type and im plants grown under normal light conditions. A magnification of 25X applies to all panels. The white sectors stain less intensely than green sectors because their plastids are deficient in internal structures. (A) wild-type. (B) green leaf sector of im. (C) white leaf sector of im. (D) adjacent green and white sectors of im. (This figure is from Aluru et al., 2001).
In addition to the regulation of transcription of nuclear photosynthetic genes, analyses of a handful of nuclear gene-induced pigment mutants, including im (Aluru et al., 2001), have led to the hypothesis that plastid signals also control cell differentiation and thereby affect tissue and organ morphogenesis (reviewed by Rodermel, 2001). These mutants include dag of Antirrhinum (Chatterjee et al., 1996), dcl of tomato (Keddie et al., 1996), and several Arabidopsis mutants, including cla1 (Mandel et al., 1996; Estévez et al., 2000), cue1 (Li et al., 1995; Streatfield et al., 1999) and pac (Reiter et al., 1994; Meurer et al., 1998). Like im, the white leaf tissues of these mutants have abnormal plastids and cells, abnormal cell sizes and/or numbers, and altered palisade and/or spongy mesophyll cell layer organizations. Because the products of the genes defined by these mutations reside in the plastid, it has been argued that these proteins are not required independently for plastid development, cell differentiation and leaf morphogenesis, but rather that the affects on cell differentiation and leaf morphogenesis are a consequence of incomplete chloroplast differentiation. The molecular details are not understood.
If cellular differentiation and development are affected by plastid-to-nucleus signaling, are plastid “developmental” signals the same as the plastid signals that regulate photosynthetic gene expression? Most of the mutants discussed above express Lhcb at markedly reduced levels in their white tissues. The one exception is pac, which has normal Lhcb mRNA accumulation (Reiter et al., 1994; Meurer et al., 1998). This suggests that plastid “developmental” signaling pathways can be separated from the plastid-to-nucleus signaling pathways involved in regulating nuclear photosynthetic gene expression.
Recent experiments in immutans have revealed that the green leaf sectors, as well as the white leaf sectors, have an aberrant anatomy (Aluru et al., 2001). In particular, the blades are thicker than normal due to a dramatic enhancement in mesophyll and epidermal cell sizes and to an increase in intercellular air space volume (Figure 10). Analyses of fluorescence-activated cell-sorter (FACS)-purified cells have demonstrated that cells from green im leaf sectors have more chlorophyll than similarly-sized cells from wild-type plants (Meehan et al., 1996). The green sectors also have significantly elevated photosynthetic rates (Aluru et al., 2001). These observations point towards a complex mechanism whereby the photosynthetic potential of the green sectors is enhanced to compensate for a lack of photosynthesis in the white sectors. It has yet to be tested whether part of this compensating mechanism involves feedback on cell differentiation by the metabolic state of the plastid.
We also found that the green im cells have significantly higher chlorophyll a/b ratios than wild-type cells under normal light conditions. High chlorophyll a/b ratios are typically found in “sun” versus “shade” plants and are indicative of smaller light harvesting complexes and/or an altered stoichiometry of PSI and PSII (reviewed by Stitt, 1991). These are typically adaptations to avoid light stress. Our working hypothesis is that a lack of IM gives rise to morphological and biochemical adaptations in the green sectors that make the leaf more “sun”-like, perhaps as a way to avoid photooxidative damage.
Mechanism of im variegation: the threshold model for phytoene desaturation capacity
Taken together with the observation that immutans plants are heteroplastidic and variegated, not albino, the finding of null immutans alleles suggests that there is a redundant function able to compensate for the absence of IMMUTANS activity in the green plastids. Computer searches and low stringency hybridizations have failed to detect IMMUTANS-related sequences in the Arabidopsis genome (Wu et al., 1999). Hence, the redundant or parallel redox component is unlikely to be another IMMUTANS-like AOX protein. Given the likelihood that IM acts as a quinol oxidase, one possibility is that the redundant function is a redox component downstream of the PQ pool.
According to our working model (Figure 11), different pathways of electron transport function in phytoene desaturation at different stages of development, and with different efficiencies, depending on which electron transport components are available (Wu et al., 1999). A fundamental assumption is that IMMUTANS is one of these components and is required for carotenoid synthesis during early chloroplast biogenesis, i.e., when thylakoid membranes are being elaborated in growing chloroplasts, following division of either progenitor proplastids in the meristem or mature chloroplasts in the expanding leaf (reviewed by Mullet, 1988). This is consistent with expression studies showing that IM is expressed ubiquitously in Arabidopsis tissues (Aluru et al., 2001). We hypothesize that PDS is unable, or only minimally able, to carry out phytoene desaturation during the early stages of thylakoid development when IMMUTANS is absent and the redundant or parallel pathway is not yet fully-functional. Phytoene would accumulate because of over-reduction of the PQ pool, causing a blockage in carotenoid synthesis. The plastids would thus be in a state vulnerable to high-light induced photooxidation by newly accumulating chlorophylls. In essence, a developmental race would occur between, on the one hand, photooxidation due to a lack of carotenoid photoprotection, and on the other hand, the development of an efficient mechanism of electron transport away from phytoene to accommodate PDS activity and the synthesis of enough carotenoids to afford photoprotection. Low light and thus lower photooxidative pressure would allow more plastids to survive the race through the vulnerable stage, accumulate chlorophylls and turn green. In the presence of a functional IMMUTANS, electron transport would not be inhibited during early development and carotenoid synthesis would proceed unhindered, thus avoiding photooxidative vulnerability.
Figure 11.
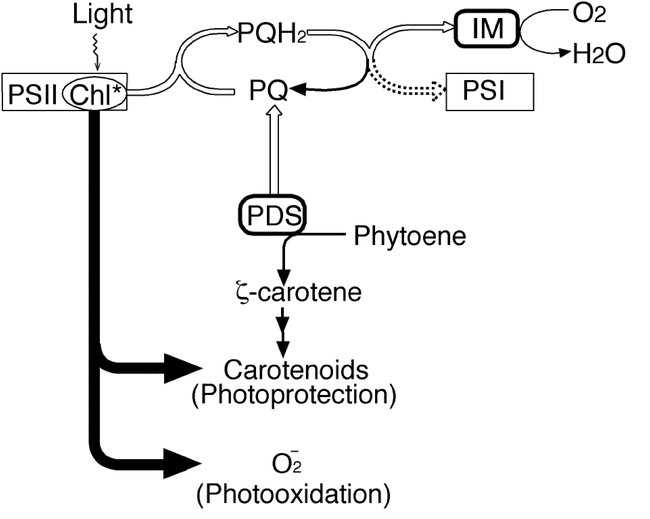
Working model of immutans variegation.Light is absorbed by the light harvesting complex of photosystem II (PSII), and energy from excited state chlorophyll can either be transferred to the reaction center or be used to form triplet chlorophyll. Unless quenched by colored carotenoids, energy from triplet chlorophyll generates oxygen radicals (O2−), which can lead to the photooxidation of the contents of the plastid if not detoxified by free radical scavengers (such as superoxide dismutase). The pathway of energy flow from triplet chlorophyll is indicated by shaded arrows, and the pathway of electron transport from PSII via the plastoquinone pool (PQ) to photosystem I (PSI) is shown as an open arrow. Electrons can also enter the PQ pool by the PDS-mediated desaturation of phytoene. In the model, PDS transfers electrons from phytoene to the PQ pool and IMMUTANS acts after this step to reduce molecular oxygen. In the absence of IMMUTANS, electron flow from the PQ pool to PSI serves as the redundant function to generate green plastids. (This figure is from Wu et al., 1999).
In sum, our model in Figure 11 invokes a threshold of electron transport capacity for phytoene desaturation that is required for carotenoid synthesis and the development of green chloroplasts (Wu et al., 1999). Below this threshold, carotenoids cannot be made in a sufficient enough quantity to prevent light-induced photodestruction. Consequently, the outcome of development in IMMUTANS-deficient plastids is either a white, photooxidized state or a fully-functional, green state. If the inner membrane structure has been destroyed along with the resident pigments then there will be no electron transport from phytoene and consequently no carotenoid synthesis (white plastids). If, on the other hand, enough electron transfer from phytoene can occur to support colored carotenoid accumulation, then wild type levels of pigments can accumulate, as observed in green immutans plastids and cells (Wetzel et al., 1994; Meehan et al., 1996). White plastids are capable of division because the requisite components are imported from the nucleus-cytoplasm (Tilney-Bassett, 1975). Therefore, the immutans mutation is effectively plastid autonomous, and each round of plastid division and differentiation carries the same risks of photooxidation.
Summary of immutans
In light of our current knowledge about im, several observations about the phenotype of im made by Rédei and co-workers in the 1960's and 1970's bear keeping in mind, particularly when considering the putative activity that compensates for a lack of IM in the green tissues of the mutant. As noted above, an acid RNase is overproduced in the white sectors of im (Rédei, 1967a). Rédei also found that two enzymes of the pyrimidine biosynthetic pathway are overproduced in the white im sectors, viz., orotidylic acid pyrophosphorylase (O5Ppase) and orotidylic acid decarboxlase (O5Pdase), but that growth on 6-azauracil, a pyrimidine analog that inhibits the activity of O5PDase, is able to partially normalize the im phenotype (Rédei, 1967b, Chung and Rédei, 1974; Chung et al., 1974). These and related observations prompted Rédei to suggest that IM is a primary regulator of RNA metabolism (Rédei, 1967a; 1975). Given the fact that IM codes for an AOX homolog, it would appear that the impact of IM on acid RNase activities and on pyrimidine biosynthesis are pleiotropic consequences of the lack of terminal oxidase activity in the plastid membrane. Yet, the partial phenotypic reversal by azapyrimidines is important and suggests that azapyrimidine-treatment might be a means of bypassing the primary lesion. This might provide a tool to understand compensating activities in the green im tissues.
Another intriguing finding by Rédei (1963) was that growth of im on cysteine promotes green sector formation and enhanced pigment accumulation. Cysteine is a precursor of glutathione, which plays a major role in oxidative stress by acting as a donor of reducing equivalents for the scavenging of reactive oxygen species (May et al., 1998). Transgenic plants with elevated levels of cysteine and glutathione are resistant to oxidative stress (Foyer et al., 1995; Wellburn et al., 1998; Blaszczyk et al., 1999; Youssefian et al., 2001). Given the possibility that cells become photooxidized in the absence of IM, one hypothesis is that cysteine acts as an antioxidant buffer to counter photooxidative pressure in immutans.
As a third example, Rédei (1967a) reported that growth on kinetin promotes pigment production in im. However, the concentrations of the hormone that were used in these experiments were very high and the treated plants were stunted. Although the impact of the hormone on sector formation and plastid morphology were not reported, these findings might be related to the observation that cytokinin is able to phenocopy some aspects of the det phenotype, viz., the development of chloroplasts in dark-grown plants (Chory et al., 1994). Cytokinin is also able to induce greening in pac2 (Grevelding et al., 1996) and atd2 (van der Graaff, 1997), two nuclear gene-induced variegations (see Table 1). Therefore, further insight into how cytokinins control plastid biogenesis might lend insight into im compensating activities.
A final observation worth noting is that heavy doses of X irradiation result in an increase in the number and size of im green sectors (Rédei, 1967c). The reason for this is unknown.
var2
Sectoring in the var2 variegation mutant of Arabidopsis is due to the action of a nuclear recessive gene (Martínez-Zapater, 1993; Chen et al., 1999). The mutant has normal-appearing cotyledons, but the first true leaves are bright yellow (Figure 12). As leaf expansion proceeds, green islands become visible and expand in size, and the yellow sectors fade to white. Sector boundaries and identity become fixed at full expansion. Whereas cells in the green leaf sectors and cotyledons of var2 contain morphologically normal chloroplasts, cells in the yellow and white sectors are heteroplastidic and contain vacuolated plastids with few, if any, lamellae (Figure 13), as well as some normal-appearing chloroplasts (Chen et al., 1999). Similar to immutans, the presence of heteroplastidic cells indicates that plastids in the mutant are affected differently by the nuclear mutation, i.e., that the phenotype of var2 is plastid autonomous. Also similar to immutans, defective plastids are not maternally inherited in var2, suggesting that the plastid defect can be “cured” during or after reproduction (Martínez-Zapater, 1993; Chen et al., 1999).
Figure 12.

The var2 mutant of Arabidopsis.The cotyledons are green and the first true leaves are yellow. The yellow variegated allele is shown (see Table 3).
Figure 13.
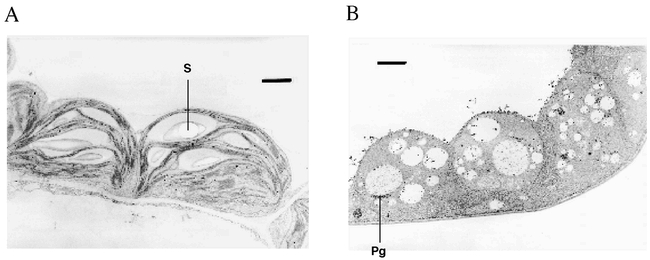
Ultrastructure of var2 plastids.Electron micrograph of representative plastids from A) a var2 green sector and B) a var2 yellow sector (Bar = 1 μm). Pg = plastoglobulae. S =starch. (This figure is from Chen et al., 1999).
Early studies showed that carotenoid and chlorophyll precursors do not accumulate in var2, indicating that the primary lesion in the mutant does not involve a blockage in pigment biosynthesis (Chen et al., 1999). We recently cloned VAR2 by map based methods and found that it codes for a 74 kDa plastid protein with high amino acid sequence similarity to the AAA protein class of Walker A/GTPases (Chen et al., 2000). AAA proteins (ATPases associated with diverse cellular activities) have one or more “AAA cassette” domains (∼200–250 amino acids) (Kunau et al., 1993; Beyer, 1997; Neuwald et al., 1999). These domains contain an ATP binding site, which is composed of Walker boxes A and B, and other conserved sequences whose functions are not understood. VAR2 shows high similarity to the “metal-dependent protease” subfamily of AAA proteins (Chen et al., 2000). The members of this family are ubiquitous among prokaryotes and eukaryotes and appear to be derived from an ancestral prokaryotic FtsH gene (Beyer, 1997). FtsH-like genes have a single AAA cassette, two transmembrane helices in their N-terminus, and a zinc binding site in their C-terminus (Beyer, 1997) (Figure 14).
Figure 14.
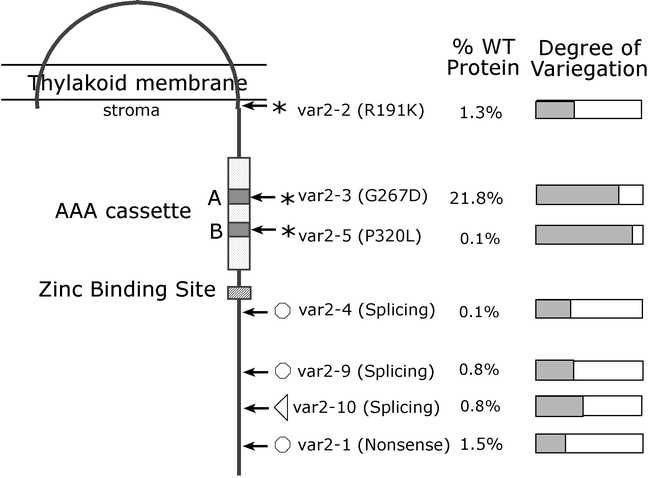
Topology of VAR2 in the thylakoid membrane.Shown are the two transmembrane domains, the AAA cassette (with Walker boxes “A” and “B”), and the zinc binding site. Sites of mutations in the various var2 alleles are illustrated (see also Table 3). Protein and pigment amounts have been reported for most of the alleles (Chen et al., 1999, 2000). Pigment contents provide a measure of the degree of variegation; hatched boxes represent the amount of pigment relative to wild type (100% green) determined on a fresh weight basis using fully-expanded first true leaves.
FtsH has been most thoroughly studied in E. coli, where it is involved in a diversity of processes, including the degradation of misfolded and excess proteins (protein quality control), the export of proteins from the cell, the integration of membrane proteins, mRNA decay, and resistance to colicins (Akiyama et al., 1994; Gottesman et al., 1997; Granger et al., 1998). The genes for several FtsH homologs have also been cloned in yeast (e.g., YME1), and these proteins are found in mitochondrial inner membranes where they associate into multimeric complexes that degrade unassembled subunits of inner membrane components (reviewed by Suzuki et al., 1997). It has been proposed that FtsH proteins are versatile because they have an intrinsic chaperone activity in addition to their protease activity (e.g., Akiyama et al., 1994; Arlt et al., 1996; Shirai et al., 1996; Gottesman et al., 1997; Akiyama et al., 1998; Leonhard et al., 1999).
Among photosynthetic organisms, four FtsH homologs have been identified in the completely sequenced genome of the cyanobacterium Synechocystis 6803 (Kaneko et al., 1996), and a small number of FtsH-like sequences have been identified in eukaryotic algae and higher plants (Beyer, 1997; Chen et al., 2000; Adam et al. 2001). In higher plants these genes are found in the nuclear genome, but in brown and red algae they are present in the plastid genome. Consistent with the endosymiotic origin of plastids from a single cyanobacterium (Doolittle, 1998), this suggests that progenitor FtsH-like sequences were transferred from the genome of the symbiont to that of the host in the lineage of photosynthetic organisms that gave rise to higher plants, but not in the lineage that gave rise to the red and brown algae (Chen et al., 2000).
Analyses of the complete Arabidopsis genome sequence have revealed that FtsH-like genes comprise a nuclear multigene family of at least 11 members (Adam et al., 2001) (Table 2). These genes have limited sequence similarity, except in the AAA cassette portion of the protein. In fact, sequences downstream from the zinc binding site (i.e., the C-terminal one-third of the protein) cannot generally be aligned for most of the family members. The lack of sequence conservation in this region is consistent with the notion that the C-terminus confers unique biochemical activities on the various family members. Seven of the Arabidopsis FtSH gene family members are predicted to have chloroplast targeting sequences and two are predicted to have mitochondrial targeting sequences. Organellar targeting sequences cannot be discerned for the other two family members (Table 2). The diversity of FtsH sequences in Arabidopsis suggests that these proteins function in multiple compartments and have varied activities.
Table 2.
Proposed nomenclature for FtsH proteases from Arabidopsis
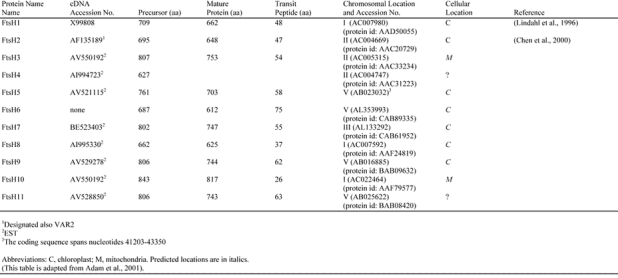
Of the Arabidopsis FtsH proteins, only VAR2 (also designated FtsH2 in Table 2) and FtsH1 have been characterized to any extent (Lindahl et al., 1996, 2000; Chen et al., 2000). Both proteins appear to be localized on stromally-exposed regions of the thylakoid membrane (Figure 14), and the expression of both genes is up-regulated during the greening of etiolated seedlings (Lindahl et al., 1996; Chen et al., 2000). These findings suggest that VAR2 and FtsH1 play a role in membrane modeling events during thylakoid biogenesis. In vitro studies have shown that FtsH1 catalyzes the proteolytic degradation of photooxidatively-damaged D1 proteins (the reaction center protein of PSII) (Lindahl et al., 2000). A proteolytic activity stimulated by zinc has also been implicated in the degradation of unassembled Rieske FeS protein (RISP) in the thylakoid membrane (Ostersetzer and Adam, 1997); however, it is not known whether this activity is caused by VAR2, FtsH1, another FtsH homolog, or a different type of metalloprotease. Although VAR2 substrate(s) have not yet been defined, it is possible that they differ from those of FtsH1 because sequences downstream from the zinc binding sites of these two proteins cannot be aligned. VAR2, at least, does not appear to be a general plastid membrane biogenesis factor inasmuch as etioplasts have a normal morphology in var2 plants (Chen et al., 2000).
Of the FtsH-like proteins described to date, VAR2 most closely resembles the red pepper Pftf protein (Hugueney et al., 1995; Chen et al., 2000). The sequences of VAR2 and Pftf can be aligned over their entirety, and the fact that both proteins share a high degree of amino acid identity (nearly 79%) suggests that they might have similar activities and functions. Pftf was isolated as a soluble factor that promotes membrane fusion and/or translocation events in an in vitro vesicle fusion assay using chromoplast membrane vesicles from red pepper fruit (Hugueney et al., 1995). Because Pftf was isolated as a soluble factor, it was thought to reside in the stroma. However, Summer and Cline (1999) have provided compelling evidence that Pftf is a membrane protein. Given the high sequence similarity between Pftf and VAR2, the finding that VAR2 is also a membrane protein (Chen et al., 2000) lends support to the conclusions of Summer and Cline (1999).
var2 allelic series
We initially became interested in the yellow variegated mutant of Arabidopsis because we thought it might be an allele of immutans (G.P. Redéi, personal communication). However, complementation analyses revealed that it is allelic to var2, not immutans (Chen et al., 1999). var2 was first characterized by Martínez-Zapater (1993), who reported two alleles of the locus (designated var2-1 and var2-2) (Table 3). We have identified five additional var2 alleles from the collection of variegation mutants in the Ohio State and Nottingham stock centers. Complementation tests were performed on candidate lines (i.e., ones with a phenotype similar to var2), and those showing allelism to var2 were sequenced. These alleles are designated var2-3 through var2-5, var2-9 and var2-10 (Chen et al., 1999, 2000; A. Manuell and S. Rodermel, unpublished findings) (Table 3). Takechi et al. (2000) have isolated an additional three var2 alleles: var2-6, a T-DNA insertion allele; var2-7, an uncharacterized line from the Ohio State stock center; and var2-8, an EMS-generated allele. As mentioned earlier, not all the lines in the stock centers define unique alleles. For instance, six lines that were identified as alleles of var2 in complementation tests were found, upon sequencing, to be identical to other var2 alleles (A. Manuell and S. Rodermel, unpublished findings) (Table 3).
We have generated a var2 allelic series on the basis of differences in the extent of white sector formation in first leaves (Chen et al., 1999, 2000). The differences between the alleles are visually striking (Figure 15). These differences have been quantitated by determining chlorophyll concentrations in first fully-expanded leaves (Figure 14): var2-1 is the most severe allele (leaves are predominantly white), whereas var2-5 is the least severe allele (leaves are nearly all-green with little white-sectoring).
Figure 15.

Alleles of var2.The plants were germinated at the same time and maintained under identical conditions (100 μmol·m−2·s−1, continuous illumination; 22°C).
The lesions in nine of the ten var2 alleles have been determined (see Figure 14 and Table 3). Two of the mutations are in the Walker ATP binding sites A and B (var2-3 and var2-5, respectively), providing in vivo evidence that these sequences are important for VAR2 activity (Chen et al., 2000). var2-1 has a nonsense mutation (Q597*) and is predicted to produce a truncated protein with nearly 100 fewer amino acids at the C-terminus, whereas var2-2 (R191K) has a conservative amino acid substitution immediately adjacent to the predicted end of the second transmembrane domain (Chen et al., 2000). It is not obvious how the latter mutation affects function and/or protein stability, but one possibility is that it influences integration of the protein into the membrane. var2-6 has a T-DNA insertion in the extreme C-terminus of the coding region, and var2-7 has a base pair deletion that causes a frameshift and the putative generation of a truncated protein (Takechi et al., 2000). No mutation has yet been found in the var2-8 allele (Takechi et al., 2000). The mutations in the remaining three alleles (var2-4, var2-9 and var2-10) are in intron splice sites (A. Manuell, V. Brendel and S. Rodermel, manuscript in preparation). Figure 14 shows that of the seven alleles examined, all have decreases in the abundance of the mutant VAR2 protein. Whereas this is consistent with the idea that the mutant proteins are unstable, there is evidence that decreases in protein abundance in some of the alleles might be due, in part, to decreases in var2 mRNA abundance (Chen et al., 2000; Takechi et al., 2000).
Mechanism of var2 variegation
One of the most intriguing aspects of var2 concerns the mechanism of variegation. Most var2 alleles have appreciable reductions in mutant VAR2 protein abundance in their green sectors, but the protein cannot be detected in the white sectors (Chen et al., 2000). Therefore, it's possible that green tissue formation in var2 requires only limited VAR2 activity, whereas a lack of VAR2 accumulation in the white sectors might be a secondary consequence of the mutation. For example, VAR2 might be turned over if it can't be assembled properly in the membrane. Alternatively, it might be degraded if it becomes photodamaged, e.g., as a consequence of photooxidation if photoprotective carotenoids fail to accumulate in the mutant white sectors. Consistent with the latter possibility, white sector formation in var2 is promoted by increased light intensity, although the effects are not as pronounced as with immutans (Martínez-Zapater, 1993; Chen et al., 1999; Takechi et al., 2000). Another possibility to explain the formation of normal-appearing green sectors in var2 plants is that there is an activity able to compensate for a lack of VAR2, at least in some cells of the mutant. It is also possible that events catalyzed by VAR2 can occur at a low rate even in its absence or near-absence.
In addition to light, sector formation in var2 is sensitive to temperature (Martínez-Zapater, 1993; Chen et al., 1999). We have also found that factors that promote green sector formation (i.e., increased temperature and decreased light) also depress the growth rate of the plant (Chen et al., 1999). Therefore, similar to immutans, we suggest that the development of a green chloroplast requires the attainment of a threshold VAR2 activity (or a VAR2 redundant activity), and that once attained, the “green” state is able to propagate itself. Accordingly, factors that promote slow growth might allow more time for threshold VAR2 activities to accumulate in developing var2 plastids. Plastids with below-threshold activities, on the other hand, might remain white due to a blockage in chloroplast biogenesis and/or to photooxidation.
chloroplast mutator (chm)
The chloroplast mutator (chm) mutant was first isolated by Rédei (1973) following EMS mutagenesis of Arabidopsis seeds. He reported that recessive mutations at the CHM locus induce the generation of white and yellow sectors in normally-green organs of the plant. Mesophyll cell differentiation and leaf morphology are also affected in chm, leading to a “rough-leaf” appearance. Rédei and Plurad (1973) found that long term maintenance of lines homozygous for chm results in the accumulation of plastids with different morphologies and the production of heteroplastidic (mixed) cells, and they suggested that the different plastid types are blocked at various steps of chloroplast biogenesis. Rédei (1973) showed that defective plastids are maternally inherited in chm, and he proposed that mutations in CHM cause the induction of mutations at various sites in the plastid genome, generating permanently-defective organelles. Stable chm-free homoplastidic lines, i.e., lines with a single type of mutant plastid, have been derived from backcrosses of the mutant and wild type (Rédei, 1973; Rédei and Plurad,1973; Mourad and White, 1992). These lines arise by the sorting out of defective plastids in chm-free progeny plants.
Rédei (1973) isolated two EMS-induced chm alleles (chm1 and chm2). Martínez-Zapater et al. (1992) isolated a third chm allele (chm3) from the progeny of T-DNA-tagged Arabidopsis regenerated from tissue culture; however, the T-DNA did not segregate with the variegation phenotype. Molecular studies showed that the mitochondrial genomes of chm3 are rearranged, and that these rearrangements cosegregate in a maternal fashion with the variegation trait (Martínez-Zapater et al., 1992). Gross chloroplast DNA polymorphisms could not be detected in these studies. The mitochondrial genomes of most higher plants are polyploid and have a multipartite organization, with subgenomic molecules arising by intramolecular recombination (reviewed by Goldschmidt-Clermont, 1998). Mitochondrial DNAs with an abnormal organization (termed “sublimons”) are also maintained at very low levels in plant mitochondria (reviewed by Leon et al., 1998). The stoichiometries of the various mitochondrial DNAs appear to be under nuclear control. One of the first demonstrations of this was the finding that cytoplasmic male sterility in common bean is associated with a mutation (pvs-orf239) on a subgenomic mitochondrial DNA molecule (Janska and Mackenzie, 1978). The dominant nuclear restorer gene, Fr, restores pollen fertility by reducing the content of this molecular species (reviewed by Leon et al., 1998).
Martínez–Zapater et al. (1992) suggested that CHM acts in a manner similar to Fr and prevents the amplification of mutant subgenomic mitochondrial DNA molecules that cause variegation. Support for this proposal has come from the observation that mutant mitochondrial DNAs are abundant in chm plants, and that these DNAs are also present in wild type plants, but at extremely low levels (Sakamoto et al., 1996). These conclusions were based on experiments with maternal distorted leaf (MDL), a mutant derived from a cross between chm1 and wild type plants; these plants lack the chm allele (Sakamoto et al., 1996). MDL plants are not variegated, but they grow slowly and have rough (distorted) leaves and aborted floral organs. The plants are homoplastidic for normal-appearing plastids, but the mitochondria have an abnormal ultrastructure. Mitochondrial DNAs in MDL are rearranged in regions of the chromosome coding for ribosomal proteins (rps3 and rpl16), and expression from these genes is reduced in the mutant. The rearranged DNAs, which are abundant in the mutant, are also present in very low amounts in the wild type, as assayed by quantitative PCR. Consistent with the suggestion of Martínez–Zapater et al. (1992), these results led Sakamoto et al (1996) to propose that the function of CHM is either to suppress the amplification of mutant mitochondrial DNAs or to maintain a high level of master mitochondrial genomes that contain the complete complement of genetic information. The isolation of CHM has yet to be reported.
If chm causes the production of permanently-defective mitochondria, why are some chm plants variegated? The reason for this appears to be that the defective mitochondria secondarily affect chloroplast function. This might not be surprising in light of the fact that mitochondria and chloroplasts exchange a variety of metabolites (Raghavendra et al., 1994). Support for such exchange comes from the non chromosomal stripe (NCS) mutants of maize (Newton and Coe, 1986). These mutants have pale-green or yellow leaf stripes, reduced growth, and sectors of aborted kernels on the ears. The morphology and function of both mitochondria and chloroplasts are defective in the pale-green and yellow sectors of the NCS mutants (Roussell et al., 1991; Gu et al., 1993). Chloroplast DNA mutations have not been detected in these mutants, but lesions in the mitochondrial DNA have been observed, e.g., deletions in the cytochrome oxidase subunit 2 gene (Cox2) in NCS5 and NCS6 (Lauer et al., 1990; Newton et al., 1990; Gu et al., 1993), a deletion in genes encoding ribosomal proteins in NCS3 (Hunt and Newton, 1991), and a chimeric nad4-nad7 gene in NCS2 (Marienfeld and Newton, 1994). The mutant and normal forms of these genes are present in the green sectors of NCS mutants, but the pale-green or yellow sectors are homoplasmic, or nearly so, for the mutant DNAs (Gu et al., 1993). NCS mutants are viable because of mitochondrial DNA heteroplasmy in the green sectors.
Although both mitochondria and chloroplasts are defective in NCS plants, the variegation phenotype cosegregates with the mitochondrial DNA defect in a maternal fashion. This suggests that the primary mutation in the mutants is the mitochondrion, and that the defective mitochondria secondarily affect chloroplast form and function, generating cells with pigment-deficient plastids. It is proposed that sectors arise during development because of somatic segregation of the mutant and normal mitochondria. Similar explanations may hold for sectoring in chm plants.
cue1
The cue (CBA underexpression) mutants of Arabidopsis were generated by mutagenesis of transgenic Arabidopsis bearing an Lhcb promoter/GUS reporter gene fusion (Li et al., 1995; López-Juez et al., 1998). Plants were selected with lower than normal levels of GUS activity, suggesting that they were also impaired in Lhcb transcription. Consistent with this idea, RNA gel blot assays revealed that the cue mutants underexpress Lhcb mRNAs, as well as transcripts from other nuclear genes for plastid proteins.
The cue mutants define seven complementation groups and fall into several phenotypic classes; only CUE1 has been cloned (Streatfield et al., 1999). Some cue mutants are allelic to known phytochrome-deficient mutants (hy1 and phyB); some are virescent (i.e., young leaves are pale, mature leaves are green), some are uniformly pale, and one (cue1) has pale-green mesophyll cells and dark-green bundle sheath cells that align the veins (a reticulate phenotype). The paraveinal regions of cue1 have normal bundle sheath cells, normal plastids and normal Lhcb mRNA expression, whereas the interveinal regions contain reduced numbers of palisade mesophyll cells, smaller than normal chloroplasts and reduced Lhcb expression (Li et al., 1995). The reductions in palisade cell number and chloroplast size explain why the interveinal regions of cue1 are pale-green.
Streatfield et al. (1999) cloned CUE1 and found that it codes for the plastid phosphoenolpyruvate/phosphate translocator (PPT). PPT is localized on the inner envelope and imports PEP in exchange for inorganic phosphate. PEP is the first substrate of the shikimate pathway, which produces aromatic amino acids and a variety of secondary metabolites, including UV light protectants, photosynthetic pigments, quinones and other phenolic redox compounds. These components are reduced in concentration in the pale-green sectors of cue1, and the mutants have reduced photosynthetic efficiencies. The fact that the mutant has normal-appearing bundle sheath cells and plastids, but abnormal mesophyll cells and plastids, indicates that there is a mesophyll cell-specific requirement for PPT in Arabidopsis leaves. Because PPT appears to be a member of a multigene family, one possibility is that expression of CUE1 is mesophyll cell-specific, while another PPT gene is bundle sheath cell-specific. Another possibility is that PPT is not required in bundle sheath cells. These hypotheses have yet to be tested.
It is interesting that reductions in Lhcb transcription in the mesophyll cells of cue1 are accompanied by decreases in plastid size and in chlorophyll content per cell, i.e., the mesophyll cells have the same number of chloroplasts but a reduced chloroplast plan area (Streatfield et al., 1999). Therefore, cue1 cells might have less Lhcb transcription because their nuclei perceive a lower than normal chloroplast plan area, perhaps via a reduction in plastid-to-nucleus signaling. An alternative hypothesis is provided by the finding that the reticulate phenotype of cue1 is light-sensitive, with growth in high light promoting the formation of white (versus pale-green) sectors. One reason for this sensitivity might be that cue1 plastids lack sufficient carotenoids to afford protection against chlorophyll-sensitized photooxidation (reviewed by Oelmüller, 1989). Lhcb expression in cue1 is also sensitive to light, with white sectors having less transcription than pale-green sectors. One hypothesis to explain these findings comes from the observation that cue1 plastids have decreased quinone concentrations and an altered redox state of the quinone pool (Streatfield et al., 1999). It has been demonstrated in several systems that the redox state of the plastoquinone pool serves as a plastid signal to regulate the transcription of nuclear photosynthetic genes (reviewed by Huner et al., 1998; Pfannnschmidt et al., 2001; Rodermel, 2001). This might also be the case for cue1.
One final comment about plastid signaling and cue1 comes from the finding that palisade cells are reduced in number in the mutant. Like other mutants perturbed in leaf morphogenesis (discussed above), this suggests that the CUE1 gene product (PPT) is required for normal palisade cell differentiation, and consequently for the normal morphogenesis of the mesophyll cell layer (Streatfield et al., 1999). In some manner, the lack of PPT disrupts one or more plastid-to-nucleus signaling pathways that regulate palisade cell division.
dov, re
In addition to cue1, other reticulate mutants have been reported in Arabidopsis, and several of these have been partially characterized. These include reticulata (re) (Rédei and Hirono, 1964) and dov1 (differential development of vascular associated cells) (Kinsman and Pyke, 1998). Both of these mutants have green vasculature on a pale-green lamina (similar to the phenotype of cue1). dov1 was selected for study from the large collection of reticulate mutants (over 200) in the Arabidopsis stock centers (Kinsman and Pyke, 1998). It is a nuclear recessive gene and is not allelic to re or cue1. dov1 has normal bundle sheath cells and chloroplasts. Mesophyll cell sizes and numbers are also normal, but the plastids are reduced in size and number. This provides an explanation for the pale-green phenotype of the interveinal regions of dov1 leaves. Chloroplast morphology is also disrupted in dov1: the mutant plastids are vacuolated and lack grana. re and dov1 have not been cloned.
pale cress
Two T-DNA-tagged alleles of the pale cress (pac) mutant have been isolated (Reiter et al., 1994; Grevelding et al., 1996). One allele is pale-green (pac1) and the other has green/white variegated leaves (pac2). Plastids in the pale-green sectors of pac1 appear to be blocked early in chloroplast biogenesis. They contain rudimentary lamellae and low levels of chlorophylls and carotenoids. Leaf morphology is also markedly altered in pac1. Early leaf development is normal, but in later stages the palisade cells fail to elongate and the air spaces increase in size. As with other mutants discussed above (e.g., im, dcl, dag), this suggests that pac is disrupted in the transmission of a plastid signal that regulates palisade cell differentiation. Ultrastructural and anatomical studies of pac2 have not been reported.
The PAC locus has been cloned in both pac1 (Reiter et al., 1994) and pac2 (Grevelding et al., 1996). In pac1, at least three unique cDNAs correspond to the locus (PAC1, 2 and 3) (Reiter et al., 1994). The three are products of alternative splicing and have the capacity to encode different proteins. By contrast, a single cDNA is found in the pac2 allele (Grevelding et al., 1996). This cDNA corresponds to the most abundant cDNA (PAC3) in pac1, and codes for a 36 kD protein of unknown function. It is not understood why PAC expression patterns differ in pac1 and pac2, but it might be related to differences in ecotype (pac1 is in the Wassilewskija background, and pac2 is in Columbia).
The putative PAC3 protein has a plastid transit sequence, and translational fusions of the N-terminus of PAC3 with the green fluorescent protein (GFP) are imported into plastids in transgenic Arabidopsis (Tirlapur et al., 1999). These findings demonstrate that PAC is a chloroplast protein. Meurer et al. (1998) found that specific plastid mRNAs are altered in abundance and in their pattern of maturation in pac plants, suggesting that PAC is a nuclear factor required for plastid mRNA maturation and accumulation.
The phenotypic differences of the pac alleles are intriguing. Particularly interesting is why one allele is variegated and the other is not. This might be related to differences in PAC expression, or to other background–specific factors. Nevertheless, the pac mutants are an ideal system to explore the molecular mechanisms of nuclear-gene induced variegation in which only some cells of the mutant plant exhibit the mutant phenotype. Of relevance in this context is the observation that cytokinin is able to bypass PAC function and to induce greening in pac plants (Grevelding et al., 1996). An understanding of how suppressors of variegation (such as cytokinin) work should also lend insight into the variegation mechanism.
var1
The var1 mutant was isolated from the progeny of an Arabidopsis thaliana plant (Columbia) regenerated from tissue culture (Martínez-Zapater, 1993). The mutant is nuclear recessive and resides on chromosome 5. Defective plastids are not maternally inherited. Variegation is limited to the leaves, and the extent of variegation is temperature-sensitive: plants grown below 20°C appear normal, but at 25°C the leaves have chlorotic sectors. Leaves that develop at the low temperature, then transferred to the higher temperature, develop chlorotic sectors in newly-differentiated tissues. By contrast, chlorotic cells, once produced, are unable to revert to a normal-appearing state, even if the temperature is reduced. var1 has not been further characterized.
atd2
The atd2 (glutamine 5-phosphoribosylpyrophosphate amidotransferase 2-deficient) mutant was generated by T-DNA mutagenesis (van der Graaff, 1997; Leon et al., 1998). It has white leaves and green cotyledons. Whereas cells in the cotyledons have normal-appearing chloroplasts, plastids in cells of the white leaves are smaller than normal, are vesiculated, and lack organized lamellae. The anatomy of the cotyledons is normal, but the palisade cells fail to expand in the white leaves. This suggests that atd2, like some of the other mutants discussed above (e.g., im, dcl, dag), is disrupted in the transmission of a plastid signal that regulates palisade cell differentiation.
The ATD2 locus codes for glutamine 5-phosphoribosylpyrophosphate amidotransferase (Atase2). Atase2 is one of two isoenzymes (the other is Atase1) that catalyzes the first step of purine biosynthesis. Because there are two Atase isoenzymes, it is possible that atd2 mutants are variegated because Atase1 is expressed in cotyledons and Atase2 is expressed in leaves. This has yet to be tested.
albomaculans
The albomaculans (am) variegation mutant was isolated by Röbbelen (1966) following X-ray treatment of Arabidopsis pollen. The mutation is inherited in a Mendelian fashion and sectoring is observed only in homozygous recessive individuals. The mutation generates permanently-defective, maternally inherited plastids. The abnormal plastids are vesiculated, lack lamellae and contain plastoglobulae (lipid bodies). Like immutans and var2, “mixed” cells are found in the white sectors of am. These cells are heteroplastidic for abnormal plastids and normal-appearing chloroplasts, and are probably products of incomplete sorting out. Tilney-Bassett (1975) suggested that albomaculans is an example of a plastome mutator (i.e, a nuclear gene that causes chloroplast DNA mutations), but this has yet to be demonstrated.
Acknowledgments
I wish to thank all the people in my lab who have contributed to research on variegation mutants. Work in my laboratory is supported by grants from the U.S. Department of Energy, Energy Biosciences panel (DE-FG02-94ER20147), and from the U.S. Department of Agriculture Competitive Research Grants Program, Biochemistry panel (#20013531810004). This review includes references through mid - 2001.
Footnotes
Citation: Rodermel S. (2002) Arabidopsis Variegation Mutants. The Arabidopsis Book 1:e0079. doi:10.1199/tab.0079
elocation-id: e0079
Published on: March 27, 2002
Table 4.
Genes cited in this review and their GenBank accession numbers.
REFERENCES
- Abdallah F., Salamini F., Leister D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 2000;51(1):141–142. doi: 10.1016/s1360-1385(00)01574-0. [DOI] [PubMed] [Google Scholar]
- Adam Z., Adamska I., Nakabayashi K., Ostersetzer O., Haussuhl K., Manuell A., Zheng B., Vallon O., Rodermel S. R., Shinozaki K., Clarke A. K. Chloroplast and mitochondrial proteases in Arabidopisis. A proposed nomenclature. Plant Physiol. 2001;1251(1):1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y., Ogura T., Ito K. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention efficiency of a cytoplasmic reporter. J. Biol. Chem. 1994;2691(1):5218–5224. [PubMed] [Google Scholar]
- Akiyama Y., Ehrmann M., Kihara A., Ito K. Polypeptide binding of Escherichia coli FtsH (HflB). Mol. Microbiol. 1998;281(1):803–812. doi: 10.1046/j.1365-2958.1998.00843.x. [DOI] [PubMed] [Google Scholar]
- Aluru M., Bae H., Wu D., Rodermel S. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiol. 2001;1271(1):67–77. doi: 10.1104/pp.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. E., Nordlund P. A revised model of the active site of alternative oxidase. FEBS Letters. 1999;4491(1):17–22. doi: 10.1016/s0014-5793(99)00376-2. [DOI] [PubMed] [Google Scholar]
- Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;851(1):875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Barak S., Nejidat A., Heimer Y., Volokita M. Transcriptional and posttranscriptional regulation of the glycolate oxidase gene in tobacco seedlings. Plant Mol. Biol. 2001;451(1):399–407. doi: 10.1023/a:1010688804719. [DOI] [PubMed] [Google Scholar]
- Bartley G. E., Scolnik P. A. Plant carotenoids: pigments for photoprotection, visual attraction, and human health. Plant Cell. 1995;71(1):1027–1038. doi: 10.1105/tpc.7.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley G. E., Viitanen P. V., Pecker I., Chamovitz D., Hirschberg J., Scolnik P. A. Molecular cloning and expression in photosynthetic bacteria of a soybean cDNA coding for phytoene desaturase, an enzyme of the carotenoid biosynthesis pathway. Proc. Natl. Acad. Sci. USA. 1991;881(1):6532–6536. doi: 10.1073/pnas.88.15.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich A. J. Why do chloroplasts and mitochondria contain so many copies of their genomes? Bioessays. 1987;61(1):279–282. doi: 10.1002/bies.950060608. [DOI] [PubMed] [Google Scholar]
- Berthold D. A., Andersson M. E., Nordlund P. New insight into the structure and function of the alternative oxidase. Biochem. Biophys. Acta. 2000;14601(1):241–254. doi: 10.1016/s0005-2728(00)00149-3. [DOI] [PubMed] [Google Scholar]
- Beyer A. Sequence analysis of the AAA protein family. Protein Science. 1997;61(1):2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer P., Mayer M., Kleinig H. Molecular oxygen and the state of geometric isomerism of intermediates are essential in the carotene desaturation and cyclization reactions in daffodil chromoplasts. Eur. J. Biochem. 1989;1841(1):141–50. doi: 10.1111/j.1432-1033.1989.tb15000.x. [DOI] [PubMed] [Google Scholar]
- Birky C. W. Relaxed cellular controls and organelle heredity. Science. 1983;2221(1):468–475. doi: 10.1126/science.6353578. [DOI] [PubMed] [Google Scholar]
- Blaszczyk A., Brodzik R., Sirko A. Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J. 1999;201(1):237–243. doi: 10.1046/j.1365-313x.1999.00596.x. [DOI] [PubMed] [Google Scholar]
- Bogorad L. Chloroplasts. J. Cell. Biol. 1981;911(1):256s–270s. doi: 10.1083/jcb.91.3.256s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T., Sears B. B. Plastome mutants. Plant Mol. Biol. Reporter. 1986;41(1):69–92. [Google Scholar]
- Carol P., Stevenson D., Bisanz C., Breitenbach J., Sandmann G., Mache R., Coupland G., Kuntz M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell. 1999;111(1):57–68. doi: 10.1105/tpc.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;51(1):174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- Chang T-L., Stoike L. L., Zarka D., Schewe G., Chiu W-L., Jarrell D. C., Sears B. B. Characterization of primary lesions caused by the plastome mutator of Oenothera. Curr. Genet. 1996;301(1):522–530. doi: 10.1007/s002940050165. [DOI] [PubMed] [Google Scholar]
- Chatterjee M., Sparvoli S., Edmunds C., Garosi P., Findlay K., Martin C. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 1996;151(1):4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Chen M., Choi Y. D., Voytas D., Rodermel S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 2000;21(1):303–313. doi: 10.1046/j.1365-313x.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- Chen M., Jensen M., Rodermel S. The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast biogenesis. J. Heredity. 1999;901(1):207–214. doi: 10.1093/jhered/90.1.207. [DOI] [PubMed] [Google Scholar]
- Chory J., Reinecke D., Sim S., Washburn T., Brenner M. A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 1994;1041(1):339–347. doi: 10.1104/pp.104.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. C., Rédei G. P. An anomaly of the genetic regulation of the de novo pyrimidine pathway in the plant Arabidopsis. Biochem. Genet. 1974;111(1):441–453. doi: 10.1007/BF00486077. [DOI] [PubMed] [Google Scholar]
- Chung S. C., Rédei G. P., White J. A. Plastid differentiation on 6-azauracil media. Experientia. 1974;301(1):92–94. [Google Scholar]
- Coe E. H., Thompson D., Walbot V. Phenotypes mediated by the iojap genotype in maize. Amer. J. Bot. 1988;751(1):634–644. doi: 10.1002/j.1537-2197.1988.tb13486.x. [DOI] [PubMed] [Google Scholar]
- Connett M. B. Mechanisms of maternal inheritance of plastids and mitochondria: developmental and ultrastructural evidence. Plant Mol. Biol. Reporter. 1987;41(1):193–205. [Google Scholar]
- Cournac L., Redding K., Ravenel J., Rumeau D., Josse E-M., Kuntz M., Peltier G. Electron flow between photosystem II and oxygen in chloroplasts of photosystem-deficient algae is mediated by a quinol oxidase involved in chlororespiration. J. Biol. Chem. 2000;2751(1):17256–17262. doi: 10.1074/jbc.M908732199. [DOI] [PubMed] [Google Scholar]
- Doolittle W. F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends in Genetics. 1998;141(1):307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- Estévez J. M., Cantero A., Romero C., Kawaide H., Jiménez L. F., Kuzuyama T., Seto H., Kamiya Y., León P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000;1241(1):95–103. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federoff N. V. Maize transposable elements. 1989;1(1):375–411. In Mobile DNA, D.E. Berg and M.M. Howe, eds (Washington, DC: American Society for Microbiology), pp. [Google Scholar]
- Foyer C. H., Souriau N., Perret S., Lelandais M., Kunert K-J., Pruvost C., Jouanin L. Overexpression of glutathione reductase but not glutathione synthetase leads to increases in antioxidant capacity and resistance to photoinhibition in poplar trees. Plant Physiol. 1995;1091(1):1047–1057. doi: 10.1104/pp.109.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 1998;1771(1):115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Wickner S., Maurizi M. R. Protein quality control: triage by chaperones and proteases. Genes and Development. 1997;111(1):815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- Granger L. L., O'Hara E. B., Wang R-F., Meffen F. V., Armstrong K., Yancey S. D., Babitzke P., Kushner S. R. The Escherichia coli mrsC gene is required for cell growth and mRNA decay. J. Bacteriol. 1998;1801(1):1920–1928. doi: 10.1128/jb.180.7.1920-1928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granick S. Die plastiden und chondriosomen. 1995;1(1):507–564. In Encyclopedia of Plant Physiology, Volume 1, W. Ruhland, ed (Berlin: Springer Verlag), pp. [Google Scholar]
- Gray J. C., Sornarajah R., Zabron A. A., Duckett C. M., Khan M. S. Chloroplast control of nuclear gene expression. 1995;1(1):543–550. In Photosynthesis: From Light to Biosphere, Volume 3, P. Mathis, ed (Dordrecht: Kluwer Academic Publishers), pp. [Google Scholar]
- Gray M. W. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 1992;1411(1):233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- Grevelding C., Suter-Crazzolara C., von Menges A., Kemper E., Masterson R., Schell J., Reiss B. Characterization of a new allele of pale cress and its role in greening in Arabidopsis thaliana. Mol. Gen. Genet. 1996;2511(1):532–541. doi: 10.1007/BF02173642. [DOI] [PubMed] [Google Scholar]
- Gu J., Miles D., Newton K. J. Analysis of leaf sectors in the NCS6 mitochondrial mutant of maize. Plant Cell. 1993;51(1):963–971. doi: 10.1105/tpc.5.8.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann R. A special type of nucleus-plastid-interactions: nuclear gene induced plastome mutants. 1986;1(1):455–466. In Regulation of Chloroplast Differentiation, G. Akoyunoglou and H. Senger, eds (New York: Alan R. Liss), pp. [Google Scholar]
- Han C-D., Coe E. H., Martienssen R. A. Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO J. 1992;111(1):4037–4046. doi: 10.1002/j.1460-2075.1992.tb05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C-D., Martienssen R. A. The iojap protein (IJ) is associated with 50S chloroplast ribosomal subunits. Maize Coop. Newsletter. 1995;691(1):32. [Google Scholar]
- Haughn G. W., Smith J., Mazur B., Somerville C. Transformation with a mutant Arabidopsis acetolactate synthase gene renders tobacco resistant to sulfonylurea herbicides. Mol. Gen. Genet. 1988;2111(1):266–271. [Google Scholar]
- Hedtke B., Wagner I., Börner T., Hess W. R. Inter-organellar crosstalk in higher plants: impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 1999;191(1):635–643. doi: 10.1046/j.1365-313x.1999.00554.x. [DOI] [PubMed] [Google Scholar]
- Hess W. R., Hoch B., Zeltz P., Hübschmann T., Kössel H., Börner T. Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell. 1994a;61(1):1455–1465. doi: 10.1105/tpc.6.10.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W. R., Linke B., Börner T. Effects of plastid differentiation on nuclear gene transcription. 1997;1(1):233–242. In Eukaryotism and Symbiosis, H.E.A. Schenk, R.G. Hermann, K.W. Jeon, N.E. Müllert and W. Schwemmler, eds (Heidelberg: Springer Verlag), pp. [Google Scholar]
- Hess W. R., Müller A., Nagy F., Börner T. Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol. Gen. Genet. 1994b;2421(1):305–312. doi: 10.1007/BF00280420. [DOI] [PubMed] [Google Scholar]
- Hess W. R., Prombona A., Fieder B., Subramanian A. R., Börner T. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast encoded RNA polymerase. EMBO J. 1993;121(1):563–571. doi: 10.1002/j.1460-2075.1993.tb05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W. R., Schendel R., Rüdiger W., Fieder B., Börner T. Components of chlorophyll biosynthesis in a barley albina mutant unable to synthesize D-aminolevulinic acid by utilizing the transfer RNA for glutamic acid. Planta. 1992;1881(1):19–27. doi: 10.1007/BF00198935. [DOI] [PubMed] [Google Scholar]
- Hugueney P., Bouvier F., Badillo A., D'Harlingue A., Kuntz M., Camara B. Identification of a plastid protein involved in vesicle fusion and/or membrane protein translocation. Proc. Natl. Acad. Sci. USA. 1995;921(1):5630–5634. doi: 10.1073/pnas.92.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huner N. P. A., Oquist G., Sarhan F. Energy balance and acclimation to light and cold. Trends Plant Sci. 1998;31(1):224–230. [Google Scholar]
- Hunt M. D., Newton K. J. The NCS3 mutation: genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 1991;101(1):1045–1052. doi: 10.1002/j.1460-2075.1991.tb08043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janska H., Mackenzie S. A. Unusual mitochondrial genome organization in cytoplasmic male sterile common bean and the nature of cytoplasmic reversion to fertility. Genetics. 1993;1351(1):869–879. doi: 10.1093/genetics/135.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse E-M., Simkin A. J., Gaffé J., Labouré A-M., Kuntz M., Carol P. A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 2000;1231(1):1427–1436. doi: 10.1104/pp.123.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;31(1):109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kanevski I., Maliga P. Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA. 1994;911(1):1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie J. S., Carroll B., Jones J. D. G., Gruissem W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 1996;151(1):4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Kinsman E. A., Pyke K. A. Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development. 1998;1251(1):1815–1822. doi: 10.1242/dev.125.10.1815. [DOI] [PubMed] [Google Scholar]
- Kirk J. T. O., Tilney-Bassett R. A. E. The Plastids, 2nd ed. 1978. Amsterdam: Elsevier/North-Holland.
- Klimyuk V. I., Nussaume L., Harrison K., Jones J. D. G. Novel GUS expression patterns following transposition of an enhancer trap Ds element in Arabidopsis. Mol. Gen. Genet. 1995;2491(1):357–365. doi: 10.1007/BF00287097. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Dresselhuys H. C., Ramulu K. S. The genetic identification of translocations in Arabidopsis. Arab. Inf. Serv. 1982;191(1):93–99. [Google Scholar]
- Kunau W-H., Beyer A., Franken T., Götte K., Marzioch M., Saidowsky J., Skaletz-Rorowski A., Weibel F. F. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;751(1):209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- Lauer M., Knudsen C., Newton K. J., Gabay-Laughnan S. J., Laughnan J. R. A partially deleted mitochondrial cytochrome oxidase gene in the NCS6 abnormal growth mutant of maize. New Biologist. 1990;21(1):179–186. [PubMed] [Google Scholar]
- Leon P., Arroyo A., Mackenzie S. Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;491(1):453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- Leonhard K., Stiegler A., Neupert W., Langer T. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 1999;3981(1):348–351. doi: 10.1038/18704. [DOI] [PubMed] [Google Scholar]
- Li H., Culligan K., Dixon R. A., Chory J. CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell. 1995;71(1):1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Spetea C., Hundal T., Oppenheim A. B., Adam Z., Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II DI protein. Plant Cell. 2000;121(1):419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Tabak S., Cseke L., Pichersky E., Andersson B., Adam Z. Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J. Biol. Chem. 1996;2711(1):29329–29334. doi: 10.1074/jbc.271.46.29329. [DOI] [PubMed] [Google Scholar]
- López-Juez E., Jarvis R. P., Takeuchi A., Page A. M., Chory J. New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998;1181(1):803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M. A., Feldmann K. A., Herrera-Estrella L., Rocha-Sosa M., León P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;91(1):649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Mackinney G., Rick C. M., Jenkins J. A. The phytoene content of tomatoes. Proc. Natl. Acad. Sci. USA. 1956;421(1):404–408. doi: 10.1073/pnas.42.7.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld J. R., Newton K. J. The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics. 1994;1381(1):855–863. doi: 10.1093/genetics/138.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Herrmann R. G. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;1181(1):9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zapater J. M. Genetic analysis of variegated mutants in Arabidopsis. J. Hered. 1993;841(1):138–140. [Google Scholar]
- Martínez-Zapater J. M., Gil P., Capel J., Somerville C. R. Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell. 1992;41(1):889–899. doi: 10.1105/tpc.4.8.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. J., Vernoux T., Sanchez-Fernandez R., Van Montagu M., Inzé D. Evidence for posttranscriptional activation of g-glutamylcysteine synthase during plant stress responses. Proc. Natl. Acad. Sci. USA. 1998;951(1):12049–12054. doi: 10.1073/pnas.95.20.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. P., Beyer P., Kleinig H. Quinone compounds are able to replace molecular oxygen as terminal electron acceptor in phytoene desaturation in chromoplasts of Narcissus pseudonarcissus L. Eur. J. Biochem. 1990;1911(1):359–363. doi: 10.1111/j.1432-1033.1990.tb19130.x. [DOI] [PubMed] [Google Scholar]
- Mayer M., Nievelstein V., Beyer P. Purification and characterization of a NADPH dependent oxidoreductase from chromoplasts of Narcissus pseudonarcissus: a redox-mediator possibly involved in carotene desaturation. Plant Physiol. Biochem. 1992;301(1):389–398. [Google Scholar]
- Mayfield S. P. Chloroplast gene regulation: interaction of the nuclear and chloroplast genomes in the expression of photosynthetic proteins. Curr. Opin. Cell Biol. 1990;21(1):509–513. doi: 10.1016/0955-0674(90)90135-2. [DOI] [PubMed] [Google Scholar]
- McKelvie A. D. Studies in the induction of mutations in Arabidopsis thaliana (L.) Heynh. Radiation Botany. 1963;31(1):105–123. [Google Scholar]
- Meehan L., Harkins K., Chory J., Rodermel S. Lhcb transcription is coordinated with cell size and chlorophyll accumulation. Plant Physiol. 1996;1121(1):953–963. doi: 10.1104/pp.112.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Grevelding C., Westhoff P., Reiss B. The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 1998;2581(1):342–351. doi: 10.1007/s004380050740. [DOI] [PubMed] [Google Scholar]
- Moore A. L., Umbach A. L., Siedow J. N. Structure-function relationships of the alternative oxidase of plant mitochondria: a model of the active site. J. Bioenerg. Biomembr. 1995;271(1):367–377. doi: 10.1007/BF02109999. [DOI] [PubMed] [Google Scholar]
- Mourad G. S., White J. A. The isolation of apparently homoplastidic mutants induced by a nuclear recessive gene in Arabidopsis thaliana. Theor. Appl. Genet. 1992;841(1):906–914. doi: 10.1007/BF00227403. [DOI] [PubMed] [Google Scholar]
- Mullet J. E. Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988;391(1):475–502. [Google Scholar]
- Neff M. M., Frankhauser C., Chory J. Light: an indicator of time and place. Genes Dev. 2000;141(1):257–271. [PubMed] [Google Scholar]
- Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;91(1):27–43. [PubMed] [Google Scholar]
- Newton K., Coe E. J. Mitochondrial DNA changes in abnormal growth mutants of maize. Proc. Natl. Acad. Sci. USA. 1986;831(1):7363–7366. doi: 10.1073/pnas.83.19.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K. J., Knudson C., Gabay-Laughnan S., Laughnan J. R. An abnormal growth mutant in maize has a defective mitochondrial cytochrome oxidase gene. Plant Cell. 1990;21(1):107–113. doi: 10.1105/tpc.2.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievelstein V., Vandekerckhove J., Tadros M. H., Lintig J. V., Nitschke W., Beyer P. Carotene desaturation is linked to a respiratory redox pathway in Narcissus pseudonarcissus chromoplast membranes. Eur. J. Biochem. 1995;2331(1):864–872. doi: 10.1111/j.1432-1033.1995.864_3.x. [DOI] [PubMed] [Google Scholar]
- Norris S. R., Barrette T. R., DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;71(1):2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R. Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochem. Photobiol. 1989;491(1):229–239. [Google Scholar]
- Ostersetzer O., Adam Z. Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell. 1997;91(1):957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald O., Martin T., Dominy P. J., Graham I. A. Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA. 2001;981(1):2047–2052. doi: 10.1073/pnas.021449998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T., Allen J. F., Oelmüller R. Principles of redox control in photosynthesis gene expression. Physiol. Plant. 2001;1121(1):1–9. [Google Scholar]
- Raghavendra A. S., Padmasree K., Saradadevi K. Interdependence of photosynthesis and respiration in plant cells: interactions between chloroplasts and mitochondria. Plant Sci. 1994;971(1):1–14. [Google Scholar]
- Rédei G. P. Somatic instability caused by a cysteine-sensitive gene in Arabidopsis. Science. 1963;1391(1):767–769. doi: 10.1126/science.139.3556.767. [DOI] [PubMed] [Google Scholar]
- Rédei G. P. Biochemical aspects of a genetically determined variegation in Arabidopsis. Genetics. 1967a;561(1):431–443. doi: 10.1093/genetics/56.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei G. P. Suppression of a genetic variegation by 6-azapyrimidines. J. Hered. 1967b;581(1):229–235. [Google Scholar]
- Rédei G. P. X-ray induced phenotypic reversions in Arabidopsis. Radiation Bot. 1967c;71(1):401–407. [Google Scholar]
- Rédei G. P. Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutation Research. 1973;181(1):149–162. [Google Scholar]
- Rédei G. P. Arabidopsis as a genetic tool. Annu. Rev. Genet. 1975;91(1):111–127. doi: 10.1146/annurev.ge.09.120175.000551. [DOI] [PubMed] [Google Scholar]
- Rédei G. P., Chung S. C., Plurad S. B. Mutants, antimetabolites and differentiation. Brookhaven Symp. Biol. 1974;251(1):281–296. [Google Scholar]
- Rédei G. P., Hirono Y. Linkage studies. Arabidopsis Inf. Serv. 1964;11(1):9–10. [Google Scholar]
- Rédei G. P., Plurad S. B. Hereditary structural alterations of plastids induced by a nuclear mutator gene in Arabidopsis. Protoplasma. 1973;771(1):361–380. [Google Scholar]
- Reiter R. S., Coomber S. A., Bourett T. M., Bartley G. E., Scolnik P. A. Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell. 1994;61(1):1253–1264. doi: 10.1105/tpc.6.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick C. M., Thompson A. E., Brauer O. Genetics and development of an unstable chlorophyll deficiency in Lycopersicon esculentum. Am. J. Bot. 1959;461(1):1–11. [Google Scholar]
- Röbbelen G. Chloroplastendifferenzierung nach geninduzierter Plastommutation bei Arabidopsis thaliana (L.) Heynh. Z. Pflphysiol. 1966;551(1):387–403. [Google Scholar]
- Röbbelen G. Genbedingte Rotlicht-Empfindlichkeit der Chloroplastendifferenzierung bei Arabidopsis. Planta. 1968;801(1):237–254. [Google Scholar]
- Rochaix J. D. Chloroplast reverse genetics: new insights into the function of plastid genes. Trends Plant Sci. 1997;21(1):419–425. [Google Scholar]
- Rodermel S. R. Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 2001;61(1):471–478. doi: 10.1016/s1360-1385(01)02085-4. [DOI] [PubMed] [Google Scholar]
- Rousell D. L., Thompson D. L., Pallardy S. G., Miles D., Newton K. J. Chloroplast structure and function is altered in the NCS2 maize mitochondrial mutant. Plant Physiol. 1991;961(1):232–238. doi: 10.1104/pp.96.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Kondo H., Murata M., Motoyoshi F. Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell. 1996;81(1):1377–1390. doi: 10.1105/tpc.8.8.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A., Ort O., Beyer P., Kleinig H. SC-0051, a 2-benzoyl-cyclohexane-1,3-dione bleaching herbicide, is a potent inhibitor of the enzyme p-hydroxyphenylpyruvate dioxygenase. FEBS Lett. 1993;3181(1):162–166. doi: 10.1016/0014-5793(93)80013-k. [DOI] [PubMed] [Google Scholar]
- Scolnik P. A., Hinton P., Greenblatt I. M., Giuliano G., Delanoy M. R., Spector D. L., Pollock D. Somatic instability of carotenoid biosynthesis in the tomato ghost mutant and its effect on plastid development. Planta. 1987;1711(1):11–18. doi: 10.1007/BF00395063. [DOI] [PubMed] [Google Scholar]
- Shirai Y., Akiyama Y., Ito K. Suppression of ftsH mutant phenotypes by overproduction of molecular chaperones. J. Bacteriol. 1996;1781(1):1141–1145. doi: 10.1128/jb.178.4.1141-1145.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow J. N., Umbach A. L. Plant mitochondrial electron transfer and molecular biology. Plant Cell. 1995;71(1):821–831. doi: 10.1105/tpc.7.7.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow J. N., Umbach A. L., Morre A. L. The active site of the cyanide-resistant oxidase from plant mitochondria contains a binuclear iron center. FEBS Lett. 1995;3621(1):10–14. doi: 10.1016/0014-5793(95)00196-g. [DOI] [PubMed] [Google Scholar]
- Stitt M. Rising CO2 levels and their potential significance for carbon flow in photosynthetic cells. Plant Cell Environ. 1991;141(1):741–762. [Google Scholar]
- Streatfield S. J., Weber A., Kinsman E. A., Häusler R. E., Li J., Post-Beittenmiller D., Kaiser W. M., Pyke K. A., Flügge U-I., Chory J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell. 1999;111(1):1609–1621. doi: 10.1105/tpc.11.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer E. J., Cline K. Red bell pepper chromoplasts exhibit in vitro import competency and membrane targeting of passenger proteins from the thylakoidal Sec and ΔpH pathways but not the chloroplast Signal Recognition Particle pathway. Plant Physiol. 1999;1191(1):575–584. doi: 10.1104/pp.119.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek R. E., Chory J. A tale of two genomes: role of a chloroplast signal in coordinating nuclear and plastid genome expression. Aust. J. Plant Physiol. 1992;191(1):387–399. [Google Scholar]
- Suzuki C. K., Rep M., van Dijl J. M., Suda K., Grivell L. A., Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. TIBS. 1997;221(1):118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- Takechi K., Sodmergen Murata M., Motoyoshi F., Sakamoto W. The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 2000;411(1):1334–1346. doi: 10.1093/pcp/pcd067. [DOI] [PubMed] [Google Scholar]
- Taylor W. C. Regulatory interactions between nuclear and plastid genomes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989;401(1):211–233. [Google Scholar]
- Terryn N., Rouzé P. The sense of naturally transcribed antisense RNAs in plants. Trends Plant Sci. 2000;51(1):394–396. doi: 10.1016/s1360-1385(00)01696-4. [DOI] [PubMed] [Google Scholar]
- Tilney-Bassett R. A. E. Genetics of variegated plants. 1975;1(1):268–308. In Genetics and Biogenesis of Mitochondria and Chloroplasts, C.W. Birky, P.S. Perlman, and T.J. Byers, eds (Columbus, OH: Ohio State University Press), pp. [Google Scholar]
- Tilney-Bassett R. A. E. The genetic evidence for nuclear control of chloroplast biogenesis in higher plants. 1984;1(1):13–50. In Chloroplast Biogenesis, R.J. Ellis, ed (Cambridge, England: Cambridge University Press), pp. [Google Scholar]
- Tilney-Bassett R. A. E. Plant Chimeras. 1986. London: Edward Arnold Publishers, Ltd.
- Tilney-Bassett R. A. E. The diversity of the structure and function of higher plant plastids. 1989;1(1):1–14. In Physiology, Biochemistry, and Genetics of Nongreen Plastids, C.D. Boyer, J.C. Shannon, and R.C. Hardison, eds (Rockville, MD: American Society of Plant Physiologists), pp. [Google Scholar]
- Tirlapur U. K., Dahse I., Reiss B., Meurer J., Oelmüller R. Characterization of the activity of a plastid-targeted green fluorescent protein in Arabidopsis. Eur. J. Cell Biol. 1999;781(1):233–240. doi: 10.1016/S0171-9335(99)80056-9. [DOI] [PubMed] [Google Scholar]
- van der Graaff E. Developmental mutants of Arabidopsis thaliana obtained after T-DNA transformation. 1997;1(1):179. Ph.D. Thesis. Leiden University. [Google Scholar]
- van der Krol A. R., Lenting P. E., Veenstra J., van der Meer I. M., Koes R. E., Gerats A. G. M., Mol J. N. M., Stuitje A. R. An antisense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature. 1988;3331(1):866–869. [Google Scholar]
- van der Krol A. R., Mur L. A., de Lange P., Gerats A. G. M., Mol J. N. M., Stuitje A. R. Antisense chalcone synthase genes in petunia: visualization of variable transgene expression. Mol. Gen. Genet. 1990;2201(1):204–212. [Google Scholar]
- Vanlerberghe G. C., McIntosh L. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;481(1):703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- Vizir I., Thorlby G., Mulligan B. Classical mutagenesis and genetic analysis. 1996;1(1):216–245. In Plant Gene Isolation: Principles and Practice, G.D. Foster and D. Twell, eds (New York: John Wiley & Sons Ltd.), pp. [Google Scholar]
- von Wettstein D., Gough S., Kannangara C. G. Chlorophyll biosynthesis. Plant Cell. 1995;71(1):1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Coe E. H. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl. Acad. Sci. USA. 1979;761(1):2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse P. M., Wang M-B., Finnegan E. J. Role of short RNAs in gene silencing. Trends Plant Sci. 2001;61(1):297–301. doi: 10.1016/s1360-1385(01)01989-6. [DOI] [PubMed] [Google Scholar]
- Wellburn F. A. M., Creissen G. P., Lake J. A., Mullineaux P. M., Wellburn A. R. Tolerance to atmospheric ozone in transgenic tobacco over-expressing glutathione synthetase in plastids. Physiol. Plant. 1998;1041(1):623–629. [Google Scholar]
- Wetzel C. M., Jiang C-Z., Meehan L. J., Voytas D. F., Rodermel S. R. Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 1994;61(1):161–175. doi: 10.1046/j.1365-313x.1994.6020161.x. [DOI] [PubMed] [Google Scholar]
- Wetzel C. M., Rodermel S. Regulation of phytoene desaturase expression is independent of leaf pigment content in Arabidopsis thaliana. Plant Mol. Biol. 1998;371(1):1045–1053. doi: 10.1023/a:1006021522259. [DOI] [PubMed] [Google Scholar]
- Wisman E., Hartmann U., Sagasser M., Baumann E., Palme K., Hahlbrock K., Saedler H., Weisshaar B. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc. Natl. Acad. Sci. USA. 1998;951(1):12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wright D. A., Wetzel C., Voytas D. F., Rodermel S. The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell. 1999;111(1):43–55. doi: 10.1105/tpc.11.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian S., Nakamura M., Orudgev E., Kondo N. Increased cysteine biosynthesis capacity of transgenic tobacco overexpressing an O-acetylserine(thiol)lyase modifies plant responses to oxidative stress. Plant Physiol. 2001;1261(1):1001–1011. doi: 10.1104/pp.126.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko M. K., Day A. Stable albinism induced without mutagenesis: a model for ribosome-free plastid inheritance. Plant J. 1998;151(1):265–271. doi: 10.1046/j.1365-313x.1998.00195.x. [DOI] [PubMed] [Google Scholar]


