SUMMARY
Chromatin immunoprecipitation (ChIP-chip and ChIP-seq) assays identify where proteins bind throughout a genome. However, DNA contamination and DNA fragmentation heterogeneity produce false positives (erroneous calls) and imprecision in mapping. Consequently, stringent data filtering produces false negatives (missed calls). Here we describe ChIP-exo, where an exonuclease trims ChIP DNA to a precise distance from the crosslinking site. Bound locations are detectable as peak-pairs by deep sequencing. Contaminating DNA is degraded or fails to form complementary peak-pairs. With the single bp accuracy provided by ChIP-exo, we show an unprecedented view into genome-wide binding of the yeast transcription factors Reb1, Gal4, Phd1, Rap1, and human CTCF. Each of these factors was chosen to address potential limitations of ChIP-exo. We found that binding sites become unambiguous and reveal diverse tendencies governing in vivo DNA binding specificity that include sequence variants, functionally distinct motifs, motif clustering, secondary interactions, and combinatorial modules within a compound motif.
INTRODUCTION
Proteins bind to specific DNA sequences to regulate genes. A fundamental and long sought goal in understanding how these interactions have evolved and their mechanism of regulation is the precise determination of where they are bound in a genome. Chromatin immunoprecipitation (ChIP) is the most widely used method to identify genomic binding locations of sequence-specific regulatory proteins (Solomon and Varshavsky, 1985). In the ChIP assay, proteins are crosslinked to their DNA binding sites in vivo, and then immunopurified from fragmented chromatin. Subsequently, the bound DNA is identified genome-wide by microarray hybridization (ChIP-chip) or deep sequencing (ChIP-seq) (Albert et al., 2007; Johnson et al., 2007; Ren et al., 2000).
Since unbound DNA contaminates the immunoprecipitate, ChIP only provides a set of statistically-enriched high-occupancy binding regions, rather than a complete and precise set of bound locations (Peng et al., 2007; Rozowsky et al., 2009; Tuteja et al., 2009). A sizeable fraction of this DNA may represent false positives (erroneous calls), and many other lower affinity sites may be missed (false negatives). Moreover, size heterogeneity of randomly sheared ChIP DNA technically limits mapping resolution, and thus cannot distinguish binding among clusters of neighboring sites.
Motif searches are insufficient to identify all in vivo binding locations for a protein because proteins recognize a wide variety of related sequences, of which only a small fraction are bound (Badis et al., 2009; Walter and Biggin, 1996). Consequently, while a consensus target motif may be extracted from data as a whole, a large fraction of putatively bound locations either lack an obvious motif or contain multiple degenerate versions of the motif (Cawley et al., 2004; Yang et al., 2006), and thus cannot be definitively assigned to a particular recognition sequence.
Protein binding microarrays have proven to be powerful in defining a DNA binding domain’s intrinsic specificity in vitro (Badis et al., 2009). However, in vivo, such specificity may be altered, prevented, or constrained in the context of the thousands of other proteins that constitute the nuclear milieu. Digital genomic footprinting can detect highly occupied binding sites at high resolution (Hesselberth et al., 2009), but identifying the source of protected genomic footprints requires a priori knowledge of which protein binds to the identified sequence. Problematically, different proteins may bind to the same sequence. Importantly, low occupancy binding is widespread in genomes (Li et al., 2008), but its physiological importance and distinction from noise has been difficult to discern by any assay thus far.
Here, we develop ChIP-exo, to precisely map a comprehensive set of protein binding locations genome-wide in any organism, and to greatly diminish both erroneous and missed calls associated with mapping. Importantly, ChIP-exo achieves near single-base resolution. The resulting maps provide a striking display of genome-wide site utilization that vividly delineates the variation in sequence recognition specificity and the underlying principles that drive specificity in vivo. From these binding events, potential mechanisms of site evolution, chromatin interplay, and genome-wide network regulation become clearer.
RESULTS
ChIP-Exo Design
We considered the possibility that a protein covalently crosslinked to DNA would block strand-specific 5′-3′ degradation by lambda exonuclease (Figure 1A), thereby creating a homogeneous 5′ border at a fixed distance from the bound protein. DNA sequences 3′ to the exonuclease block remain intact and are sufficiently long to uniquely map to a reference genome, after identification by deep sequencing (Figure S1A). Uncrosslinked nonspecific DNA is largely eliminated by exonuclease treatment, as evidenced by the repeated failure to generate a ChIP-exo library from a negative control BY4741 strain. Therefore, use of an exonuclease makes comparisons to input DNA or mock IP moot, in that such DNA is destroyed.
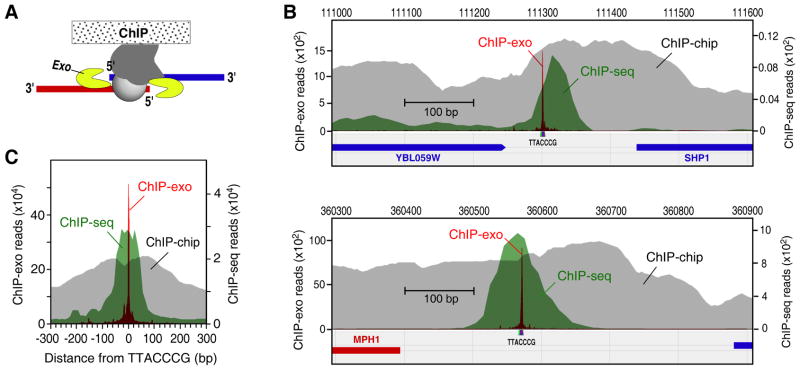
Figure 1. Single Base-Pair Resolution of ChIP-Exo.
(A) Illustration of the ChIP-exo method. ChIP DNA is treated with a 5′ to 3′ exonuclease while still present within the immunoprecipitate. The 5′ ends of the digested DNA are concentrated at a fixed distance from the sites of crosslinking, and are detected by deep sequencing (see also Figure S1).
(B) Comparison of ChIP-exo to ChIP-chip and ChIP-seq for Reb1 at specific loci. The gray, green, and magenta filled plots, respectively show the distribution of raw signals, measured by ChIP-chip using Affymetrix microarrays having 5 bp probe spacing (Venters and Pugh, 2009), ChIP-seq, and ChIP-exo. Sequencing tags on each strand were shifted towards the 3′ direction by 14 bp so as maximize opposite-strand overlap.
(C) Aggregated raw Reb1 signal distribution around all 791 instances of TTACCCG in the yeast genome. The ChIP-seq and ChIP-exo datasets included 2,938,677, and 2,920,571 uniquely-aligned tags, respectively.
ChIP-exo Improves Genome-wide Mapping Accuracy and Sensitivity
We initially focused on the yeast Reb1 protein, which has a clear DNA recognition site (TTACCCG) that can be used for independent validation (Badis et al., 2008; Harbison et al., 2004). Reb1 is involved in many aspects of transcriptional regulation by all three yeast RNA polymerases, and promotes formation of nucleosome-free regions (NFRs) (Hartley and Madhani, 2009; Raisner et al., 2005). It is also found at telomeres. We compared ChIP-exo to ChIP-chip and standard sonication-based ChIP-seq.
The unfiltered ChIP-exo signal was highly focused across the genome at TTACCCG sequences (Figure 1B,C). ChIP-chip and ChIP-seq displayed broader signals. When converted to peak-pair calls (described below), ChIP-exo displayed a standard deviation (σ of 0.3 bp (Figure S1B), which indicates that ChIP-exo of Reb1 has single-base accuracy. In comparison, ChIP-seq displayed more than 90-fold greater mapping variability (σ = 24 bp). ChIP-exo also displayed lower raw background. The raw signal-to-noise ranged from 300- to 2800-fold (Table S1). Subsequent employment of noise filters produced a comprehensive set of bound locations. In contrast, ChIP-chip and ChIP-seq had 7- and 80-fold raw signal-to-noise, respectively. ChIP-exo retained its quantitative properties, in that occupancy levels correlated with that from ChIP-seq (Figure S1C), and peak-pair intensities correlated (Figure 2A).
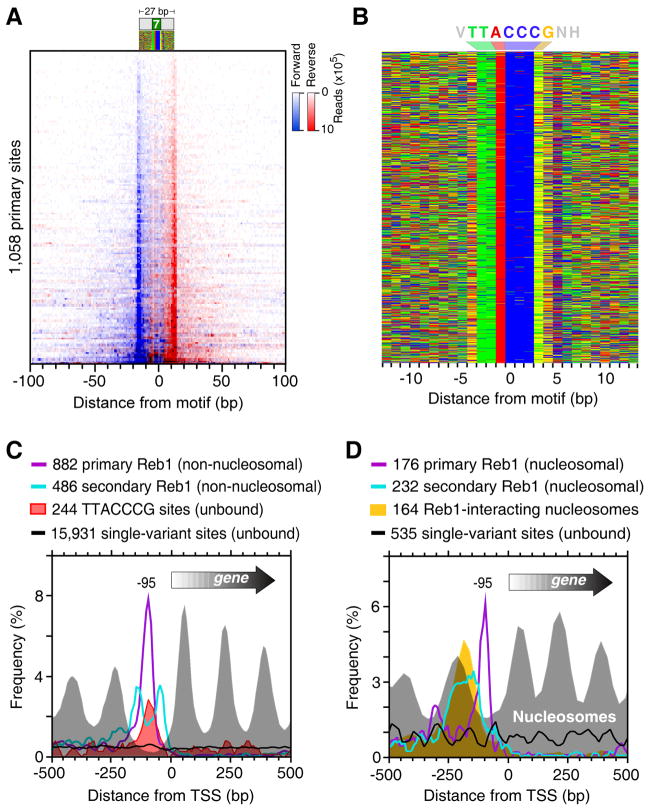
Figure 2. Genome-Wide Identification of Reb1-Bound Locations.
(A) Raw sequencing tag distribution around 1,058 primary Reb1-bound locations (rows). Blue and red indicate the 5′ ends of forward (left border) and reverse strand tags (right border), respectively, centered by the motif midpoint, and sorted by Reb1 occupancy level.
(B) Color chart representation of 27 bp of DNA sequence located between each Reb1 peak-pair, and centered by the motif midpoint. Each row represents a bound sequence ordered as in panel A. Red, green, yellow, and blue indicates A, T, G, and C. The Reb1 consensus sequence is indicated as VTTACCCGNH (V=A/C/G, H=A/T/C) (see also Discussion).
(C) Distribution of non-nucleosomal primary (purple trace) and secondary (cyan trace) Reb1 bound locations and respective nucleosome dyads (gray fill) around the TSS. Locations that were within 100 bp of a nucleosome midpoint (Figure S2I) were removed and plotted in panel D. Distribution traces of all unbound (<2% of average occupancy) TTACCCG sites and single nucleotide variants are shown by the red fill and black traces, respectively.
(D) Distribution of nucleosomal primary (purple trace) and secondary (cyan trace) Reb1 bound locations and respective nucleosome dyads (gray fill) around the TSS. The distribution of previously determined Reb1-bound nucleosome dyads is shown by the orange fill (Koerber et al., 2009). Distributions of unbound single nucleotide variants for those genes are shown by the black trace.
Reb1 Has Multiple Highly Organized Secondary Interactions at Promoters
The 5′ end of ChIP-exo tags (as well as peaks) located on one strand were largely at a fixed distance (~27 bp) from another tag or peak on the other strand, corresponding to the two exonuclease barriers formed by Reb1 (Figures 2A and S2A,B). A total of 1,776 Reb1 peak-pairs were identified (Supplemental Data File 1). Importantly, these peak-pairs were not pre-selected based upon the presence of any DNA sequence motif, although a motif was present in nearly all cases.
Of the peak pairs, 60% (1,058/1,776) were classified as primary locations, and 40% (718/1,776) as secondary. Secondary locations were defined as less-occupied locations within 100 bp of a more-occupied location. Thus, most Reb1 locations were found in clusters. Nearly all (92%) primary locations contained the TTACCCG Reb1 recognition site or a single nucleotide variant centered between its borders (Figures 2A,B and S2C). Increased deviations from TTACCCG were associated with lower occupancy levels (Figure S2D), which reflects low affinity. Such binding was clearly distinguishable from background. Surprisingly, Reb1 predominantly utilized TTACCCT at telomeres, which indicates that functionally distinct genomic regions may utilize particular site variants.
Compared to isolated sites, Reb1 had ~10-fold higher occupancy levels at clustered sites than would have been expected based upon sequence information (Figure S2D). This might reflect cooperative stabilization between primary and secondary locations. Secondary binding likely represents the same type of binding as primary binding, rather than incidental contact that is captured by crosslinking, because secondary locations tended to have canonical peak-pair distances, were reproducible from multiple biological replicates, and had centrally positioned, albeit degenerate, Reb1 motifs (Figure S2C). Remarkably, secondary sites were concentrated about 40 bp from a primary site (Figures 2C and S2E,F). Such resolution of individual binding locations within a cluster was not obtainable by standard ChIP-seq or ChIP-chip. Such a concentration of binding at a fixed distance from a primary bound location is unlikely to have arisen by chance, which suggests that even lowly-occupied secondary locations have biological relevance.
Depending on filtering criteria, only 34–72 percent of Reb1-bound regions identified in other studies were confirmed by ChIP-exo (Figure S2G). Those not identified, were not statistically enriched with Reb1 motifs and were not identified in any of the other studies, which indicates that they represent false positives in those datasets. We examined the false positive rate in our ChIP-exo dataset by searching for 48 randomized versions of the Reb1 motif located between peak pairs, and found on average <0.05% having a scrambled motif or a single nucleotide variant (Table S2). Thus, few if any, of the ChIP-exo detected Reb1-bound locations were in error (false positives).
The higher resolution afforded by ChIP-exo substantially increased the number and accuracy of Reb1-bound locations, making genome-wide ontologies more comprehensive (Figure S2H) and binding patterns more evident. For example, Reb1-bound locations were tightly positioned 95 bp upstream of the transcriptional start site of 778 annotated genes (14% of all genes, Figure 2C), well within the NFRs that they have been implicated in maintaining. Unoccupied or lowly occupied TTACCCG sites were enriched at the same location (Figure 2C), indicating that they are likely to be functionally important. These sites were nucleosome-free (not shown), indicating that a continued presence of Reb1 is not necessary to maintain these NFRs.
Reb1 also interacts with nucleosomal DNA in vivo where it binds at the NFR edge of the “−1” nucleosome (Koerber et al., 2009). ChIP-exo detected relatively strong Reb1 binding at such locations (Figure S2I). The ChIP-exo properties (peak-pair distance and motif distance) of nucleosomal and non-nucleosomal Reb1 were identical (Figures 2D and S2J), indicating that the presence of a nucleosome did not compromise Reb1 detection. Reb1-bound nucleosomes, however, did contain a higher density of secondary Reb1 locations, and unlike non-nucleosomal Reb1 these secondary locations were almost entirely located on the nucleosomal-side of primary Reb1 locations (i.e., upstream relative to the transcription start site or TSS). This would place secondary locations on the opposite side of the physical nucleosome from where primary locations are found (Figure S2K). A provocative possibility is that an intervening “−1” nucleosome appositions primary and secondary Reb1 sites in a way that promotes cooperative Reb1 binding. Conceivably this may provide one mechanism of locking down the position of the −1 nucleosome. Note in Figure 2D that the composite position of Reb1-bound −1 nucleosomes is shifted towards the NFR compared to the bulk −1 nucleosome population at the same loci, which might reflect directed positioning of the −1 nucleosome by Reb1.
Evidence of Single Nucleotide Exclusion in Gal4 Sites
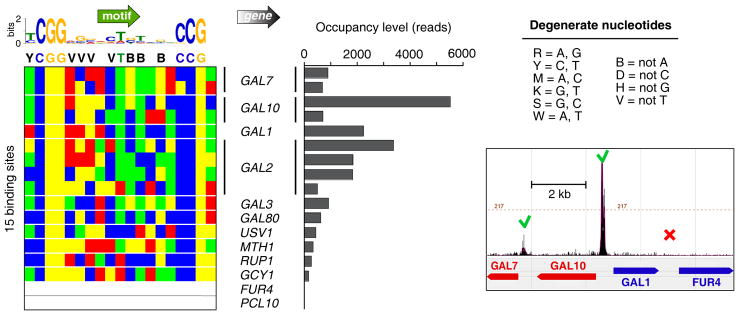
Transcription factor Gal4 reportedly binds to and regulates only ten genes related to the yeast galactose regulon (Ren et al., 2000). We identified 15 Gal4 peak-pair binding locations at eight of those genes in galactose-induced conditions, but not at the previously reported FUR4 and PCL10 loci (Figures 3 and S3). Prior low resolution detection at FUR4 (uracil permease) may have been due to contamination from adjacent GAL1, which binds Gal4 strongly; PCL10 (Pho85 cyclin) had the weakest signal of the ten regions in the prior study. Low-levels of Gal4 were detected at two new loci: USV1, which encodes a transcriptional regulator of genes involved in growth on non-fermentable carbon sources, and RUP1-SFL1, which was also detected at a low-confidence interval in another study (Harbison et al., 2004). SFL1 encodes a stress-response transcriptional activator, and RUP1 is involved in regulation of ubiquitin ligase. Thus, ChIP-exo comprehensively detected a small set of bound locations, resulting in a more accurate delineation of the Gal4 regulon. Our analysis revealed an in vivo sequence preference of 11 bp between each Gal4 half site, consistent with in vitro studies (Liang et al., 1996; Marmorstein et al., 1992). Many of these positions were limited or biased to three of the four possible nucleotide choices. This might reflect a selective negative interaction of the excluded nucleotide with Gal4, directly or indirectly, that does not occur with the other three nucleotides. This type of exclusion was also seen with other factors examined here.
Figure 3. Genome-Wide Identification of Gal4-Bound Locations.
The left panel shows a color chart representation of 18 bp of sequence located between each Gal4 peak-pair. Sites are oriented such that the nearest TSS is on the same strand. A MEME output logo is shown, along with a single-letter degenerate code of the surmised consensus (definition of the code is to the right). The bar graph shows the occupancy level at these sites. Also shown is a browser shot of Gal4 ChIP-exo tags around the contiguous GAL7, 10, 1, - FUR4 region. Sequencing tags on each strand were shifted towards the 3′ direction by 13 bp.
Transcription Factor Phd1 Recognizes Distinct Motifs
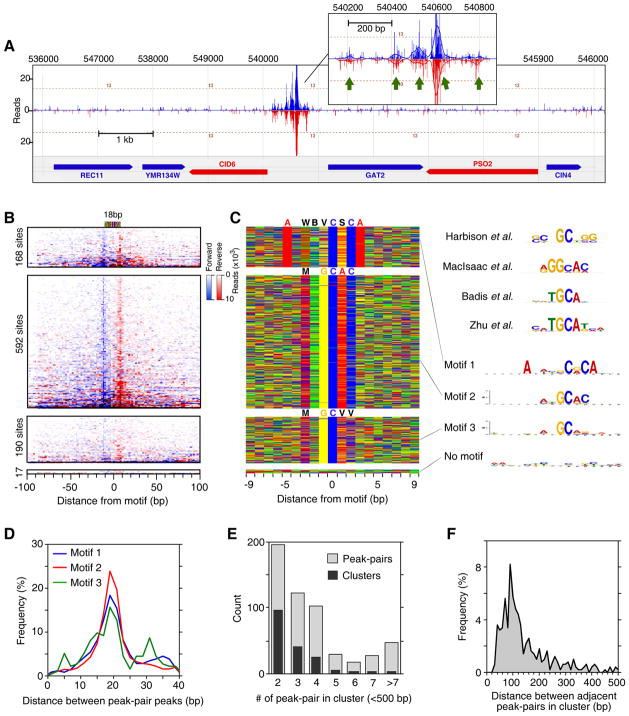
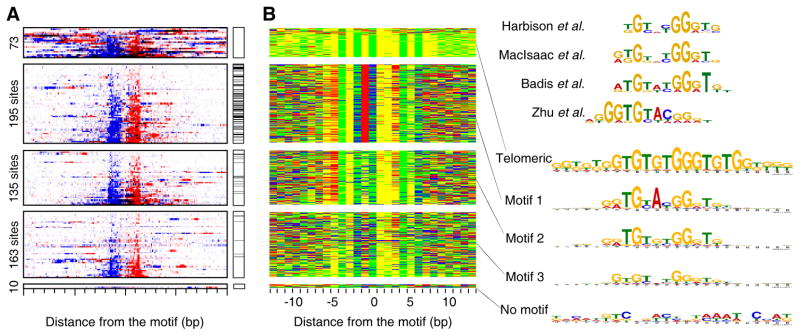
In an effort to evaluate whether ChIP-exo could define the specificity of a protein whose consensus site has been reported to be ambiguous (Badis et al., 2008; Harbison et al., 2004; MacIsaac et al., 2006; Zhu et al., 2009), we examined Phd1. Phd1 is a transcriptional activator of genes involved in yeast pseudohyphal growth (Gimeno and Fink, 1994). Its consensus site varies widely in different studies (Figure 4C, right). Figure 4A shows an example of robust Phd1 binding, where five distinct Phd1 binding locations within a 600 bp GID6-GAT2 intergenic region were resolved. ChIP-exo identified 967 Phd1 peak-pairs (Figure 4B). MEME analysis detected three motifs (Figures 4C and S4A) (Bailey et al., 2009). Motif 1 and 2 were distinct, although not entirely, which may explain its site ambiguity. Motif 3 was a degenerate version of motif 2. All motifs had the same 19 bp region protected from exonuclease digestion centered precisely over each motif (Figure 4B,D), which suggests that Phd1 binds these sites with the same interaction boundaries, despite the sites having distinct sequences at many positions.
Figure 4. Genome-Wide Identification of Phd1-Bound Locations.
(A) Example of Phd1 binding at the GID6-GAT2 locus. Green arrows indicate Phd1 motifs. Vertical blue and red bars demarcate the 5′ ends of forward and reverse strand tags, respectively, shifted in the 3′ direction by 10 bp.
(B) Raw sequencing tag distribution around 967 Phd1-bound locations. Blue and red indicate the 5′ ends of forward and reverse strand tags, respectively, centered by the motif midpoint. Rows were divided into four groups based upon the type of motif shown in panel C, and sorted by Phd1 occupancy level. The additional tags distributed distal to the main peaks reflect multiple Phd1 peak-pairs residing near each other. See Figure S4C for Gene Ontology analysis.
(C) Color chart representation of 19 bp of sequence located between each Phd1 peak-pair, and centered by the motif midpoint. Each row represents a bound sequence ordered as in panel B. MEME logos for each group are shown to the right. The upper four logos were reprinted from the indicated references.
(D) Frequency distribution of Phd1 peak-pair distances for groups defined by motifs 1–3.
(E) Multiple Phd1 peak-pairs reside in clusters. Black bars indicate the number of Phd1 clusters found having the indicated number of peak-pairs within 500 bp of each other. Light bars indicate the total number of peak-pairs present in those clusters.
(F) Frequency distribution of distances between adjacent peak-pairs in clusters defined in panel E.
The median number of tags associated with motif 1 and 2 were the same (Figure S4B), indicating that Phd1 has similar affinity for both motifs in vivo. The more degenerate motif 3 was associated with only moderately less occupancy. This contrasts with Reb1, where site divergence was associated with lower occupancy. This relative insensitivity of Phd1 occupancy to the underlying sequence and the rather low complexity of the Phd1 motifs suggest that Phd1 might rely more on additional interactions to gain specificity and affinity. Indeed, most (548/967) Phd1-bound locations were found in clusters having two to four sites (Figure 4E, dark bars), which might reflect such interactions. Locations within clusters were spaced ~100 bp apart (Figure 4F), suggesting that Phd1 binding has a restricted spatial organization at promoters. This analysis of Phd1 illustrates two principles of genome-wide protein-DNA specificity: 1) A protein may recognize a variety of related motifs in vivo with similar affinity, as demonstrated in vitro (Badis et al., 2009), and 2) binding specificity and affinity may be distributed across multiple sites.
Selective Motif Usage by Rap1 at Ribosomal Protein Genes and Telomeres
Rap1 is a yeast sequence-specific repressor and activator protein that regulates telomeres and ribosomal protein (RP) genes (Lieb et al., 2001; Moehle and Hinnebusch, 1991). ChIP-exo identified 73 Rap1 peak-pairs in telomeric regions and 503 peak-pairs elsewhere (Figure 5A). MEME analysis reported four motifs, which were not distinguished in prior large-scale studies (Figure 5B). Rap1 at telomeric regions had a broader recognition site (GTGTGTGGGTGTGG) with higher apparent occupancy than the three other motifs (Figure S5A). Telomeric Rap1 had a broader region of protection (by 4–7 bp) from exonuclease digestion compared to the three other motifs (Figure S5B), which may indicate that Rap1 is bound differently to telomeric DNA compared to other places in the genome.
Figure 5. Genome-Wide Identification of Rap1-Bound Locations.
(A) Raw sequencing tag distribution around 576 Rap1-bound locations. Blue and red indicate the 5′ ends of forward and reverse strand tags, respectively, centered by the motif midpoint. Rows were divided into four groups based upon the type of motif shown in panel B, and sorted by Rap1 occupancy level. The additional tags distributed distal to the main peaks reflect multiple Rap1 peak-pairs residing near each other. Black horizontal lines indicate the distribution of ribosomal protein (RP) genes having a TSS between 100 bp upstream and 700 bp downstream of a Rap1 location.
(B) Color chart representation of 27 bp of sequence located between each Rap1 peak-pair, and centered by the motif midpoint. Each row represents a bound motif ordered as in panel A. MEME logos are shown to the right. The upper four motifs were reprinted from the indicated references.
Rap1 motif patterns 1, 2 and 3 were 12–13 bp in length, and were centered between Rap1 peak pairs that were spaced by 24 bp. This calculates to a ~5–6 bp “headroom” between the exonuclease cleavage site and the edge of the Rap1 motif, which is a value that we frequently observe for other proteins. Compared to Reb1 and Phd1, Rap1 binding sites displayed substantially more sequence heterogeneity. However, this heterogeneity was spread over more nucleotide positions, indicating that Rap1 devotes less binding energy to each nucleotide position, while still attaining an equivalent level of specificity. Increased sequence divergence from the already degenerate pattern appeared to be only modestly associated with lower occupancy (Figure S5A, also compare between and within motifs in Figures 5A,B). As with Reb1 and Phd1, Rap1 was found in clusters (Figure S5C,D). Where Rap1 clusters occurred, they typically consisted of two sites that were separated by 20–100 bp.
Most Rap1-bound genes are involved in protein synthesis (Figure S5E). Rap1 bound to 92 of 121 RP genes (76%, p = 9.0 × 10−130), and 388 of 4671 non-RP genes (8%). While six of the non-RP genes (ENO1, GPM1,2, TDH3, PFK2, PGI1) were associated with the main glycolysis pathway (Figure S5F), Rap1 was not broadly associated with other glycolysis-related genes.
Of the RP genes, 77% (71/92) employed motif 1 (Figure 5A). Only three RP genes contained Rap1 in clusters, whereas 29 were expected by chance (p = 5 × 10−9). Thus, in rich media RP genes selectively utilize a single copy of motif 1, which is the stronger consensus. In contrast, other Rap1-bound genes had a greater tendency to use clusters of Rap1 motifs that had a weaker consensus. This difference may reflect distinct mechanisms by which Rap1 regulates RP genes versus other genes, for example, using a single strong consensus versus two weak ones to achieve similar occupancy levels.
Rap1 binds nucleosomal DNA in vivo and in vitro (Koerber et al., 2009; Rossetti et al., 2001). Consistent with this, Rap1 locations were enriched near the edge of previously-defined Rap1-bound nucleosomes (Figure S5G,H). As with Reb1, nucleosomal Rap1 displayed relatively high site occupancy compared to non-nucleosomal Rap1 (Figure S5I), which may be due to stronger consensus sites that are associated with nucleosomal Rap1 (Figure S5J). Moreover, the presence of nucleosomes did not alter the detection properties of Rap1 (Figure S5K), which further demonstrates that histones do not interfere with detection by ChIP-exo. Conceivably, the wrap of the Rap1 DNA site on the nucleosome surface may enhance Rap1 affinity (to the extent that crosslinking provides an equivalent measure of occupancy in the two putative types of interactions). Ultimately, such binding may lead to nucleosome eviction in that nucleosomes at Rap1-bound regions generally had low occupancy (Figure S5G).
Human CTCF Mapping Supports the Notion of a CTCF “Code”
We applied ChIP-exo to human CTCF, as a more complex biological system. CTCF is a sequence-specific DNA binding protein that plays important roles in metazoan gene regulation and three-dimensional chromatin organization in a wide variety of cell types (Ohlsson et al., 2010; Phillips and Corces, 2009). Where CTCF binds in a genome and its DNA recognition sequence has been the subject of some dispute (Bao et al., 2008; Barski et al., 2007; Cuddapah et al., 2009; Jothi et al., 2008; Kim et al., 2007). Estimates range from 13,000 to 29,000 binding locations across mammalian genomes (Cuddapah et al., 2009; Dion et al., 2007). We detected 35,161 CTCF-bound locations in HeLa cells (Figures 6A,B and S6A). Nearly all CTCF-bound regions (93%), determined previously in HeLa cells by ChIP-seq (Cuddapah et al., 2009), were verified by ChIP-exo (Figure S6B), although another ~17,000 locations were missed.
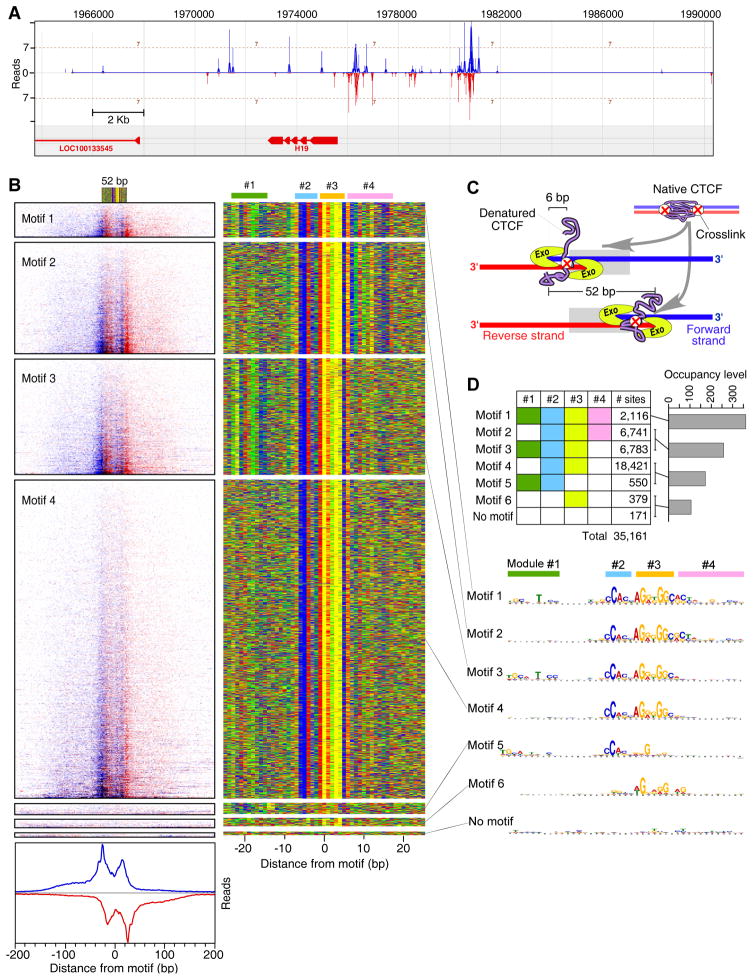
Figure 6. Genome-Wide Identification of Human CTCF-Bound Locations.
(A) Example of CTCF binding at the H19 locus. Vertical blue and red bars demarcate the 5′ ends of forward and reverse strand tags, respectively.
(B) The left panel shows raw sequencing tags distributed around 8,578 CTCF-bound locations present on chromosomes 1, 2, and 3. Blue and red indicate the 5′ ends of forward and reverse strand tags, respectively, centered by the motif midpoint. Rows were divided into six groups based upon the type of motif shown in the right panel. Summed tag counts are shown on the bottom of the left panel. The right panel shows a color chart representation of 52 bp sequence located between the most 5′ borders on each strand, and centered by the motif midpoint. Each row represents a bound motif ordered as in the left panel. Locations of four CTCF site modules are drawn on the top of the right panel.
(C) Model explaining the presence of four exonuclease blockage sites (two peak-pairs) for each CTCF-bound location. CTCF is illustrated as a native protein (purple) during the crosslinking. Two crosslinks are shown to represent a population distribution. However, any one CTCF molecule is likely to contain 0–1, and rarely 2, crosslinks. CTCF is shown in its denatured state during the exonuclease treatment, with one crosslink occurring at either site.
(D) Table colored to demarcate the combinatorial usage of the four CTCF site modules. Corresponding median tag counts for the specified motifs having different number of modules are shown as a bar graph. MEME logos for each motif and corresponding CTCF site module are shown below.
About 6% (2,106/35,161) of CTCF-bound locations were enriched in core promoters, ~85 bp upstream of a TSS, of ~10% (3,016/29,943) of all annotated genes (Figure S6C). This precise positioning is reminiscent of the positioning seen by yeast Reb1, both of which have been implicated in organizing flanking nucleosomes (Cuddapah et al., 2009; Hartley and Madhani, 2009).
Surprisingly, CTCF-bound locations tended to have four exonuclease-derived borders, 12–14 and ~52 bp apart, instead of the two seen thus far in yeast (Figures 6B and S6D). We surmise that they represent one binding event, in agreement with footprinting studies (Boyle et al., 2011; Ohlsson et al., 2001), but with two distinct sites of DNA crosslinking, each having a border on both strands (Figure 6C). This can be achieved if only a small fraction of either site becomes crosslinked in a population of molecules. The exonuclease would then proceed to either the first or second stop, depending on the location of the crosslink. This would likely require that CTCF be denatured (promoted by SDS during ChIP) such that the digestion block occurs at the site of crosslinking rather than where CTCF folds onto the DNA surface.
Strikingly, >99.5% of the CTCF locations contained parts of a single compound consensus sequence that spanned ~41 bp (Figures 6B and S6E). We identified four combinatorial modules within a compound consensus sequence (Figure 6D). Roughly half of all CTCF-bound locations utilized only modules 2 and 3, constituting the core consensus to which four of CTCF’s eleven zinc fingers bind (Ohlsson et al., 2010). The other CTCF-bound locations employed three to four modules (mainly 1–3, 2–4, and 1–4). In ~1% of the cases only module #3 was used. Sites employing more modules tended to have a higher CTCF occupancy (Figure 6D, bar graph), suggesting that modules enhance affinity. Since the exonuclease barriers were similarly arranged regardless of the modules present, modules do not alter the boundaries to which CTCF binds DNA, although details of its footprint differ (Boyle et al., 2011). A prevailing model is that CTCF uses four of its eleven zinc fingers to bind modules 2 and 3 (Filippova et al., 1996; Ohlsson et al., 2010). It then uses different combinations of its remaining fingers (“multivalency”) to recognize alternative sequences, termed a “CTCF code”. Our finding of combinatorial use of modules supports this notion.
DISCUSSION
ChIP-exo provides a comprehensive and high-resolution (to within a few bp) view of transcription factor-DNA interactions across a genome. It detects low-level binding to the point where typically 2–4 fold more binding locations are discovered. With this precision, cognate DNA binding sequences become unambiguous, thereby revealing the complexity of site-specific DNA recognition. Detection by ChIP-exo is not compromised by the presence of other bound proteins, including histones. Since only a small fraction of proteins become crosslinked to DNA, neighboring proteins are stripped away by stringent SDS detergent washes in the ChIP procedure. ChIP-exo not only resolves adjacent binding events that are indiscernible by other methods, but also resolves multiple crosslinking sites within a given bound location.
ChIP-chip and ChIP-seq Have Substantial False Discovery Rates
For the five proteins examined here, >98% of all peak-pair binding locations determined by ChIP-exo contained a recognizable DNA binding motif. The remaining ~2% had very low occupancy, and may contain highly degenerate motifs. If generally true, then many sequence-specific DNA binding proteins may not make high affinity primary contacts with nonspecific DNA as some studies have suggested. Instead such putative binding events might represent false positives or have substantially degenerate DNA recognition elements that cannot be readily discerned with the current resolution of standard ChIP-chip and ChIP-seq technology. As such, standard methods may obscure the true degeneracy intrinsic to site-specific DNA recognition in vivo. Nevertheless, many regulatory proteins might gain DNA binding specificity through protein-protein bridging interactions in lieu of sequence-specific binding (Welboren et al., 2009; Zhao et al., 2010).
The false positive and false negative rates associated with ChIP-chip and ChIP-seq vary depending upon chromatin fragmentation heterogeneity, ChIP-efficiency, background contamination, limitations imposed by the detection platform, and bioinformatic filtering/thresholding of the data. From our analysis, we suspect that as much as 50% of factor-bound locations determined by ChIP-chip and 30% by ChIP-seq could be false positives. False negatives represent a much higher percentage. Higher data thresholding produces fewer false positives, but more false negatives. While these false discovery rates associated with ChIP-chip and ChIP-seq appear high, they are sufficiently low to draw robust statistical conclusions. However, the validity of any one selected binding location may have substantially more uncertainty, which may be diminished with ChIP-exo.
Diversity and Complexity in Site Recognition In Vivo
From the five proteins examined here, we observed diverse and complex strategies for achieving sequence-specific DNA binding (Figure 7). These proteins likely represent a small sampling of this diversity. Several predominant well-known themes arise. First, each nucleotide position within a binding site has a characteristic biased usage of the four possible nucleotides. Certain positions may be essentially invariant, whereas others accept any nucleotide with equal frequency. Between these extremes, some positions are biased towards two or three nucleotides. Usage of three of four possible nucleotides at a position might indicate that the fourth causes a negative interaction, rather than the three providing a positive interaction. These position-specific tolerance profiles form the basis of a consensus. Variations from the consensus may serve to alter binding affinity, the magnitude of which may be dependent on the type of nucleotide present at other positions within the site, and/or cooperative or competitive binding with other factors (discussed below).
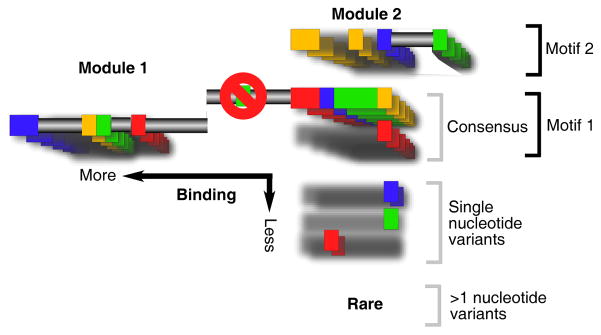
Figure 7. Themes in Genomic Binding Site Organization.
Shown is an amalgamation of hypothetical DNA recognition sequences. Each color represents one of four nucleotides (A, C, T or G) that constitute a recognition site. The core sequence is shown as a series of color boxes. For simplicity, alternative nucleotides to the core sequence are represented as additional boxes, with constant nucleotides represented as a gray shadow. Frequently used alternatives form a consensus motif, as indicated. Collectively, single-nucleotide variants are shown to be common, although any particular variant occurs infrequently. Also shown are alternative motifs, and compound motifs that are built up combinatorially from modules. Multiple motifs may exist in clusters (not shown). A position that excludes a certain nucleotide is shown as a red circle with a line drawn through. Regions where any nucleotide would suffice are drawn as a gray horizontal bars. Variants having >1 nucleotide variation from the core are designated as rare, but this should be qualified to short, well-defined motifs. Long motifs tend to have many degenerate positions.
Since site variation may occur throughout a consensus sequence, any particular deviation from the consensus may be rare. Collectively, however, deviations from the consensus appear to be quite common. As such, there may not be a clear demarcation between a consensus sequence and site variants. It may therefore be useful to think of each position in a site as a four-setting non-linear rheostat, of which some positions provide coarse tuning of affinity whereas others provide fine-tuning.
A particular level of occupancy may be needed to regulate a set of functionally related genes, in which case a single motif version may be employed. For example, Rap1 regulates a large set of ribosomal proteins genes, and these genes selectively utilize one version of the Rap1 consensus. Rap1 is also found at telomeres where a different version of the consensus is employed. The same is seen for Reb1. This phenomenon of selective motif version utilization might explain some of the reported discrepancies in consensus sequences defined in different studies that may have been derived from different subsets of binding locations.
It almost seems paradoxical that a more comprehensive set of bound locations would necessarily yield a more degenerate consensus. However, this finding is consistent with the idea that low affinity binding sites are low affinity because their sequences are farthest from the consensus. Therefore a technique with greater detection sensitivity would, by definition, yield an abundance of low affinity interactions occurring at degenerate motifs.
Physiological Importance of Lowly Occupied Sites
At what point does a sequence impart so little specificity/occupancy that it ceases to be biologically meaningful? Biological networks have been generally thought of as being discrete, meaning that a factor either regulates or does not regulate particular genes in the network. However, an alternative view is of a continuum, where a factor’s regulatory potential on a gene scales with its occupancy level (Li et al., 2008). A continuum of occupancy levels renders the concept of false negatives as somewhat meaningless, except in an operational sense. Thus, while protein binding might be detected at more than a thousand locations in a genome, only the binding of the most highly occupied sites might be rationalized. The rest may form a continuum or increasingly more subtle regulation as site occupancy decreases, which would make network definition seemingly less vivid. Thus, even with perfect data, the set of bound locations would not be definable in an absolute sense, but only at a specified occupancy threshold.
The low affinity/occupancy locations reported in this study show evidence of being real (i.e., not false positives) and functional. First, such locations are reproducibly detected in multiple biological replicates. Second, with an uncertainty of less than a few bp, such locations are almost always centered over a sequence with similarity, albeit degenerate, to a high affinity site. Third, peak-pair distances are nearly identical to distances of high occupancy locations. Fourth, and most importantly, such locations are not random in the genome, but instead are concentrated at fixed distances from genomic features. For example, isolated low and high occupancy Reb1 locations are concentrated 95 bp upstream of the TSS, and clustered low and high occupancy locations are concentrated ~40 bp from each other. When Reb1 is bound to the −1 nucleosome, the lowly occupied secondary locations selectively reside in the upstream flanking region bound by the nucleosome. In contrast, the genome is awash with equivalent sites that are intrinsically low affinity, but no binding is detected. Taken together, none of these properties are consistent with the notion of physiological irrelevance or nearby incidental contact due to looping or chromatin compaction. Such weak interactions might have little measurable regulatory potential on gene expression, but may be sufficiently important for fine-tuning to be evolutionarily maintained.
An alternative view of lowly occupied sites is a hit-and-run mechanism (Biddie et al., 2011; Voss et al., 2011), whereby the dwell time of a protein on a DNA site may be rather short, but is sufficiently long to catalyze downstream events (e.g. chromatin remodeling) that may be more long-lived and ultimately functional. As such, low occupancy sites may be functionally important.
Multiple Mechanisms by which Transcription Factors Bind Chromatin
The effective concentration of DNA binding proteins and DNA sites in the nucleus may far exceed the KD of DNA binding, and as such factors may be DNA-bound (specifically or nonspecifically) most of the time (Lin and Riggs, 1975). This raises the question as to the exact pathway of site-specific DNA binding in vivo, whether factors exist in an unbound pool or are directly transferred from other DNA sites (von Hippel et al., 1974). Our finding that isolated high affinity sites may be lowly occupied in vivo, while many intrinsically low-affinity sites have higher occupancy, suggests that intrinsic affinity is not the sole determinant of occupancy in vivo. Rather a combination of effects, including high local concentrations, direct and indirect cooperativity, and competitive binding derived from other factors including nucleosomes will likely impose additional constraints. The contribution of any constraint may vary from one location to another.
For example, Reb1 not only binds in the middle of nucleosome-free regions and has been implicated in NFR formation, but it also binds quite strongly and selectively to nucleosomes. Rap1 binds to nucleosomes as well, but such nucleosomes seem to have low occupancy, which might reflect Rap1 binding followed by nucleosome eviction, rather than simple competitive binding. Certainly, many other sequence-specific binding proteins might recognize their site only after nucleosome eviction, and thus would be mutually competitive.
Each of the yeast proteins examined here had clustered binding locations. Clustered sites had substantially higher occupancy than isolated sites, perhaps owing to mutually cooperative binding through direct or indirect interactions or through cooperative exclusion of competing proteins. Site clustering might also give rise to the perception of non-orthologous site evolution. It is well known that cis regulatory elements have a conserved presence but not necessarily a conserved position in promoter regions (Birney et al., 2007; Dermitzakis and Clark, 2002; Moses et al., 2006). Conceivably, each site in a cluster of sites might evolve back and forth from high affinity (recognizable) to low affinity (unrecognizable). As such, two sites that appear at nonorthologous locations might also have degenerate orthologous equivalents that are undetectable by consensus matching.
Summary
To our knowledge ChIP-exo is the first technique that has the potential to reveal essentially a comprehensive and unambiguous set of genomic binding locations for a protein at near single bp accuracy. Moreover, improved mapping accuracy and background reduction substantially reduce the number of tags needed to unambiguous identify a bound location, and provides a much greater range of occupancy levels that can be detected. This allows for a more complete assessment of regulatory networks, the repertoire of binding sites, their evolutionary turnover, and the context in which they interact with other factors.
EXPERIMENTAL PROCEDURES
Standard chromatin immunoprecipitation (ChIP) was performed as previously described (Venters and Pugh, 2009). While still on the sepharose resin, the immunoprecipitated DNA was polished, ligated with P2 adaptors, and digested with lambda (λ) exonuclease. λ exonuclease-digested DNA was eluted from the resin. Crosslinks were reversed and proteins degraded with Proteinase K at 65°C. DNA samples were precipitated with ethanol and denatured at 95°C. DNA samples were primer-extended using a P2 primer. P1 adaptors were ligated to the λ exonuclease-digested end. The resulting DNA was PCR-amplified, gel purified, and sequenced using the SOLiD genome sequencer (AppliedBiosystems). The genomic distribution of sequence tags was used to identify peaks on the forward and reverse strand separately using the peak-calling algorithm in GeneTrack (Albert et al., 2008). Full methods are available in the Supplemental Information. Final binding locations are reported in Supplemental Data File 1. Direct binding to defined motifs was confirmed by Protein Binding Microarray Data (Table S3).
Supplementary Material
Acknowledgments
We thank Gue Su Chang and Cizhong Jiang for bioinformatic support, Mark Biggin (Lawrence-Berkeley Laboratory) for discussions on the concept of continuous networks, and members of the Pugh lab and the Penn State Center for Eukaryotic Gene Regulation for scientific discussions. Sequencing was performed at the Penn State Genomics Core Facility. This work was supported by NIH grant ES13768.
Footnotes
ACCESSION NUMBERS
Raw sequencing data are available at NCBI Sequence Read Archive (accession number: SRA044886).
Supplemental Information includes Extended Supplemental Methods, two tables, seven figures, and one data file.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Albert I, Wachi S, Jiang C, Pugh BF. GeneTrack - a genomic data processing and visualization framework. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Zhou M, Cui Y. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011 doi: 10.1101/gr.112656.110. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biology. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SD, Marmorstein R, Harrison SC, Ptashne M. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol Cell Biol. 1996;16:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Lin S, Riggs AD. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975;4:107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rossetti L, Cacchione S, De Menna A, Chapman L, Rhodes D, Savino M. Specific interactions of the telomeric protein Rap1p with nucleosomal binding sites. J Mol Biol. 2001;306:903–913. doi: 10.1006/jmbi.2001.4458. [DOI] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Revzin A, Gross CA, Wang AC. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proc Natl Acad Sci U S A. 1974;71:4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic Exchange at Regulatory Elements during Chromatin Remodeling Underlies Assisted Loading Mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Biggin MD. DNA binding specificity of two homeodomain proteins in vitro and in Drosophila embryos. Proc Natl Acad Sci U S A. 1996;93:2680–2685. doi: 10.1073/pnas.93.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. The EMBO journal. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Zhao C, Gao H, Liu Y, Papoutsi Z, Jaffrey S, Gustafsson JA, Dahlman-Wright K. Genome-wide mapping of estrogen receptor-beta-binding regions reveals extensive cross-talk with transcription factor activator protein-1. Cancer Res. 2010;70:5174–5183. doi: 10.1158/0008-5472.CAN-09-4407. [DOI] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.