Abstract
Rationale
Accurate and comprehensive de novo transcriptome profiling in heart is a central issue to better understand cardiac physiology and diseases. Although significant progress has been made in genome-wide profiling for quantitative changes in cardiac gene expression, current knowledge offers limited insights to the total complexity in cardiac transcriptome at individual exon level.
Objective
To develop more robust bioinformatic approaches to analyze high-throughput RNA sequencing (RNA-Seq) data, with the focus on the investigation of transcriptome complexity at individual exon and transcript levels.
Methods and Results
In addition to overall gene expression analysis, the methods developed in this study were used to analyze RNA-Seq data with respect to individual transcript isoforms, novel spliced exons, novel alternative terminal exons, novel transcript clusters (i.e., novel genes) and long non-coding RNA genes. We applied these approaches to RNA-Seq data obtained from mouse hearts following pressure-overload induced by trans-aortic constriction. Based on experimental validations, analyses of the features of the identified exons/transcripts, and expression analyses including previously published RNASeq data, we demonstrate that the methods are highly effective in detecting and quantifying individual exons and transcripts. Novel insights inferred from the examined aspects of the cardiac transcriptome open ways to further experimental investigations.
Conclusions
Our work provided a comprehensive set of methods to analyze mouse cardiac transcriptome complexity at individual exon and transcript levels. Applications of the methods may infer important new insights to gene regulation in normal and disease hearts in terms of exon utilization and potential involvement of novel components of cardiac transcriptome.
Keywords: RNA-Seq, transcriptome profiling, hypertrophy, heart failure
Regulation of gene expression has a critical role in normal cardiac function and pathogenesis of heart failure. A global change in cardiac transcriptome from normal to one with characteristics of “fetal-like” profile is a major part of the pathological remodeling in failing hearts1-3. Although much insight has been learnt from transcriptome profiling studies using microarray technologies, limitations in coverage and sensitivity still leave a significant part of the cardiac transcriptome landscape un-explored, especially concerning expression and variation at single exon resolution. Recent advances in high-throughput sequencing technologies are enabling a new way to study transcriptomes: massively parallel sequencing of short reads derived from mRNAs (RNA-Seq)4, 5. Compared to microarray technologies, RNA-Seq was shown to enable more accurate quantification of gene expression levels6, 7. More importantly, RNA-Seq does not require a priori annotation of gene and transcript structures. It allows not only in-depth studies of expression changes in known genes and alternative isoforms, but also unbiased characterization of novel exons and novel transcript clusters. It also enables investigation of long non-coding RNA (lncRNA) genes, which are not usually targeted by alternative transcriptome profiling methods, such as microarrays. Thus, RNA-Seq opens the way to de novo transcriptome reconstruction and discovery of novel transcripts of any mammalian cell. Indeed, recent reports using RNA-Seq to profile transcriptome in mouse heart have revealed interesting new insights in cardiac transcriptional and signaling networks in genetic models of heart failure7-10.
In this study, we developed bioinformatic methods to identify transcript structures and analyze transcriptome complexities with a particular emphasis on quantification of RNA splicing variants at single exon resolution using RNA-Seq data of normal and failing murine hearts. The methods take full advantage of the strength of RNA-Seq. We show that they allowed in-depth profiling and quantification of alternative mRNA structures, novel exons, novel transcript clusters (NTCs) and long non-coding RNA genes. The results open ways to direct experimental investigation of these novel transcriptome features and highlight the power of RNA-Seq to provide a comprehensive bioinformatic delineation of disease-specific transcriptomes.
Methods
RNA-Seq data generation and mapping
Left ventricular tissues were collected from male C57BL/6 mice after 1 week (hypertrophy stage, HY) and 8 weeks post trans-aortic constriction (TAC) procedure (heart failure stage, HF) and their corresponding Sham controls (Sham-HY, Sham-HF) (Online Supplement and Online Table I). To conduct RNA-Seq analysis, total RNAs from six TAC and Sham-operated mice at the HY stage and four TAC and corresponding Sham mice at the HF stage were obtained. Paired-end RNA-Seq reads (2×72nt or 2×76nt long) were mapped to the mouse Ensembl transcript sequences (release 56) using Bowtie11 and BLAT12. Mapping was first carried out for individual reads without considering the read-pairing information. Next, all read pairs were inspected for correct pairing by considering whether they map to the same chromosome, potentially in the same gene and with correct orientation relative to each other (Online Supplement). The pair of reads was considered as uniquely mapped if and only if one unique pair of mapped locations was identified.
Analysis of gene and transcript isoform expression
Levels of gene and exon expression were quantified using the RPKM measure13 and a minimum RPKM value of 3 (~ 1 copy per cell) is required for expressed genes/isoforms (see Results for justification of this cutoff). Gene expression differences were evaluated using Fisher's exact test after normalizing by the total number of mapped reads in each lane using the upper-quartile normalization method14. The resulted p-values were corrected via the Benjamini and Hochberg method. Differentially expressed genes were defined as those with changes of at least 1.5 fold between a pair of samples at a false discovery rate (FDR) of 5% for genes expressed at ≥ 3 RPKM in ≥ 1 sample. The Cufflinks software (v0.9.2)15 was used to estimate expression levels of individual isoforms of an Ensembl gene, which allowed identification of isoform-specific expression changes due to alternative transcription start site (ATSS) or alternative splicing (AS). To further assess overall isoform expression dissimilarity in two samples (A and B), a dissimilarity score was defined based on the Morisita-Horn similarity index as follows:
where Pi(A) and Pi(B) represent the expression of isoform i normalized by overall gene expression in the sample A or B. We only considered genes expressed at ≥ 3 RPKM in this analysis.
Transcriptome reconstruction
We developed two different methods for the reconstruction of transcript isoforms, 1) Guided Transcriptome Reconstruction: a method to reconstruct isoforms and discover novel exons within known genes; 2) De novo Reconstruction: a method to reconstruct completely new isoforms independent of known gene annotations. Details of the two approaches are presented in Online Supplement. Briefly, the following steps are common to both approaches: 1) define expressed sequence fragments (seq-frags); 2) identify connections between seq-frags based on reads mapped to spliced junctions; 3) generate a directed graph using seq-frags and their connections for each gene or each chromosome and 4) construct the isoforms by finding all possible paths in the graph. In Guided Transcriptome Reconstruction, novel seqfrags can represent novel exons or extended regions of known exons. Identification of exon boundaries (novel or known) depends on the presence of reads mapped to spliced junctions. Thus, to reduce possible false positive isoforms, we required at least two junction reads as evidence of a splicing event. In De novo Reconstruction, NTCs were identified in intergenic regions and clustered together on each chromosome. The boundaries of NTCs were decided by confirming the absence of spliced junctions or expressed seqfrags. Filters for minimum expression levels of seq-frags and canonical splicing signals were applied. The coding potential of NTCs was evaluated using the Coding Potential Calculator software16.
Statistical and computational methods
For gene expression analysis, the statistical significance was assessed by Fisher's exact test as described above. Pearson correlation coefficients for gene expression validation were calculated in R. For GO analysis, empirical p-values were estimated based on 10,000 randomized simulations (Online Supplement) and the Bonferroni cutoff was used to determine significant p-values. All other computational procedures including transcript isoforms reconstructions were carried out using in-house programs written in Python, Perl and R.
Results
Mapping of RNA-Seq reads
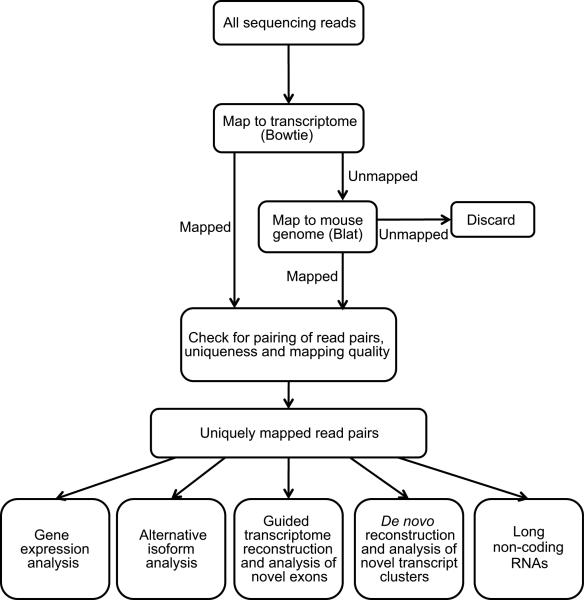
We obtained a total of 168 million pairs of reads using the standard paired-end RNA-Seq protocol on the Illumina GA II sequencer. Table 1 shows the number of reads in each TAC/Sham group and the mapping results. Our mapping procedure (Figure 1) ensures that reads generated by both known genes and novel transcribed regions were identifiable. In addition, it enabled detection of novel and known spliced junctions that connect two or more exons intervened by long introns. The usage of paired-end sequencing brings the advantage of improved mapping performance. We estimated that 4% of all the original reads were mapped nonuniquely as singletons but uniquely as pairs. Ambiguous mapping results can be removed by examining the pairing of reads. For example, 12% of all reads were categorized as unmapped in the paired-end mode but mapped uniquely as singletons (possible mapping errors in the single-end mode). Only pairs of reads that mapped uniquely to the transcriptome and/or the genome were retained for further analyses (Figure 1). Among the 168 million pairs of reads obtained in our study, ~95 million (57%) were uniquely mapped in total, which covered 72% of the known exon-exon junctions, 82% of exon bodies and 77% of known genes.
Table 1.
Number of RNA-Seq reads obtained for each type of samples and mapping results (percentages shown are relative to the number of final mapped reads).
| # of reads | HY | Sham-HY | HF | Sham-HF | Total |
|---|---|---|---|---|---|
| Total | 83,718,570 | 83,009,180 | 100,602,620 | 68,442,422 | 335,772,792 |
| Unmapped | 15,325,416 | 12,007,844 | 36,768,368 | 23,961,060 | 88,062,688 |
| Non-uniquely paired | 10,483,514 | 11,559,470 | 6,689,996 | 5,398,104 | 34,131,084 |
| Wrong pairing | 6,405,098 | 6,194,896 | 6,470,068 | 4,359,302 | 23,429,364 |
| Total uniquely paired (i.e., final mapped) | 51,504,542 | 53,246,970 | 50,674,188 | 34,723,956 | 191,049,656 |
| Mapped within exons | 37,882,946 (73%) | 39,198,116 (73%) | 32,479,491 (64%) | 22,977,952 (66%) | 132,538,505 (70%) |
| Mapped to exon-exon junctions | 9,570,514 (19%) | 9,977,970 (19%) | 10,627,444 (21%) | 7,442,858 (21%) | 37,618,786 (20%) |
| Mapped to introns | 1,619,022 (3%) | 1,555,896 (3%) | 3,058,379 (6%) | 1,688,736 (5%) | 7,922,033 (4%) |
| Mapped to intergenic regions | 2,432,060 (5%) | 2,514,988 (5%) | 4,508,874 (9%) | 2,614,410 (8%) | 12,070,332 (6%) |
“Unmapped” reads refer to those that were not mappable to the genome or transcriptome using the defined mismatch thresholds (Online Supplement). “Non-uniquely paired” means that the pair of reads mapped non-uniquely as a pair. “Wrong pairing” means that the pair of reads did not pass the filters for correct pairing (Online Supplement).
Figure 1.
Overview of the methods and procedures to analyze RNA-Seq data.
Analysis of gene expression levels via RNA-Seq
RNA-Seq has been demonstrated to be an effective approach for gene expression profiling in mouse heart7-9. One advantage of this method is its ability to provide quantitative read-out of the mRNA expression levels in one sample, in contrast to microarrays that just permit comparative analyses without absolute expression values. Consistent with the previous studies7-9, we also observed a wide dynamic range of expression values varying from ~ 1 copy per cell (3 RPKM) for the Ankrd12 gene to 8,048 copies per cell for the mt-Co1 gene in the Sham hearts. In this work, we use 3 RPKM as the minimum cutoff to filter for expressed genes13 considering its biological relevance and the fact that some genes (e.g., Kcnd2, Kcnd3) with heart-related function are expressed at ~3RPKM in Shams. Other RNA-seq studies also showed that low abundant transcripts expressed at ~1 copy per cell include transcriptional factors and other functionally important genes for cardiac regulation7-9. In addition, in a PubMed search of published abstracts, we found that genes with ≥ 3 RPKM expression levels are about twice as often associated with the keywords “heart” and “cardiac” than those expressed at < 3 RPKM. Altogether, 9,833 genes (29% of all Ensembl genes) are expressed at ≥ 3 RPKM in at least one sample in our study.
Another advantage of the RNA-Seq method over microarrays is the improved quantification of differential gene expression between samples7. We validated expression changes of 42 genes using real-time PCR (Online Figure I, Online Table II). This validation demonstrated that the results from RNASeq and real-time PCR are highly concordant (r = 0.90, Pearson correlation). To establish biologically meaningful criteria to determine significant differential gene expression, we examined the levels of genes known to be altered in failing hearts, including Myh7, Egr1, Nppb, Pln, and Actb. We confirmed that all the above genes had expression changes of at least 1.5 fold between the HF and Sham-HF samples in the real-time PCR or RNA-Seq. Thus, we used the following criteria to identify differentially expressed genes: (1) gene expression level ≥ 3 RPKM in either Sham or HY/HF or both; (2) change in expression level ≥ 1.5 fold; and (3) Fisher's exact test (see Methods) FDR < 5%. Altogether, 97 and 1,435 genes passed the above filters between Sham-HY and HY, Sham-HF and HF, respectively (Online Table III). The number of genes and their magnitudes of changes are larger at the HF stage, consistent with the expected higher degrees of cardiac remodeling in HF contributed by vascular remodeling, inflammatory response and fibrosis in the myocardium. In addition to the well-known HF or HY-related genes, we found genes with significant alteration in expression, but have never been implicated in heart failure or cardiac hypertrophy, such as Prc1, E2f1, Birc5, Iqgap3, Cdc20 and Cdca8. Our results suggest that RNASeq may provide novel insights in overall gene expression as demonstrated previously7-9.
Analysis of alternative transcript isoforms via RNA-Seq
One of the main voids in our current knowledge of cardiac transcriptome is the genome-wide profile of mRNA splicing variants, a very challenging task not readily accomplishable by most microarray platforms. For this purpose, we used the package Cufflinks15, which was shown to effectively capture isoform-specific expression and alteration in RNA-Seq data. Although Cufflinks has a number of modules for different purposes, we focused on its usage to infer expression levels and differential expression of individual transcript isoforms. As inputs to Cufflinks, we used our read mapping results described above and the set of Ensembl-defined genes and their spliced variants. The novel exons and NTCs identified in our study were not included because the nature of the short sequencing reads limits the accuracy in predicting complete structures of spliced variants that are needed to estimate their expression levels.
We first examined the absolute isoform expression estimated by Cufflinks. For genes with multiple transcript isoforms, we detected a total of 7,811 isoforms with expression level ≥ 3 RPKM in at least one sample. The most abundant isoform was from the gene Myl2 (Myosin regulatory light chain 2) in the Sham-HF sample. The isoforms of other heart-related transcripts such as Actc1, Atp2a2, Myh6,Tnnt2 and Tpm1 were also highly expressed. We observed spliced variants for numerous genes (e.g., Atp2a2, Cacna1c, Slc6a8 and Ank2) known to undergo alternative splicing in cardiac tissues7. For the gene Camk2d, We confirmed that its neuronal-specific isoform is much less expressed than the heart-specific isoforms (2.96 vs. 22.11 RPKM) in the Sham-HF sample. These findings suggest that RNA-Seq can readily detect isoforms of a gene due to alternative RNA splicing.
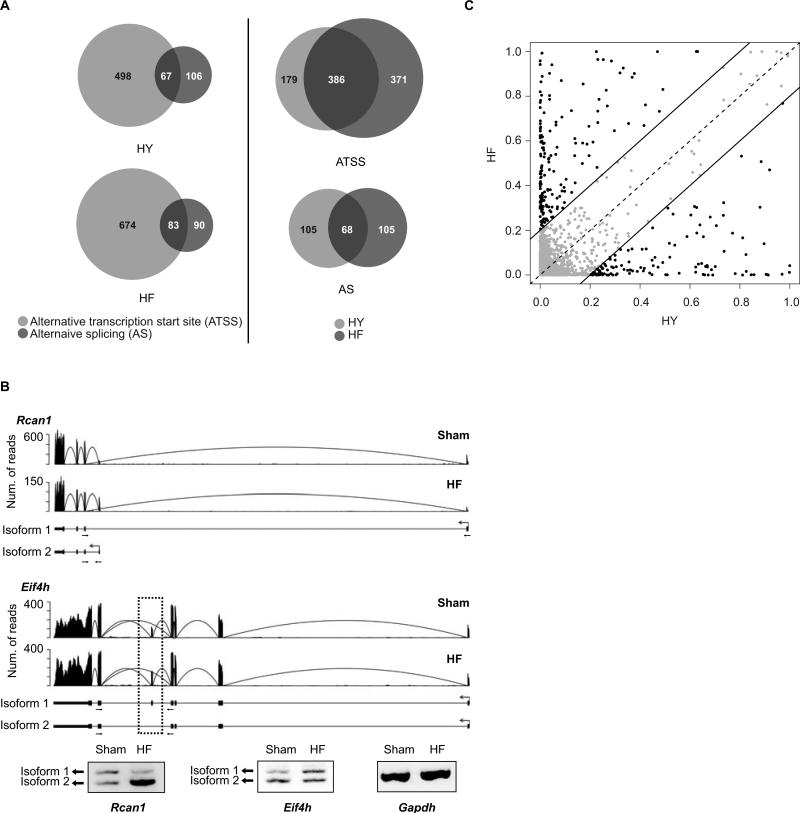
We next analyzed the differential expression patterns of individual transcript isoforms. Cufflinks analysis identified 1,087 genes (mostly protein-coding genes) with significant isoform-specific expression changes (q-value < 0.05) due to ATSS or AS or both (Figure 2A & Online Table IV). As examples, Figure 2B shows two genes (with ATSS and AS, respectively), their reads distributions and RT-PCR validation results. If the same criteria were applied as for determining differential gene expression (q-value < 0.05, fold change ≥ 1.5, and expression level ≥ 3 RPKM), a total of 720 isoforms in 475 genes were identified as differentially expressed. Overall, genes with ATSS and AS are both enriched in biological processes related to muscle function and ATP synthesis (Online Table V).
Figure 2. Expression and isoform changes of known genes in heart failure.
(A) Number of genes with expression changes of alternative isoforms due to alternative transcription start sites (ATSS) or alternative splicing (AS). Left panel: genes classified into HY and HF stages; right panel: genes classified into the ATSS and AS categories. (B) Read distribution for two genes with differential isoform expression due to ATSS (Rcan1) or AS (Eif4h), and the corresponding RT-PCR validation results (with primer locations illustrated by small arrows). Arcs represent reads mapped to exon-exon junctions. Ensembl-annotated isoforms are illustrated below read distributions. In the Eif4h gene, the skipped exon is highlighted by a dotted box. (C) Distribution of dissimilarity scores to quantify the overall isoform difference of genes in the two stages relative to their Sham samples.
Similar to our findings in overall gene expression changes, more genes were found to have altered isoform expression in the HF stage than the HY stage. To further compare the two stages, we calculated a dissimilarity score that quantifies the overall isoform difference of a gene between a pair of samples (Methods). This measure is independent of gene expression levels. Figure 2C shows that most genes have similar scores between the HY and HF stages (data distributed close to the diagonal line). However, a significant number of genes (n > 250) have dissimilarity scores differing by more than 0.2, suggesting a significant change in isoform usage at different stages of heart failure. Interestingly, among these genes, many (e.g., Garnl1, Sipa1, Rgs12, Rin2 and Rabgap1) are known to be involved in processes well-studied in heart failure. In addition, genes involved in chromatin and histone modifications (such as Hdac7, Ezh1 and Aof2) also demonstrated stage-specific isoform expression changes. Therefore, the quantitative gene isoform analysis suggests a global change in exon utilization due to alternative RNA splicing that can potentially impact functionally important genes in failing hearts.
Guided transcriptome reconstruction for novel transcripts of known genes
Another major advantage of the RNA-Seq approach is the capability to discover previously unknown transcript isoforms. We thus developed a guided transcriptome reconstruction method to enable identification of novel isoforms in known genes (Online Supplement). In this method, RNA-Seq reads that mapped inside or in the vicinity of Ensembl genes were examined. Reads that do not support the known Ensembl transcript structures (e.g., those in the intronic regions) may suggest existence of novel transcripts. However, such reads may also arise from other sources such as incompletely processed transcripts, degradation intermediates of introns or mapping errors. In order to reduce false positives, we implemented two additional requirements to define novel transcripts. First, we applied stringent filters for expression levels of the novel fragments (details in Online Supplement). Second, since novel exons should be spliced to other exons, we required the existence of at least two spliced junction reads flanking each end of the novel exon. When the detected novel fragments were identified to be extensions of known internal exons or terminal exons, they would indicate new exon splicing pattern, or alternative transcriptional initiation or termination events.
For the 10,061 multi-exon genes (defined by Ensembl and/or our isoform reconstruction) with an expression level of ≥ 3 RPKM, 5,112 (51%) were detected with novel isoforms (with novel exons or novel splicing patterns among known exons) as a result of alternative splicing, 1,651 (16%) as a result of alternative initiation or termination, and 830 (8%) with both types of novel isoforms. Novel transcript structures were identified across a broad range of expression levels (Online Figure II), with more isoforms detected for higher expressed genes (most likely due to higher read coverage). Therefore, our findings suggest a significant deficiency in the current mouse transcriptome annotation (Ensembl v56 used in this analysis) and deep RNA-Seq combined with guided transcriptome reconstruction can provide a much more comprehensive profile of the total complexity in transcript structures.
Evaluation of novel spliced exons identified by guided transcriptome reconstruction
Although a large number of genes were identified with novel spliced variants, it is possible that many of them were resulted from random errors or noise in the process of splicing or transcription detected by the highly sensitive RNA-Seq method. Thus, we analyzed in detail the novel exons in the spliced variants to determine if our method leads to findings with potential biological significance. A total of 1,873 novel exons were identified corresponding to different types of alternative splicing events (Table 2 and Online Table VI). Since the Ensembl v56 database was used as a reference to define known genes and exons and updated databases now exist, we examined whether the above novel exons are annotated as known exons in the new Ensembl v61 database (April, 2011), or the UCSC, RefSeq databases. Indeed, 26% of the novel spliced exons identified from our original analysis are now “known” to one or more of the above databases (Table 2). This serves as a validation of our approach for identifying the novel exons and we refer to those exons that remain to be not annotated in the above databases (1,384 in total) as “updated novel exons”. We validated 29 of the randomly selected updated novel exons via RT-PCR and the expression of 97% of them was confirmed (Online Table VII).
Table 2.
Novel alternative splicing (AS) events identified in Ensembl genes (ENSGs) and novel transcript clusters (NTCs). Exons are categorized according to the type of AS events.
| Alternative splicing events | SE* | RI† | A5E‡ | A3E§ | MXE∥ |
|---|---|---|---|---|---|
| Original novel exons in ENSG | 968 | 691 | 268 | 332 | 15 |
| % of total (1,873) | 52% | 37% | 14% | 18% | < 1% |
| Updated novel exons in ENSG | 623 | 629 | 197 | 235 | 8 |
| % of total (1,384) | 45% | 45% | 14% | 17% | < 1% |
| Novel AS exons in NTCs | 223 | 0 | 138 | 109 | 7 |
| % of total (421) | 53% | 0 | 33% | 26% | 1% |
“Original”: novel exons identified relative to Ensembl v56. “Updated”: novel exons identified relative to the most recent databases including Ensembl v61, UCSC KnownGenes and RefSeq genes. Note that one exon may be associated with multiple types of AS. Such exons are counted into all applicable types. (Thus, the % values for all categories may not sum to 100%.)
SE: Skipped exon
RI: Retained intron
A5E: Alternative 5' splice site exon
A3E: Alternative 3' splice site exon
MXE: Mutually exclusive exon
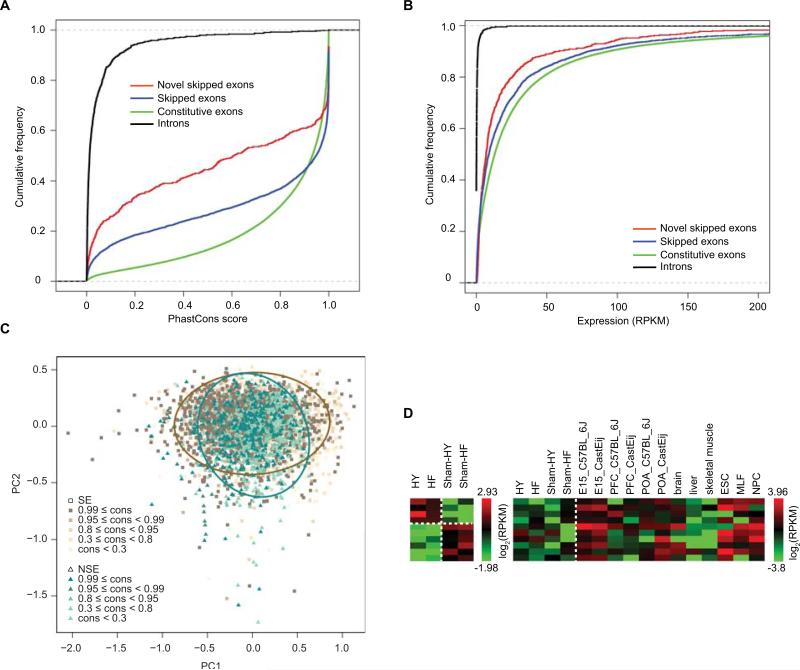
To provide further evaluation on the identified “updated novel exons”, we performed additional bioinformatic analyses on these novel exons that were alternatively skipped, the most common type of alternatively spliced exons. We first examined whether the novel exons have features that resemble those of known skipped exons. The following features were considered: evolutionary conservation, expression level, exon length and splice site strength. The conservation level and expression level of the novel exons, although lower than those of the known skipped exons, are significantly higher than intronic regions with matched GC content and length (Figures 3A and 3B). About 54% of the novel skipped exons have a length that is multiples of three, significantly higher than expected (p-value = 3.1e-07, Chi-Square test). This observation implies the existence of strong selection on the alternative protein products derived from these exons. Approximately 96% of the novel skipped exons are flanked by GT-AG in the immediate intronic regions, representing consensus splice site sequences. Figure 3C shows the result of principal component analysis of the above features in the novel and the known exon populations respectively, which suggests that the two groups of exons are largely similar. The similarities of the novel exons to the known alternative exons suggest that they are likely authentic exons with biological function.
Figure 3. Novel exons in known genes identified via RNA-Seq.
(A) Cumulative distribution functions of conservation levels of novel skipped exons, introns, known skipped exons and constitutive exons. (B) Cumulative distribution functions of expression levels of the same exons/introns in (A). (C) Principal component analysis (PCA) and clustering of novel (NSE) and known skipped exons (SE) based on their expression level, exon length and splice site strength. Conservation level (cons) of the exonic regions is represented in varying shades of colors. The representative ellipses for novel and known skipped exons were obtained by fitting the principal components with Gaussian distributions (Online Supplement). (D) Heatmaps of expression levels (log2RPKM; reads per kilobase of exon per million mapped reads) of novel skipped exons. Left panel: 10 example novel exons with differential expression in HY or HF compared to Sham controls. Right panel: 10 example novel exons lowly expressed in our data, but highly expressed in one or more of the other published mouse RNA-Seq data sets. C57BL_6J and Cast/Eij are mouse strains used in 17. E15: embryonic day 15 (E15) whole brain; PFC: adult male and female medial prefrontal cortex; POA: adult male and female preoptic area17. Brain, liver and skeletal muscle data from13; ESC: embryonic stem cells, MLF: lung fibroblasts, NPC: neural progenitor cells18.
To further evaluate their biological significance, we then analyzed the expression patterns of the novel exons in more detail. If a novel exon exists due to nonfunctional random splicing noise, its absolute expression level is most likely low and similar across different samples. However, we found about 72% of the novel exons had an expression level of ≥ 3 RPKM in at least one of the samples used in our study. In examining the expression difference of the novel exons in the HY/HF and Sham controls, we observed that a substantial fraction (682 exons, 68% of all with ≥ 3 RPKM in absolute expression) had an expression difference of at least 1.5 fold (10 examples shown in Figure 3D, left panel). Furthermore, we computed the expression of the novel exons in other mouse tissues or cell types based on available RNASeq data13, 17, 18. Interestingly, many novel exons with relatively low abundance in the mouse heart had much higher expression levels in one or more of other tissues (Online Figure III, examples in Figure 3D). Thus, our result suggests that the novel exons identified by RNA-Seq even with low expression level in heart may be authentic exons with biological roles in other tissues.
Finally, we analyzed the impact of these novel exons on the predicted protein products. A large fraction (60%) of them will introduce premature termination codons or are expected to induce nonsense-mediated decay, suggesting that many novel exons may have a major impact on the final protein expression (Online Supplement). Furthermore, we found evidence of translation for 174 of the detected novel exons in public proteomic databases, suggesting that these exons may contribute to the overall complexity of the proteome (Online Supplement).
Alternative initiation and termination events
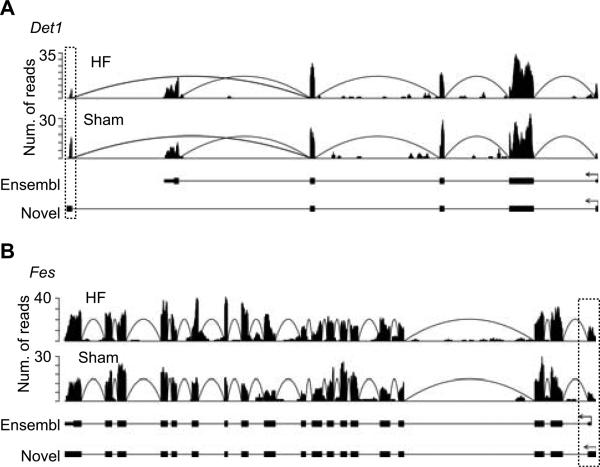
Based on the reconstructed transcript structures, we identified novel alternative 5' terminal exons that differ from the Ensembl annotations. Two different categories of such events were defined: (1) 5' terminal exon overlapping the annotated first exon but with extended regions or alternative splice sites supported by junction reads; (2) 5' terminal exon not overlapping any annotated exon and occurring upstream of the annotated 5' start sites. To be conservative and avoid the complication of incomplete transcript reconstruction, we excluded 5' terminal exons whose start sites are downstream of the annotated 5' start sites. Similar analyses were carried out to identify alternative 3' terminal exons. Altogether, we identified 1,613 exons (in 1,535 genes expressed at ≥ 3 RPKM) with novel alternative initiation or termination events that are not annotated in the most recent databases including UCSC, RefSeq and Ensembl (v61) (Online Tables VIII, IX). Figure 4 shows two examples of such events. Among these exons, about 469 differ in expression by at least 1.5 fold between HY and Sham or HF and Sham. In addition, the corresponding genes significantly overlap the list of differentially expressed genes (162 genes in common, p = 4.2e-13). Interestingly, 39% of the alternative 3' terminal exons contain predicted target sites of known miRNAs expressed in mouse heart (Online Supplement), suggesting that the alternative terminal events may be functionally involved in gene regulation.
Figure 4. Examples of novel alternative terminal exons identified via RNA-Seq.
Read distribution plots defined similarly as in Figure 1. Novel terminal exons are highlighted in dotted boxes. The Ensembl-annotated isoforms (Ensembl) and the novel isoforms (Novel) reconstructed from RNA-Seq are shown. (A) Novel alternative 3' terminal exon in the gene Det1. (B) Novel alternative 5' initiation exon in the gene Fes.
Methods to identify and evaluate novel transcript clusters
In addition to the guided transcriptome reconstruction, we also conducted de novo transcript identification for those reads that match to genomic regions with no annotated genes (Methods). Here we focused on NTCs corresponding to genes with multiple exons only. We refer to an NTC as a cluster of all possible transcript isoforms. It is possible that an NTC contains false positive isoforms because the nature of the short reads in RNA-Seq does not allow identification of complete transcript structures. In addition, complete profile of all isoforms may not be identifiable due to low read coverage. Nevertheless, each NTC suggests the possible existence of a novel gene with multiple transcript isoforms.
Using these criteria, 1,884 NTCs were identified that do not overlap any Ensembl genes (v56), all of which were multi-exon transcripts (Online Table X). Among the 5,869 exons within these clusters, 8% had spliced junction reads suggesting alternative splicing, with the most prevalent type being alternatively skipped exons (Table 2). Of all NTCs, 863 (46%) are now annotated as known genes in the most recent databases of UCSC, Refseq and Ensembl (v61), supporting the validity of our method in identifying novel genes. We validated the expression patterns of 7 NTCs (3 newly annotated and 4 remain novel) using real-time PCR in the same mouse samples as used for RNA-Seq (Online Figure I and Online Table II). The results confirmed the expression of all 7 NTCs and also showed that RNA-Seq and real-time PCR gave highly consistent measures in gene expression changes (r = 0.87, Pearson correlation).
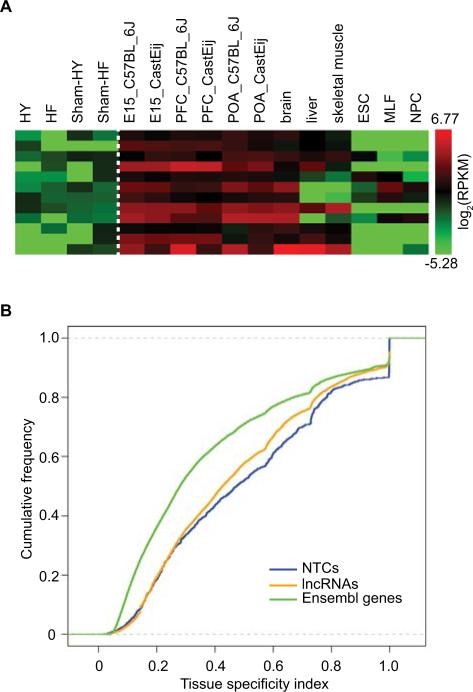
Next, we focused on the 1,021 NTCs that are still not annotated in the newest databases. To infer biological significance, we analyzed their expression patterns in our samples and the other mouse RNASeq data sets mentioned above13, 17, 18. Remarkably, many of these NTCs with low abundance in the heart had much higher expression levels in other tissues (examples shown in Figure 5A). A total of 199 NTCs passed the minimum expression level cutoff of 3 RPKM in at least one sample. When examined for differential expression using the same criteria as for known genes, 195 NTCs were differentially expressed between at least one pair of samples (34 between HF/HY and Sham controls). In addition, we found that the NTCs may be more tissue-specific than annotated genes evaluated by an entropy-based tissue specificity index (Online Supplement, Figure 5B). These results suggest that many NTCs may be actively regulated and may have functional ramifications in specific tissues.
Figure 5. Analysis of novel transcript clusters (NTCs) and long non-coding RNA (lncRNA) genes identified via RNA-Seq.
(A) Heatmap of expression levels (log2RPKM) of 12 example NTCs in our data and other published RNA-Seq data sets, see Figure 3 for details of the published data sets. (B) Cumulative distribution functions of the tissue-specificity index of NTCs, all novel and known lncRNA genes and Ensembl protein-coding genes.
We next classified the NTCs into coding and non-coding clusters. Among all 1,021 NTCs, 105 clusters (containing 315 possible transcript isoforms) were found to have significant coding potential16. Interestingly, the coding clusters had significantly higher expression and conservation levels than the non-coding clusters (Online Figure IV), consistent with the existence of stronger selection pressure on protein-coding sequences as observed in known genes. Indeed, we found that 46% (48 out of 105) of the putatively coding NTCs had sequence matches in public proteomic databases (Online Supplement). These results suggest that our de novo transcript identification method can effectively identify novel transcripts from RNA-seq data sets, and the novel genes identified in heart contributes to the total complexity of cardiac transcriptome and proteome.
Inference of potential function of novel transcript clusters
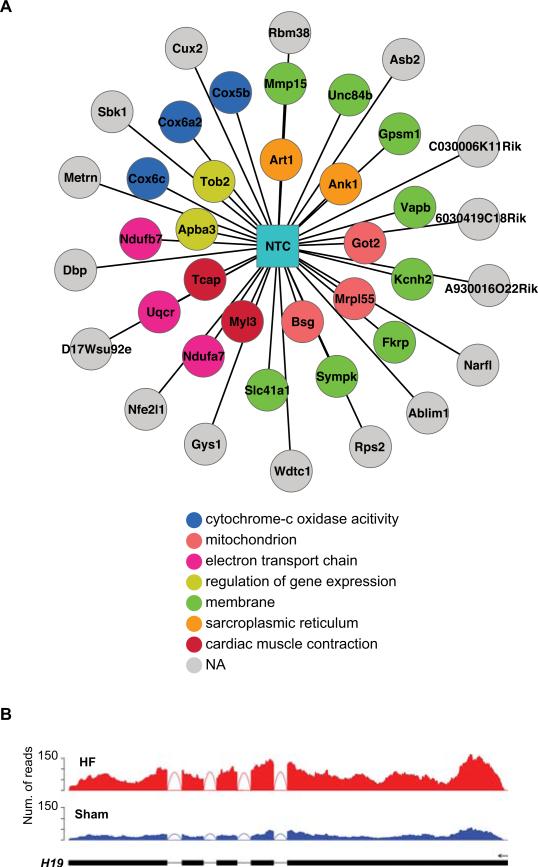
Functionally related genes involved in the same biological pathways or protein interaction networks are often regulated by similar transcription factors or other gene regulators. Thus, one approach to infer the potential function of novel genes is by determining whether their expression patterns correlate with those of known genes of certain function based on co-expression analysis19. Note that such analyses only provide tentative functional indications of a gene. However, they may help to formulate hypotheses for further experimental studies. We applied this scheme to examine the potential functions of NTCs. Using the WGCNA method20, we constructed co-expression networks encompassing all known genes and 98 NTCs discovered in the heart samples (≥ 3 RPKM). We identified 52 network modules in total and focused on 20 that are enriched with differentially expressed genes between HY/HF and the corresponding Shams. Genes in these modules are significantly associated with GO categories related to heart and muscle functions (Online Table XI). A total of 58 NTCs were included in the 20 significant modules. Among them, we analyzed the 10 most highly connected NTCs (i.e., with highest connectivity) in each module that had at least 10 neighboring known genes. Fifteen NTCs from 6 modules were chosen in this way (example shown in Figure 6A). A complete list of GO categories related to each of the 15 NTCs is included in Online Table XII.
Figure 6. Novel transcript clusters (NTCs) and long non-coding RNA (lncRNA) genes potentially involved in disease mechanisms.
(A) An example NTC and its neighboring genes in the gene co-expression networks. Non-grey nodes correspond to genes that are associated with GO categories enriched among all the neighboring genes of the NTC. Otherwise, the nodes are denoted in grey. (B) Read distributions of a differentially expressed lncRNA gene (H19) in HF and Sham RNA-Seq data.
Long non-coding RNA (lncRNA) genes
Among the novel multi-exon transcript clusters, a large fraction (81%) had low coding potential (Online Table X) and thus most likely belongs to the category of lncRNA genes. LncRNAs were recently recognized to have diverse functions in gene regulation and may contribute to disease etiology21. To assess the expression of lncRNAs in mouse heart failure, we combined our non-coding NTCs with the Ensembl and other mouse lncRNA data sets18, 22 to constitute a comprehensive repository of 5,767 lncRNAs. A total of 703 lncRNAs were expressed at levels ≥3 RPKM in our RNA-Seq data of at least one sample. Interestingly, for lncRNAs expressed below 3 RPKM, 62% of them had an expression level of at least 3 RPKM in one or more of the publically available mouse RNA-Seq data13, 17, 18. Figure 5B shows that lncRNAs have higher tissue specificity in their expression than protein-coding genes on average. Among all expressed lncRNAs (≥ 3 RPKM in ≥ 1 of our samples), 15 and 135 are differentially expressed (> 1.5 fold change, FDR < 5%) between HY and Sham-HY and between HF and Sham-HF, respectively. Intriguingly, the well-known H19 gene demonstrated significant up-regulation in the HF stage compared to the corresponding Sham controls (Figure 6B). This gene is highly expressed during embryogenesis and was shown to have tumor-suppressor activity23. However, its role in heart failure is not yet clear.
Discussion
Accurate and de novo transcriptome profiling is a central issue in disease research. Here we describe methods and applications of transcriptome analysis via RNA-Seq in normal and failing murine hearts. As clearly demonstrated by Matkovich et al7, RNA-Seq has both accuracy and sensitivity to detect transcripts with low abundance (such as those encoding transcription factors), so new gene expression networks associated with heart failure can be established (such as transcriptional networks). Our work highlights the power of RNA-Seq in de novo transcriptome profiling given its accuracy and the independence of a priori knowledge of transcripts to be analyzed. We used newly developed bioinformatic tools, namely a combination of guided transcriptome reconstruction and de novo reconstruction approaches, to identify novel exons and transcripts in normal and diseased mouse hearts. As demonstrated by our validation studies, the vast majority of the identified novel exons or transcripts are confirmed by RT-PCR for their expression in heart. We showed that the sequence and conservation features of the novel exons are generally similar to those of the known alternatively spliced exons. In addition, both novel exons and NTCs were found to be expressed in a tissue-specific manner. These properties suggest the potential functional significance of the novel isoforms and novel transcripts. They also suggest that the RNA-Seq technology, combined with bioinformatic analysis, is a sensitive tool to comprehensively profile the transcriptome complexity at individual exon resolution.
It is somewhat unexpected to us that cardiac transcriptome encompasses such a large number of novel transcripts and exons with potential biological significance that have never been annotated despite extensive genome-wide profiling of cardiac transcriptome in the past decade. These findings open the way to further experimental investigations of their relevance in the pathogenesis of heart failure. Yet, our study does have limitations. Since the findings are mainly based on bioinformatic observations, their functional relevance would need to be further established at molecular and cellular levels experimentally. In addition, the poly-A selection procedure during RNA-Seq library preparation may introduce a positional bias in the coverage of the entire transcript (favoring the 3' end) and not all expressed RNAs can be included in the library. In this study, we focused on the pressure-overload induced heart failure and whether the findings are general to other types of heart failure models is unclear. Nevertheless, our analyses revealed previously uncharacterized complexity in the cardiac transcriptome as well as their dynamic changes during heart failure. This study highlights the need to employ both RNA-Seq and bioinformatic tools to re-evaluate cardiac transcriptome in other heart disease models as well as in human heart failure. Since the bioinformatic approaches developed in our study are generally applicable to any RNA-Seq data sets, we suggest that application of these new tools will vastly expand our current knowledge of transcriptome architecture and dynamics in general.
Supplementary Material
Novelty and Significance.
What is known?
Accurate and de novo transcriptome profiling is a central issue in studying the mechanisms of cardiovascular development and diseases.
Understanding of the global cardiac transcriptome landscape is currently limited concerning expression and variation at single exon resolution.
Whole-transcriptome sequencing (RNA-Seq) offers a new way to study transcriptomes.
What new information does this article contribute?
We present bioinformatic methods to identify transcript structures and to analyze transcriptome complexities with a particular emphasis on quantification of RNA splicing variants at single exon resolution using RNA-Seq data of normal and failing murine hearts.
We validate the effectiveness and accuracy of our bioinformatic approaches based on experimental confirmation and cross-database analyses.
We show that the bioinformatic analyses of RNA-Seq allow in-depth profiling and quantification of alternative mRNA structures, novel exons, novel transcript clusters and long non-coding RNA genes in mouse heart.
Transcriptome profiling offers detailed insight in understanding gene regulation in health and diseases. Previous technologies (e.g., microarrays) for this purpose are limited in coverage and sensitivity. RNASeq using massively parallel next-generation sequencing platforms can potentially enable de novo and unbiased characterization of the transcriptome at individual exon resolution. However, as a result of the massive amount of raw data, RNA-Seq poses new challenges to data analysis and interpretation. Here, we present bioinformatic methods that enable effective analysis of RNA-Seq data either integrating or independent of known gene annotations. We demonstrate that such analyses can provide a more comprehensive profile of the mouse cardiac transcriptome. Indeed, a large number of novel transcripts and exons with potential biological significance were found from this study that had never been annotated previously. These findings open the way to further experimental investigations of their relevance in the pathogenesis of heart failure. Our study highlights the need to employ both RNA-Seq and bioinformatic tools to achieve comprehensive evaluation of cardiac transcriptome in heart disease models as well as in human heart failure.
Acknowledgments
Sources of Funding
The work was supported in part by NIH grant R21DA027039, Alfred P. Sloan Foundation Research Fellowship and Research Grant No. 5-FY10-486 from the March of Dimes Foundation to X.X, an American Heart Association Postdoctoral Fellowship to JH Lee and NIH grants HL088640, HL070079, HL103205, HL098954 to Y.W.
Non-standard Abbreviations and Acronyms
- HY
hypertrophy
- HF
heart failure
- Sham-HY
corresponding sham control for HY
- Sham-HF
corresponding sham control for HF
- NTC
novel transcript cluster
- LncRNA
long non-coding RNA
- RPKM
reads per kilobase of exon per million mapped reads
- ATSS
alternative transcription start site
- AS
alternative splicing
Footnotes
Disclosures
None.
Data availability. RNA-Seq data are available at the Gene Expression Omnibus with ID GSE29446.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson EN, Backs J, McKinsey TA. Control of cardiac hypertrophy and heart failure by histone acetylation/deacetylation. Novartis Found Symp. 2006;274:3–12. discussion 13-19, 152-155, 272-156. [PubMed] [Google Scholar]
- 3.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358(13):1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8(6):469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 6.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW., 2nd Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq. Circ Res. 2010;106(9):1459–1467. doi: 10.1161/CIRCRESAHA.110.217513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Matkovich SJ, Duan X, Gold JI, Koch WJ, Dorn GW., 2nd Nuclear effects of g-protein receptor kinase 5 on histone deacetylase 5-regulated gene transcription in heart failure. Circ Heart Fail. 2011;4(5):659–668. doi: 10.1161/CIRCHEARTFAILURE.111.962563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Matkovich SJ, Duan X, Diwan A, Kang MY, Dorn GW., 2nd Receptor-independent protein kinase C alpha (PKCalpha) signaling by calpain-generated free catalytic domains induces HDAC5 nuclear export and regulates cardiac transcription. J Biol Chem. 2011;286(30):26943–26951. doi: 10.1074/jbc.M111.234757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang SY, Vanhoutte D, Del Re DP, Purcell NH, Ling H, Banerjee I, Bossuyt J, Lang RA, Zheng Y, Matkovich SJ, Miyamoto S, Molkentin JD, Dorn GW, 2nd, Brown JH. RhoA protects the mouse heart against ischemia/reperfusion injury. J Clin Invest. 2011;121(8):3269–3276. doi: 10.1172/JCI44371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–349. doi: 10.1093/nar/gkm391. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329(5992):643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 20.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.