Abstract
There are 272 cytochrome P450 genes (including 26 pseudogenes) in the Arabidopsis genome. P450s thus form one of the largest families of proteins in higher plants. This explosion of the P450 family is thought to have occurred via gene duplication and conversion, and to result from the need of sessile plants to adapt to a harsh environment and to protect themselves from pathogens and predators. P450s sometimes share less than 20% identity and catalyze extremely diverse reactions. Their biological functions range from the synthesis of structural macromolecules such as lignin, cutin or suberin, to the synthesis or catabolism of all types of hormone or signaling molecules, the synthesis of pigments and defense compounds, and to the metabolism of xenobiotics. In despite of a huge acceleration in our understanding of plant P450 functions in the recent years, the vast majority of these functions remain completely unknown.
Introduction
One of the surprising disclosures of the complete Arabidopsis sequence was the extraordinary diversity of the superfamily of cytochrome P450 proteins. The final P450 count in the almost complete genome is 246 genes and 26 pseudogenes, which makes of P450s one of the largest family of proteins in higher plants. Lists of A. thaliana P450 genes and accession numbers are available on several web sites: http://www.biobase.dk/P450, http://drnelson.utmem.edu/CytochromeP450.html.
Nomenclature
A common system for the nomenclature of P450 genes from all organisms has been set up on the basis of protein sequence identity and phylogeny (Figure 1; Nelson et al., 1996). P450s in a same family usually share at least 40% identity. In a same subfamily they normally share at least 55% identity. This identity rule has some exceptions, especially in plants, where gene duplication and shuffling sometimes makes a straightforward nomenclature difficult. In this case, family assignment is based on phylogeny and gene organization. Sequence identity among P450s from Arabidopsis can be less than 20%.
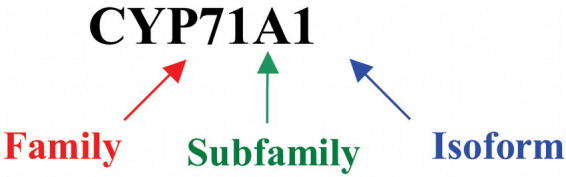
Figure 1: Naming a P450 protein.
P450s are named in chronological order of submission to a nomenclature committee (David Nelson: DNELSON@utmem1.utmem.edu). Plant P450s have been assigned names from CYP71A1 to CYP99XY, then from CYP701A1 and above.
What's a P450, what is it doing?
P450s are enzymes found in all organisms from bacteria to humans (Nelson, 1999). Reactions they catalyze are extremely diverse but usually based on activation of molecular oxygen with insertion of one of its atoms into the substrate and reduction of the other to form water (Mansuy, 1998; Werck-Reichhart and Feyereisen, 2000). They are hence classified as monooxygenases.
Reduced P450 can bind carbon monoxide instead of O2. CO coordination results in a shift of the maximum of absorbance of the heme to 450 nm (Omura and Sato, 1964). This property is characteristic of P450 enzymes. P450 actually stands for Pigment absorbing at 450 nm. CO is bound with high affinity and prevents binding and activation of O2. The result is an inhibition of P450 activity which can be reversed by light, with maximal efficiency at 450 nm.
P450s all share a common catalytic center, heme with iron coordinated to the thiolate of a conserved cysteine, and a common overall topology and tridimensional fold (http://p450terp.swmed.edu/Bills_folder/billhome.htm) (Graham and Peterson, 1999; Werck-Reichhart and Feyereisen, 2000). Such common features imply a few conserved characteristics in the primary structure and scattered signature motifs.
Bacterial P450s are soluble proteins, but all plant P450s described so far are bound to membranes, usually anchored on the cytoplasmic surface of the endoplasmic reticulum through a short hydrophobic segment of their N-terminus, and possibly a hydrophobic loop of the protein (Williams et al., 2000). Analysis of the Arabidopsis, and other plant P450 sequences, however predicts potential signal peptides that should target some of them to the plastids or to the mitochondria. To be active, P450s need to be coupled to electron donating proteins, cytochrome P450 reductases or cytochrome b5, also anchored via their N- or C-terminus to the surface of the E.R. For more on P450 reductases: http://www.uky.edu/pharmacy/ps/porter/cpr.htm. Schematic or model pictures of P450-reductase interactions can be seen at: http://p450terp.swmed.edu/Bills_folder/billhome.htm, http://ibmp.u-strasbg.fr/dep_stress/Dany/P450gris.html, or http://rex.nci.nih.gov/RESEARCH/basic/lm/fkf.htm. Other electron donors would be expected to operate in plastids or mitochondria.
In canonical P450s, heme-bound O2 is activated by the successive transfer of two electrons from NADPH via the NADPH-cytochrome P450 reductase. In some cases, the second electron can be provided more efficiently by an alternative electron transport chain involving NADH, and NADH cytochrome b5 reductase and cytochrome b5 (De Vetten et al., 1999). More on P450 redox cycle and mechanism can be found in (Werck-Reichhart and Feyereisen, 2000). The result of P450 catalysis is very often an hydroxylated substrate, but the reaction can be more complex, involve different reduction states of the heme iron or lead to rearrangements of intermediates or final products (Mansuy, 1998). Examples of such complex reactions can be found in the list of the Arabidopsis P450 functions given below.
The natural substrates of plant P450 enzymes include precursors of membrane sterols and of structural polymers such as lignin, cutin or suberins. P450s also contribute to the homeostasis of hormones and signaling molecules by controlling their biosynthesis (gibberellins, auxin, brassinosteroids, jasmonate…) and their catabolism (brassinosteroids, abscissic acid). They are involved in the biosynthesis of pigments, antioxidants and defense compounds, including flavonoids, phenolic esters, coumarins, glucosinolates, cyanogenic glucosides, benzoxazinones, isoprenoids, alkaloids…(Kahn and Durst, 2000). In addition to their physiological substrates, cytochromes P450 metabolize and usually detoxify exogenous molecules such as pesticides and pollutants (Werck-Reichhart et al., 2000).
Phylogeny and gene organization
The P450s constitute a superfamily of highly divergent sequences. To authenticate a true evolutionary relationship is not simple. Bootstrapped phylogenetic trees based on careful multiple alignments of primary amino acid sequences and characteristics such as intron position and phase, as well as a comparison of unique amino acid substitutions in highly conserved domains, are needed for the validations. Introns are lost and gained through evolution. Accordingly, intron position and phase are fossils of the evolutionary relationship that may be used as diagnostic fingerprints to validate the phylogeny predicted from the primary sequences (Long et al., 1995; Stoltzfus et al., 1997). Only a few domains are highly conserved in the primary amino acid sequence of P450s (Figure 2): the proline rich membrane hinge, the sequence surrounding the cysteine which is the axial ligand to the heme, the I-helix involved in oxygen binding, and the E-R-R triade using the Glu and Arg of the K-helix consensus (KETLR) and the Arg in the “PERF” consensus. The E-R-R triad is generally thought to be involved in locking the heme pocket into position and to assure stabilization of the conserved core structure (Hasemann, 1995). Of these conserved domains, only the E-R-R triad and the cysteine in the heme-binding domain are conserved in all plant P450 sequences.
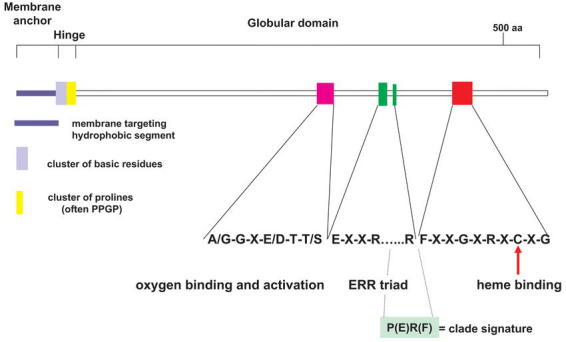
Figure 2: Conserved structures and sequences in P450 proteins.
Plant P450s are generally classified in two main clades: the A-type and the non-A type (Durst and Nelson, 1995; Paquette et al., 2000). The A-type clade is specific to plants. P450s involved in the biosynthesis of secondary metabolites or natural products are found in this group. In contrast, the non-A type clade is a much more divergent group of sequences consisting of several individual clades that often show more similarity to non-plant P450s than to other plant P450s (Paquette et al., 2000). It was initially proposed by Durst and Nelson (1995) that the A-type P450s originate from a single common ancestral gene. A previously published detailed phylogenetic analysis of 135 Arabidopsis sequences coupled to a comparison of the gene organization confirmed this proposal (Paquette et al., 2000).
The sequencing of the Arabidopsis genome has revealed that 17% of the genes are organized in tandem arrays. In addition, around 60% of the genome has arisen by duplication of large chromosomal segments (TAGI, 2000). Particularly within the A-type P450 family such gene duplication events during the evolutionary history have generated expanded families. Members of the P450 (sub) families like CYP71A, CYP71B, CYP705, and CYP81 are thus organized as tandem arrays repeated on different chromosomes (Figure 3). Plants are sessile organisms that synthesize a vast number of secondary metabolites to adapt to abiotic and biotic stresses as well as to secure communication among plants. The biosynthesis of these compounds often involves A-type P450s. This may explain the explosion of especially A-type P450s in plant. Also within the non-A type P450 families, duplication of e.g. CYP72A has resulted in 8 CYP72A genes and one pseudogene all located in a tandem array on the upper arm of chromosome III.

Figure 3: Genetic locations of cytochromes P450 in the Arabidopsis genome. Chromosome Maps were made by BLAST of deduced amino acid sequence against whole chromosome sequences. Arrows indicate the relative gene orientation. Putative pseudogenes are shown in italics.
Originally the A-type P450s were proposed to catalyze reactions specific for plants: e.g. A-type P450s like CYP73, CYP75, CYP84, and CYP98 are involved in the phenylpropanoid pathway and CYP79s and CYP83s are involved in glucosinolate biosynthesis. The non-A type P450s were thought to have maintained essential biochemical functions that evolved before the radiation of plants (Durst and Nelson, 1995): e.g. the non-A type P450s CYP86 and CYP94 are involved in fatty acid metabolism.
With the accumulating knowledge of biochemical functions of individual P450s from Arabidopsis and from other plant species, the original division of A and non-A type P450s should be reevaluated. The general picture is now that A-type P450s are primarily involved in biosynthesis of secondary metabolites and in plant specific pathways, and the non-A type are primarily involved in metabolizing house-hold functions, and in conserved functions essential for development and signaling. However, several examples of non-A P450s involved in biosynthesis of secondary metabolites are known. The non-A type CYP72A1 from Catharanthus roseus is involved in biosynthesis of terpene indole alkaloids (Irmler et al., 2000). Other non-A type P450s are involved in biosynthesis of taxol (Schoendorf et al., 2001). Both terpene indole alkaloids and taxol are species specific secondary metabolites. Conversely, P450s in the biosynthesis of hormones such as CYP701 (gibberellin biosynthesis) or CYP79B (biosynthesis of auxin) are found in the group A. The functional distinction between the two groups is thus not strict, and it is obvious that, during evolution, P450s resulting from gene duplication events have been recruited for use in new pathways. As more biochemical and genetic knowledge is gained from Arabidopsis and from other plant species a more refined understanding of the non-A and A P450 clades should emerge.
The example of the biosynthesis of giberellins shows that cases of convergent evolution have to be expected. Members of the CYP68 family in the fungus Gibberella fujikuroi (Rojas et al., 2001) and two very phylogenetically distinct plant P450s CYP701 (A-type) (Helliwell et al., 1998) and CYP88 (non-A group) (Helliwell et al., 2001) are all involved in gibberellin biosynthesis catalyzing similar reactions but they have significant differences in their genetics and biochemistry.
The Arabidopsis genome-sequencing project has revealed the presence of 26 P450 pseudogenes. These pseudogenes have been annotated based on the absence of a complete open reading frame due to segmented genes and to the presence of in-frame stop codons or frameshifts. Additional pseudogenes may be present in the genome that have not been found using these criteria.
The alignments (including marked intron positions and phase) behind the trees presented in Figures 4 and 5, as well as un-rooted trees can be found at http://www.biobase.dk/P450. This website also offers a local BLAST facility that enables database searching of Arabidopsis P450 EST, cDNA, genomic, and deduced amino acid sequences, as well as against whole chromosomes.
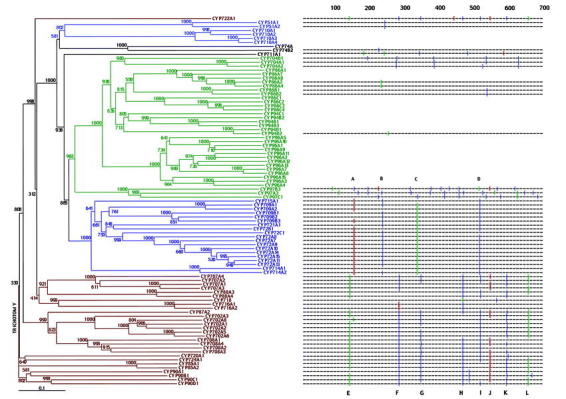
Figure 4: Neighbor-join bootstrap tree and intron map of the non-A type P450s. P450 groups thought to comprise individual clades are colored. Bootstrap values are out of 1000 trials. Phase 0 introns: |; Phase 1 introns: [; Phase 2 introns: ]. Phase 1 introns are positioned between two codons, phase 2 introns are positioned after the first base in the codon, and phase 3 introns after the second base of the codon. P450s lacking an intronmap are intronless.
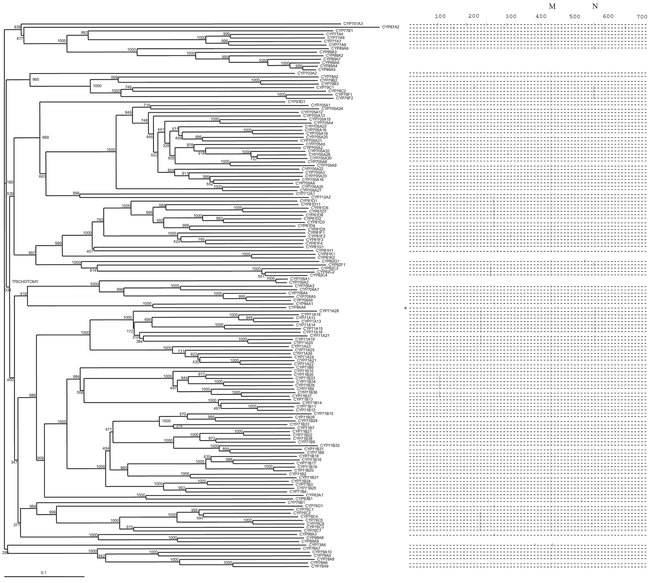
Figure 5: Neighbor-join bootstrap tree and intron map of the A-type P450s. Phase 0 introns: |; phase 1 introns: [; phase 2 introns: ]. P450s lacking an intronmap are intronless. Bootstrap values are out of 1000 trials.
Non-A type P450s:
There are 93 non-A P450s and 7 putative non-A pseudogenes. (Figure 4). In the non-A P450 tree published in 2000 by Paquette et al., four branch-points were identified, based on bootstrap values and intron conservation, but the tree could not resolve all of the deeper branches. The completion of the Arabidopsis genome has provided 32 additional non-A sequences, and now enables a more complete analysis of the paralogous non-A Arabidopsis P450s.
Members of the CYP94 and CYP86 families have been shown to be fatty acid hydroxylases. They group with the CYP96 and CYP704 families whose function is currently not known. CYP96 and CYP94 family members are generally intronless, whereas the CYP86 and CYP704 family members contain unique introns. The generally highly conserved PERF motif has a Phe to Trp substitution, so that members of the families CYP86, CYP94, CYP96, and CYP704 read the consensus PERW. The evolutionary relatedness of these sequences suggests that CYP96 and CYP704 may also take fatty acids as substrates, just as CYP86 and CYP94. CYP97 groups with a strong bootstrap value of 963/1000 to the fatty acid clade indicating that it belongs to the fatty acid clade as well. However, the CYP97 family is characterized by having up to 13 introns that are not shared with any other Arabidopsis P450. In addition, CYP97C1 and CYP97B3 contain the PERF consensus as opposed to the PERW consensus generally seen in the fatty acid clade. Biochemical analysis of recombinant CYP96, CYP97, and CYP704 or mutant studies should help to reveal the evolutionary relatedness of this expanded clade, and clarify if they all are fatty acid hydroxylases.
The CYP90 and CYP85 families form a clade with the CYP87, CYP720, CYP724, CYP702, and CYP708 families. Bootstrap values, conservation of intron positions, and the change of the PERF consensus into P(W/S)RW validates this clade. The position of families CYP88, CYP707, and CYP722 in the tree is not as clear as for members of the CYP90 containing group. However, taking intron position and phase into consideration, these families are more related to the clade containing the CYP90s than to any other non-A type Arabidopsis clade. Of all the members of this highly divergent clade, only the function of CYP90s, CYP85, and CYP88s are known. CYP90 and CYP85 are involved in brassinolide biosynthesis and use sterols as substrates. CYP88s are involved in biosynthesis of gibberellins. Thus the known members of this clade are all involved in biosynthesis of large terpenoids. Interestingly, the P450s involved in biosynthesis of the diterpenoid taxol from Taxus are related to this clade (Schoendorf et al., 2001). Several of the enzymes that catalyze steps in brassinolide- and phytosterol biosynthesis are currently unknown. Members of this clade could catalyze them. Functional characterization of additional members of this clade should clarify if their role is confined to the synthesis of large terpenoids.
CYP72, CYP714, CYP721, CYP709, CYP715 form a third clade characterized by four highly conserved introns, A, B, C, and D. Intron position and phase are highly conserved in this clade and bootstrap values are generally high.
CYP710 and CYP51 appear to form a fourth clade, but the intron position of these two families is not conserved. The CYP51s have a single intron and the CYP710s lack introns. It could be argued that the intron present in CYP51 has been lost in CYP710. However, when non-plant eukaryotic P450s are included in the phylogenetic analysis, CYP710 and CYP51 appear to be only distantly related (Paquette et al., 2000) (Figure 1) and probably should be regarded as two independent clades.
The CYP74 family constitutes its own clade. The primary sequence of CYP74 differs significantly from other plant P450s and hence the position of CYP74 in the tree is not strongly fixed. CYP74 members do not utilize molecular oxygen or a redox partner. Oxygen, reducing equivalents and protons needed for the reaction are all brought by the fatty acid hydroperoxide which is used as a substrate. Accordingly, it is not surprising that the otherwise conserved residues in the oxygen-binding domain are absent and only the cysteine residue in the heme-binding domain is present. The E-R-R triad is conserved in the CYP74 family.
Families CYP711, CYP718, CYP716, and CYP722 are difficult to place in the phylogenetic trees presented. CYP711, CYP718, and CYP722 are single-member families whereas there are two CYP716 family members that share 83 % identity on the amino acid level. Such divergent single-family members are generally difficult to place in phylogenetic tree using the Neighbor-Join method employed by the ClustalW program (Thompson et al., 1994). CYP722A1 has the conserved E, F, G, I, J, K, and L introns shared with the clade containing CYP90, arguing that it should be contained in this clade. The CYP716s contain the conserved H intron present in the CYP90 containing clade, whereas the intron in CYP718 is positioned in the same position but in a different phase. The intron at this position in CYP718 should be regarded as a separate intron, as intron sliding, if it occurs at all, is considered negligible (Stoltzfus et al., 1997). The phase and position of the introns in the CYP711 family are not shared by any of the other clades suggesting that it is a separate clade. In unrooted trees (http://www.biobase.dk/P450), CYP722A1, CYP716, and CYP718 group with the CYP90 containing clade, though the branches are very deep. In contrast, CYP711 does not group with any other family in the unrooted trees either. Biochemical function as well as sequences from other plant genomes may help to further authenticate the position of these families in phylogenetic trees.
A type P450s:
There are 153 A-type P450s, and 19 putative A pseudogenes in Arabidopsis (Figure 5). These A-type P450s are generally characterized by having a single highly conserved intron, M, which is not found in any of the non-A type P450s. Within the different A-type families and subfamilies, some additional conserved introns are seen. However, the number of introns tends to be less than those seen in the non-A P450s. Generally, the nomenclature of the individual families follows the phylogenetic grouping with the exception of the CYP83 family. The CYP71 family is the most expanded P450 family with 52 members in the Arabidopsis genome and encloses the CYP83 family. Truly, the CYP83 family should be regarded as belonging to the larger CYP71 clade. It was decided not to rename CYP83A1 and CYP83B1 to avoid nomenclature ambiguities in the literature (http://drnelson.utmem.edu/CytochromeP450.html).
Members of the CYP79, CYP703, and CYP71A families have a Phe to His substitution in the PERF domain. Whereas CYP71A's and CYP79's are clearly not closely related, CYP703 clusters with a bootstrap value of 960/1000 to the CYP79s. The branch is deep indicating that CYP79's and CYP703's may be distantly related phylogenetically.
Four families, CYP73, CYP77, CYP89, and CYP701 are difficult to place in the phylogenetic trees. These families contain PERF and heme sequences that more resemble the consensus of core A-type P450 members such as the CYP705 subfamily and the CYP71 subfamilies, rather than the consensus found in non-A type clades. In comparison with non plant and other plant P450s sequences they are phylogenetically closer to the A-type than to the non-A types (Paquette et al 2000). CYP73 is the cinnamic acid 4-hydroxylase and only a single CYP73 family member exists in the Arabidopsis genome. CYP73 lacks the conserved M intron and does not share its two introns with any other A-type P450. The low bootstrap value of 295/1000 indicates that ClustalW places CYP73 more or less randomly in the tree. The members of families CYP77 and CYP89 form a strong cluster with a bootstrap value of 1000/1000 and they are generally characterized by being intron less, thus lacking the conserved A-type M intron. CYP701A3 is an unusual A-type P450 as it contains 6 unique introns and lacks the conserved M intron. Despite their intron organization, these four families are closer related to the A type P450 than to the non-A type P450 and non-plant P450s, thus arguing that they belong to the A type P450s.
P450 functions and regulation
Seven plant P450 families are not represented in Arabidopsis. Those are CYP80, CYP92, CYP99, CYP719, CYP723, CYP725 and CYP726. Some of them, such as CYP80 or CYP725, may represent very specialized functions in taxa-specific pathways. Others, such as CYP92, may still come up from unsequenced fragments of the genome or have been possibly lost during evolution.
CYP51
Two CYP51 copies are present in Arabidopsis. CYP51 is the only P450 family conserved across evolution from bacteria to fungi, animals and plants. Its function, 14α-demethylation of sterols, is conserved as well, although substrates and pathway structures differ in fungi, animal and plants. The substrate in plants is obtusifoliol (Figure 6). The gene function was investigated in vitro with the recombinant enzymes of wheat and sorghum (Cabello-Hurtado et al., 1997; Bak et al., 1997).
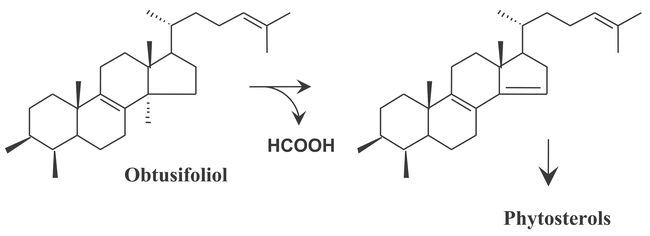
Figure 6: The reaction catalyzed by CYP51 genes in higher plants. CYP51 is an obtusifiliol 14α-demethylase and catalyzes three successive oxidations of the carbon 32, leading to C-C rupture and release of the methyl in C-14 as formic acid to form 4αmethyl-5αergosta-8,14,24(241)-trien-3β ol.
CYP51 is considered as an essential gene in all organisms since it controls the biosynthesis of sterols for membranes, needed in low amounts for meiosis and cell cycle activation. The two CYP51 proteins in Arabidopsis are 72% identical and probably non redundant since CYP51A1 is predominantly expressed in roots while CYP51A2 is evenly expressed in all tissues (Kim et al., 2000).
CYP71
CYP71 emerges as the largest P450 family in all plant species, showing large clusters of duplicated genes. There are 17 CYP71As and 37 CYP71Bs in the genome of Arabidopsis. CYP71Bs form the largest cluster of 13 P450s on chromosome III.
The function of CYP71As is so far obscure, and this despite of the fact that CYP71A1 was the first plant P450 gene to be isolated from ripening avocado (Bozak et al., 1990). Glandular trichome localization and in vitro activity of CYP71As from Nepeta racemosa suggest a possible involvement in the metabolism of isoprenoids (Hallahan et al., 1994). Herbicide metabolism by the CYP71A10 from soybean has also been reported (Siminsky et al., 1999).
The PAD3 mutation in Arabidospis, causing a defect in the biosynthesis of camalexin was associated with the CYP71B15 gene (Zhou et al., 1999). This suggests that CYP71B15 metabolizes an indole derived molecule. Consistent with a role in camalexin biosynthesis CYP71A15 transcripts accumulate in infected or salicylic acid treated plants, but not in mutants affected in the regulation of camalexin biosynthesis.
More is known about fuctions of CYP71Cs, CYP71Ds and CYP71Es. But those are quite distantly related to CYP71As and CYP71B subfamilies, close from having a family status and completely absent from the Arabidopsis genome.
CYP72
The CYP72 family is formed of three subfamilies in Arabidopsis, 9 CYP72A genes, one CYP72B and one CYP72C. CYP72As form a large cluster of 8 genes and one pseudogene on chromosome III. The function of Arabidopsis CYP72As and CYP72C is completely unknown. CYP72A1 from Catharanthus roseus is a secologanin synthase, the ring-opening enzyme in the biosynthesis of the seco-iridoid unit of terpene indole alkaloids (Irmler et al., 2000). This possibly hints to the involvement of other P450s of the CYP72A subfamily in the biosynthesis of monoterpenes or indole alkaloids.
CYP72B1 is a brassinolide 26-hydroxylase, involved in the catabolism of brassinosteroid hormones and regulation of light perception (Neff et al., 1999). Its activity and substrate specificity were not yet confirmed using recombinant protein, but it seems likely that brassinosteroids upstream from brassinolide are also substrates of the enzyme. CYP72B1, also termed BAS,1 was one of the first plant P450 gene to be characterized by activation tagging. It was isolated as an activation-tagged suppressor of the phyB-4 allele, a missense mutation in the gene of the photoreceptor phytochrome B. Transgenic lines with reduced CYP72B1 expression have hypocotyls with enhanced responses to brassinolide and reduced responses to light. Overexpression of CYP72B1 in tobacco results in a dwarf phenotype indicative of brassinosteroid deficiency (Figure 7). CYP72B1 expression is regulated in a tissue-specific manner, and seems to be a control point between multiple photoreceptor signal transduction pathways and brassinosteroid signaling.
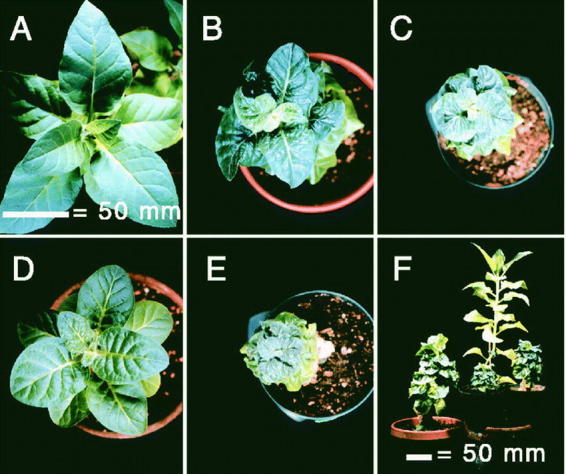
Figure 7: Dwarf phenotype induced by BAS1 overexpression in tobacco (Neff et al., 1999).Five-months-old, primary tobacco transformants. Seeds from wild-type (A), two lines transformed with the entire CYP72B1 ORF in the context of the four enhancer elements of the activation tagging system (B and C), or two lines transformed with the CAMV35S::BAS1 cDNA (D and E) were grown for 9 days in the dark. A side view compared to wild type (tallest) is shown in F.
CYP73
CYP73A5 is the cinnamic acid 4-hydroxylase (C4H), and the only CYP73 family member in Arabidopsis. The enzyme catalyzes the second step of the phenylpropanoid pathway (Figure 8). CYP73s were the first plant P450 genes to be associated with a specific biochemical and physiological function in plants.
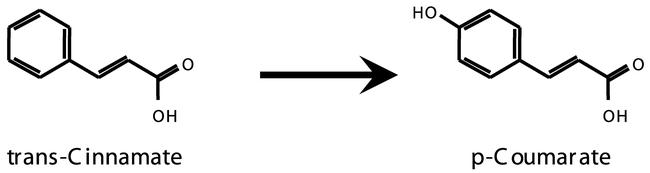
Figure 8: The reaction catalyzed by CYP73A5.
The function of CYP73A5 from Arabidopsis was confirmed by expression of the cDNA in insect cells (Mizutani et al., 1997). The CYP73A5 gene and its promoter were isolated by two groups (Bell-Lelong et al., 1997; Mizutani et al., 1997). The coding sequence contains two unique introns conserved in most CYP73 sequences reported so far. Colombia and Lansdberg erecta alleles differ in 13 positions of the coding sequences, 12 are silent, the last results in a H92 to L substitution in the Columbia ecotype (Bell-Lelong et al., 1997). One of the silent modifications also results in the loss of a StyI site in the Columbia allele.
Both groups investigated CYP73A5 regulation and showed that its expression pattern was essentially the same as that of other genes in the upstream phenylpropanoid pathway, such as those coding for phenylalanine ammonia-lyase and 4-coumarate CoA ligase. Message was detected in all plant tissues, but was highest in stems, roots and siliques. Transcripts increased within 1 hour of light exposure or wounding. In agreement with these data, various boxes (P, A, G, H, L, AC-I, AC-III, 1, 2 and 3) previously identified in other genes of the phenylpropanoid pathway were also found in the promoter of CYP73A5. Tissue-specific expression, analyzed with GUS translational fusions, detected strong expression in all vascular tissues from stems, roots and leaves, in young shoot meristems and in reproductive tissues (Bell-Lelong et al., 1997). Expression was induced by wounding of mesophyll and parenchyma cells from mature leaves (Figure 9).
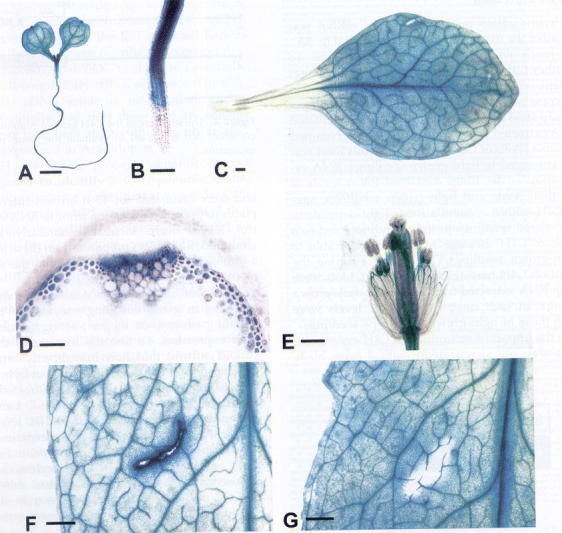
Figure 9: In vivo GUS staining in CYP73A5 promoter:GUS transformants (Bell-Lelong et al., 1997).A: Ten-day-old seedlings; B: 10-day-old seedling root; C: mature leaf; D: rachis transverse setion; E: flower; F: mature leaf stained 48h after wounding; G: mature leaf stained immeditely after wounding. A, C, E, F, G, Bar = 500 mm. B, D, Bar = 10 mm.The CYP73A5 promoter was successfully used to drive the expression of the coniferaldehyde 5-hydroxylase (CYP84A1) gene in the vascular tissues from Arabidopsis and tobacco (Franke et al., 2000; Meyer et al., 1998).
Overall CYP73A5 expression was not significantly altered in tt, fah1-2 or sng1-1 mutant backgrounds from 8 days old plants, except in the case of the tt8 mutant (Bell-Lelong et al., 1997). Strong perturbations in the phenypropanoid metabolism thus does not seem to result in large changes in the expression of CYP73A5. A more recent study provides evidence of a regulation of CYP73A5 dissociated from that of other genes of the phenylpropanoid pathway in response to UV-B irradiation (Jin et al., 2000). This study demonstrates that CYP73A5 is the principal target of AtMYB4, a transcription repressor down-regulated in plants exposed to UV-B. Regulation of the synthesis of sinapate ester sunscreens may thus occur at the level of CYP73A5.
CYP74
CYP74s are the most atypical P450s in Arabidopsis and all plant species. They do not activate molecular oxygen but catalyze the conversion of already oxygenated fatty acid hydroperoxides into allene epoxides or volatile cleavage products. As a result, they significantly differ from other P450s in their primary sequences. The residues involved in oxygen binding and activation, or electron transfer, are absent from their sequences which also have unusual N-termini with putative chloroplast transit peptides. The sequence alteration in the oxygen-binding region results a reduced affinity of these P450s for carbon monoxide. The two CYP74A and B subfamilies which seems to be present in most, if not all, plant species each have a single member in the Arabidopsis genome.
The CYP74A function was extensively studied in Arabidopsis. The recombinant enzyme expressed in E. coli was shown to catalyze the dehydration of both 13-hydroperoxylinoleic and 13-hydroperoxylinolenic acids into allene oxides (Figure 10) (Laudert et al., 1996), reactions leading to the synthesis of the signaling molecules jasmonates and their active precursors such as 12-oxo phytodienoic acid (12-OPDA). These compounds, collectively referred to as octadecanoids, mediate the accumulation of antimicrobial defense metabolites, and can substitute to mechanical stimulus in mechanoreactions, for example tendril coiling in Bryonia dioica.
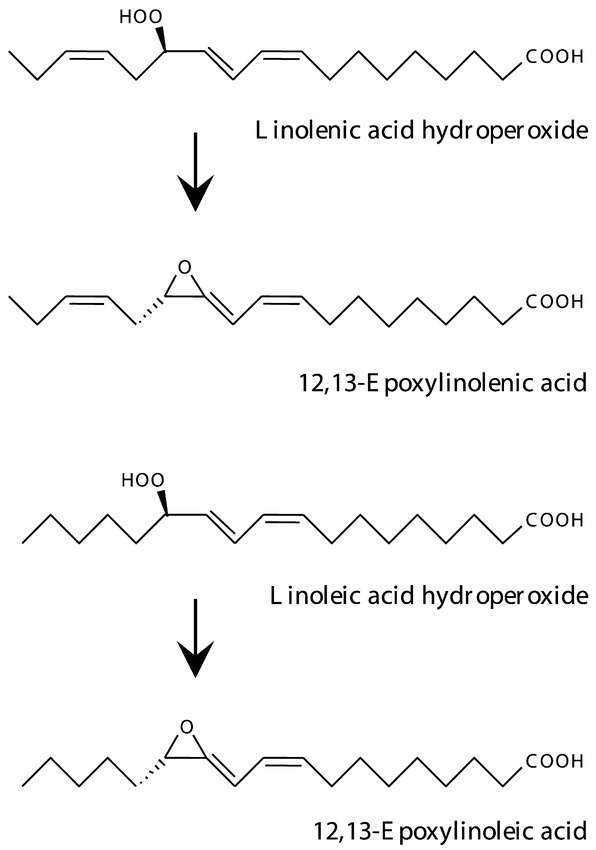
Figure 10: CYP74A is an allene oxide synthase (AOS)
CYP74A catalyzes the first step of the octadecanoid pathway. The protein is predominantly found in leaves and flowers of the mature A. thaliana plant (Laudert and Weiler, 1998). The single and intronless gene was isolated from Arabidopsis, and its expression studied using promoter GUS translational fusions (Kubigsteltig et al., 1999). Developmental control of CYP74A includes an expression confined to veins in young leaves, and spreading throughout leaves and cotyledons in older plants (Figure 11). Transient expression is also observed in flower organs: weak and transient in developing carpels, very strong in maturing pollen and at the basis of anther filaments. The latter patterns of expression was taken as indication of the involvement of octadecanoids in floral organ abscission and pollen maturation.
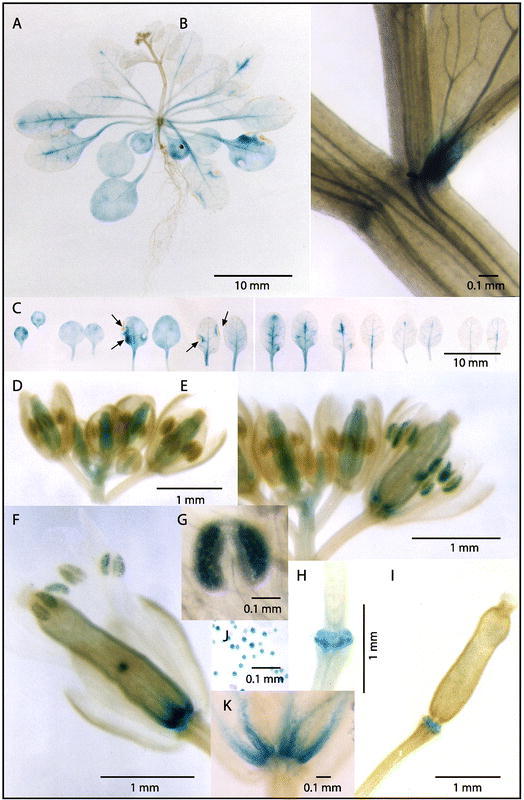
Figure 11: Developmental control of CYP74A analyzed in A. thaliana (A-I, K) or Nicotiana tabacum (J) plants grown under sterile conditions and harboring the translational CYP74A promoter:uidA fusion (Kubigsteltig et al., 1999).A: Overview of a 3-week old plant. B: Leaf base and stipule of a bract of the inflorescence axis. C: Detached leaves of the plant shown in A arranged in the order of development (from left to right: cotyledons to younger leaves), necrotic areas marked by arrows. D, E: Developing flower buds. F: Flower at the stage of fertilization. G: Close-up of anther. H,I: Abscission zone scars after shedding of flower organs. J: Pollen grains of transgenic tobacco. K: Close-up of the bases of floral organs.
A fast increase in CYP74A transcripts, protein, catalytic activity and jasmonic acid is induced in wounded leaves (Laudert et al., 2000; Laudert and Weiler, 1998). A similar increase is also observed, although smaller and delayed, in the intact leaves of the wounded plants. Treatment of the leaves with octadecanoids (12-OPDA and jasmonic acid methyl esters, coronatine), salicylic acid, or the ethylene releasing agent etephon, also induces an increase in message and protein accumulation in leaf tissues (Laudert and Weiler, 1998). In agreement with the increase in transcripts and protein, the AOS activity is also induced. Kinetics of induction and the octadecanoids produced by the plant however differ, depending on the nature of the inducer. The modes of action and impact on the plant of the effectors are thus expected to be different. Stress, salycilate and ethylene-responsive elements are found in the CYP74A promoter which drives wound, jasmonate, 12-OPDA and coronatine-induced expression of GUS. A interesting topological analysis of this induction can be found in (Kubigsteltig et al., 1999). The most striking aspect of CYP74A regulation is its feedback amplification by downstream metabolites that leads to an increased output of the jasmonate pathway capacity.
CYP74B2 uses the same substrate as CYP74A, but instead of dehydrating the CI8 lipid hydroperoxides generated from free fatty acids by lipoxygenases, cleaves them to form a C6-aldehyde and a 12 carbon oxo acid (Figure 12). The C12 product leads to the formation of traumatin implicated in wound signaling. The C6-aledhydes are the basis of the “green note” flavor from disrupted plant tissues, and are an important determinant of fresh fruit and vegetable flavor. The described impacts of the C6 volatiles on plants include antimicrobial effect, induction of phytoalexin accumulation, and inhibition of seed germination (Bate et al., 1998). The first 74B cDNA was isolated from pepper bell, but the Arabidopsis cDNA was also isolated (Bate et al., 1998). The protein expressed in E. coli was shown to catalyze the cleavage of 13S-hydroperoxy-9(Z),11(E),15(Z)-octadecatrienoic acid 10 times more efficiently than that of 13S-hydroperoxy-9(Z),11(E)-octadecadienoic acid.
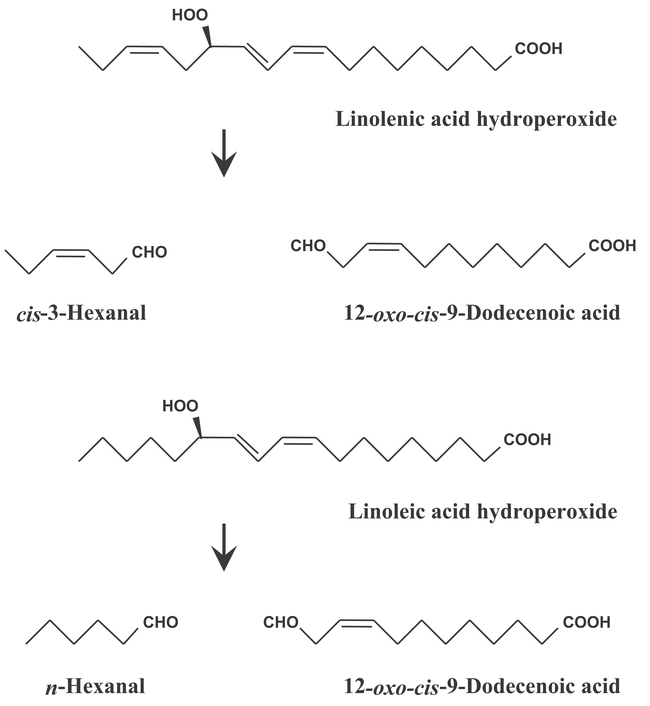
Figure 12: CYP74B2 is an hydroperoxide lyase. Recombinant CYP74B2 catalyzes the cleavage of 13S-hydroperoxy-9(Z),11(E),15(Z)-octadecatrienoic acid (linolenic acid hydroperoxide) 10 times more efficiently than that of 13S-hydroperoxy-9(Z),11(E)-octadecadienoic acid (linoleic acid hydroperoxide).
The CYP74B2 gene is highly expressed in flowers, siliques and roots (Bate et al., 1998). Levels of transcripts detected in green leaves are low, but their accumulation is induced within 30 minutes after wounding. Plant treatment with 10 mM methyl-jasmonate induces CYP74A expression but has no effect on the expression of CYP74B2.
CYP75
A single gene belonging to the CYP75 family is found in the Arabidopsis genome. CYP75B1 maps at the top of chromosome V and corresponds to the tt7 mutation. The tt7 mutants have a transparent testa phenotype, pale-brown seeds, due to the lack of tannins in the seed coats, reduced anthocyanins in the whole plant, and accumulate 4'-hydroxylated flavonoids, pelargonidin and kaempferol, instead of the 3′,4′-hydroxylated flavonoids which are present in the wild plants (Koornneef, 1982; Sheahan, 1993). It was thus assumed that the TT7 (or AtF3′H) gene encoded a flavonoid 3′-hydroxylase. This assumption was recently confirmed by the expression of the cDNA in yeast and the demonstration that recombinant CYP75B1 catalyzes the 3′-hydroxylation of the B-ring of flavonoids such as naringenin and dihydrokaempferol (Figure 13) (Schoenbohm et al., 2000). Sequencing of the tt7 allele then showed that the mutation leads to premature protein termination after 113 amino acids (Schoenbohm et al., 2000). Comparison of the Col and Ler sequences also revealed the presence of a BstXI restriction polymorphism.
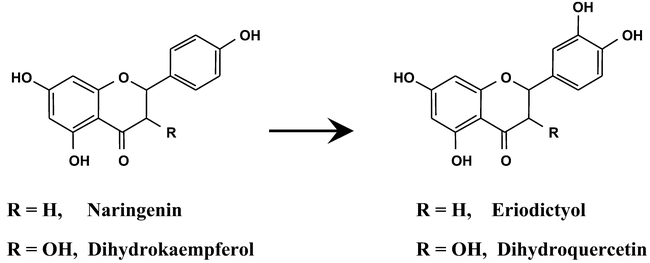
Figure 13: CYP75B1 is a flavonoid 3′-hydroxylase. CYP75B1 catalyzes the 3′-hydroxylation of the ring B of naringenin and dihydrokaempferol to form eriodictyol and dihydroquercetin, respectively.
CYP75B1 is highly expressed in siliques, flowers and senescent leaves, in agreement with the distribution of 3′,4′-hydroxylated flavonoids in the A. thaliana plant. Transcripts accumulate after UV light treatment of cultured cells which accumulate 3′,4′-hydroxylated flavonoids in the vacuole.
The CYP75A subfamily that catalyzes the flavonoid 3′,5′-hydroxylation is not represented in Arabidopsis.
CYP76
There are nine CYP76 genes in the Arabidopsis genome, eight belong to the CYP76C subfamily (including one pseudogene), a single one to the subfamily CYP76G. The function of these P450s is not known. CYP76C1 to C4 form a small cluster on chromosome 2, and appear to have arisen by gene duplication. The expression of two genes of the cluster has been compared to that of other Arabidopsis P450s in two independent studies (Godiard et al., 1998; Mizutani and Ohta, 1998). CYP76C1 was shown to be highly expressed in flowers, and at lower levels in stem, siliques and leaves (Mizutani et al., 1998). Its expression is decreased by wounding. It gradually increases upon illumination and drops when plants are returned to the dark. CYP76C2 expression was associated with stress response, being very low in young tissues, but induced during hypersensitive response to pathogens, senescence, after wounding, heat-shock, or lead nitrate treatment (Godiard et al., 1998).
CYP77
Five CYP77As and one CYP77B are present in the Arabidopsis genome. No information about their function or expression is available so far.
CYP78
The six Arabidopsis CYP78 genes are scattered over the whole genome. Like CYP78 members from other plant species, they all belong to the CYP78A subfamily. The catalytic function of the Arabidopsis CYP78 proteins is not known. The pattern of expression and in vivo impact of the related genes suggest their possible involvement in the biosynthesis of some unknown type of plant growth regulator (Ito and Meyerowitz, 2000; Zondlo and Irish, 1999). Homozygous insertional mutants of CYP78A6, which is ubiquitously expressed in all plant tissues, showed no mutant phenotype (Ito and Meyerowitz, 2000). The other CYP78 genes have been described as flower or meristem specific in their expression.
CYP78A5, was isolated from a differential screening for genes expressed in early flower development (Zondlo and Irish, 1999). In situ hybridization showed that it is strongly expressed in the peripheral region of the vegetative and reproductive shoot apical meristems, defining a boundary between the central meristematic zone and the developing organ primordia. Expression was detected throughout floral development, in the organ primordia, on the adaxial side of the pedicels and floral organs, in the developing ovules. Constitutive overexpression of the gene results in plants with twisted and kinked stems with an irregular surface. Flowers are very slow to open, with stunted petals and stamens, and disorganized tissues. They lead to very few viable seeds. Cell morphology is altered with abnormal expansion of epidermal tissues, defects in cell size and shape in internal tissues. Most striking defects are observed in tissues which undergo a directional cell elongation. Accordingly, it was proposed that CYP78A5 is involved in regulating directional cell growth.
CYP78A9 was isolated from an activation tagging line showing large seedless fruits and altered cell growth (Ito and Meyerowitz, 2000). The mutant phenotype initially characterized in the apetala2-1 background, was slightly altered in a wild-type background, with fruits more elongated than wide (Figure 14). Overexpression of CYP78A9 allows fruit growth independently of fertilization. Flower modifications are similar to those observed in CYP78A5 overexpressors, with shortened stamens and reduced aborted ovules, which suggests that both genes may be involved in the same pathway. CYP78A9 however differs from CYP78A5 by its very strict tissue-specific expression in funiculi (the stalks of the developing ovules) during late flower development (anthesis). It was proposed that one of the functions of CYP78A9 is to produce a signal that activates or enhances fruit development.
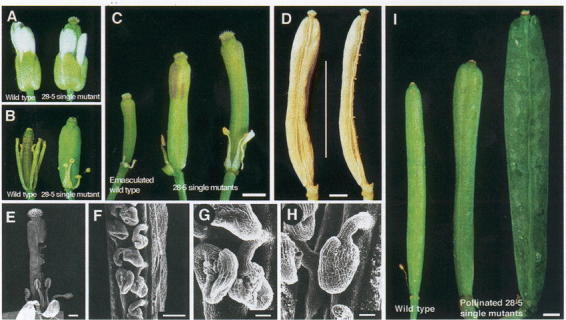
Figure 14: Phenotype resulting from the over-expression of CYP78A9 (Ito and Meyerowitz, 2000).(A) and (B) Wild-type (left) and mutant flowers (right). The length of the sepals and petals of mutant flowers did not differ from wild-type. However stamens of mutants were approx. 50% shorter than those of the wild type. The single mutant pistils were longer and wider than those of the wild type.(C) Wild type siliques 14 days after emasculation (left), mutant siliques 3 days (middle), and 5 days (right) after anthesis. The siliques of mutants continued to elongate without fertilization. Stigmatic papillae of elongating siliques were still intact.(D) Unpollinated dried mutant silique. The silique of the mutant showed a parthenocarpic phenotype. One carpel was removed to view the inside (right). No seeds were produced.(E) SEM of mutant pistil. Part of one carpel was removed to view the ovules inside.(F) to (H) Close-up view of the ovules. Most of the ovules were shriveled around the region where the embryo sac would be in the wild type (G). However, a few ovules showed normal morphology (H).(I) Self-pollinated wild-type silique (left) and pollinated mutant silique with wild type pollen (middle). Pollinated silique elongated to as much as 18 mm in one extreme case (right).Bars in (C), (D) and (I) =1 mm; bar in (E) 500 mm; bar in (F) = 200 mm; bars in (G) and (H) = 50 mm.
CYP79
CYP79s are multifunctional P450s catalyzing two consecutive N-hydroxylations of amino acids followed by a dehydration and decarboxylation reaction resulting in the formation of the corresponding aldoxime (Halkier et al., 1995; Sibbesen et al., 1995). There are seven CYP79 genes and six pseudogenes in Arabidopsis covering 4 subfamilies. Except for CYP79C1, CYP79C2, and CYP79F2, of which the function is currently unknown, all the Arabidopsis CYP79s metabolize amino acids or their chain elongated forms into the corresponding aldoximes in the biosynthesis of glucosinolates (Hull et al., 2000; Mikkelsen et al., 2000; Wittstock and Halkier, 2000). Arabidopsis contains at least 24 different glucosinolates derived from tryptophan, homophenylalanine, and chain elongated homologs of methionine. The CYP79 homologs are highly substrate-specific and are thought to dictate the specific glucosinolate profiles in a plant.
CYP79B2 and CYP79B3 catalyze the conversion of tryptophan to indole-3-acetaldoxime (Hull et al., 2000; Mikkelsen et al., 2000) (Figure 15). Indole-3-acetaldoxime is a shared intermediate between biosynthesis of indole glucosinolates and the main growth regulator auxin (indole-3-acetic acid (IAA)). Accordingly, ectopic overexpression of CYP79B2 cDNA results in a significant overproduction of indole glucosinolates, as well as in plants that exhibit reduced growth (Mikkelsen et al., 2000). IAA levels were not measured in the CYP79B2 overexpression lines, but it is plausible that the phenotype observed can be related to an increase in free IAA. In a parallel study, overexpression of CYP79B2 did not alter the phenotype (Hull et al., 2000). However, neither indole glucosinolate levels nor IAA levels were measured in these transgenic lines.
Figure 15.
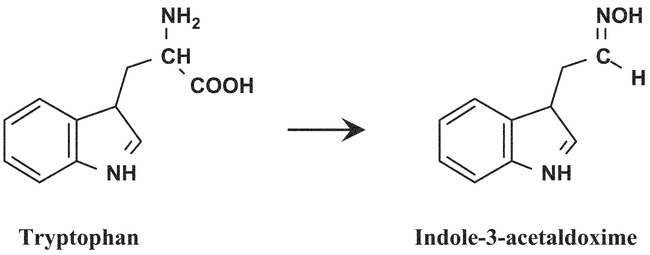
Conversion of tryptophan to indole-3-acet-aldoxime by CYP79B2/B3.
Recombinant CYP79A2 catalyzes the conversion of phenylalanine, but not homophenylalanine. In good agreement with these data, ectopic overexpression of CYP79A2 cDNA results in accumulation of phenylalanine derived benzylglucosinolate, and not of homophenylalanine derived phenylethylglucosinolate (Wittstock and Halkier, 2000). Such results were unanticipated since phenylethylglucosinolates, but not benzylglucosinolates, are usually detected in the Arabidopsis ecotype Columbia.
CYP79F1 and CYP79F2 share 88% amino acid sequence identity. Their genes are both located on chromosome 1, separated by less than 1600 bp. A knock-out of CYP79F1 by En-1 transposon tagging was designated the bus1-1 mutation. The bus1-1 mutants exhibit a short bushy phenotype with crinkled leaves and retarded vascularization (Figure 16). They are depleted in short chain methionine-derived glucosinolates (Reintanz et al., 2001). Auxin as well as indole glucosinolate levels are elevated in bus1-1. Similarly, co-suppression of CYP79F1 results in bushy plants with lowered short chain methionine-derived glucosinolates, increased expression of CYP79B2, and increased indole glucosinolates (Hansen et al., 2001). The Ds transposon null-mutant supershoot sps is allelic and phenotypically similar to bus1-1 (Tantikanjana et al., 2001). In the sps mutants, cytokinine and auxin levels are both elevated. This indicates the involvement of SPS/CYP79F1 in regulation of hormone homeostasis. As expected, recombinant CYP79F1 metabolizes short chained methionine analogs into the corresponding aldoximes precursors of glucosinolate biosynthesis (Hansen et al., 2001). This study demonstrates the pleotropic effects that can occur when studying metabolic mutants.
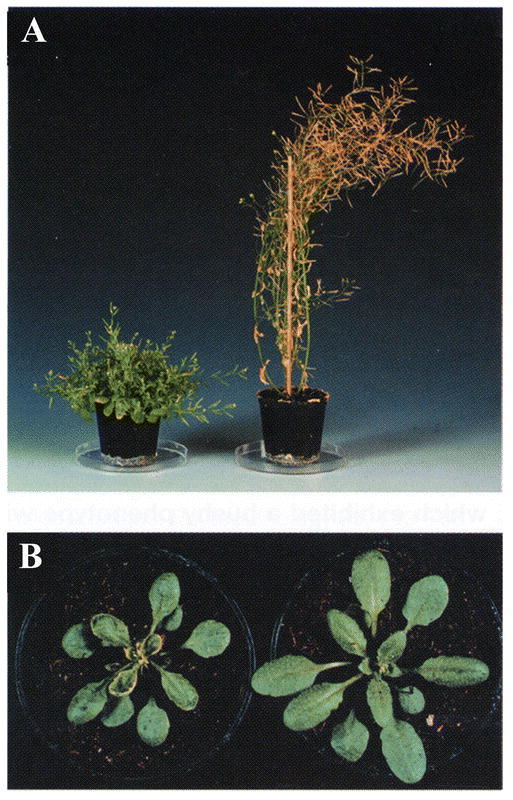
Figure 16: The bus1-1 phenotype induced by En-1 insertion in CYP79F1 (Reintanz et al., 2001).(A) The mature bus1-1 mutant exhibits a small bushy phenotype. bus1-1 is shown on the left, wild type on the right. (B) Under normal greenhouse conditions, the bus1-1 mutant has smaller rosette leaves (3 weeks old).
CYP81
There are eight CYP81Ds, four CYP81Fs, single CYP81H and G, and two CYP81Ks, forming small clusters in the Arabidopsis genome. The function and expression of these genes has so far not been investigated.
CYP82
No information is available concerning the function of the cluster of the three CYP82Cs, and of the single CYP82F and CYP82G genes present in Arabidopsis.
CYP83
CYP83 succeed CYP79 in glucosinolate biosynthesis. There are two CYP83s in the Arabidopsis genome, CYP83A1 and CYP83B1. The two P450s share 65% sequence identity on the amino acid level, and group between the CYP71A and CYP71B subfamilies in phylogenetic trees. Nevertheless, they have retained their initial naming. In the presence of thiol compounds, CYP83B1 catalyzes the initial conversion of indole-3-acetaldoxime to the corresponding S-alkyl-thiohydroximate in indole glucosinolate biosynthesis (Bak et al., 2001, Bak and Feyereisen, 2001) (Figure 17). In the absence of thiol compounds, the product of the enzymatic reaction inactivates the enzyme. Indole-3-acetaldoxime is an intermediate in tryptophan dependent IAA biosynthesis (reviewed by Normanly and Bartel, 1999). Accordingly, the CYP83B1 knockout mutant, rnt1-1 (Figure 18), is characterized by a high IAA phenotype (apical dominance) and low indole glucosinolate levels. Conversely, ectopic overexpression of CYP83B1 induces a low IAA phenotype (reduced apical dominance) and elevated indole glucosinolate levels (Bak et al., 2001, Bak and Feyereisen, 2001). rnt1-1 is allelic to the well-characterized auxin mutant sur2, known to hyperaccumulate free IAA (Barlier et al., 2000; Delarue et al., 1998). The CYP83B1 mutants and overexpression analysis thus concur to demonstrate that CYP83B1 occupies the metabolic branch-point between IAA and indole glucosinolate biosynthesis (Bak et al., 2001, Bak and Feyereisen, 2001). Ectopic expression of the CYP83A1 cDNA in rnt1-1 rescues the high IAA phenotype, and restores indole glucosinolate levels back to wild type levels, which suggests that CYP83A1 is functionally redundant to CYP83B1 (Bak et al., 2001). However, CYP83A1 has a 50 fold higher Km for indole-3-acetaldoxime than CYP83B1. Under normal conditions, CYP83A1 is thus not functionally redundant to CYP83B1 (Bak et al., 2001). In addition to the tryptophan-derived indole-3-acetaldoxime, CYP83B1 also metabolizes the tyrosine-derived p-hydroxyphenylacetaldoxime and phenylalanine derived phenylacetaldoxime to the corresponding S-alkyl-thiohydroximates, but with lower affinity than for indole-3-acetaldoxime (Bak et al., 2001; Hansen et al., 2001). Conversely, p-hydroxyphenylacetaldoxime is a preferred substrate for CYP83A1 as compared to CYP83B1. CYP83A1 and CYP83B1 thus have overlapping but not identical substrate specificities (Bak et al., 2001). The substrate specificity of CYP83A1 and CYP83B1 towards aliphatic oximes is currently not known.
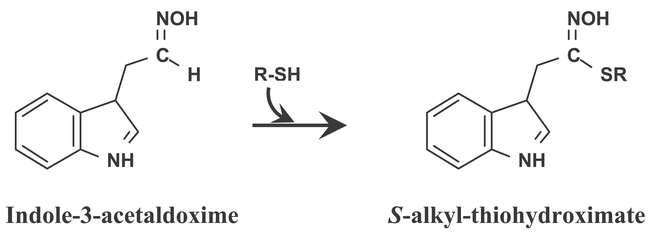
Figure 17: CYP83B1 catalyzes the first committed step of indole glucosinolate biosynthesis. In the presence of thiol compounds (R-SH) CYP83B1 form S-alkyl-thiohydroximate adducts. Formation of the adducts proceeds non-enzymatically outside the active site. In the absence of a thiol compound the highly electrophilic product of CYP83B1 catalysis inactivates the enzyme.
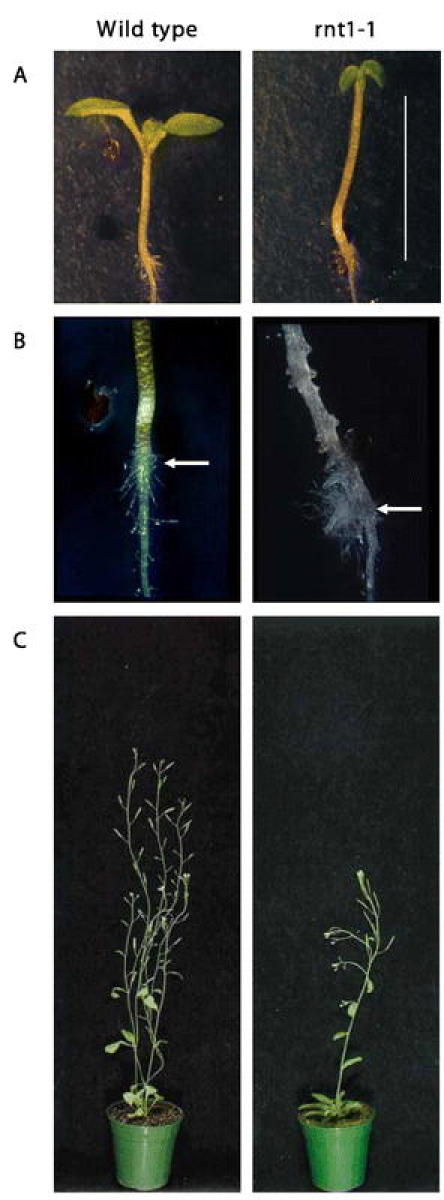
Figure 18: The rnt1-1 phenotype induced by knockout of CYP83B1 (Bak and Feyereisen, 2001).(A) 1 week old rnt1-1 seedlings have increased hypocotyl length and epinastic cotyledons (bar = 5 mm). (B) After two weeks, exfoliation of the hypocotyl begins at the root-hypocotyl junction (arrow). Secondary roots initiate from the hypocotyl, and there is an enhanced formation of secondary roots and root hairs. (C) 6 weeks old rnt1-1 plants have increased apical dominance due to elevated IAA levels. Typically, the plants have a reduced height, an increased number of epinastic rosette leaves and a single inflorescence.
CYP84
CYP84A1 is the the most extensively studied P450 gene in Arabidopsis. A forward genetics approach based on the screening of EMS mutants for a defect in sinapic esters accumulation has enabled the isolation of mutants at a locus designated SIN1 (Chapple et al., 1992). The sin1 mutant phenotype included an altered lignin composition with absence of synringyl units, lack of blue fluorescence due to sinapoyl malate in the leaf epidermis, and lack of sinapoyl choline in seeds (Figure 19). A biochemical analysis of the mutants, and complementation assays, revealed that the block in sinapoyl malate biosynthesis affected the 5-hydroxylation step of the phenolic ring. The SIN1 locus was then re-named FAH1 (for ferulic acid hydroxylase).
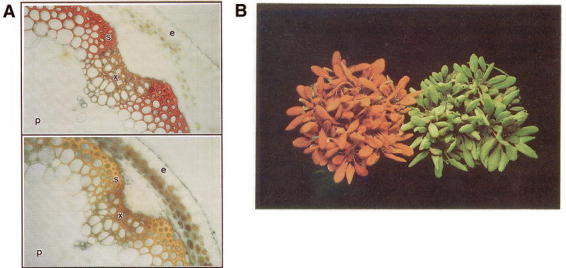
Figure 19: The sin1 (fah1) mutant phenotype (Chapple et al., 1992).A. Impact of the mutation on the staining of lignin with the Maüle reagent which produces a red coloration in the presence of syringyl units. Upper panel: wild type. Lower panel: sin1 mutant. The position of the pith (p), sclerified parenchyma (s), xylem (x), and epidermis (e) are indicated.B. Wild-type and sin1 mutant photographed under UV light. Wild-type (right) and sin1 mutant (left) plants were photographed using a 365 nm transilluminator as a light source and a pale yellow barrier to remove reflected UV light. The green color of the wild-type plants is due to the fluorescence of sinapoyl malate in the leaves' upper epidermis and appears green rather than blue because of the filter employed during photography. The red color of the mutant is due to the UV-induced chlorophyll fluorescence that is revealed in the absence of sinapoyl malate.
Additional fah1 alleles were isolated from a T-DNA tagged library on the basis of the red fluorescence of the mutants under UV-light. The interrupted sequence was isolated and shown to encode CYP84A1 (Meyer et al., 1996). A genomic CYP84A1 sequence complemented the fah1 mutation. The CYP84A1 gene is expressed in a tissue specific manner, only in the oldest rachis internodes, with developed sclerified parenchyma and increased syringyl lignin (Meyer et al., 1998). Overexpression of CYP84A1 in the mutant fah1 plants devoid of syringyl lignin, suppresses the tissue-specificic expression of the FAH gene, and leads to plants with increased syringyl lignin compared to wild-type. Thus, the temporal and tissue-specific expression of the CYP84A1 gene seems to control the composition of the lignin polymer in Arabidopsis. Interestingly, this study showed that the CaMV 35S promoter fails to drive high levels of CYP84A1 expression in tissues undergoing lignification. Much higher levels of expression are attained using the tissue-specific promoter of CYP73A5 which leads to the formation of lignin derived solely from syringyl units (Marita et al., 1999). CYP84A1 is thus a target of choice for the manipulation of lignin composition in higher plants (Franke et al., 2000).
Other experiments with the fah1 mutants demonstrated that, in Arabidopsis, sinapoylmalate is an important UV-B screen, more powerful than flavonoids, (Landry et al., 1995). Complementation of the mutation with different constructs, including the genomic sequence and different alternative promoters, indicated that CYP84A1 expression in leaves and de novo synthesis of sinapoyl choline is dependent on a regulatory domain that is located 3′ of the stop codon (Ruegger et al., 1999). This region is not required for expression of CYP84A1 in the embryo and accumulation of sinapoyl choline in the seeds. The same study showed that CYP84A1 is regulated distinctly from other phenylpropanoid genes, and strongly dependent on light in young seedlings. CYP84A1 expression, however, is not rate-limiting for the biosynthesis of sinapate esters in seedlings and developing siliques.
The catalytic function of CYP84A1 was assumed from the mutant analysis, but not confirmed with recombinant enzyme. The recent expression of the protein in yeast has led to a revision of this catalytic function and of the lignin biosynthesis pathway, revealing that ferulic acid is a very poor substrate, while coniferaldehyde and coniferyl alcohol are hydroxylated with a much higher efficiency (Humphreys et al., 1999). The efficiency of the metabolism of coniferaldehyde or coniferyl alcohol is about 1000 times that of ferulic acid which is probably not a substrate in vivo. The revised biochemical function of CYP84A1 (Figure 20) provides an explanation to many results considered as enigmatic on the basis of the previous model.
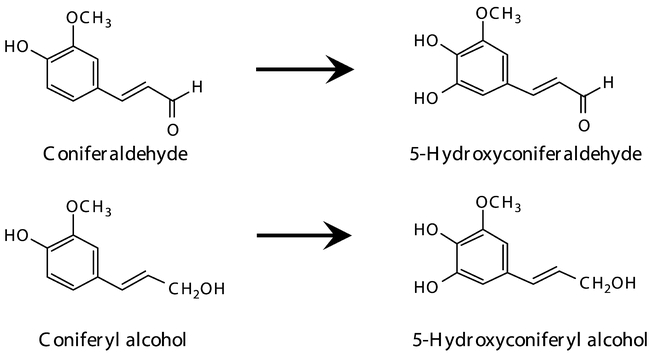
Figure 20: Reactions catalyzed by CYP84A1.
A second member of the same familly, CYP84A4, was found in the last stages of the Arabidopsis genome sequencing. CYP84A4 is 64% identical in amino acids to CYP84A1 and its gene has one intron less. No ESTs are reported for CYP84A4 and there is so far no indication concerning its possible function.
CYP85
There are two P450 genes (CYP85A and CYP85A2) belonging to the CYP85 family in Arabidopsis. Because, transposon tagging of CYP85A1 in tomato lead to a dwarf phenotype, it was assumed that this gene is involved the synthesis of hormonal compounds (Bishop et al., 1996). Expression of recombinant enzymes in yeast then demonstrated that the tomato (Bishop et al., 1999), and Arabidopsis CYP85A1s (Shimada et al., 2001) both catalyze the two successive C-6 oxidation steps in the synthesis of the steroidal plant hormones, brassinosteroids (Figure 21). A transient CYP85 expression was detected in a small patch of cells in leaf primordia which suggests a role of brassinolide in early leaf development (Pien et al., 2001). The role of CYP85A2 was not yet investigated, but it is likely to catalyze early or late C6 oxidations of brassinosteroids.
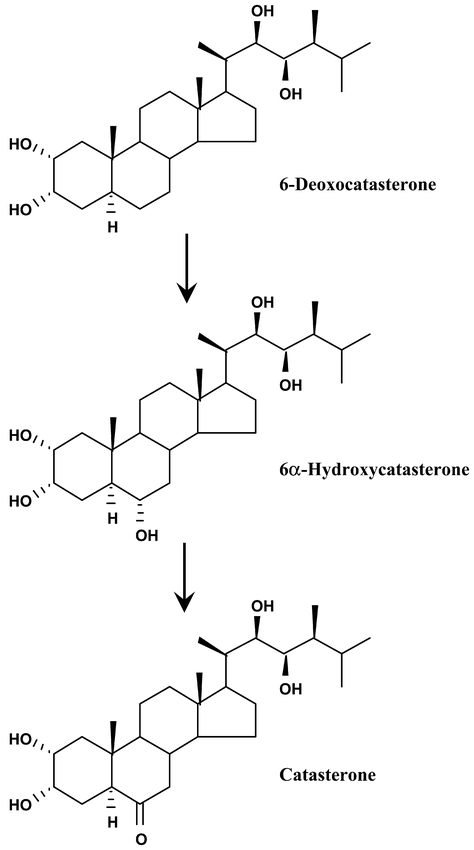
Figure 21: CYP85A1 catalyzes two successive C6 oxidations in the synthesis of brassinosteroids. The same sequence of reactions can be catalyzed on all 22,23-hydroxylated precursors: 6-deoxoteasterone, 3-dehydro-6-deoxoteasterone, 6-deoxotyphasterol and 6-deoxocastasterone (Shimada et al., 2001).
CYP86
There are five CYP86As, one CYP86B, four CYP86Cs scattered in the Arabidopsis genome. Two genes initially classified as CYP86Ds have been renamed CYP94D. CYP86s are typical non-A P450s, showing significant sequence homology with the fatty acid and alkane metabolizing CYP4s and CYP52s from mammals and fungi. The Arabidopsis CYP86A1 cDNA was isolated on the basis of this sequence homology, and the protein shown to catalyze the ω-hydroxylation of C12 to C18 fatty acids (Benveniste et al., 1998). The sequence conservation thus reflected a conservation of enzyme function. A transposon tagged Arabidopsis mutant, showing various developmental abnormalities, including postgenital organ fusions (lacerata or lcr) was then described (Wellesen et al., 2001). As other fusion mutants, the lcr plants support pollen germination on the leaf surface. The LCR gene encodes a P450, CYP86A8, which catalyzes ω-hydroxylation of fatty acids when expressed in yeast. Fatty acids ranging from C12 to C18:1 are substrates of the enzyme. Thus, it seems likely that CYP86A8 is implicated in the biosynthesis of cutins in the epidermis for preventing postgenital organ fusion. The lcr mutation is pleiotropic. Other characteristics of the lcr plants include abnormalities in cell morphology, retarded formation of trichomes, delayed senescence, lower height and increased number of shoots which seems indicative of reduced apical dominance. The molecular basis of these deffects which might be related to the absence of some lipid derived signal is however not understood.
CYP87
The function of the single CYP87 gene in Arabidopsis is currently unknown.
CYP88
There are two CYP88As (A3 and A4) in Arabidopsis, belonging to the non-A class of P450 genes. CYP88A3 and CYP88A4 are only 76 % identical, but both catalyze the conversion of ent-kaurenoic acid to GA12 when expressed in a yeast recombinant system (Helliwell et al., 2001) (Figure 22). CYP88A3 and CYP88A4 seem to be at least partially functionally redundant, being expressed in all aerial parts of the plants with higher expression in inflorescence stem, inflorescence and siliques. Mutations of the CYP88 gene in barley (grd5) and maize (dwarf3) leads to a dwarf phenotype as expected for a pertubation in the gibberellin biosynthesis. Such dwarf mutants have not been described in Arabidopsis, possibly due to gene redundancy.
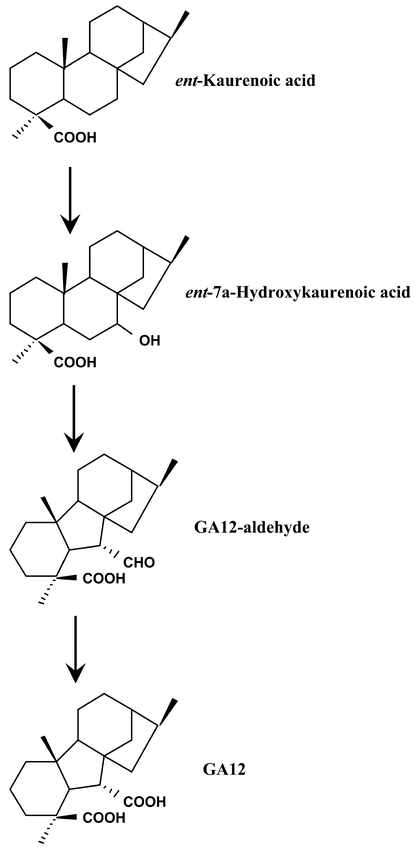
Figure 22: CYP88s are ent-kaurenoic acid oxidases (KAO). These enzymes catalyze the three successive oxidations at C-7 in the gibberellin hormone biosynthesis pathway.
CYP89
The function of the 7 CYP89A genes, four of which form a small cluster on the chromosome I of Arabidopsis, is not known.
CYP90
CYP90s are clearly related to the plant, fungal and mammalian enzymes involved in sterol biosynthesis, and are typical examples of non-A P450s. Each of the four CYP90s in Arabidopsis belongs to a different subfamily. Mutants in the Arabidopsis CYP90 genes have helped to define the role of brassinosteroids in higher plants, two of them catalyzing different steps in the biosynthesis of this class of steroid hormones.
CYP90A1 was the first P450 to be functionally characterized by reverse genetics in Arabidopsis. During the screening of a T-DNA insertional mutant collection for plants defective in hypocotyl or root elongation, a recessive mutation causing constitutive photomorphogenesis and dwarfism (cpd) was identified (Szekeres et al., 1996). In the dark, the cpd mutant developed a short hypocotyl, no apical hook, open cotyledons and extended leaf primordia (Figure 23 A). These phenotypic traits indicated derepression of photomorphogenesis and de-etiolation. When grown under white light, the size of the mutant was 20 to 30-fold smaller than that of the wild-type (Figure 23 H and I). Various histological differences were also observed between cpd and the wild-type. The mutant phenotype included male sterility. Sequencing of the T-DNA borders showed that its insertion knocked out CYP90A1. CYP90A1, but also brassinolide and all of its 23-hydroxylated precursors, successfully complemented the mutant phenotype, which led to the conclusion that CYP90A1 catalyzed the 23α-hydroxylation step in the biosynthesis of brassinosteroids (Figure 24).
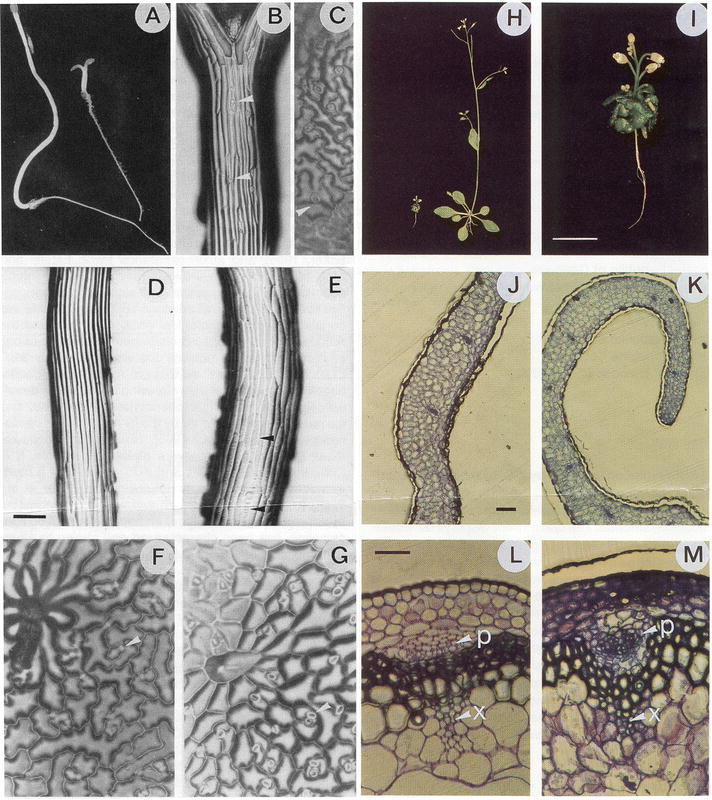
Figure 23: Phenotype of the cpd mutant (Szekeres et al., 1996).In the dark the cpd mutant (right in [A]) exhibits short hypocotyl and open cotyledons, whereas the hypocotyl is elongated and the hook of the cotyledon is closed in the wild-type (left in [A]). Unusual cell division and guard cell differentiation in the hypocotyl epidermis (B) and closely spaced stomata in the cotyledon epidermis C) are seen in the cpd mutant. In contrast with wild type (D), the length of epidermal cells is reduced in the cpd mutant (E), and their surface is covered by transverse cellulose microfibrils (indicated by arrowheads). In comparison with the wild-type (F), the adaxial leaf epidermis of the cpd mutant (G) shows straight cell walls and duplicated stomatal structures. In the light (H), the cpd mutant (left) is smaller than the wild-type (right), owing to inhibition of longitudinal growth in all organs. (I) Close-up of the mutant. Cross-sections of wild-type (J) and cpd mutant (K) leaves show difference in size and elongation of mesophyll cells. Comparison of the organization of phloem (p) and xylem (x) cell files in stem cross-sections of wild-type (L) and cpd mutant (M) plants. (D) and (E), (F) and (G), (J) and (K), and (L) and (M) are identical magnifications. Scale bar represents 200 mm in (D) and 100 mm in (J) and (L).
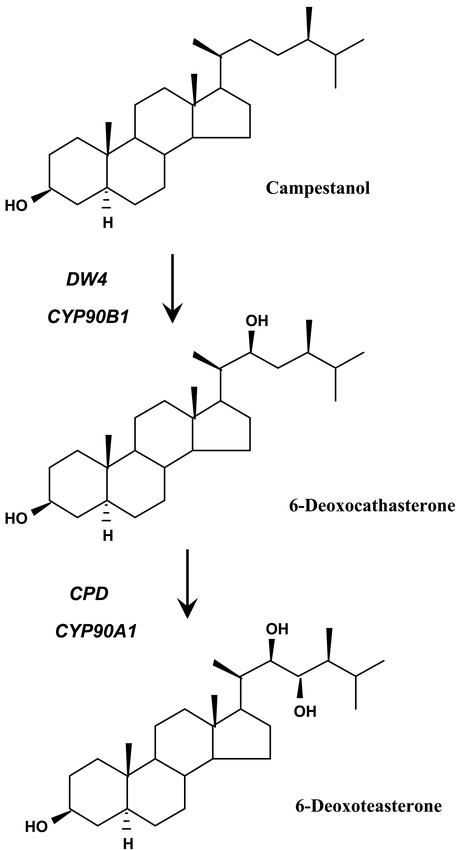
Figure 24: Reactions catalyzed by CYP90A1 and CYP90B1. The 6-oxidized precursors 6-oxocampestanol and cathasterone are also substrates of CYP90B1 and CYP90A1, respectively.
Comparable amounts of CYP90A1 transcripts are found in roots, leaves and flowers, but the gene seems expressed at lower levels in inflorescence stems and siliques. Expression of the CYP90A1 gene is confined to cotyledons and leaf primordia in etiolated seedlings, and detectable in the adaxial parenchyma of expanding leaves in light-grown plants (Mathur et al., 1998). Its transcription is modulated by external signals such as light, sucrose, or cytokinin, and repressed by brassinosteroids.
A similar approach was used for the characterization of CYP90B1. A dwarf (dwf4) mutant was isolated from a T-DNA mutagenized collection, showing a phenotype similar, but slightly different from cpd (Azpiroz et al., 1998). Among the main differences were a reduced apical dominance and less severe size reduction in dwf4. Sterility of the dwf4 mutants was simply due to the shortness of stamens, and could be reversed by manual pollination. The major defect of the dwf4 mutants was a reduced cell elongation. As for cpd, dwarfism was rescued by application of brassinosteroids (Choe et al., 1998). Only 22-hydroxylated compounds rescued the phenotype. Compounds not hydroxylated at C-22 had no effect. Sequence analysis of the DWF4 gene showed that it encoded a P450, CYP90B1. It was thus concluded that CYP90B1 catalyzed the 22α-hydroxylation step in brassinosteroid biosynthesis (Figure 24).
CYP90B1 is a gene expressed at very low levels, and was proposed to constitute the rate-limiting step in the pathway. In support of this hypothesis, steady-state levels of CYP90B1 transcripts are very significantly increased in mutants affected in brassinosteroid perception or biosynthesis (Noguchi et al., 2000). CYP90B1 is a target of brassinazole, a synthetic triazole P450 inhibitor that induces dwarfism in Arabidopsis and other plants species (Asami et al., 2000; Asami et al., 2001; Asami and Yoshida, 1999).
Like CYP90A1 and CYP90B1, CYP90C1 seems to have a signaling function. The CYP90C1 null mutants (rot3) exhibit a defect in the polar elongation of leaves and floral organs, without abnormalities in internodes and roots (Kim et al., 1998). The result is an alteration in leaf shape (rotundifolia phenotype), but the defect in cell elongation does not results in dwarfism and is not complemented by exogenous brassinosteroids. The gene is expressed in all organs, leaves, stems, floral organs and roots and in all leaf cell layers. In leaves, its expression is higher in distal than in the basal region (Kim et al., 1999). Over-expression of CYP90C1 results in plants with longer leaves, petioles and floral organs than parent plants, without change in leaf width. The impact of CYP90C1 activity thus is on control of cell elongation, but restricted to leaves and leaf-based organs.
No information is available concerning the function of CYP90D1.
CYP91
The genes previously classified as CYP91A1 and CYP91A2 have been renamed CYP81D1 and CYP81F1.
CYP93
There is a single representative of the CYP93 family in Arabidopsis. All other genes of this family described so far in other plant species are involved in the biosynthesis of flavonoids, usually in the modification of the C2-C3 bond of ring C. The Arabidopsis CYP93D1 protein is however less than 50% identical to the other P450s of the same family, and its function might be different.
CYP94, CYP96 and CYP97
There are six CYP94s (three 94Bs, one CYP94C and two CYP94Ds), twelve CYP96A genes and one pseudogene, and three CYP97s (each belonging to a different subfamily: CYP97A, CYP97B and CYP97C) in Arabidopsis. The three families have in common to be non-A P450s related to fatty acid and alkane oxidizing enzymes from fungi or animals. Involvement in fatty acid ω-hydroxylation has been confirmed for CYP94s from other plant species (e.g. Tijet et al., 1998). Such P450s might be involved in the biosynthesis of cutin monomers or defense reactions.
CYP98
There are three CYP98 genes in Arabidopsis. CYP98A8 and CYP98A9 are 69% identical and clustered on chromosome I. They probably result from a recent gene duplication, and are only 52% identical to the third gene, CYP98A3. The frequency of the CYP98A3-related ESTs found in many plant species, especially in lignifying tissues, has drawn attention on this gene. CYP98A3 was found expressed in all organs from Arabidopsis, but at higher levels in inflorescence stems, roots and siliques (Schoch et al., 2001). Immunolocalization of the protein on tissue prints indicated that its expression was confined to tissues undergoing active lignification in stems and roots. The site and high expression level of CYP98A3 thus suggested its involvement in the 3-hydroxylation of p-coumaric acid. Recombinant CYP98A3 does not metabolize free p-coumaric acid nor its CoA or glucose conjugates, but very actively hydroxylated its shikimimate and quinate esters in the 3 position of the phenolic ring (Figure 25). Isomers of the substrates are also converted. The products of CYP98A3 metabolism can be converted back into CoA esters before being methylated by caffeyl CoA O-methyltransferase. CYP98A3 is thus involved in the biosynthesis of chlorogenic acid, and probably also in that of the lignin monomers.
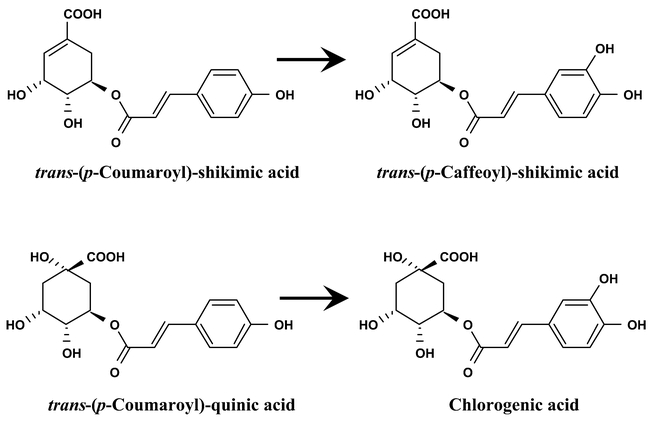
Figure 25: Reactions catalyzed by CYP98A3. 5-O-(4-Coumaryol)-shikimate is metabolized 4 times more efficiently than 5-O-(4-coumaryol)-D-quinate and is probably the favored substrate in vivo.
CYP701
CYP701A3 is involved in the biosynthesis of gibberellin hormones, like CYP88A3 and CYP88A4. There are however striking differences between the two families of enzymes. The CYP701A3 gene is not redundant, and a single member of the CYP701 family is found in Arabidopsis. CYP701 and CYP88s catalyze very similar enzymatic reactions and two successive steps in the same pathway, but CYP701A3 belongs to the class A of P450 enzymes while CYP88s belong to the non-As, being related to P450s in the biosynthesis of brassinosteroids. This example shows that the rule that P450s involved in the same pathway or catalyzing similar reactions are phylogenetically related does not hold in all cases, and that genes for a same pathway can sometimes be recruited in very distant families.
The ga3 mutants of Arabidopsis was identified as a non-germinating, gibberellin-responsive dwarf after EMS mutagenesis (Figure 26). The mutation was recessive, and complemented by application of ent-kaurenoic acid and GA. ent-Kaurene accumulated in the mutants. It was thus postulated that the mutants were blocked in the three successive oxidation steps converting ent-kaurene to ent-kaurenoic acid (Helliwell et al., 1998). These steps were known to be catalyzed by a P450 enzyme. CYP701A3 was isolated on the basis of its mapping to the GA3 locus and complementation of the ga3 mutation. The CYP701A3 coding sequence was then expressed in yeast to confirm the function of the protein that catalyzed as expected the conversion of ent-kaurene to ent-kaurenoic acid (Helliwell et al., 1999) (Figure 27).
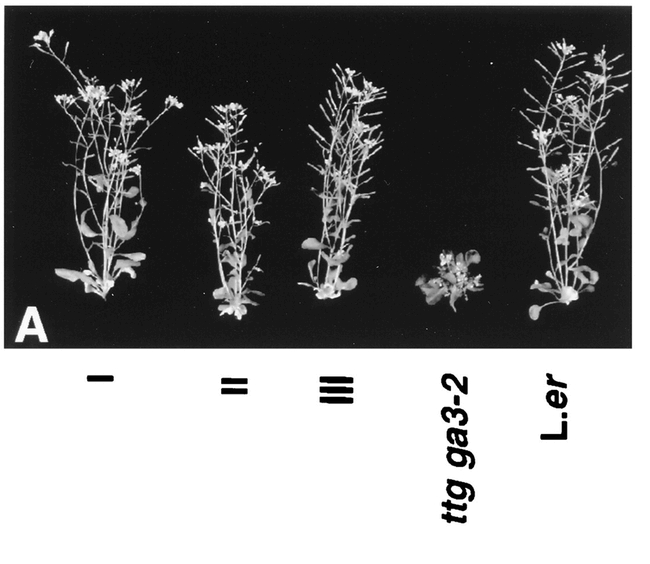
Figure 26: The ga3 mutant phenotype and complementation of the mutation with CYP701A3 (Helliwell et al., 1998).(I–III) Three kanamycin resistant plants from the transformation of ga3-2 with a genomic CYP701A3 clone. (ttg ga3-2): The mutant ga3-2 line. (L.er): The wild-type Landsberg erecta line.
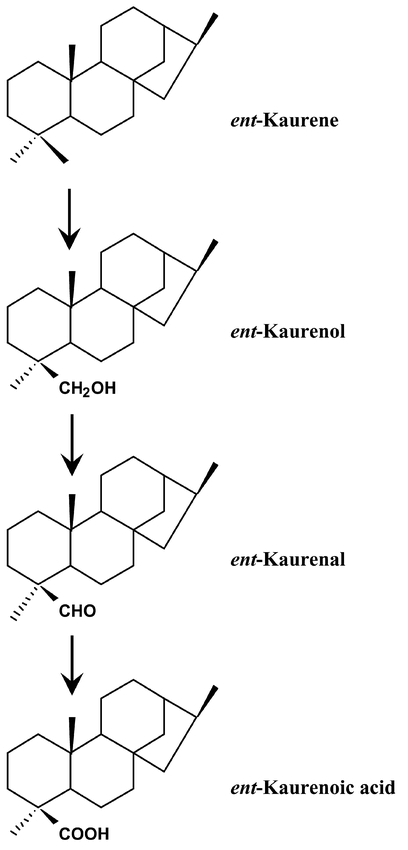
Figure 27: CYP701A3 is an ent-kaurene oxidase.The enzyme catalyzes the first P450-dependent step in the biosynthesis of gibberellins, the three successive oxidations at C-19.
The CYP701A3 gene is expressed in young, fast growing, seedlings (Helliwell et al., 1998). In older plants, expression is detected in leaves, but is higher in elongating stems and inflorescence. The gene does not seem to be subject to feed-back inhibition. Isolation of a bZIP transcriptional activator from tobacco (RSG for repression of shoot growth activator) has shown that one of its important targets is CYP701 (Fukazawa et al., 2000).
CYP702 to CYP724
The function of the P450s from the families CYP702 to CYP724 is completely unknown. Several of these families, especially CYP705, seem to result from repeated gene duplications and form large clusters in the Arabidopis genome. The functional significance of such duplications is currently not understood.
Conclusion
The first plant P450 gene was isolated in 1990 from avocado (Bozak et al., 1990) and its function is still unknown. In the last ten years, there was a huge acceleration in plant P450 gene discovery and in our understanding of their diversity and function. In despite of this acceleration, the function of more than 80% of the P450s found in the Arabidopsis genome is still completely unknown. When information is available on their biochemical function, understanding of the role of these P450s in planta is often missing. On the contrary, several null mutants were described which show interesting phenotypes, but no explanation of the phenotypes were provided. A huge field of investigation is thus opened, that should lead to the discovery on new plant pathways for the synthesis of secondary metabolites and signaling compounds. It is extremely likely that systematic approaches, such as screening of plant P450 functions and expression, by reverse genetics (Winkler et al., 1998) or extensive transcriptome analysis (Xu et al., 2001) are going to reveal unsuspected defense mechanisms and new chemical mediators or regulation mechanisms in the control of plant development.
Footnotes
Citation: Werck-Reichhart D., Bak S., and Paquette S. (2002) Cytochromes P450. The Arabidopsis Book 1:e0028. doi:10.1199/tab.0028
elocation-id: e0028
Published on: April 4, 2002
References
- Asami T., Min Y. K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;1231(1):93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T., Mizutani M., Fujioka S., Goda H., Min Y. K., Shimada Y., Nakano T., Takatsuto S., Matsuyama T., Nagata N., Sakata K., Yoshida S. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J. Biol. Chem. 2001;2761(1):25687–25691. doi: 10.1074/jbc.M103524200. [DOI] [PubMed] [Google Scholar]
- Asami T., Yoshida S. Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 1999;41(1):348–353. doi: 10.1016/s1360-1385(99)01456-9. [DOI] [PubMed] [Google Scholar]
- Azpiroz R., Wu Y., LoCascio J. C., Feldmann K. A. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;101(1):219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Feyereisen R. The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 2001;1271(1):108–118. doi: 10.1104/pp.127.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Kahn R. A., Olsen C. E., Halkier B. A. Cloning and expression in Escherichia coli of the obtusifoliol 14 alpha-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14 alpha-demethylases (CYP51) from fungi and mammals. Plant J. 1997;111(1):191–201. doi: 10.1046/j.1365-313x.1997.11020191.x. [DOI] [PubMed] [Google Scholar]
- Bak S., Tax F. E., Feldmann K. A., Galbraith D. W., Feyereisen R. CYP83B1, a Cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;131(1):101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I., Kowalczyk M., Marchant A., Ljung K., Bhalerao R., Bennett M., Sandberg G., Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci U S A. 2000;971(1):14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N. J., Sivasankar S., Moxon C., Riley J. M., Thompson J. E., Rothstein S. J. Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiol. 1998;1171(1):1393–400. doi: 10.1104/pp.117.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Lelong D. A., Cusumano J. C., Meyer K., Chapple C. Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 1997;1131(1):729–738. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste I., Tijet N., Adas F., Philipps G., Salaün J. P., Durst F. CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem. Biophys. Res. Commun. 1998;2431(1):688–693. doi: 10.1006/bbrc.1998.8156. [DOI] [PubMed] [Google Scholar]
- Bishop G. J., Harrison K., Jones J. D. The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell. 1996;81(1):959–969. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G. J., Nomura T., Yokota T., Harrison K., Noguchi T., Fujioka S., Takatsuto S., Jones J. D., Kamiya Y. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA. 1999;961(1):1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozak K. R., Yu H., Sirevag R., Christoffersen R. E. Sequence analysis of ripening-related cytochrome P-450 cDNAs from avocado fruit. Proc. Natl. Acad. Sci. USA. 1990;871(1):3904–3908. doi: 10.1073/pnas.87.10.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Hurtado F., Zimmerlin A., Rahier A., Taton M., DeRose R., Nedelkina S., Batard Y., Durst F., Pallett K. E., Werck-Reichhart D. Cloning and functional expression in yeast of a cDNA coding for an obtusifoliol 14α-demethylase (CYP51) in wheat. Biophys. Biochem. Res. Commun. 1997;2301(1):381–385. doi: 10.1006/bbrc.1996.5873. [DOI] [PubMed] [Google Scholar]
- Chapple C. C S., Vogt T., Ellis B. E., Somerville C. R. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell. 1992;41(1):1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Dilkes B. P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K. A. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;101(1):231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vetten N., Ter Horst J., Van Schaik H. P., De Boer A., Mol J., Koes R. A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA. 1999;961(1):778–783. doi: 10.1073/pnas.96.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Prinsen E., Onckelen H. V., Caboche M., Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;141(1):603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Durst F., Nelson D. R. Diversity and evolution of plant P450 and P450-reductases. Drug Metab. Drug Interact. 1995;121(1):189–206. doi: 10.1515/dmdi.1995.12.3-4.189. [DOI] [PubMed] [Google Scholar]
- Franke R., McMichael C. M., Meyer K., Shirley A. M., Cusumano J. C., Chapple C. Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000;221(1):223–234. doi: 10.1046/j.1365-313x.2000.00727.x. [DOI] [PubMed] [Google Scholar]
- Fukazawa J., Sakai T., Ishida S., Yamaguchi I., Kamiya Y., Takahashi Y. Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell. 2000;121(1):901–915. doi: 10.1105/tpc.12.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiard L., Sauviac L., Dalbin N., Liaubet L., Callard D., Czernic P., Marco Y. CYP76C2, an Arabidopsis thaliana cytochrome P450 gene expressed during hypersensitive and developmental cell death. FEBS Lett. 1998;4381(1):245–249. doi: 10.1016/s0014-5793(98)01309-x. [DOI] [PubMed] [Google Scholar]
- Graham S. E., Peterson J. A. How similar are P450s and what can their differences teach us. Arch. Bochem. Biophys. 1999;3691(1):24–29. doi: 10.1006/abbi.1999.1350. [DOI] [PubMed] [Google Scholar]
- Halkier B. A., Sibbesen O., Koch B., Møller B. L. Characterization of cytochrome P450TYR, a multifunctional haem-thiolate N-hydroxylase involved in the biosynthesis of the cyanogenic glucoside dhurrin. Drug Metab. Drug Interact. 1995;121(1):285–297. doi: 10.1515/dmdi.1995.12.3-4.285. [DOI] [PubMed] [Google Scholar]
- Hallahan D. L., Lau S. M., Harder P. A., Smiley D. W., Dawson G. W., Pickett J. A., Christoffersen R. E., O'Keefe D. P. Cytochrome P-450-catalysed monoterpenoid oxidation in catmint (Nepeta racemosa) and avocado (Persea americana); evidence for related enzymes with different activities. Biochim. Biophys. Acta. 1994;12011(1):94–100. doi: 10.1016/0304-4165(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Hansen C. H., Du L., Naur P., Olsen C. E., Axelsen K. B., Hick A. J., Pickett J. A., Halkier B. A. CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol Chem. 2001;2761(1):24790–24796. doi: 10.1074/jbc.M102637200. [DOI] [PubMed] [Google Scholar]
- Hansen C. H., Wittstock U., Olsen C. E., Hick A. J., Pickett J. A., Halkier B. A. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 2001;2761(1):11078–11085. doi: 10.1074/jbc.M010123200. [DOI] [PubMed] [Google Scholar]
- Hasemann C. A., Kurumbail R. G., Boddupalli S. S., Peterson J. A., Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of thee crystal structures. Structure. 1995;31(1):41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- Helliwell C. A., Chandler P. M., Poole A., Dennis E. S., Peacock W. J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA. 2001;981(1):2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Poole A., Peacock W. J., Dennis E. S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999;1191(1):507–510. doi: 10.1104/pp.119.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Sheldon C. C., Olive M. R., Walker A. R., Zeevaart J. A., Peacock W. J., Dennis E. S. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA. 1998;951(1):9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull A. K., Vij R., Celenza J. L. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA. 2000;971(1):2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys J. M., Hemm M. R., Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA. 1999;961(1):10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmler S., Schroder G., St-Pierre B., Crouch N. P., Hotze M., Schmidt J., Strack D., Matern U., Schroder J. Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 2000;241(1):797–804. doi: 10.1046/j.1365-313x.2000.00922.x. [DOI] [PubMed] [Google Scholar]
- Ito T., Meyerowitz E. M. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell. 2000;121(1):1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. Embo J. 2000;191(1):6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R., Durst F. Function and evolution of plant cytochrome P450. Recent Adv. Phytochem. 2000;341(1):151–189. [Google Scholar]
- Kim G. T., Tsukaya H., Saito Y., Uchimiya H. Changes in the shapes of leaves and flowers upon overexpression of cytochrome P450 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;961(1):9433–9437. doi: 10.1073/pnas.96.16.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. T., Tsukaya H., Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;121(1):2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Park J. H., Qi C., Ma J., Van Etten H., Galbraith D., Feldmann K., Feyereisen R. Isolation of Arabidopsis CYP51 mutants using systematic reverse genetic and molecular genetic analysis. 2000. In Fifth International Symposium on P450 Biodiversity (Copenhagen), C4.
- Koornneef M. A gene controling flavonoid 3′-hydroxylation in Arabidopsis. Arabidopsis Information Service. 1982;191(1):113–115. [Google Scholar]
- Kubigsteltig I., Laudert D., Weiler E. W. Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta. 1999;2081(1):463–471. doi: 10.1007/s004250050583. [DOI] [PubMed] [Google Scholar]
- Landry L. G., Chapple C. C., Last R. L. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995;1091(1):1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert D., Pfannschmidt U., Lottspeich F., Hollander-Czytko H., Weiler E. W. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 1996;311(1):323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Laudert D., Schaller F., Weiler E. W. Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta. 2000;2111(1):163–165. doi: 10.1007/s004250000316. [DOI] [PubMed] [Google Scholar]
- Laudert D., Weiler E. W. Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998;151(1):675–684. doi: 10.1046/j.1365-313x.1998.00245.x. [DOI] [PubMed] [Google Scholar]
- Long M., Rosenberg C., W. G. Intron phase correlation and the evolution of the intron/exon structures of genes. Proc. Natl. Acad. Sci. USA. 1995;921(1):12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy D. The great diversity of reactions catalyzed by cytochrome P450. Comp. Biochem. Physiol. Part. C. 1998;1211(1):5–14. doi: 10.1016/s0742-8413(98)10026-9. [DOI] [PubMed] [Google Scholar]
- Marita J. M., Ralph J., Hatfield R. D., Chapple C. NMR characterization of lignins in Arabidopsis altered in the activity of ferulate 5-hydroxylase. Proc. Natl. Acad. Sci. USA. 1999;961(1):12328–12332. doi: 10.1073/pnas.96.22.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J., Molnar G., Fujioka S., Takatsuto S., Sakurai A., Yokota T., Adam G., Voigt B., Nagy F., Maas C., Schell J., Koncz C., Szekeres M. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;141(1):593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Meyer K., Cusumano J., Somerville C., Chapple C. Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monoxygenases. Proc. Natl. Acad. Sci. USA. 1996;931(1):6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Shirley A. M., Cusumano J. C., Bell-Lelong D. A., Chapple C. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in arabidopsis. Proc. Natl. Acad. Sci. USA. 1998;951(1):6619–6623. doi: 10.1073/pnas.95.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M. D., Hansen C. H., Wittstock U., Halkier B. A. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 2000;2751(1):33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Mizutani M., Ohta D. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 1998;1161(1):357–367. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Ohta D., Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997;1131(1):755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Ward E., Ohta D. Cytochrome P450 superfamily in Arabidopsis thaliana: isolation of cDNAs, differential expression, and RFLP mapping of multiple cytochromes P450. Plant Mol. Biol. 1998;371(1):39–52. doi: 10.1023/a:1005921406884. [DOI] [PubMed] [Google Scholar]
- Neff M. M., Nguyen S. M., Malancharuvil E. J., Fujioka S., Noguchi T., Seto H., Tsubuki M., Honda T., Takatsuto S., Yoshida S., Chory J. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;961(1):15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. R. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 1999;3691(1):1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Koymans L., Kamataki T. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;61(1):1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Tax F. E., Yoshida S., Feldmann K. A. Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 2000;1241(1):201–209. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Bartel B. Redundancy as a way of life - IAA metabolism. Curr. Opin. Plant Biol. 1999;21(1):207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- Omura T., Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964;2391(1):2370–2378. [PubMed] [Google Scholar]
- Paquette S. M., Bak S., Feyereisen R. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000;191(1):307–317. doi: 10.1089/10445490050021221. [DOI] [PubMed] [Google Scholar]
- Pien S., Wyrzykowska J., Fleming A. J. Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the apical meristem. Plant J. 2001;251(1):663–674. doi: 10.1046/j.1365-313x.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., Godde M., Uhl R., Palme K. bus, a Bushy arabidopsis CYP79F1 knockout mutant with abolished synthesis of short chain aliphatic glucosinolates. Plant Cell. 2001;131(1):351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M. C., Hedden P., Gaskin P., Tudzynski B. The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc. Natl. Acad. Sci. USA. 2001;981(1):5838–5843. doi: 10.1073/pnas.091096298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Meyer K., Cusumano J. C., Chapple C. Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 1999;1191(1):101–110. doi: 10.1104/pp.119.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch G., Goepfert S., Morant M., Hehn A., Meyer D., Ullmann P., Werck-Reichhart D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001;2761(1):36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- Schoenbohm C., Martens S., Eder C., Forkmann G., Weisshaar B. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem. 2000;3811(1):749–753. doi: 10.1515/BC.2000.095. [DOI] [PubMed] [Google Scholar]
- Schoendorf A., Rithner C. D., Williams R. M., Croteau R. Molecular cloning of a cytochrome P450 taxane 10b-hydroxylase cDNA from Taxus and functional expression in yeast. Proc. Natl. Acad. Sci. USA. 2001;981(1):1501–1506. doi: 10.1073/pnas.98.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan J. J. Differential visualization of transparent testa mutants in Arabidopsis thaliana. Anal. Chem. 1993;651(1):961–963. [Google Scholar]
- Shimada Y., Fujioka S., Miyauchi N., Kushiro M., Takatsuto S., Nomura T., Yokota T., Kamiya Y., Bishop G. J., Yoshida S. Brassinosteroid-6-oxidases from arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;1261(1):770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbesen O., Koch B., Halkier B. A., Moller B. L. Cytochrome P-450TYR is a multifunctional heme-thiolate enzyme catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J. Biol. Chem. 1995;2701(1):3506–3511. doi: 10.1074/jbc.270.8.3506. [DOI] [PubMed] [Google Scholar]
- Siminsky B., Corbin F. T., Ward E. R., Fleischmann T. J., Dewey R. E. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc. Natl. Acad. Sci. USA. 1999;961(1):1750–1755. doi: 10.1073/pnas.96.4.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A., Logsdon J. M J., Palmer J. D., Doolittle W. F. Intron “sliding” and the diversity of intron positions. Proc. Natl. Acad. Sci. USA. 1997;941(1):10739–10744. doi: 10.1073/pnas.94.20.10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Nemeth K., Koncs-Kalman Z., Mathur J., Kauschmann A., Altmann T., Redei G. P., Nagy F., Schell J., Koncs C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450 controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;851(1):171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- TAGI Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;4081(1):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Tantikanjana T., Yong J. W., Letham D. S., Griffith M., Hussain M., Ljung K., Sandberg G., Sundaresan V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2001;151(1):1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;221(1):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N., Helvig C., Pinot F., Le Bouquin R., Lesot A., Durst F., Salaun J. P., Benveniste I. Functional expression in yeast and characterization of a clofibrate-inducible plant cytochrome P-450 (CYP94A1) involved in cutin monomers synthesis. Biochem J. 1998;3321(1):583–589. doi: 10.1042/bj3320583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K., Durst F., Pinot F., Benveniste I., Nettesheim K., Wisman E., Steiner-Lange S., Saedler H., Yephremov A. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid w-hydroxylation in development. Proc. Natl. Acad. Sci. USA. 2001;981(1):9694–9699. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D., Feyereisen R. Cytochromes P450 : a success story. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-reviews3003. REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D., Hehn A., Didierjean L. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 2000;51(1):116–123. doi: 10.1016/s1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Cosme J., Sridhar V., Johnson E., Mc Ree D. E. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell. 2000;51(1):121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- Winkler R. G., Frank M. R., Galbraith D. W., Feyereisen R., Feldmann K. A. Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Isolation of mutations in the cytochrome P450 gene superfamily. Plant Physiol. 1998;1181(1):743–750. doi: 10.1104/pp.118.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U., Halkier B. A. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. Catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 2000;2751(1):14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]
- Xu W., Bak S., Decker A., Paquette S. M., Feyereisen R., Galbraith D. W. Microarray-based analysis of gene expression in very large gene families: the cytochrome P450 gene superfamily of Arabidopsis thaliana. Gene. 2001;2721(1):61–74. doi: 10.1016/s0378-1119(01)00516-9. [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T. L., Glazebrook J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell. 1999;111(1):2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondlo S. C., Irish V. F. CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 1999;191(1):259–268. doi: 10.1046/j.1365-313x.1999.00523.x. [DOI] [PubMed] [Google Scholar]


