Abstract
Mitochondria represent the powerhouse of cells through their synthesis of ATP. However, understanding the role of mitochondria in the growth and development of plants will rely on a much deeper appreciation of the complexity of this organelle. Arabidopsis research has provided clear identification of mitochondrial components, allowed wide-scale analysis of gene expression, and has aided reverse genetic manipulation to test the impact of mitochondrial component loss on plant function. Forward genetics in Arabidopsis has identified mitochondrial involvement in mutations with notable impacts on plant metabolism, growth and development. Here we consider the evidence for components involved in mitochondria biogenesis, metabolism and signalling to the nucleus.
INTRODUCTION
Over the past 50 years there have been many contributions to understanding the form and function of mitochondria in plants. Here we have not attempted to review this wider literature, but to focus on the contribution of Arabidopsis to our understanding of mitochondria in plants. As in many areas of biology, the advances in mitochondrial research facilitated by Arabidopsis have centred on the availability of genome sequence data (The Arabidopsis Genome Initiative, 2000). This has provided clear identification of mitochondrial components, allowing wide-scale analysis of gene expression, and has aided reverse genetic manipulation to test the impact of mitochondrial component loss on plant function. Forward genetics in Arabidopsis has identified mitochondrial involvement in mutations with notable impacts on plant metabolism, growth and development.
DIVISION AND BIOGENESIS
Mitochondria form and structure has been investigated in Arabidopsis using green fluorescent protein (GFP) to image mitochondria in living cells. These studies conclude that mitochondria in Arabidopsis are dynamic organelles that move rapidly and vary in size and shape. Mitochondria are not evenly distributed in cells and move, with oscillations of a few hundred nanometres up to larger, rapid movements of micrometres. Both fission and fusion events are frequent, with the rate of each varying with cell type and environmental conditions implying they are regulated processes (Logan and Leaver, 2000; Sheahan et al., 2005). The production of stably transformed mitochondrial targeted GFP plants provides a useful experimental tool to investigate mitochondria fission, fusion and segregation between cells and identify mutants in these processes (Logan and Leaver, 2000; Arimura et al., 2004b; Feng et al., 2004). Mitochondrial biogenesis can be defined as an increase in mitochondrial numbers and/or mass. As mitochondria contain lipids, nucleic acids and proteins, biogenesis requires synthesis of all three of these components. The expression of the mitochondrial genome and synthesis and import of nuclear-encoded proteins has been the most intensively studied aspects of biogenesis, largely because they are most amenable to experimental investigations. However, studies of genome replication and lipid synthesis are on the increase.
Mitochondrial division
The semi-autonomous nature of mitochondria means that they must divide to be passed on to subsequent generations and this is accomplished by fission, in much the same fashion as in many prokaryotes. Due to the endosymbiotic origin of mitochondria, and from findings in some lower algae, it might be expected that mitochondria utilise the machinery inherited from their bacterial progenitor, a key component being a GTPase protein FtsZ (McFadden and Ralph, 2003). However, in contrast to plastids, where an ortholog to FtsZ has been shown to be involved in division, no mitochondrial orthologs of FtsZ are encoded in the genomes of Arabidopsis, humans, mouse, C. elegans or yeast. Instead, mitochondria appear to employ an alternative fission process, which in yeast requires a complex of the dynamin-related protein Dnm1p, the TPR protein Fis1p and the WD repeat protein Mdv1p (Okamoto and Shaw, 2005). Using homology searches with the yeast dynamin-like proteins, Dnm1p and Drp1, which have been shown to co-localise with sites of mitochondrial constriction and fission, it was demonstrated that the GTPase proteins ADL2a (DRP3A) and ADL2b (DRP3B) play a crucial role in mitochondrial division in Arabidopsis (Arimura and Tsutsumi, 2002; Arimura et al., 2004a; Logan et al., 2004). As well as being localised at the sites of constriction and the tips of mitochondria, knock-out or dominant negative mutants result in abnormal mitochondria that are elongated, bulging and fewer in number. ADL2a/DRP3A is also required for peroxisomal fission (Mano et al., 2004). Arabidopsis also has a homologue of yeast Fis1p, and mutants in this gene (BIGYIN) display reduced numbers of large mitochondria with abnormal morphology (Scott et al., 2006). It seems clear now that plant mitochondrial division has much in common with the process in animal and fungal mitochondria.
Two other Arabidopsis dynamin-like proteins ADL1C (DRP1C) and ADL1E (DRP1E) play a crucial role in maintaining mitochondrial morphology. They appear to have overlapping functions as ADL1C (DRP1C) can compensate for the loss of ADL1E (DRP1E) in some tissues (Jin et al., 2003). Interestingly, DRP1E has also been implicated in pathogen-induced cell-death (Tang et al., 2006). Using a chemical mutagenesis approach with Arabidopsis expressing mitochondrial targeted GFP, a series of mutants have been isolated with altered mitochondrial morphology (Logan et al., 2003). One mutant, termed friendly mitochondria (fmt), was characterised by clusters of mitochondria in cells, although some mitochondria also appeared normal. FMT is homologous to the CluA gene in Dictyostelium in which mutants also cause clustering of mitochondria. Homologues are present in all eukaryotic cells, with greatest homology observed in the tetratricopeptide (TPR) domain. The subcellular localisation of the protein product has not been determined and its role in maintaining mitochondrial morphology is not clear. It is possible that it interacts with the cytoskeleton.
Mitochondrial genome structure and replication
Even before the complete sequence of the Arabidopsis mitochondrial genome was completed, it was clear that the majority of the components required for transcription and translation were encoded in the nucleus and needed to be imported into mitochondria (Unseld et al., 1997). What became evident with the complete sequencing of both the mitochondrial and nuclear genomes was the added complexity and mixed evolutionary origin of the components involved in expressing the mitochondrial genome. The basic replication machinery involves at least a DNA polymerase (Christensen et al., 2005), a DNA gyrase (Wall et al., 2004) and a DNA ligase (Sunderland et al., 2004, 2006). Plant mitochondrial genomes are organised as multiple circular molecules that undergo a high frequency of inter- and intramolecular recombination (Lonsdale et al., 1984; Mackenzie and McIntosh, 1999). Mitochondrial homologues of bacterial proteins involved in recombination and repair such as RecA and MutS have been identified in Arabidopsis (Abdelnoor et al., 2003; Khazi et al., 2003; Shedge et al., 2007). Amongst other probable functions, these proteins appear to be involved in suppression of ectopic recombination. Mutations in CHM, encoding a MutS homologue (Abdelnoor et al., 2003), and other genes encoding proteins probably involved in recombination, replication or segregation such as the single-strand DNA binding protein OSB1 (Zaegel et al., 2006), lead to a process named ‘sub-stoichiometric shifting’ (Abdelnoor et al., 2003) whereby rare rearrangements of the genome become predominant (Shedge et al., 2007). These shifts are often reproducible and can lead to phenotypic abnormalities through incorrect expression of mitochondrial genes. Our understanding of the structure, replication, segregation and transmission of plant mitochondrial genomes remains limited. Genetic studies in Arabidopsis currently offer the best hope for progress.
Expression of the mitochondrial genome–transcription, RNA processing, translation
Plant mitochondrial genes are transcribed by phage-type single subunit RNA polymerases (Hedtke et al., 1997; Cahoon and Stern, 2001). Arabidopsis contains three RNA polymerases in this group, which arose from a single progenitor by two independent duplications. RpoT1 is targeted to mitochondria, RpoT3 is targeted to plastids, and RpoT2 is targeted to both mitochondria and plastids. The relative functions of these different polymerases are not clear. RpoT1 and RpoT2 show overlapping expression patterns (Emanuel et al., 2006), and mutants lacking expression of the RpoT2 protein show no obvious defects in mitochondrial gene expression (Baba et al., 2004). Expression of both RpoT1 and RpoT3 is affected in these mutants, so the apparent lack of effect may be due to compensatory processes. Unlike the case for the phage proteins, additional transcription factors are required in yeast and animals for efficient and specific initiation on mitochondrial promoters. It is not clear whether this is also true in plants. RpoT1 and RpoT3 initiate correctly on known organelle promoters in vitro (Kuhn et al., 2007), implying additional factors may not be required. The Arabidopsis nuclear genome contains homologues of many proteins proposed to act as mitochondrial transcription factors in other systems, but as yet little functional characterisation has been carried out and the location and role of such proteins in mitochondria have not been defined. The human mitochondrial transcription factor, mtTFB, is related to RNA adenine methyltransferases. Three Arabidopsis homologues of these proteins exist, at least one of which may function in mitochondria (Gagliardi and Gualberto, 2004), although there is no evidence that this one is a transcription factor. Some of the transcription factor-like proteins of the Whirly family are predicted to be targeted to mitochondria or plastids, with some experimental support (Krause et al., 2005). As yet, though, no function within the organelles has been ascribed to any of them, and they may be involved in retrograde signalling rather than organelle gene expression (Schwacke et al., 2007).
The mitochondrial RNA polymerases recognise rather poorly defined promoter sequences. The core motif of the promoter is CRTA that in dicots usually can be expanded to CRTAAGAGA, transcription initiating at the underlined G residue (Brennicke et al., 1999; Binder and Brennicke, 2003). These promoter motifs are found upstream of rRNA genes, protein coding and tRNA genes, although only 29 of the 57 genes encoded in the Arabidopsis mitochondrial genome have this sequence upstream (Unseld et al., 1997). After transcription the primary transcript is modified in several different ways, including editing, splicing, and maturation by 5′ and 3′ processing. Many genes probably rely on co-transcription with upstream genes followed by subsequent processing of the polycistronic RNAs to generate translatable mRNAs. In the Arabidopsis mitochondrial genome, about 40 genes are arranged in clusters of two or more genes, with the clusters having no apparent functional link as the resulting proteins are not found in the same protein complex or biochemical pathway. Other promoter types probably remain to be identified to account for the transcription of all 57 genes in the Arabidopsis mitochondrial genome. Mapping of all Arabidopsis mitochondrial transcripts by circular RT-PCR has given a much clearer view of the mitochondrial transcriptome and the processes involved in generating mature transcripts (Forner et al., 2007). Most 5′ ends are generated by endonucleolytic cleavage, and 3′ ends by endonucleolytic cleavage and/or exonucleolytic trimming. So-called t-elements, derived from tRNA-like sequences, are often found close to transcript termini and are likely to be sites of endonucleolytic cleavage by RNase P or RNase Z (Forner et al., 2007).
The transcription rate of Arabidopsis mitochondrial genes appears to have only a small role in transcript abundance. Minor differences in the rate of transcription are overshadowed by much larger differences in RNA degradation rates and therefore post-transcriptional processes appear to play the crucial role in determining final RNA abundance. Direct evidence for this has come from experiments where Arabidopsis mitochondrial DNA was introduced into a Brassica nuclear background via protoplast fusion. Despite large differences in transcription rates and RNA processing patterns, the final steady state levels of coding transcripts were relatively conserved in these cybrid plants (Leino et al., 2005). Degradation of RNA is initiated at the 3′ end of the molecule. It is now clear that polyadenylation of transcripts is a signal for rapid 3′-5′ exonucleolytic degradation by polynucleotide phosphorylase (PNPase) (Perrin et al., 2004a). A study of polyadenylated transcripts from mutants lacking mitochondrial PNPase has shown accumulation of a surprisingly wide diversity of RNAs normally rapidly turned over (Holec et al., 2006). The functions of these (often non-coding) RNAs is not clear. Some of them may have regulatory roles.
The exonucleolytic processing of transcripts is not only a degradation pathway, but also a normal step in the production of stable, translatable RNAs. Primary transcripts for some mitochondrial genes may be thousands of nucleotides longer than the stable, accumulated RNA. The initial trimming appears to be a function of PNPase, whilst a second stage of 3′-5′ exonucleolytic trimming may be carried out in some cases by a second enzyme, a mitochondrial homologue of bacterial RNase II (Perrin et al., 2004b).
The ends of transcripts are not the only parts that are modified. Twenty-three group II introns are present in protein-coding mitochondrial genes of Arabidopsis that undergo cis or trans splicing (Bonen, 2008). Many proteins are likely to be involved in these splicing processes, but very few have been identified. Intron 4 of nad1 encodes a probable maturase (MatR) but it is not known which introns, if any, require its activity for splicing. There are also genes encoding proteins homologous to MatR in the Arabidopsis nuclear genome. One of them has been shown to be required for efficient splicing of the mitochondrial nad4 transcript (Nakagawa and Sakurai, 2006). The first factor identified as being required for splicing of a plant mitochondrial trans-spliced intron is a PPR protein, OTP43 (Falcon de Longevialle et al., 2007). Many other plant mitochondrial splicing factors remain to be discovered by analogy with the plethora of factors known from better studied organelles such as yeast mitochondria or plastids.
RNA editing (the post-transcriptional alteration of the RNA sequence by a site-specific mechanism that modifies cytosine bases to uracils) is an important and prevalent part of mitochondrial (and plastid) gene expression (reviewed recently in Shikanai, 2006). Biochemical studies in other species indicate that the modification is probably a simple deamination reaction. In Arabidopsis, 456 C to U changes have been defined in sequenced mitochondrial mRNA (Giege and Brennicke, 1999). The degree of editing between genes varies greatly with some genes displaying no editing while others contain over 30 edited sites. Editing can also vary between Arabidopsis ecotypes, which provides a means of genetically identifying some of the factors involved (Bentolila et al., 2005). The protein machinery involved in RNA editing has not yet been experimentally identified in plant organelles. Current models are based on a requirement for editing specificity factors (likely to be very numerous, given the little conservation around editing sites (Cummings and Myers, 2004; Mulligan et al., 2007)) and an enzymatic activity likely to resemble a cytidine deaminase. The large family of PPR proteins (Figure 1) make good candidates as specificity factors (Lurin et al., 2004; Delannoy et al., 2007) and two Arabidopsis chloroplast PPR proteins have been experimentally shown to have this role (Kotera et al., 2005; Okuda et al., 2007). More recently, it has been proposed, based on sequence analysis and circumstantial phylogenetic evidence, that at least some PPR proteins carry a catalytic domain that could carry out RNA editing (Salone et al., 2007). As in many other areas of plant research, definitive answers are likely to stem from the study of Arabidopsis mutants, and the description of a new method for screening for editing mutants is timely (Chateigner-Boutin and Small, 2007). As yet, no Arabidopsis mutants affected in the editing of specific mitochondrial sites have been published.
Figure 1.
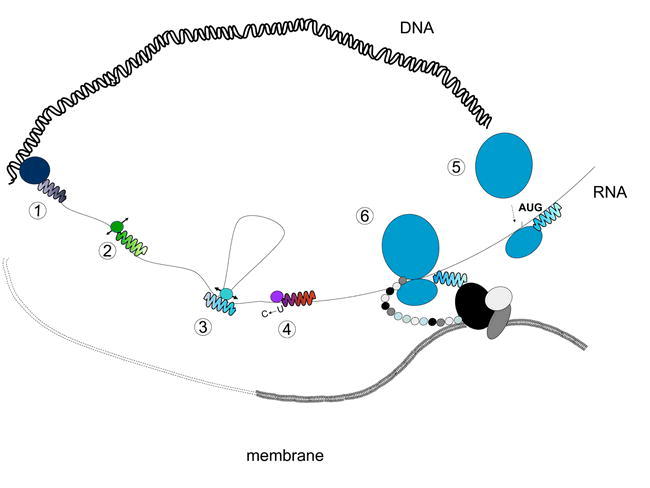
Roles for pentatricopeptide (PPR) proteins in mitochondrial gene expression.
PPR proteins have been found associated with every known stage of gene expression between transcription and translation. 1: PPR proteins have been found associated with the mitochondrial RNA polymerase; 2: PPR proteins have been implicated in RNA cleavage; 3: PPR proteins have been implicated in splicing; 4: PPR proteins have been implicated in editing; 5: PPR proteins are strongly thought to play a role in translation initiation; 6: PPR proteins are thought to be associated with ribosomes and to in some cases to tether the translation machinery to the mitochondrial inner membrane to facilitate insertion of newly synthesized polypeptides into the correct complex. Figure reproduced from Andres et al. (2007).
There is enough data already to confidently predict that several hundred different RNA-binding proteins must be postulated to perform all the known splicing, editing and 5′ and 3′ processing events. Very few of these proteins have been identified as yet. However, emerging evidence from several sources suggests that many of them are likely to be members of the large family of proteins defined by the PPR motif that interacts with nucleic acids (Small and Peeters, 2000). PPR proteins are thought to be sequence-specific RNA-binding proteins (reviewed in Delannoy et al., 2007) and likely to perform many of the activities associated with organelle transcription and processing (Andres et al., 2007; Saha et al., 2007) (Figure 1). These proteins are predicted to constitute up to 15 % of the soluble proteome in Arabidopsis mitochondria (Small et al., 2004), although direct proteomic approaches have identified only 10 PPR proteins in Arabidopsis mitochondria to date (Heazlewood et al., 2004) suggesting this large array of proteins are probably present at very low concentrations. Increasingly, mutations in genes encoding PPR proteins are being linked to mitochondrial dysfunction (Lurin et al., 2004; Kocabek et al., 2006), although the exact molecular defects are often not known. A good example is LOVASTATIN INSENSITIVE 1 (LOI1), a mitochondrial PPR protein of unknown molecular function whose absence leads to an up-regulation of the activity of cytosolic 3-hydroxy-3-methylglutaryl coenzyme A reductase (Kobayashi et al., 2007). This enzyme catalyzes the first committed step in isoprene biosynthesis leading to synthesis of, amongst other important compounds, ubiquinone, a mobile electron carrier of the mitochondrial electron transport chain. It is tempting to speculate that this PPR protein is affecting expression of a mitochondrial protein that in some way generates a signal regulating isoprenoid synthesis.
The research in Arabidopsis on PPR proteins has helped drive our rapidly increasing understanding of the complex phenomenon known as cytoplasmic male sterility (CMS). CMS occurs when expression of a mitochondrial gene leads to flower abnormalities or pollen abortion such that the affected plants are male sterile (Leino et al., 2008). Such sterility genes can be naturally selected in wild populations of many plants (but not as far as we know in self-pollinated Arabidopsis populations where the trait would be extremely disadvantageous). CMS is widely used in plant breeding to facilitate the production of hybrid seed, but for grain crops such as rice or canola, this requires the CMS trait to be suppressed in the hybrid plants by so-called ‘restorer’ genes (Chase, 2007). Most of the restorer genes so far identified are PPR proteins, starting with Rf592 from petunia (Bentolila et al., 2002) and including similar proteins from radish (Desloire et al., 2003; Koizuka et al., 2003; Brown et al., 2003), rice (Wang, et al., 2006; Komori, et al., 2004; Akagi et al., 2004) and sorghum (Klein et al., 2005). Restorer proteins presumably act by binding to the transcript encoding the sterility-causing mitochondrial protein (Gillman et al., 2007), thus preventing its expression, although the mechanism by which this is achieved is not always clear and may vary in different CMS systems. Although Arabidopsis does not exhibit CMS, and therefore, strictly speaking, has no restorer proteins, the Arabidopsis genome does contain very similar PPR genes at chromosomal loci that are very close to restorer loci in related brassicas (Desloire et al., 2003) (Geddy & Brown, 2007), leaving open the possibility that the molecular function of restorer proteins could be addressed by Arabidopsis molecular genetics in the future.
Other RNA-binding proteins are known to be present in Arabidopsis mitochondria that may play more general roles as RNA chaperones facilitating many processes. An experimental approach identified a plant specific family of mitochondrial single-stranded nucleic acid binding proteins (Vermel et al., 2002). These proteins contain a typical RNA recognition motif in the N-terminal region but differ considerably at the C-terminal end. They are induced by cold treatment, which may indicate a chaperone function to inhibit the formation of inappropriate secondary structures (Vermel et al., 2002). RNA helicases are also important for unravelling RNA structure during many post-transcriptional processes, particularly at low temperatures. The first observations on cold-induced complexes containing DEAD-box helicases have been published recently (Matthes et al., 2007).
The hundreds of genes and proteins needed for the myriad aspects of mitochondrial genome maintenance, transcription and RNA processing all serve only one purpose – to allow the synthesis of the 33 polypeptides encoded by the Arabidopsis mitochondrial genome. Although this is only a small number, these polypeptides are essential for mitochondrial (and thus cellular) function. A block in mitochondrial translation is lethal at very early stages in either embryo development, as shown by an analysis of mutations in genes encoding mitochondrial aminoacyl-tRNA synthetases (Berg et al., 2005) or in gametophyte development, as shown by two mutants lacking mitochondrial ribosomal proteins (Portereiko et al., 2006). The lethality is much more precocious than for similar mutants blocked in plastid translation (Berg et al., 2005). The absolute requirement for mitochondrial translation in plants and the continuing inability to reconstitute translation in mitochondrial extracts has greatly hampered research in this field.
Currently, studies are largely limited to describing the components of the translation system. The Arabidopsis mitochondrial genome only encodes 22 tRNA molecules that constitute 18 different tRNA types. This is not sufficient to decipher the entire set of codons used in the protein coding genes (Marienfeld et al., 1999). It is presumed that the five tRNA types that are completely missing are imported from the cytoplasm in Arabidopsis, as has been reported in studies of other plant species (Dietrich et al., 1996). In fact, considering overlapping specificities, it is estimated that 13 tRNA molecules must be imported from the cytoplasm in Arabidopsis (Unseld et al., 1997; Duchene and Marechal-Drouard, 2001). Six of the 18 tRNA types encoded in the Arabidopsis mitochondrial genome are of plastid origin, transferred to the mitochondrion via an RNA, or more likely, DNA intermediate. However, two of these chloroplast-like tRNA genes are not expressed in Arabidopsis in contrast to homologues in other plant species examined (Duchene and Marechal-Drouard, 2001). Almost all of the Arabidopsis mitochondrial aminoacyl-tRNA synthetases that recognise these tRNAs have been identified (Duchene et al., 2005), and the majority are shared with the plastid compartment. This sharing of the aminoacylating enzymes probably explains the ease with which plastid tRNAs can be incorporated into the mitochondrial translation system. The ribosome itself also contains subunits of mixed phylogenetic origin, although no complete analysis of plant mitochondrial ribosome protein composition has been undertaken, and ribosomal biogenesis in plant mitochondria has been scarcely studied at all. Some steps of ribosomal RNA processing have been examined (Perrin et al., 2004b), as well as a mitochondrial homologue of a bacterial GTPase involved in ribosomal large subunit assembly (Hill et al., 2006). A total of 7 ribosomal proteins are encoded in the mitochondrial genome and the remainder are encoded in the nucleus and imported into mitochondria. The full set of nuclear-encoded mitochondrial ribosomal proteins awaits characterisation, but already some surprises have been noted. Unlike animal and yeast mitochondria or bacteria, four different variants of RPL12 are present in Arabidopsis mitochondria (Delage et al., 2007). Analysis of two other nuclear-encoded subunits indicates that RPS8 has been substituted by its cytosolic counterpart (Adams et al., 2002) and that RPS13 has been substituted by its plastid counterpart (Adams et al., 2002; Mollier et al., 2002). These evolutionary exchanges between compartments are found at all levels of organelle gene expression. Many of the proteins involved in the metabolism of DNA, RNA and proteins are dual-targeted to mitochondria and plastids (reviewed in Peeters and Small, 2001; Millar et al., 2006). It appears that the complex machinery involved in gene expression in Arabidopsis mitochondria is a genetic mosaic derived from the ancestral sequences of the three genomes present in the cell.
Protein import from the cytosol
The biogenesis of mitochondria requires the synthesis and import of hundreds if not thousands of different proteins from the cytosol (Figure 2). Although protein import into plant mitochondria was first reported in 1987 (Boutry et al., 1987), it was not until the complete genome sequence from Arabidopsis, together with its associated technologies, became available that the actual components of the protein import apparatus could be investigated in detail (Figure 3). One approach to identify components of the protein import apparatus in Arabidopsis is by comparative analysis to the intensively studied model system of Saccharomyces cerevesiae. Although it was initially believed that two major import pathways existed, i.e. the general and carrier import pathways, studies in yeast have revealed two additional pathways, the sorting and assembly machinery for outer membrane proteins (SAM), (also called topogenesis of β-barrel proteins (TOB)), and the intermembrane space import and assembly machinery (MIA) (Neupert and Herrmann, 2007; Bolender et al., 2008). The general import pathway is responsible for the import of proteins that contain N-terminal cleavable targeting signals, and the carrier import pathway which functions to import protein of the inner membrane involved in metabolite and other transport functions. A single translocase of the outer membrane (TOM) contains the receptors for both pathways and passes the proteins to the translocases of the inner membrane (TIM). Proteins imported via the general import pathway are passed to TIM17:23 and imported into or across the inner membrane. Once in the matrix the targeting signal is removed, proteins are re-directed across or into the inner membrane, or assembled into a functional complex in the matrix. Proteins imported via the carrier import pathway interact with small TIM proteins of the intermembrane space and are inserted into the inner membrane via TIM22 (Pfanner and Geissler, 2001). The SAM complex on the outer mitochondrial membrane is responsible for the import and/or assembly of both β-barrel and α-helical outer membrane proteins such as Tom40 and Tom20 respectively (Neupert and Herrmann, 2007; Stojanovski et al., 2007). The MIA machinery is responsible for the import of a variety of small intermembrane space proteins (Hell, 2008; Bolender et al., 2008).
Figure 2.
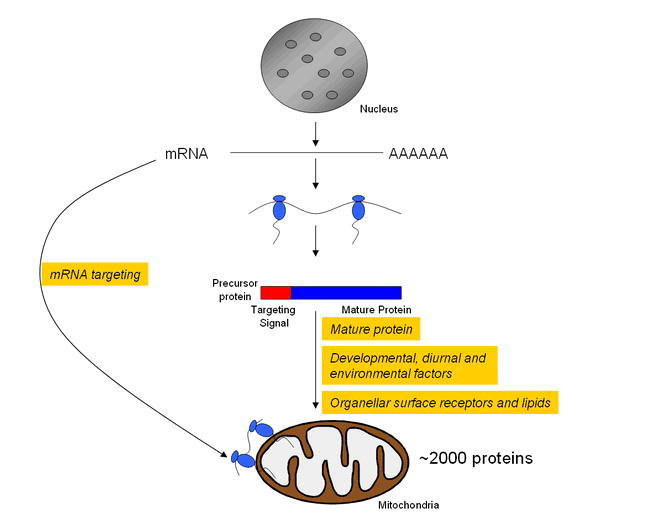
Proposed model for sorting nuclear-encoded proteins to mitochondria in Arabidopsis.
A given targeting signal directs nuclear-encoded precursor proteins to mitochondria. It is proposed that to obtain targeting specificity, the targeting of chloroplast precursor proteins to mitochondria must be prevented. Current experimental data exists to explain how this might be achieved at a variety of levels, including the involvement of the mature protein, synthesis of mitochondrial proteins at the mitochondrial surface via mRNA targeting, and additional factors such as the unique composition of organellar surface receptors and lipids, development, diurnal rhythm and environmental conditions.
Figure 3.
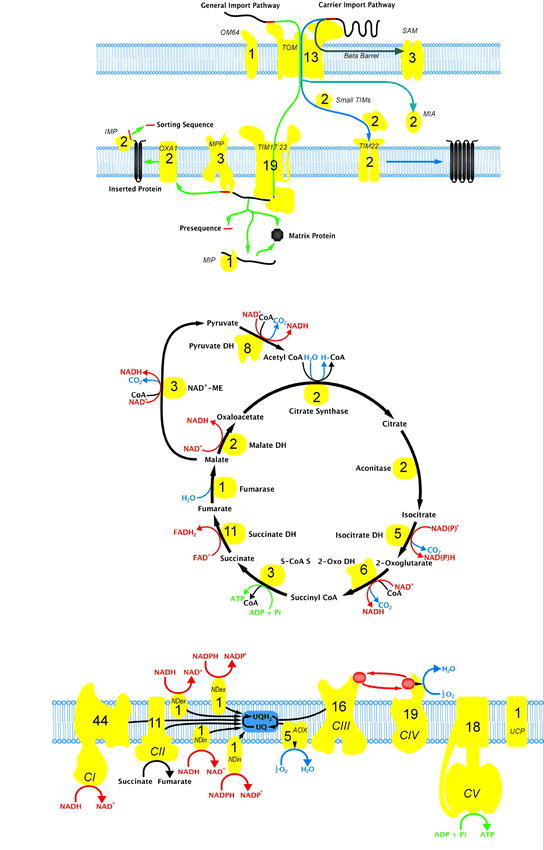
Major mitochondrial protein complexes of the pathways responsible for protein import, organic acid oxidation (TCA cycle) and oxidative phosphorylation.
The number of genes that encode polypeptide components involved in each complex in Arabidopsis are annotated.
Although it is often stated that the protein import apparatus is well conserved between species based on sequenced comparison alone (Lister et al., 2003), functional studies reveal significant differences, especially evident with outer membrane components that act as the gatekeepers to define what enters mitochondria (Figure 3). Several studies of outer membrane components reveal that Arabidopsis, and likely all plants, do not contain any orthologues of the three primary protein receptors present in yeast and mammals, i.e. Tom20, Tom70 and Tom22. Tom22 has been defined as a central and essential component of the TOM complex in yeast (van Wilpe et al., 1999). Arabidopsis does not contain any orthologues of Tom22, but does contain a smaller protein called Tom9, that encodes a protein that is similar to Tom22 in that it contains an α-helical transmembrane region and an intermembrane space domain (Macasev et al., 2000). Tom22 from yeast contains an additional domain exposed in the cytosol that act as a receptor domain for precursor proteins, similar in recognition specificity to Tom20 (Yamano et al., 2008). If the coding sequence of Arabidopsis Tom9 is fused to the receptor domain of yeast Tom22 it can complement a yeast tom22 mutant (Macasev et al., 2004), indicating that Arabidopsis Tom9 carries out some of the functions of yeast Tom22, such as assembly of the TOM complex, but lacks the receptor domain. In the case of Tom20, although initial studies suggested the existence of a Tom20 protein in Arabidopsis (and all plants) (Werhahn and Braun, 2002; Werhahn et al., 2003), the protein is not orthologous to yeast or mammalian Tom20. The plant protein is anchored in the outer membrane in the opposite orientation, the animal and fungal Tom20 proteins are anchored in the membrane by a transmembrane region in the N-terminal region of the protein whereas Arabidopsis Tom20 is anchored in the membrane via a C-terminal located transmembrane region (Lister and Whelan, 2006; Perry et al., 2006). Structural studies on one Arabidopsis Tom20 isoform reveal that it is remarkably similar to that of rat Tom20, and thus this represents a case of convergent evolution (Perry et al., 2006). In the case of Tom70, a variety of studies conclude that there is no orthologue in Arabidopsis (Lister et al., 2005; Chan et al., 2006), and biochemical approaches also have failed to identify any Tom70 type proteins from a variety of plants including Arabidopsis (Werhahn et al., 2003).
The finding in Arabidopsis that plants lacking Tom20 have no major phenotype, despite having significantly lower rates of protein import for some precursor proteins, indicates that other receptor components are present that can compensate (Lister et al., 2007). Two additional outer membrane protein characterised in Arabidopsis appear to play a role in mediating recognition and/or transport across the outer membrane. Direct proteomic approaches in Arabidopsis identified a metaxin homologue in Arabidopsis mitochondria (Heazlewood et al., 2004). Metaxin was originally shown in mammalian mitochondria to play a role in the import of preadrenodoxin (Armstrong et al., 1997). Subsequently Metaxin has been identified as a subunit of the SAM complex in yeast, and called SAM37 (Bolender et al., 2008). Deletion of Metaxin has severe phenotypic effects in Arabidopsis, that are likely to be related to the fact that the SAM complex is required for the insertion of ß-barrel proteins into the outer membrane, including Tom40 (Neupert and Herrmann, 2007; Bolender et al., 2008). However several independent protein-protein interaction assays suggest that Arabidopsis Metaxin interacts with a variety of precursor proteins directly (Lister et al., 2007), suggesting that it plays a role in the recognition of precursor proteins and may act as a receptor for precursor proteins, in addition to playing a role in the insertion of β-barrel proteins into the outer membrane.
Direct proteomic approaches have also identified a protein on the mitochondrial surface with high sequence identity to a chloroplast protein import receptor (Chew et al., 2004). The translocase of the outer envelope of chloroplasts contains a component of 64 kDa (Toc64), that is proposed to act as a receptor for the cytosolic guidance complex characterised for chloroplast protein import. Arabidopsis contains three genes with high sequence identity to Toc64. Investigations show that Toc64-III is targeted to chloroplasts but that Toc64-V is targeted to and located on the outer mitochondrial membrane (Chew et al., 2004) (Figure 3). There are some differences in the literature concerning the proposed role of Toc64 as a receptor for chloroplast proteins (Rosenbaum Hofmann and Theg, 2005; Qbadou et al., 2006; Aronsson et al., 2007; Qbadou et al., 2007). The mitochondrial located Toc64-V, now called mtOM64 (Chew et al., 2004), appears to play a role in the import of at least some proteins. Inactivation of the gene encoding mtOM64 (At5g09420) results in a decreased rate of protein import for one precursor protein, and additionally protein-protein interaction assays indicate that it can act with a variety of precursor proteins (Lister et al., 2007).
In summary, it appears that there are a variety of proteins that can act as receptor proteins in Arabidopsis mitochondria, namely TOM20, Metaxin and mtOM64. Inactivation of any of the genes encoding these proteins leads to an increase in the abundance of at least one of the others at a protein level (Lister et al., 2007), This suggests a number of proteins can act as receptors with overlapping specificity. Also given that none of these proteins are orthologous to the receptor components in yeast it is possible that additional receptor components may also exist.
TIM components in Arabidopsis display significantly higher sequence similarity with their counterparts in other organisms than those of the TOM complex, but still retain many plant specific features. All components of the TIM17:23 complex are well conserved compared to yeast, but for the TIM22 complex, sub-components TIM12, 18, and 54 appear to be absent (Lister et al., 2003; Murcha et al., 2003). The absence of TIM12 in mammals, and the absence of TIM54 and 18 in all other organisms studied to date, suggests that the mechanism of carrier protein import via the TIM22 translocase may differ significantly between organisms. Experimental studies using plant mitochondria indicate that direct insertion of carrier proteins into the inner membrane can take place and is stimulated by small intermembrane space proteins (Lister et al., 2002). In yeast, on the other hand, a physical link between the TOM and TIM22 translocase during carrier protein import has been proposed (Truscott et al., 2003).
However, there are several significant differences in the gene families encoding the TIM components that suggest functional divergence even in the components that are apparently common to yeast and Arabidopsis. Specifically, there is an expansion in the small gene family that encodes components of the TIM complexes and some contain additional domains (Murcha et al., 2007). Functional analysis of TIM components has only being carried out for genes encoding TIM17, 23 and 22. TIM17 in Arabidopsis has three orthologues that encode proteins with predicted molecular masses of 26, 23 and 14 kDa. The last of these is truncated compared to yeast, but the other two contain C-terminal extensions of approximately 80 amino acids. None of these proteins can complement a yeast mutant for tim17, but the longer TIM17 protein from Arabidopsis can complement a tim17 mutant from yeast if the additional C-terminal extensions are removed (Murcha et al., 2003). Detailed analysis of one of the TIM17 isoforms, TIM17-2, revealed that this protein spanned both the outer and inner membranes (Murcha et al., 2005). Removal of the intermembrane space located domain by mild protease treatment of outer membrane ruptured mitochondria slows down the rate of protein import into outer membrane ruptured mitochondria, suggesting that this domain plays a role in transferring proteins from the TOM to the TIM17:23 complex.
The TIM23 protein of TIM17:23 does not contain the outer membrane exposed domain found in yeast and the most prominently expressed isoform does not contain the preprotein and amino acid transporter signature sequence (PRAT). Complementation of yeast mutants with the Arabidopsis proteins can only be achieved if a PRAT domain is inserted in TIM23 (Murcha et al., 2003). Phylogenetic analysis alone has not identified TIM22 in Arabidopsis, however complementation of a yeast tim22 mutant identifies the probable orthologue, which exists as a recent tandemly duplicated gene in Arabidopsis (Murcha et al., 2007).
Thus in Arabidopsis, even components that are orthologous to the well studied components of the yeast mitochondrial protein import apparatus cannot be assumed to be functionally equivalent. Additionally the role of three other PRAT type transporters that are target to mitochondria is not known. They may play roles in protein import or act as metabolite carriers similar to some of the outer envelope proteins of chloroplasts that also belong to this family (Murcha et al., 2007).
Lipid synthesis and biogenesis of membrane composition and structure
A complex set of lipids are present in plant mitochondrial membranes. It has long been known that due to the limited ability of mitochondria to synthesize lipids, they can import their lipid content (Daum and Vance, 1997). It now appears that there is a combination of denovo synthesised lipids from mitochondriallocalised machinery, and the incorporation of lipids, presumably via lipid vesicles, into mitochondrial membranes from elsewhere in the cell.
De novo fatty acid synthesis can occur in plant mitochondria via a type II fatty acid synthase (FAS) enzyme complex (Olsen et al., 2004). The substrate specificity of condensing enzymes is a predominant factor determining the nature of fatty acyl chains incorporated by FAS. The specificity of the mitochondrial Beta-ketoacyl-[acyl carrier protein] (ACP) synthase (KAS) has been analysed using C(4)-C(18) fatty acids, and it exhibits a bimodal distribution with peaks at C(8) and C(14)-C(16) (Yasuno et al., 2004), which is very reminiscent of the biomodel distribution found in mitochondrial lipid extracts. Thus this enzyme could theoretically be responsible for most of the lipid species found in mitochondrial membranes. However, a lot of evidence suggests this is not the case and that much of the lipid is imported.
An Arabidopsis phosphatidylglycerophosphate synthase was identified, functionally characterized in yeast (Muller and Frentzen, 2001) and shown to be dual-targeted to plastids and mitochondria in Arabidopsis (Babiychuk et al., 2003). Examination of a knockout line revealed an Arabidopsis plant with pigment-deficiency and lack of thylakoid membrane biosynthesis. This gene was essential for the biosynthesis of phosphatidylglycerol in plastids, but mitochondria still accumulated phosphatidylglycerol and its derivative cardiolipin. This suggested that mitochondria, unlike plastids, could either import phosphatidylglycerol from the endoplasmic reticulum or were making it (Babiychuk et al., 2003). Subsequently a cardiolipin synthase has been identified in Arabidopsis, which catalyses CL synthesis from CDP-diacylglycerol and phosphatidylglycerol, and this protein is targeted to mitochondria. Thus is appears the mitochondrial CL is synthesized within the organelle (Katayama et al., 2004).
Phosphatidylethanolamine (PE) is an abundant lipid in mitochondria, but it also found in other cellular membranes. In Arabidopsis three genes encode phosphatidylserine decarboxylases (PSD) involved in PE synthesis. The PSD1 protein is localized to mitochondria, while PSD2 and PSD3 are found in the endomembrane system. Insertional mutagenesis in each gene separately showed few phenotypes. But the triple mutant was devoid of PSD activity and showed significant loss of PE in mitochondria, while maintaining wildtype PE levels of other cellular membranes (Nerlich et al., 2007). It thus appears that both mitochondrial and extra-mitochondrial PSD contribute to mitochondrial membrane lipid biosynthesis. These data also point to the fact that specific phospholipid transfer from the endoplasmic reticulum to mitochondria exists in plants, as it does in yeast and animals.
Arabidopsis mutants deficient in omega-6-oleate desaturase have mitochondria with an oleic acid content of more than 70% of the total fatty acids, and a greatly increased lipid/protein ratio (Caiveau et al., 2001). Oxygen consumption rate and inner membrane proton permeability were decreased, and the temperature response of respiration increased in these mutants, suggesting a changed lipid composition altered lateral mobility of lipids and the bioenergetic capabilities of these mitochondria (Caiveau et al., 2001).
Transfer of lipids has also been detected from chloroplasts to mitochondria. During phosphate deficiency in plants, phospolipids are converted to glycolipids in chloroplasts to release Pi, but these glycolipids synthesized on the plastid envelope can also be detected in mitochondrial membranes. This transfer does not apparently involve the endomembrane system but appears to be dependent upon physical contacts between plastids and mitochondria (Jouhet et al., 2004).
Proteases
Proteolysis is crucial for mitochondrial biogenesis and function, and regulated proteolysis plays a central role in the assembly of various protein complexes in mitochondria (Tatsuta and Langer, 2008). Additionally, mitochondrial targeting peptides due to their amphiphatic nature have uncoupling properties (Hugosson et al., 1994), and the failure to remove targeting signals from mitochondrial proteins results in them being targeted for degradation (Mukhopadhyay et al., 2007). In yeast bioinformatic and experimental data suggest that mitochondria contain approximately 40 proteases (Esser et al., 2002; Koppen and Langer, 2007). It is likely that a similar situation exists in Arabidopsis, but with only a few notable exceptions there is little experimental characterization of proteases in Arabidopsis mitochondria. Mitochondria contain proteases involved in removal of targeting signals (mitochondrial processing peptidase), a protease involved in degrading targeting signals after they have been removed (mitochondrial intermediate peptidase), ATP dependent proteases called AAA proteases (AT-Pases Associated with a number of cellular Activities), Lon, Clp and Rhomboid type proteases. The mitochondrial processing peptidase (MPP) has been extensively characterized in potato and spinach mitochondria, and is integrated into the cytochrome bc1 complex, whereas in yeast it is located in the matrix. Extensive reports exist on the function of plant MPP (Glaser and Dessi, 1999; Glaser and Whelan, 2007).
The most extensively characterized mitochondrial protease from Arabidopsis is the presequence peptidase called PreP (Stähl et al., 2002; Bhushan et al., 2003; Stahl et al., 2005; Johnson et al., 2006). Two genes exist in Arabidopsis encoding proteins called AtPreP1 and AtPreP2 that display 93% sequence similarity. Both PreP isoforms are dual targeted to mitochondria and chloroplasts. PreP is a metalloendopeptidase belonging to the pitrilysin subfamily containing an inverted zinc-binding motif. Although the two isoforms display high sequence similarity, cleavage site recognition differs. To date there are no reports of inactivation of one or both proteases to determine if they play a unique role or whether their functions can be compensated by a variety of other proteases.
There are few details available for other proteolytic activities from Arabidopsis mitochondria. An emerging theme with many types of organelle proteases is their dual targeting to mitochondria and chloroplasts (Adam et al., 2001; Glaser and Whelan, 2007).
CENTRAL PROCESSES IN MITOCHONDRIAL FUNCTION
Carriers and Transporters
The double membrane system of the mitochondrion allows for relatively non-specific transport of small molecules from the cytosol into the intermembrane space, and more selective transport of small molecules across the inner membrane to the matrix space (Mannella, 1992; Mannella et al., 2001). This allows a complex set of carrier functions to have a large influence on the catabolic, biosynthetic and anaerpleotic functions of mitochondria (Laloi, 1999). Transport across the outer membrane is dominated by the voltage dependent anion channels (VDAC), or porins, that form pores for the movement of solutes up to 1000 kDa (Mannella and Tedeschi, 1987). Six genes encoding putative isoforms of these porins exist in Arabidopsis and of these, four have been shown to be expressed and present in mitochondria (Clausen et al., 2004; Heazlewood et al., 2004).
A clearly defined family of mitochondrial inner membrane carriers operate for the transport of metabolically important compounds into mitochondria, including organic acids, amino acids, inorganic phosphate and adenine di- and tri-nucleotides. This family, most of which are 30 kDa, are basic proteins with six transmembrane domains (Laloi, 1999). In Arabidopsis, a family of ~50 genes can be readily identified from genome sequence data. A sub-set of ~10 are clear orthologs of yeast mitochondrial carriers of known function, while a further ~20 still do not have even a putative function (Millar and Heazlewood, 2003; Picault et al., 2004). Eight different members of this family have been directly identified by mass spectrometry in Arabidopsis mitochondrial preparations (Millar and Heazlewood, 2003; Heazlewood et al., 2004).
A carrier previously annotated as a specific oxoglutarate/malate transporter, has been experimentally analysed and shown to be a general carrier able to transport both di- and tricarboxylic acids (Picault et al., 2002). Two basic amino acid carriers (BAC1 and BAC2) have also been identified and functionally characterised by two groups independently (Catoni et al., 2003b; Hoyos et al., 2003). Detailed work on the kinetics of these carriers as recombinant proteins in liposomes shows they both transport various basic L-amino acids, but that BAC2 transported citrulline while BAC1 did not, and that BAC2 had less stereospecificity than BAC1 and an altered pH optimum (Palmieri et al., 2006a). Early expression of BAC1 and BAC2 is consistent with the delivery of arginine, released from seed reserves, to mitochondrial arginase and the export of ornithine. Increase of BAC2 transcript levels later in seedling development is consistent with roles in NO, polyamine or proline metabolism—processes involving arginine, citrulline and/or ornithine (Palmieri et al., 2006a).
The Arabidopsis mitochondrial succinate-fumarate carrier homolog has also been identified by complementation of the yeast acr1 mutant (Catoni et al., 2003a). Key work on the inorganic phosphate carrier (Takabatake et al., 1999; Hamel et al., 2004), the uncoupling protein (Maia et al., 1998; Watanabe et al., 1999) and the adenine di- and tri-nucleotide carriers (Haferkamp et al., 2002) has also been undertaken in Arabidopsis.
The activation of uncoupling protein by ROS (originally demonstrated with mammalian uncoupling protein and subsequently with plant uncoupling protein from potato) indicates an important biochemical control mechanism of this protein that needs to be considered when measuring activity or assigning function (Echtay et al., 2002; Considine et al., 2003). Analyses of an insertional knockout of the classical Arabidopsis UCP (AtUCP1) shows that absence resulted in localized oxidative stress but it did not impair the ability of the plant to withstand a wide range of abiotic stresses. However, absence of UCP1 resulted in restriction in photorespiration with a decrease in the rate of oxidation of photorespiratory glycine in the mitochondrion and reduced photosynthetic carbon assimilation rate (Sweetlove et al., 2006). This suggests that the main physiological role of UCP1 in Arabidopsis leaves is related to maintaining the redox poise of the mitochondrial electron transport chain to facilitate photosynthetic metabolism (Sweetlove et al., 2006). The claims that there are up to 4 more UCPs in Arabidopsis other than UCP1 (Vercesi et al., 2006) have been recently challenged as a number of these carriers have been shown to be organic acid transporters in artificial bilayer experiments (Palmieri et al., 2008).
S-adenosylmethionine (SAM) in an essential metabolite in the methylation of DNA, RNA and proteins. Arabidopsis homologs of the yeast and human mitochondrial SAM transporters have been identified. Transport studies with recombinant proteins show both SAMC1 and SAMC2 display a very narrow substrate specificity confined to SAM and its closest analogs (Palmieri et al., 2006b). GFP expression suggested SAMC1 was located in mitochondria, while proteomic studies suggest localization in plastids. Given the need for SAM in both organelles these findings suggest that the provision of cytosolically synthesized SAM to mitochondria and possibly also to plastids is mediated by SAMC1 according to the relative demands for this metabolite in the organelles (Palmieri et al., 2006b)
Other major classes of carriers also exist on the mitochondrial inner membrane, including half-ABC transporters. One of these (ATM1) is known to be an Fe-S transporter and mutation of this gene product leads to dwarfism and chlorosis in Arabidopsis (Kushnir et al., 2001). These data support the view that plant mitochondria possess an evolutionarily conserved Fe/S cluster biosynthesis pathway, which is linked to intracellular iron homeostasis by ABC transporters (Kushnir et al., 2001). Recent detailed analysis of this Arabidopsis family of three half-transporters by expression in yeast lacking the Fe-S transporter shows that each is transported to mitochondria, but that while AtAMT1 and AtAMT3 complement the yeast mutant to varying degrees, AtAMT2 is lethal to yeast, precluding its functional analysis (Chen et al., 2007). However, these transporters may have wider roles that Fe-S export as overexpression of AtATM3 enhances Cd tolerance and knockouts were more sensitive to Cd (Kim et al., 2006a). Based on the correlation of changes in glutathione synthesizing pathways and the redox poise of mutants and wildtype, it is speculate that glutathione-Cd(II) complexes formed in the mitochondria may be exported by AtATM3 (Kim et al., 2006a).
Bioenergetic Functions
The electron transport chain of plant mitochondria has been studied in a variety of species over the last few decades, most notably in potato, spinach and soybean. A large part of this research has focussed on the plant specific bypasses of the classic respiratory chain, the rotenone-insensitive NADH dehydrogenases (NDs of the classes NDA, NDB and NDC) and the alternative oxidase (AOX). A variety of biochemical purifications in several plant species have also provided insight into the components of the classical electron transport chain, but the absence of genes and full-length protein sequences in these other species has limited the degree to which these preparations could be compared to the well-described complexes in mammals and fungi. Arabidopsis research has allowed a detailed and direct insight into the components of the respiratory chain and the plant specific bypasses (Figure 3).
Complex I or NADH-UQ oxidoreductase is a 30–46 subunit complex in fungi and mammals. It is made from nuclear and mitochondrial-encoded proteins, and genome coordination is required to form an active complex with the correct stoichiometry of subunits (Rasmusson et al., 1998). In plants, up to 30 subunits can be separated by SDS-PAGE in potato, broad-bean and wheat. In Arabidopsis, a set of 35 putative proteins with high similarity to 29 of the 46 mammalian enzyme I components are encoded in the nuclear or mitochondrial genome (Rasmusson et al., 1998; Heazlewood et al., 2003a). Blue native PAGE separation of complex I components followed by mass spectrometry have identified most of these. This approach has also highlighted a range of novel complex I proteins in plants raising the total number of proteins associated with this complex to 44 (Heazlewood et al., 2003a; Sunderhaus et al., 2006; Meyer et al., 2008). Looking back to N-terminal sequences from potato and bean complex I reveals that some of these novel components were also apparent in the earlier plant complex I preparations. The newly identified components include a series of gamma carbonic anhydrase-like proteins (Parisi et al., 2004), several small proteins of unknown function and, interestingly, the final enzyme in ascorbate synthesis pathway in plants, galactonolactone dehydrogenase (Heazlewood et al., 2003a). This plant specific protein was known to be associated with mitochondrial membranes (Bartoli et al., 2000), but its physical association with complex I was first identified in Arabidopsis. Subsequently, it was found that ascorbate synthesis by Arabidopsis mitochondria is sensitive to complex I function as both are strongly inhibited by rotenone (Millar et al., 2003). The long-held notion that acyl carrier proteins (ACP), that are part of complex I in N. crassa, were also subunits of complex I in animals and plants, appears less likely based on recent investigations in both bovine (Cronan et al., 2005) and Arabidopsis (Meyer et al., 2007). Several Arabidopsis mutants diminished in or lacking complex I have been reported through the knockout of nuclear-encoded components such as the plant specific carbonic anhydrases (Perales et al., 2005) and the knockout of mitochondrial components via mutations in a mutase (Nakagawa and Sakurai, 2006) and in a PPR (Falcon de Longevialle et al., 2007) involved in the correct generation of mitochondrial transcripts for NAD4 and NAD1, respectively.
Complex II, succinate dehydrogenase, is an enzyme of both the citric acid cycle and the respiratory electron transport chain (Figure 3). It is classically comprised of four subunits: a flavoprotein (SDHI), an iron-sulphur subunit (SDH2) and two membrane anchor subunits (SDH3 and SDH4). Figueroa et al. (2002) reported the successful import of the Arabidopsis SDH polypeptides into isolated plant mitochondria. Notably, while the Arabidopsis SDHI and SHD2 proteins are highly conserved when compared to their counterparts in other organisms, SDH3 and SDH4 share little similarity with non-plant homologues. SDH2 is encoded by a single-copy nuclear gene in mammals and fungi and by a mitochondrial gene in some protists. In Arabidopsis, the SDH2 orthologs are encoded by three nuclear genes, each containing the essential conserved sequences required for function (Figueroa et al., 2001). Subsequently, using blue native-PAGE, Eubel et al. (2003) successfully separated an intact SDH complex from Arabidopsis mitochondrial membranes. This complex contained SDH1-3 as well as four other proteins that co-migrate with the SDH complex. All four co-migrating proteins have been identified by mass spectrometry as proteins of unknown function in the Arabidopsis genome (Eubel et al., 2003; Millar et al., 2004). Thus a novel composition of SDH seems very likely in plants, but to date, no function for the additional subunits has been determined.
Complex III, ubiquinone-cytochrome c oxidoreductase, consists of 10 subunits including the bifunctional matrix processing peptidase proteins, 16 genes encode these subunits in Arabidopsis (Figure 3). In BN-PAGE separations from Arabidopsis mitochondria, most of these subunits have been identified and linked back to a set of mostly single copy genes in the nuclear genome (Kruft et al., 2001; Werhahn and Braun, 2002; Meyer et al., 2008). However, the Arabidopsis homolog of the so-called Hinge subunit first identified in potato (Braun et al., 1994) has yet to be identified as a subunit of the complex in Arabidopsis. One subunit of this complex, cytochrome b (COB), is encoded in the Arabidopsis mitochondrial genome (Unseld et al., 1997).
Complex IV, cytochrome c oxidase, in mammals contains 13 non-identical subunits. Three of these are mitochondrial-encoded and ten are nuclear-encoded. Early purifications from plants found only seven or eight subunits in the plant complex (Peiffer et al., 1990), but more recently, Eubel et al. (2003) and Millar et al. (2004) separated a COX complex containing 10–12 protein bands from Arabidopsis, and in the latter study identified eight proteins homologous to known COX subunits from other organisms and a further six proteins that may represent plant-specific COX subunits. A comparative study of cytochrome c from Arabidopsis and human provides the first systematic analysis of its redox behavior and the kinetics of both their reduction by flavin semiquinones (lumiflavin, riboflavin, and FMN) and oxidation by cytochrome c oxidase (Rodriguez-Roldan et al., 2006). This work shows intriguing differences between the way Arabidopsis and human cytochrome c interacts with cytochrome c oxidase which could be in response to variations between the plant and animal COX structure (Rodriguez-Roldan et al., 2006). Arabidopsis homologs of the yeast metal chaperone Cox19p, involved in cytochrome c oxidase biogenesis, have also been studied (Attallah et al., 2007). AtCOX19-1 is able to restore growth on non-fermentable carbon sources when expressed in a yeast cox19 null mutant. AtCOX19 transcript levels increase by treatment with copper or compounds that produce reactive oxygen species, by leaf wounding and Pseudomonas infection, suggesting a role of this protein in the biogenesis of cytochrome c oxidase in Arabidopsis to replace damaged forms of the enzyme (Attallah et al., 2007).
Alternative oxidase (AOX), which appears to play an antioxidant role in plant mitochondria, is encoded by five genes in Arabidopsis, four AOX1 type and one AOX2 type (Considine et al., 2002) (Figure 3). The expression of the different genes has been shown to be both tissue and development specific (Saisho et al., 1997; Saisho et al., 2001; Thirkettle-Watts et al., 2003). AOX1a is the predominantly expressed isoform in most tissues, with AOX1c expressed in young cotyledons and, more strongly, in floral tissue. AOX1b, 1d and AOX2 are expressed but at much lower levels. Microarray studies have revealed co-expression of AOX1a and NDB2 observed under a number of treatments suggested co-regulation that may be directed by common sequence elements arranged hierarchically in the upstream promoter regions of these genes (Clifton et al., 2005). The stress-induced nature of AOX in a variety of plants is further supported in Arabidopsis as AOX1a and AOX1d are amongst the most stress responsive genes amongst the hundreds of known genes encoding mitochondrial proteins. Analysis of genes on microarrays co-expressed with AOXs from studies of responses to various stress treatments altering mitochondrial functions and/or from plants with altered Aox levels reveal that: 1) AOXs are co-expressed with genes that typically function outside the mitochondrion; 2) that several pathways for AOX induction exist and there is a difference in the magnitude of the induction in each pathway; 3) that the magnitude of induction depends on the endogenous levels of AOX, and 4) that while induction can be oxidative stress-dependent, it can also be oxidative stress-independent depending on the Aox gene member investigated and the tissue analysed (Clifton et al., 2006). An overall role for AOX in re-programming cellular metabolism in Arabidopsis in response to a changing environment encountered by plants is proposed.
Transgenic studies assessing the impact of cold treatment revealed that anti-sense and overexpressing lines for AOX1a showed, reduced or increased leaf area and rosette size, respectively (Fiorani et al., 2005). The observed phenotypes were correlated with the amount of total shoot anthocyanin at low temperature and with the transcription of the flavonoid pathway genes PAL1 and CHS, suggesting AOX plays a role in shoot acclimation to low temperature in Arabidopsis, and that it can function to prevent excess reactive oxygen species formation in whole tissues under stressful environmental conditions but also influence extramitochondrial metabolism through more pervasive effects (Fiorani et al., 2005). In the companion study by Umbach et al 2005 antisense and overexpression lines for AOX1a were used to show the role of AOX in planta in preventing ROS production when the cytochrome pathways was inhibited. A study analyzing the affect of knocking out AOX1a via insertional inactivation has also showed that aox1a plants display a distinct phenotype of accumulating anthocyanins in the leaves under combined light and drought stress. Further analysis of aox1a plants under these conditions revealed there was an increase in non-photochemical quenching, and an increase in ROS that likely originates in chloroplasts (Giraud et al., 2008). Additionally studies from both groups analysing the effect of altering AOX1a in Arabidopsis indicated that the transcript abundance of genes encoding proteins involved in ROS metabolism were changed (Umbach et al., 2005; Giraud et al., 2008). This is also consistent with studies in tobacco (Amirsadeghi et al., 2006) where a lack of AOX results in lower cellular levels of ROS. However the mechanism(s) through which AOX suppresses the synthesis of ROS are still unclear. Although studies in tobacco show that the presence of AOX appears to dampen the production of mitochondrial generated ROS (Maxwell and McIntosh 1999; Yip and Vanlerberghe 2001), the studies in Arabidopsis reveal that changes in transcript abundances for genes encoding proteins not located in mitochondria were greater than alterations in transcripts for genes encoding mitochondrial proteins (Umbach et al., 2005; Giraud et al., 2008). This suggests that AOX may alter ROS equilibrium throughout the cell indirectly by altering REDOX balance.
Rotenone-insensitive NADH dehydrogenases that bypass complex I, are increasingly being studied in Arabidopsis. These alternative NDH-DHs are closely related to the type 2 NAD(P)H dehydrogenases found in bacteria and their activity is easily distinguished from that of complex I by its insensitivity to the complex I inhibitor rotenone. Studies in various other plant species had demonstrated a complex set of NAD(P)H oxidative activities (pointing to at least four different enzymes), induced by various conditions and associated with proteins of varying molecular mass. Using sequence searches based on NDH-DHs identified in yeast and potato, seven putative NDH-DH genes appear to exist in Arabidopsis and these fall into three distinct classes (Michalecka et al., 2003).
Using radiolabelled precursor import experiments, NDA1, NDA2 and NDC1 are considered to be located on the inside of the inner membrane, while NDB1, NDB2 and NDB4 are located on the outside of the inner membrane (Elhafez et al., 2006). Circadian regulation for the NDA1 gene has been suggested (Michalecka et al., 2003), and was later confirmed and clustered with the gene encoding the P-subunit of glycine decarboxylase in microarray analyses, suggesting that NDA1 plays a role in integrating metabolic activities of chloroplasts and mitochondria during photorespiration (Elhafez et al., 2006). These enzymes have very high turnover numbers and can be present in very small amounts despite there significant impact on the respiratory chain (Menz and Day, 1996). However, two proteins, NDB3 and NDB2, have been found in proteomic analyses of mitochondria isolated from Arabidopsis cell cultures (Heazlewood et al., 2004).
Functional studies to confirm that all these genes encode NAD(P)H dehydrogenases are underway in several laboratories. In the first study reported using an Arabidopsis knock-out, the gene encoding NDA1 (At1g07180) was shown to be a rotenone insensitive NAD(P)H dehydrogenase activity that is located on the inside of the inner membrane (Moore et al., 2003). Subsequently NDB2 and NDB4 were found to functionally complemented an E. coli mutant deficient in endogenous type I and type II NADH dehydrogenases (Geisler et al., 2007). Recombinant NDB1 and NDB2 enzymes were also found to bind Ca2+ and under physiologically relevant conditions, the recombinant NDB1 acted as a Ca2+ -dependent NADPH dehydrogenase, while NDB2 and NDB4 were NADH-specific dehydrogenases. The observed activity profiles of the NDB-type enzymes provide a foundation for understanding the mitochondrial system for direct oxidation of cytosolic NAD(P)H in plants and explain many aspects of the observed NADH, NADPH specificity and Ca2+ dependency of NAD(P)H-dependent respiration of plant mitochondria (Geisler et al., 2007).
ATP synthesis
Complex V is the membrane bound F1FO-type H+ ATP synthase of mitochondria which catalyses the terminal step in oxidative respiration, converting the electrochemical gradient into ATP for cellular biosynthesis (Figure 3). The general structure and the core subunits of the enzyme are highly conserved in both prokaryotic and eukaryotic organisms. The structure consists of two distinct parts, the hydrophilic F1 which contains the nucleotide-binding site and the FO which channels protons through the membrane. The plant mitochondrial ATP synthase also displays the classical F1 five subunit structure. Searches of the Arabidopsis nuclear and organellar genomes identify homologs for all five of the F1 subunits. The βsubunit is encoded in a small multigene family of three members on chromosome 5 and these contain over 98% sequence identity at an amino acid level. The FO consists of three subunits in the E. coli enzyme, a, b and c in a 1:2:12 stoichiometry. In mammals and yeast the FO contains a central core of three subunits analogous to the E. coli proteins in a 1:1:9-12 stoichiometry and a series of associated proteins involved in F1-FO interactions and as components of the second or ‘stator’ stalk. A number of associated subunits varies between mammals and yeast. In Arabidopsis, orthologs for subunit 6 and 9 components of the FO core, OSCP and d-subunit peripheral components are present in the genome. Using blue native PAGE (BN-PAGE) and mass spectrometry, Heazlewood et al. (2003b) attempted to characterize the protein components of the F1FO ATP synthase complex and link these directly back to specific gene products in Arabidopsis. In this process, two mitochondrial-encoded ORFs were identified for putative proteins not previously characterized as associated with the ATP synthase complex. These are likely to represent the plant equivalents of the FO components, AL6 (or subunit 8) and b subunit (or subunit 4), and are encoded by orfB and orf25 respectively in the Arabidopsis mitochondrial genome. Thus five subunits of the ATP synthase are mitochondrial-encoded in plants, four being the core components of the FO complex. These identifications were confirmed by the report that orfb in sunflower was ATP8 (Sabar et al., 2003) and identification of ORF ymf39 (an orf25 otholog) as a probable ATP4 subunit in protists (Burger et al., 2003). Subsequently, Eubel et al. (2003) has noted 13 protein bands in ATP synthase separated from Arabidopsis by BN-PAGE, and tentatively identified a putative g subunit (At4g29480). Meyer et al (2008) has identified the highly hydrophobic mitochondrial encoded ATP6 and a number of other peripheral subunits using BN-PAGE followed by a new diagonal SDS-PAGE technique.
Organic acid metabolism
Members of the tricarboxylic acid (TCA) cycle dominate the carbon metabolising enzymes present in plant mitochondria. The number of genes identified as encoding TCA cycle components in Arabidopsis are noted in Figure 3. Several enzymes and associated pathways have attracted considerable attention in Arabidopsis, namely the pyruvate dehydrogenase complex, citrate synthase and isocitrate dehydrogenase and the TCA cycle associated GABA shunt pathway.
The pyruvate dehydrogenase complex uses E1 (2-oxo acid dehydrogenase), E2 (acyltranaferase) and E3 (lipoamide dehydrogenase) enzymes and five cofactors (TPP, CoA, lipoic acid, FAD, NAD) in its catalytic cycle. Active-site coupling is used to catalyse decarboxylation of pyruvate (E1), esterification of aldehydes to CoA (E2) and reduction of NAD to NADH (E3). The regulation of this complex is facilitated by a specific kinase and phosphatase that act on a subunit of E1 to activate or inactivate this primary step in the catalysis. The subunits of the Arabidopsis complex were systematically sequenced in the 1990s. Leuthy et al. (1994 Leuthy et al. (1995) sequenced the E1αand E1βsubunits and Guan et al. (1995) sequenced the first E2 subunit. A smaller single lipoyldomain E2 subunit was identified much latter (Thelen et al., 1999) followed by the E1 alpha subunit kinase (Thelen et al., 2000). Antisense repression of the kinase in Arabidopsis leads to increased mtPDC activity, increased leaf and seed respiration rates and altered vegetative growth with reduced accumulation of vegetative tissues, early flower development and a shorter generation time (Zou et al., 1999; Marillia et al., 2003). This provides direct evidence of the importance of this regulatory site in influencing both plant metabolism and development.
Citrate synthase catalyses the condensation of oxaloacetate and acetyl CoA to form a short-lived intermediate, citroyl CoA, which hydrolyses to form citrate and CoA. The mitochondrial citrate synthase has been purified from pea leaves and shown to consist of a single polypeptide of 50 kDa apparent molecular mass. Full-length cDNAs were isolated from Arabidopsis (Unger et al., 1989) and, based on this sequence, from a variety of other plants (La Cognata et al., 1996). Antisense inhibition in potato leads to male sterility (Landschutze et al., 1995), while over-expression in Arabidopsis allows enhanced growth under low phosphorous conditions, putatively due to enhanced citrate excretion from the roots, which acts as a phosphate chelator (Koyama et al., 2000).
Sequencing of Arabidopsis genes encoding the other TCA cycle enzymes, such as fumarase and isocitrate dehydrogenase, has allowed the generation of antisera to over-expressed proteins that are now used widely in plant research on mitochondria (Behal and Oliver, 1997, 1998). NAD-dependent isocitrate dehydrogenase (IDH) catalyses the oxidation of isocitrate to form 2-oxoglutarate In Arabidopsis five genes encode IDH subunits, two ‘catalytic’ and three ‘regulatory’ subunits according to their homology with yeast. Complementation of yeast idh mutants with the different Arabidopsis IDHs indicate a combination of a single catalytic and regulatory subunit was sufficient to restore acetate growth of the yeast idh double mutant. Four of the IDH genes are expressed in all plant organs, while one gene (At4g35650) was mainly expressed in the pollen (Lemaitre and Hodges, 2006). Knockout of several of the mitochondrial IDHs in Arabidopsis have surprisingly mild phenotpyes especially given that IDH is considered an important enzyme for the synthesis of 2-oxoglutarate that is essential as the primary carbon skeleton for nitrogen assimilation in plants. But while they don't appear to be rate limiting for nitrogen assimilation, their loss does impact on steady state abundance of a broad range of organic acids and amino acids (Lemaitre et al., 2007).
In bacteria, a GABA shunt around the TCA cycle exists, bypassing the steps from 2-oxoglutarate to succinate. However, in plants there has been comparatively little discussion of this pathway. One of its constituents, succinic semialdehyde dehydrogenase (SSADH), was first identified in Arabidopsis (Busch and Fromm, 1999). A detailed analysis of the kinetics of the purified Arabidopsis protein by surface plasmon resonance and fluorescence spectroscopy supported a model of feedback regulation of SSADH, and therefore the entire GABA shunt, by the energy status of the mitochondria (Busch et al., 2000). A number of the proteins of the GABA shunt pathway have been directly identified in Arabidopsis mitochondria by mass spectrometry (Millar et al., 2001b), except for the GABA transaminase. The functional identification of such a transaminase from Arabidopsis was published the following year (Van Cauwenberghe et al., 2002), rapidly followed by the direct identification of this protein in Arabidopsis mitochondria (Sweetlove et al., 2002). Knockout mutation of SSADH in Arabidopsis has a dramatic effect in white light, containing UV-B, under which mutants appear dwarfed with necrotic lesions. Analysis of this mutant suggests that the GABA shunt is required to prevent the accumulation of reactive oxygen intermediates and cell death, especially during environmental stress (Bouche et al., 2003). How it does this is yet to be determined.
Amino acid metabolism
Cysteine metabolism has received significant attention in Arabidopsis mitochondria. Cysteine represents not only an essential amino acid for protein structure and function, but also is an interface point between sulfur and nitrogen metabolism in plants. Through gene sequencing and functional analysis, the Arabidopsis cysteine synthase pathway involving serine acetyltransferase (SAT) and O-acetylserine (thiol) lyase (OAS-TL) had been uncovered. The genes responsible for the mitochondrial pathway have been differentiated from the chloroplast and cytosolic pathways (Noji et al., 1998; Jost et al., 2000). Structural analysis of Arabidopsis mitochondrial SAT has also revealed insights into its C-terminal bifunctional domain involved in both catalysis and the binding of the OAS-TL (Wirtz et al., 2001). Sulfur based metabolism in mitochondria links cysteine to Fe-S cluster formation and possible pathways of cyanide metabolism. A series of researchers have investigated the controversial roles of mercaptopyruvate- and thiosulfate-sulfurtransferases and beta-cyanoalanine synthase in Arabidopsis, providing clear evidence for mitochondrial forms of these enzymes (Hatzfeld et al., 2000; Hatzfeld and Saito, 2000; Papenbrock and Schmidt, 2000). However, their role in vivo still remains unresolved, despite the exploitation of knockout mutants to probe function (Nakamura et al., 2000). One member of the family of sulfurtransferases in Arabidopsis, which catalyze the transfer of a sulfur atom from suitable sulfur donors to nucleophilic sulfur acceptors, has been experimentally localised to mitochondria (Bauer et al., 2004).
Proline accumulation occurs via rapid synthesis from glutamate and can protect plants from osmotic stress. Accumulated proline is rapidly oxidized back to glutamate when osmotic stress is relieved. The first step of this process is catalyzed by a mitochondrial proline dehydrogenase. Studies in Arabidopsis have shown rapid transcript response of proline oxidase to changing osmotic pressure, making it a regulatory link in proline homeostatis in plants (Kiyosue et al., 1996; Verbruggen et al., 1996). A subsequent step in this pathway is catalysed by delta-1-pyrroline-5-carboxylate dehydrogenase. The expression of this mitochondrial enzyme is regulated by osmotic stress in Arabidopsis (Deuschle et al., 2001).
Glycine metabolism, which is central to the photorespiratory cycle, occurs within the matrix of mitochondria. Sommerville and Ogren (1982) highlighted the potential of Arabidopsis mutants to investigate this pathway through the presentation and analysis of a glycine decarboxylase mutant over 25 years ago. Detailed assessment of the coordinated expression of genes for mitochondrial photorespiratory enzymes and their co-expression with Calvin cycle enzymes in Arabidopsis, has helped to elucidate both light responsive promoter elements (Srinivasan and Oliver, 1995) and circadian expression patterns (McClung et al., 2000). Through recent analysis of the genomes of Arabidopsis and rice it appears that the genes for glycine decarboxylase and serine- hydroxymethyltransferase are similarly organized, even in very distantly related plant species, suggesting a strong linkage in both function and inheritance (Bauwe and Kolukisaoglu, 2003).
The intracellular location of branched-chain amino acid catabolism in plants has been controversial, with early evidence suggesting a peroxisomal location (Gerhardt, 1992) as opposed to the well established mitochondrial location in mammals and yeast (Graham and Eastmond, 2002). The three branched-chain amino acids, Val, Leu and Ile, are initially transaminated to their respective branched-chain α-keto acids by the branched-chain amino-transferase (BCAT). This reversible reaction is also the final step of the biosynthesis of these amino acids. A series of BCAT genes are present in Arabidopsis and GFP-targeting studies have shown that separate members are targeted to chloroplasts and mitochondria (Diebold et al., 2002). Following this transamination, the α-keto acids are decarboxylated and esterified to CoA by the branched chain α-keto acid dehydrohenase complex (BCKDC). Early reports suggested BCKDC activity was located in peroxisomes (Gerhardt, 1992), but, when genes for the Arabidopsis prtoeins were sequenced, it was found that each has an N-terminal extension, indicative of mitochondrial targeting (Fujiki et al., 2000). The CoA esters are then oxidised by an acyl-CoA dehydrogenase. An Arabidopsis mitochondrial acyl-CoA dehydrogenase (isovaleryl-CoA dehydrogenase (IVD)), with activity towards both isovaleryl-CoA (from Leu) and isobutyryl-CoA (from Val), has been identified using gene cloning, in vivo targeting studies and biochemical analysis in mitochondria (Daschner et al., 2001). Following this step the three pathways diverge to a series of separate reactions leading to propionyl-CoA in the case of Ile and Val metabolism, and to acetyl-CoA and acetoacetate in the case of Leu metabolism. Molecular analysis in Arabidopsis has been critical in showing the mitochondrial location of this pathway. Work on the induction of BCKDC genes using LUC-constructs linked to their promoters has shown that sugar-starvation drives expression in cell cultures (Fujiki et al., 2002). In this system, induction of the BCKDC gene promoters was influenced by protein kinase and phosphatase inhibitors, indicating the presence of a phosphorylation-based signal transduction pathway in the perception and transduction of the sugar-starvation response (Fujiki et al., 2002). Similar work on 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis has complemented the BCKDC work, indicating the coordinated regulation of the leucine catabolism pathway (Che et al., 2002). Overall, these data suggest that branched-chain amino acids promote their own catabolism, but only when cells are sugar starved. Recently, the mitochondrial components of this pathway were directly identified in Arabidopsis by mass spectrometry, pinpointing the members of gene families responsible for the mitochondrial pathway and also revealing a gap in the pathways for Leu and Ile that are likely to be localized in peroxisome rather than the mitochondrion (Taylor et al., 2003a).
The electron-transfer flavoprotein:ubiquinone oxidoreductase (ETFQO) and electron-transfer flavoprotein (ETF) work in sequence to accept electrons from catabolism of these branched chain amino acids and enter them into the mitochondrial electron transport chain. Analysis of Arabidopsis ETFQO and ETFbeta knockouts revealed a dramatic reduction in their ability to withstand extended darkness, resulting in early senescence and death (Ishizaki et al., 2005; Ishizaki et al., 2006). In dark-treated leaves a significant accumulation of an intermediate of Leu catabolism was observed. A strikingly accumulation of phytanoyl-CoA also imply a novel role for these mitochondrial proteins in the chlorophyll degradation pathway activated during dark-induced senescence and sugar starvation (Ishizaki et al., 2005; Ishizaki et al., 2006).
Biosynthetic Functions
The pathways for the biosynthesis of many vitamins have been elucidated at the molecular level in Arabidopsis revealing the role mitochondria play in the synthesis of folate (vitamin B9), biotin (B7), pantothenate (B5), ascorbate (C), thiamin (B1) and lipoic acid (LA). The production of some of these cofactors is regulated by developmental cues, and perhaps more surprisingly, by environmental signals such as high light and salinity.
Lipoic acid is an essential cofactor in a number of decarboxylating, dehydrogenase reactions and is made in mitochondria in plants. The first plant mitochondrial lipoic acid synthase was identified, cloned and analysed in Arabidopsis (Yasuno and Wada, 1998). The synthesised lipoic acid is fixed to the acyl carrier protein (Shintani and Ohlrogge, 1994) and then transferred to the dehydrogenase complexes by a mitochondrial lipoyltransferase (Wada et al., 2001). Subsequently, an additional Arabidopsis lipoic acid synthase isoform was identified in plastids (Yasuno and Wada, 2002), showing that both mitochondria and plastids contain this cofactor assembly pathway. Recent study of a mutant mapped to the gene of the mitochondrial beta-ketoacyl-[acyl carrier protein (ACP)] synthase (mtKAS), showed reduced lipoylation of glycine dehydrogenase and PDC and OGDC in leaves, but still high levels in roots, suggesting this pathway of lipoate biosynthesis found in leaves may occur by another pathway in root mitochondria (Ewald et al., 2007).
The cofactor biotin is also synthesised in mitochondria. Biochemical work in pea and potato has been complemented by molecular cloning and expression of Arabidopsis components of the plant mitochondrial biotin synthesis pathway (Baldet et al., 1997). This lead to the identification of the Arabidopsis biotin synthase (BIO2) gene (Picciocchi et al., 2001) and the subsequent identification of three other mitochondrial proteins, adrenodoxin, adrenodoxin reductase and cysteine desulfurase, as essential accessory proteins of this biosynthesis pathway (Picciocchi et al., 2003). Attempts to complement a bio2 mutant with a truncated version of BIO2 lacking the region encoding the mitochondrial targeting sequence failed, even with provision of the substrate dethiobiotin (DTB), suggesting biochemical constraints, and the apparent close connection with the mitochondrial Fe-S machinery may account for the reaction being retained within mitochondria (Arnal et al., 2006). The second and third enzymes in the biotin biosynthetic pathway (BIO1 and BIO3) are encoded in adjacent genes and are expressed in both single and chimeric BIO3-BIO1 transcripts. One of the fused transcripts is monocistronic and encodes a bifunctional fusion protein. Biotin biosynthesis in Arabidopsis thus provides an intriguing example of a bifunctional locus that catalyzes two sequential reactions in the same metabolic pathway. This complex locus exhibits several unusual features that distinguish it from biotin operons in bacteria and from other genes known to encode bifunctional enzymes in plants (Muralla et al., 2008).
Thiamine synthesis in plants was thought to be plastid specific, while in yeast it is a mitochondrial process. Identification of the Arabidopsis thiamine synthase gene showed that it encodes a precursor protein that drives dual-localisation of GFP to both chloroplast and mitochondria in vivo (Chabregas et al., 2001). Further analysis revealed two translation products from this transcript, leading to a longer form destined for chloroplasts and a shorter form addressed to mitochondria (Chabregas et al., 2003). However, functional evidence for thiamine synthesis in plant mitochondria is lacking to date, to our knowledge.
Folate is required for formation of the cofactor tetrahydrofolate involved in one-carbon (C1) metabolism. Identification and functional characterization in Arabidopsis revealed that dihydrofolate synthetase (DHFS) is exclusively located in the mitochondria, making this compartment the sole site of synthesis of folate in the plant cell (Ravanel et al., 2001). Several isoforms of enzymes involved in isoprenoid biosynthesis have also been localised to mitochondria in Arabidopsis, including a farnesyl-diphosphate synthase (Cunillera et al., 1997) and a geranylgeranyl pyrophosphate synthase (Zhu et al., 1997). However, the isoprenoid end-products in mitochondria are poorly defined.
Anti-oxidant defence systems
The mitochondrial electron transport chain generates superoxide by reaction of molecular dioxygen with ubiquinol radicals in the inner membrane. To detoxify this superoxide a series of pathways are present in plant mitochondria. Much of the dissection of these pathways has been performed in Arabidopsis. A Manganese-dependent superoxide dismutase (Mn-SOD) is the primary defense in the matrix compartment. Knockdown of this activity in Arabidopsis leads to altered TCA cycle operation and a more oxidised redox poise in the mitochondrial matrix (Morgan et al., 2008). In the intermembrane space, a Cu-Zn-like SOD has been previously reported in yeast mitochondria but has not been confirmed or identified in Arabidopsis to date. The hydrogen peroxide generated by these SODs can be degraded to water via a series of reactions, including—peroxiredoxins (Sweetlove et al., 2002) and an ascorbate-glutathione cycle (Chew et al., 2003a). Knockout of the peroxiredoxin in Arabidopsis leads to a root phenotype under stress conditions induced by CdCl2 or salicylhydroxamic acid (an inhibitor of AOX respiration), but it could be largely compensated for by other peroxidases in plant mitochondria (Finkemeier et al., 2005). The ascorbate-glutathione cycle in mitochondria largely consists of dual-targetted enzymes that form the same cycle in the plastid stroma (Chew et al., 2003a). A matrix localised glutathione peroxidise has been measured in some plant mitochondria (Moller, 2001) but has yet to be characterised in Arabidopsis. Lipid peroxidation by superoxide in mitochondria yields peroxidation products like the lipid aldehyde HNE which can selectively modify and damage mitochondrial proteins (Winger et al., 2005; Winger et al., 2007). Detoxification of lipid aldehydes could be achieved by the thioredoxin-dependent peroxiredoxin (Finkemeier et al., 2005), but this has yet to be proven in the knockout pxn background that this enzyme is a major component in this detoxification pathway.
Activation of programmed cell death
Mitochondria play a key role in the regulation of programmed cell death (PCD), triggering a controlled destruction of the cell in response to environmental stress and internal damage. In animal cells, the central role of the mitochondrion in this process is well established (Newmeyer and Ferguson-Miller, 2003; Bras et al., 2005). Essentially, proteins that activate the PCD process are stored in the intermembrane space of the mitochondria and are released into the cytosol where they activate a complex of other proteins that trigger the cell death pathway. Intracellular signals (such as ROS) cause the opening of a pore in the outer mitochondrial membrane, releasing the mitochondrial proteins into the cytosol and activating the death pathway. The most widely studied of these proteins is cytochrome c, which binds to a cytosolic protein, APAF-1, causing oligomerisation with a protease, pro-cas-pase-9. This sets in motion a protease cascade in which sequential cleavage and activation of other caspases ultimately activates downstream targets such as DNAses that initiate dismantling of the cell (Reed, 2000). Other proteins in the mitochondrial intermembrane space in animals may also be involved in cell death, including an endonuclease (Li et al., 2001), and a flavoprotein called apoptosis activating factor. Upon release from the mitochondrion, these proteins move to the nucleus where, in concert with DNases, they cause chromatin condensation and inter-nucleosomal DNA cleavage.
PCD is a process conserved widely in eukaryotes and the molecular events underlying PCD including the involvement of mitochondria are also assumed to be conserved in plants, albeit with significant differences from those identified in animal cells (Lam et al., 2001). For example, while there is evidence that release of mitochondrial proteins can trigger cell death in plants, the role of cytochrome c may not be as central as it is in animals (Lam et al., 2001). Balk et al (2003), using an in vitro model system, showed that rupture of plant mitochondria caused internucleosomal cleavage of DNA in isolated nuclei, but the active mitochondrial component was identified as an endonuclease. Likewise, Yao et al. (2004) found that cytochrome c release did not always accompany cell death in Arabidopsis. Thus while cytochrome c release may be a convenient marker of mitochondrial outer membrane rupture (Balk and Leaver, 2001; Rikhvanov et al., 2007), it may not be the main player in plant PCD.
The mechanism of release of death proteins from mitochondria has been widely studied in animals but remains a subject for debate. The current consensus is that two mechanisms operate. One involves the formation of a transient pore, the permeability transition pore (PTP), which is a transient complex of the voltage-dependent anion channel (VDAC) on the outer membrane, the adenine nucleotide transporter on the inner membrane, and cyclophilin D in the matrix. Rapid flux of water and solutes through this channel dissipates the membrane potential, uncoupling oxidative phosphorylation from electron flux, and leading to swelling of the matrix, eventually causing rupture of the outer membrane and release of intermembrane proteins (Green and Reed, 1998). Importantly, pore formation is stimulated by thiol oxidants (Fortes et al., 2001), possibly providing a link between redox signalling and PCD, but genetic experiments in mice have cast doubt on the role of this pore in apoptosis (Nakagawa et al., 2005). The second mechanism, very definitely involved in PCD, uses only VDAC when pro-apoptotic members of the Bcl-2 family (e.g. Bax) bind to the channel and modify its permeability (Shimizu et al., 1999). This does not cause swelling of the mitochondrion. The immunosup-pressive drug cyclosporin A (CsA) has been used to discriminate between the two mechanisms. CsA specifically binds cyclophilin D in the matrix, preventing the formation of the PTP and, thus, the release of cytochrome c (Halestrap and Davidson, 1990).
The Bcl-2 family of proteins is critically important in controlling cell death. Some Bcl2 proteins promote cell survival (Bcl-SL, Bcl-2,) while others promote cell death (Bax, Bak). A homologue of a Bax inhibitor protein (Bl-1) has been found in plants (Kawai-Yamada et al., 2004) and Arabidopsis Bl-1 suppresses cell death triggered by expression of Bax in protoplasts. Further, it has been shown that the cdf1 gene from Arabidopsis causes Bax-like cell death when expressed in yeast (Kawai-Yamada et al., 2005). Direct genetic homologues of mammalian Bcl-2 proteins have not been identified in the Arabidopsis genome or those of other plants, but it is possible that evolutionarily divergent, but structurally conserved, proteins that fulfil the same function as Bcl-2 proteins in mammals, do exist in plants. On the other hand, there is reasonably good experimental evidence that a permeability transition pore can be induced in mitochondria from various plants (Fortes et al., 2001; Arpagaus et al., 2002). Whether it contains the same components as that in animals (see above), has not been proven yet, but sensitivity to cyclosporin A has been observed in some cases (Contran et al., 2007). In animals, the toxic lipid ceramide has been shown to trigger apoptosis in cells. This compound has also been implicated in the permeability transition in plant mitochondria and may be involved in the hypersensitive response of plants to pathogens (Townley et al., 2005). It is also possible that release of death proteins by mitochondrial swelling and subsequent outer membrane rupture could be mediated by other proteins such as the K+ATP channel (Casolo et al., 2005), but it should be borne in mind that, whatever the mechanism, permeability transition pore formation has yet to be conclusively demonstrated to lead to PCD in plants.
Mitochondria appear to be integral to PCD in plants in a more indirect fashion also, since their electron transport and related metabolism influence the susceptibility of plant cells to induction of PCD. In particular, AOX seems to help prevent PCD. In tobacco cells lacking AOX, PCD could be triggered by the addition of cysteine, which suppressed expression of cytochrome chain components, but in cells containing Aox, this did not occur (Vanlerberghe et al., 2002). These same transgenic tobacco cells were more sensitive to various death-inducing compounds (hydrogen peroxide, salicylic acid and a protein phosphatase inhibitor) and underwent a PCD-like cell death (Robson and Vanlerberghe, 2002). AOX may ameliorate PCD by maintaining a functional electron transport chain when the cytochrome pathway is compromised, thereby preventing over-reduction of respiratory chain components involved in ROS formation. More recently, Kim et al. (2006b) showed that silencing of the gene encoding the mitochondrial-associated hexokinase induced PCD, while, over-expression of the enzyme bestowed resistance to PCD. Since addition of purified hexokinase to isolated mitochondria prevented cytochrome c release and loss of membrane potential, it seems that it may act to prevent permeabilisation of the mitochondrial outer membrane. It is probably no coincidence that hexokinase can be tethered to the outer membrane by an association with VDAC. This role of hexokinase is intriguing, since it also participates in sugar-sensing in Arabidopsis. Thus the mitochondria are well equipped to monitor cell carbon and energy status and feed this information into the cell death pathways.
ANALYSIS OF MITOCHONDRIAL PROTEIN COMPOSITION
Predicting the Mitochondrial Proteome
The accurate, sequence-based prediction of the subcellular localisation of a protein has been a goal of researchers for a number of years. There are a variety of subcellular prediction programmes that can be used for this endeavour, falling into two groups based on their prediction method. The programmes PSORT (Nakai and Horton, 1999), iPSORT (Bannai et al., 2002) and MitoProt II (Claros and Vincens, 1996) utilise defined experimental rules to determine protein localisation while the programmes TargetP (Emanuelsson et al., 2000), and SubLoc (Hua and Sun, 2001) use computer-learning techniques to drive their predictions. Using a variety of training sets, the authors of these prediction programmes report relatively high levels of success in their overall prediction rates, typically ranging from 80 to 90%. Typically an end-user provides a single or small number of protein sequences for analysis, and the outputs are commonly attributed an 80–90% accuracy in the literature. The fully annotated nuclear genome of Arabidopsis has allowed a direct analysis of the targeting prediction of a whole plant genome. Using TargetP as a model predictor, the Arabidopsis Genome Initiaitive provided a series of large numbers representing the likely proteomes of different compartments in the cell. The mitochondrial set contained some 2897 members (The Arabidopsis Genome Initiative, 2000). However, it has been very difficult to define a pertinent but rigorously unbiased test to evaluate the performance of the various predictors proposed, and more recent programs claim better results than TargetP, with some justification. Predotar (Small et al., 2004) uses neural networks while LOCtree (Nair and Rost, 2005) and MultiLoc (Hoglund et al., 2006) primarily use support vector machines. Comparison of different prediction programmes shows that while many predict sets of 3000–4000 proteins as destined for mitochondria, these programmes do not overlap highly on the actual set of proteins predicted. Common sets between any two prediction programmes are normally 1500–2000, and between any three prediction programmes are typically only 500–1000 in size (Richly et al., 2003; Heazlewood et al., 2004). This range and disagreement between prediction tools, many of which are based on very similar set of known-location proteins either for defining rules or as training-sets for machine-learning, raises significant doubt about the current accuracy of this approach. Only about 40% of the experimentally identified Arabidopsis mitochondrial proteins are predicted to be mitochondrial by all the main bioinformatic prediction tools (Target P, Predotar). This subset is heavily skewed to energy and metabolism protein functional categories (Figure 4), largely reflecting the training sets used to build the prediction tools. As noted earlier, the dual-targeted subset of proteins in Arabidopsis that enter both chloroplasts and mitochondria, are skewed in the opposite direction, away from metabolic functions and towards the low abundance functional groups of DNA, RNA and protein synthesis/regulation and organelle stress defense (Figure 4). A comparative analysis of different programmes that calculated their false positive and false negative rates against an experimentally defined set of mitochondrial proteins has been used to calculate a predicted mitochondrial proteome for Arabidopsis of about 2000 proteins (Millar et al., 2006). None of the current tools predict integral membrane proteins with any accuracy, the predictions being based on the characteristics on the cleavable N-terminal targeting peptides that are a feature of most, but not all, soluble matrix proteins. The subsets of Arabidopsis proteins predicted to be mitochondrial by a wide range of popular prediction programmes can be accessed via the SubCellular Protein Location in Arabidopsis Database (SUBA, http://www.suba.bcs.uwa.edu.au)
Figure 4.
Comparison of the functional group breakdown of the Arabidopsis mitochondrial proteome (543) with the subset of proteins containing clear N-terminal mitochondrial targeting sequences (207) and the set of experimentally determined dual-mitochondrial and chloroplast targeted proteins (35).
Functional breakdown was obtained from SUBA. Subset with N-terminal sequences defined as those predicted to be mitochondrial by both Predotar and TargetP. Dual-targeted sets were defined predominantly by GFP-tagging and were all gained from published literature searches.
Isolation of Plant Mitochondrial Proteins
The principles of isolation, fractionation and electrophoretic separation of plant mitochondrial proteins have been reviewed several times over many years (Douce et al., 1972; Neuburger, 1985; Millar et al., 2001a). Importantly, the introduction and refinement of sucrose and Percoll density gradients has allowed the extraction of high purity mitochondrial samples from plant tissues (Douce et al., 1972; Neuburger et al., 1982; Day et al., 1985). Arabidopsis has dominated recent studies of the plant mitochondrial proteome. Combining differential centrifugation and Percoll gradient density gradients allows isolation of mitochondria with only a few percent contamination by cytosol, peroxisomes, plastids and other membranes (Kruft et al., 2001; Millar et al., 2001b; Werhahn et al., 2001; Heazlewood et al., 2004). A recent addition to mitochondrial isolation protocols has been the use of free-flow electrophoresis to separate mitochondria from other organelles in cell extracts based on the specific surface change of the organelles (Eubel et al., 2007).
Experimental display and identification of proteins
A range of techniques have been used to profile the protein contents of these mitochondrial preparations. Denaturing 1D SDS-PAGE and 2D IEF/SDS-PAGE followed by mass spectrometry, N-terminal sequencing and immunoreactivity, have been used to identify many of the well known abundant proteins of this organelle (Kruft et al., 2001; Millar et al., 2001b; Millar and Heazlewood, 2003). Additionally, blue native (BN) PAGE and 2D BN-PAGE/SDS-PAGE have been used to look at native complexes and their constituents in Arabidopsis mitochondria (Werhahn and Braun, 2002), providing new insights to both supercomplex formation in mitochondrial membranes (Eubel et al., 2003) and the components of electron transport chain and import apparatus complexes in plants (Werhahn et al., 2001; Heazlewood et al., 2003a; Heazlewood et al., 2003b; Werhahn et al., 2003; Millar et al., 2004; Meyer et al., 2008). Affinity purification from mitochondrial lysates has also revealed the set of ATP binding proteins in Arabidopsis mitochondria (Ito et al., 2006). Nongel proteomics relying on microbore HPLC separation and mass spectrometry have also been used to further analyse the contents of Arabidopsis mitochondria (Heazlewood et al., 2004). Together these approaches have identified a wide range of proteins involved in many aspects of mitochondrial function (Figure 4).
Experimental approaches to the mitochondrial proteome can also be error prone. Sub-cellular proteomics is only as good as the purity of the preparation of the sub-compartment. The sensitivity of mass spectrometry means that contamination of the order of 10% by other compartments will lead to the identification of a large number of contaminants. In earlier times, such contamination levels were considered reasonable for many applications in plant mitochondrial research, such as assessment of respiratory function and immunoreaction studies (Douce et al., 1972; Neuburger, 1985; Millar et al., 2001a). But contamination of not much more than 1% is required to avoid a large number of false-positive assignments in mitochondrial proteome analysis (Kruft et al., 2001; Millar et al., 2001b; Sickmann et al., 2003; Taylor et al., 2003b). Despite this, relative to algorithm prediction, the size and nature of the errors in experimental identifications are both more definable and typically much smaller in magnitude. Further, the advent of new approaches to further purify mitochondria (Eubel et al., 2007) and the use of quantitative tools to measure the enrichment of mitochondria and loss of contaminating proteins through the separation process (Dunkley et al., 2004; Dunkley et al., 2006; Eubel et al., 2007; Hartman et al., 2007) can further reduce false-positive calls by mass spectrometry.
To date, nearly 550 non-redundant proteins have been identified in Arabidopsis mitochondrial samples by mass spectrometry (Heazlewood et al., 2005; Heazlewood et al., 2007) (Figure 4). This analysis has involved both peptide mass fingerprinting and tandem MS/MS spectra for pattern matching to predicted peptide spectra from Arabidopsis genome sequence. Broadly, this analysis has provided three things to mitochondrial research in plants. Firstly, it provides detailed profiles of the putative functions and biochemical operations of mitochondria in plants. This is vital for understanding the full roles of these organelles in plants and is a prerequisite for comparative analysis with the functions of the same organelles in other eukaryotic lineages. Secondly, specific gene for protein matches allow forward genetics to be undertaken with a mitochondrial focus and reverse genetic results to be understood in the context of the protein destination of mapped gene products. Finally, we now have a genomic context for identified mitochondrial proteins within complex gene families and we can analyse global expression data (microarray, MPSS, SAGE) from a organelle destination perspective. Full lists of the Arabidopsis mitochondrial proteome can be accessed via the SubCellular Protein Location in Arabidopsis Database (SUBA, http://www.suba.bcs.uwa.edu.au)
The physical attributes of mitochondrial proteins identified solely by isoelectric focusing followed by SDS-PAGE, often appear skewed compared to those of total proteomes, due to an inherent inability to detect large and small proteins, and low detection rates of basic, hydrophobic and low abundance proteins. Utilisation of LC-MS/MS has greatly aided in the identification of large (>80 kDa) and hydrophobic proteins (Heazlewood et al., 2004). The current set of over 550 identified Arabidopsis mitochondrial proteins cover an isoelectric point range of 4 to 11, a molecular mass (kDa) range of 6 to 428, and a GRAVY range of −1.6 to +1.0 (GRAVY; Grand Average of hydropathicity. -GRAVY=hydrophilic, +GRAVY=hydrophobic). The representation of proteins in EST databases allows an assessment of transcript abundance and thus probable protein abundance for different gene products. Most interestingly, there is clear evidence of identification of proteins that are poorly represented in EST sequencing programmes. This strongly suggests that a significant depth of the proteome has been uncovered by analysis in Arabidopsis to date, identifying key low abundance components. Many of these are in important areas of electron transfer processes, regulation of enzyme function and signalling.
It is also interesting to compare the LC-MS/MS proteomes of human (Taylor et al., 2003b), yeast (Sickmann et al., 2003) and Arabidopsis. This provides an overview of the functional characteristics of mitochondria in these widely different model organisms. At least 20% of these proteomes are comprised of proteins whose function is currently unknown. These unknown sets are quite distinct in the three eukaryotes (Karlberg et al., 2000; Richly et al., 2003; Heazlewood et al., 2004) and, therefore, presumably have different functions in these organisms.
GFP and in vitro import as tools for proteome confirmation
The use of fluorescence of epitope tagged constructs for in vivo visualisation of protein targeting has been widely used in Arabidopsis. This more direct method of testing a particular product of interest complements the shot-gun approach of mass spectrometry sequencing of proteomes. Traditionally, this tagging approach has been restricted to small numbers of proteins, especially dual targeted proteins (Peeters and Small, 2001; Silva-Filho, 2003), a variety of higher throughput studies have commenced in model organisms including yeast (Ross-Macdonald et al., 1999; Kumar et al., 2002) and, more recently, Arabidopsis (Hilson et al., 2003). A variety of groups have used GFP-constructs to confirm localisations predicted from either bioinformatic programmes (Elo et al., 2003) or defined by mass spectrometry in proteomic analyses (Chew et al., 2003a; Chew et al., 2004). However, the trend towards using GFP-chimeric location as the only means to determine compartmentalisation is fraught with potential errors. Firstly, co-localisation with mitochondria does not necessarily indicate that a protein is imported into the organelle and may lead to false positives. For example, glycolytic enzymes were localised to mitochondria by GFP tagging, and mass spectrometry indicated mitochondrial localisation, but these are, in fact attached to the outer surface of mitochondria and not imported (Giege et al., 2003). Secondly, although GFP localisation is often used with proteins that contain N-terminal targeting signals, it is not known if proteins that contain internal or C-terminal targeting signals can function with such fusions (Lee et al., 1999) and this may thus lead to false negatives. Even with proteins with clear N-terminal targeting signals, sub-cellular localisation with GFP fusions does not always give the complete picture. Glutathione reductase is a dual targeted protein to mitochondria and plastids and although the targeting signal for pea glutathione reductase can target phosphinothricin acetyl transferase to mitochondria and plastids, it cannot target GFP to mitochondria (Chew et al., 2003b). In contrast, the Arabidopsis glutathione reductase can target GFP to both organelles (Chew et al., 2003a). This suggests that the cargo protein to be transported can affect targeting ability for a subset of targeting signals. Further, it has been shown that the ability of three different dual targeting signals to direct proteins to a particular location is dependent on the nature of the mature protein (Zhou and Weiner, 2001; Rial et al., 2002; Chew and Whelan, 2003; Dessi et al., 2003). All three signals were able to target proteins to mitochondria or chloroplasts, or both, but the location depended on the nature of the passenger protein. The characterisation of the yeast mitochondrial proteome also demonstrated that proteins from various mitochondrial compartments were transported to other cellular organelles when tagged, but only to mitochondria when “un-tagged” (Sickmann et al., 2003). In the case of GFP, such problems may arise because of its ability to dimerise. It is possible that dimerisation may slow down or inhibit import into mitochondria and thus overexpressed protein can be mis-targeted or sequestered elsewhere in the cell. Such a situation is readily observed in Arabidopsis where “novel” flourescesent organelles have been identified as ER located GFP aggregates (Hawes et al., 2001; Brandizzi et al., 2002). The use of in vitro uptake assays can be used in conjunction with direct proteomics and in vivo approaches to confirm localisation (Rudhe et al., 2002). An added advantage of this approach is that it can be readily used to determine intra-organelle location of proteins (Chew et al., 2003a). However, care must be taken to use appropriate controls with in vitro assays, as protease resistant fragments, by-pass import or mis-targeting can also occur with these systems (Whelan and Schleiff, 2004). Full lists of the Arabidopsis proteins localised to mitochondria using GFP fusions can be accessed via the Sub-Cellular Protein Location in Arabidopsis Database (SUBA, http://www.suba.bcs.uwa.edu.au)
Dual-targeting of proteins to mitochondria and other organelles
Dual targeting of proteins to mitochondria and chloroplasts was first reported for pea glutathione reductase but the availability of the Arabidopsis genome has allowed the identification of many more dual targeted proteins (Peeters and Small, 2001). Dual targeting is potentially a powerful means of coordinating organelle biogenesis because it acts as a feed forward mechanism, whereby signals from one organelle can induce protein expression in another organelle. Because metabolic pathways, as well as stress response pathways, encompass several organelles, such a communication network allows a strategic pre-positioning of resources and links biogenesis of one organelle to that of another. The dual-targeting of proteins, together with retrograde signalling to the nucleus, provide effective cross-talk between organelles. Although dual targeting is best characterised for mitochondria and plastids, examples for dual located proteins are know for the cytosol and mitochondria, mitochondria and endoplasmic reticulum, and mitochondria and peroxisomes (Silva-Filho, 2003; Whelan and Schleiff, 2004). The mechanisms that direct dual targeting have been reviewed elsewhere (Small et al., 1998). The relatively large number of proteins shown to be dual targeted in Arabidopsis underscores how important this form of inter-organellar communication is in plants. Enzymes involved in DNA replication and repair, RNA transcription and translation, lipid synthesis, and various metabolic pathways (including enzymes of the ascorbate-glutathione cycle), illustrate the fundamental nature of the proteins that are dual targeted (Figure 4).
COORDINATION OF NUCLEAR AND MITOCHONDRIAL GENE EXPRESSION
The regulation of nuclear located genes encoding mitochondrial proteins can be broadly divided into anterograde and retrograde signalling pathways (Leister, 2005). Anterograde regulation is a top-down approach, where factors external to the organelle such as environmental and or developmental cues affect the expression of genes encoding mitochondrial proteins. Retrograde signals are those that originate within the organelle, these can effect gene expression by their own independent mechanisms but can also interact with anterograde signals to modify gene expression and thus affect the metabolic circumstances of the cell.
Coordination during development
The expression of several nuclear-encoded mitochondrial genes has been examined during development in Arabidopsis. As is evident in other plant species, there is a general increase in these transcripts in inflorescence organs. The expression of three complex I genes and the elements controlling this expression have been extensively studied. The 55 kDa NADH binding subunit, the 22 kDa PSST subunit and a 28 kDa FeS protein all display a common theme. Their promoters are organised as bi-partite structures with the region immediately upstream of the transcriptional start site, approximately −100 bp, responsible for constitutive expression throughout the plant. Upstream of this region is a strong enhancer region, approximately −100 to −250, which drives strong expression in anthers and pollen. A pollen box motif, with high sequence identity to one found in tomato, is located in all three promoters. Placement of the pollen box region in front of a disarmed 35 CaMV promoter resulted in a gain of function with strong expression in anthers and pollen indicating that this region contained all the elements required for the high level of anther and pollen specific expression (Zabaleta et al., 1998; Binder and Brennicke, 2003). Further investigations indicated that this region could bind a response regulator ARR2, which is part of a two-component signalling system that contains a sensor kinase and a response regulator (Lohrmann et al., 2001). The response regulators in Arabidopsis have been classified into A class that respond to cytokines and B class that do not. ARR2 falls into the class B and has been shown to bind within the region of the pollen box at two sites. The perturbation of complex I either by direct inhibition with rotenone or by knock-out mutants results in an increase in mitochondrial biogenesis or an inability to respond to cold stress respectively (Lee et al., 2002; Lister et al., 2004). Additionally, an increase in the expression of nuclear encoded complex I genes caused by mitochondrial dysfunction induced by engineering male sterility in Arabidopsis, points to it being an important sensor for mitochondrial function; coordinating gene expression for mitochondrial and non-mitochondrial genes (Gomez-Casati et al., 2002).
It has been proposed that site II elements (5′ TGGGCC/T 3′ or reverse complement) may play a role in co-ordinating the expression of nuclear located genes encoding mitochondrial proteins with cell growth and division (Welchen and Gonzalez, 2005, 2006). Site II elements were first characterised to be function in the promoter of the proliferating cell nuclear antigen (PCNA), c cyclin gene and in several promoters of genes that encode proteins of the cytosolic located ribosome. Site II elements bind to TCP proteins (Teosinte binding 1 cycloides PCF), a plant specific family of transcription factors encoded by 24 genes in Arabidopsis (Kosugi et al., 1995; Tremousaygue et al., 2003; Navaud et al., 2007). Site II elements are over-represented in the promoter regions of genes encoding proteins located in the mitochondrial electron transport chain (Welchen and Gonzalez, 2006). Functional analysis indicates that site II elements play some role in the expression of AOX1c (Ho et al., 2007), Cytc-1 and genes encoding several subunits of the cytochrome c oxidase complex (Welchen and Gonzalez, 2005; Gonzalez et al., 2007). However site II elements are diverse in nature, they often occur in pairs in various orientations and can bind up to 24 potential TCP proteins. Therefore, it cannot be concluded that Site II elements play a role in co-regulating the expression of nuclear genes encoding mitochondrial proteins, albeit they likely play some role in the expression of some genes encoding mitochondrial proteins. Notably, the reported co-expression of genes encoding proteins involved in oxidative phosphorylation did not correlate with the presence of site II elements (Gonzalez et al., 2007).
Studies of the expression of components of the respiratory chain complexes, metabolite carriers, branched chain amino acid catabolism pathway, the photorespiratory pathway and the pyruvate dehydrogenase complex, all show that nuclear-encoded mitochondrial protein gene expression responds to various metabolic signals. While components of the respiratory chain such as cytochrome c and cytochrome c oxidase are induced by an increase in sucrose or light (Ohtsu et al., 2001; Figueroa et al., 2002; Welchen et al., 2002; Curi et al., 2003), enzymes involved in branched chain amino acid catabolism are induced by low sugar levels (Fujiki et al., 2001; Fujiki et al., 2002). Circadian, tissue and developmental parameters all affect the expression of various genes with different members of small gene families responding differently, either in terms of the magnitude of inductions, or in varying spatial or developmental responses (McClung et al., 2000; Drea et al., 2001; Lutziger and Oliver, 2001; Lu et al., 2003). A good example of this is seen with the two genes that encode basic amino acid carriers on the inner mitochondrial membrane (Catoni et al., 2003b; Catoni et al., 2003a; Hoyos et al., 2003). AtmBAC1 is prominently expressed in seedlings where it likely plays a role in mobilising amino acids that act as an important nitrogen store that is crucial in germination. In contrast AtmBAC2 is more prominently expressed in older tissues and flowers.
Coordination during environmental stress
The availability of large microarray datasets allows a detailed analysis of which genes encoding mitochondrial proteins are responsive to environmental stresses. Direct inhibition of the mitochondrial electron transport chain with antimycin A, an inhibitor of the cytochrome bc1 complex, only induced the expression of cytochrome c and no other components of the cytochrome chain, although AOX was induced (Yu et al., 2001). In this study, which analysed changes in expression of greater than 11,000 genes when plants were treated with antimycin A, it was observed that many of the changes in gene expression clustered with various biotic and abiotic stresses, such as aluminium and cadmium stress, viral infection and the cell death response induced by hydrogen peroxide treatment. This may indicate that the plant stress response is at least in part mediated via mitochondria involving unidentified mitochondrial-nuclear communication pathways. More in depth analysis of larger datasets reveal that overall transcripts encoding subunits of cytochrome electron transport chain are not responsive to various stresses, while genes encoding AOX or NDs are highly responsive (Clifton et al., 2005; Clifton et al., 2006).. Upon a variety of stress treatments it is primarily AOX1a and NDB2 that are increased in abundance (Clifton et al., 2005) (Figure 5). The induction of both AOX1a and NDB2 under a variety of conditions strongly suggests they are co-regulated, but this has yet to be experimentally verified (Clifton et al., 2006).
Figure 5.
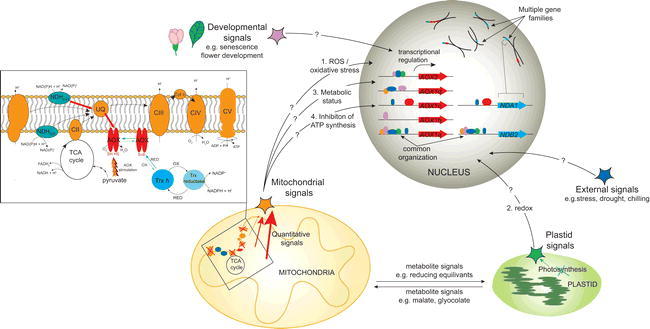
Overview of the signalling pathways that regulate expression of AOX genes in Arabidopsis.
This diagram depicts the potential signalling pathways that influence the expression of AOX in a gene-specific manner. Signals originating within the cell from mitochondrial or plastidic function interact with external signals from organ, developmental or environmental stimuli to activate expression of AOX. These inputs are processed via the promoters of genes where transcription factors involved in these signal transduction pathways converge to activate gene expression. The components involved in transducing the signals which ultimately result in altered expression are unknown. These pathways may interact to affect the magnitude and/or Aox gene induced. Abbreviations: CI = complex I, CII = complex II, CIII = complex III, CIV = Complex IV, CV = Complex V, AOX = alternative oxidase, NDH = alternative type II NAD(P)H dehydrogenase, INT = internal, EXT = external, ROS = reactive oxygen species.
A number of other genes encoding mitochondrial protein are also up-regulated in transcript abundance in response to stress. A protein with unknown function encoded by At2g21640 has been defined as one of five genes that can be considered as hallmarks of oxidative stress (Gadjev et al., 2006). A number of other genes encoding small heat shock proteins and inner membrane proteins belonging to the mitochondrial carrier family are also responsive to a wide variety of stresses (Clifton et al., 2006). As yet experimental evidence that any of these proteins are essential for a normal stress response, including AOX1a and NDB2, has not been reported.
Signalling pathways (mitochondrion-nucleus signalling, anterograde and retrograde pathways)
One over-arching question concerning mitochondrial biogenesis is, how is its timing coordinated with other organelles in the cell and how it is controlled? While there are many possible ways of coordinating organelle biogenesis in cells, currently three potential modes have been elucidated: retrograde regulation, dual targeting of proteins to various locations, and common transcription responses to external stimulae.
Retrograde regulation of gene expression, in the context of organelle biogenesis, refers to regulation of nuclear gene expression by metabolic changes or signals originating in the organelle. This mode of regulation was initially elucidated by Butow and colleagues in yeast using mitochondrial citrate synthase mutants, but is thought to occur in all eukaryotes (Liao and Butow, 1993). Retrograde regulation is well described for plastids in Arabidopsis where several GUN mutants (genomes uncoupled) have been created, which disrupt the normal communication pathway. Some of the molecular components that mediate this signalling have been identified (Larkin et al., 2003; Strand et al., 2003; Koussevitzky et al., 2007; Woodson and Chory 2008). Several reports also indicate that retrograde regulation plays a role in controlling synthesis of mitochondrial proteins in plants. The addition of mitochondrial electron transport chain inhibitors or various metabolites (notably citrate), or the imposition of environmental and oxidative stresses, to cells or tissue pieces, results in the induction of a number of nuclear genes encoding mitochondrial proteins (Vanlerberghe and McLntosh, 1996). The induction of these genes is blocked by protein kinase or phosphatase inhibitors, depending on the genes and signals involved (Vanlerberghe and McLntosh, 1996; Djajanegara et al., 2002; Vanlerberghe et al., 2002). These studies have largely been performed with tobacco or soybean but are now being extended in Arabidopsis. Studies in Arabidopsis, using the AOX1a promoter driving expression of luc, confirmed biochemical and molecular studies that induction of AOX1a takes place via a number of signal transduction pathways (Zarkovic et al., 2005), but to date none of these components has been identified (Figure 5). Further analysis of the AOX1a promoter identified a 93 bp region that was necessary for induction by a variety of treatments (Dojcinovic et al., 2005). This region contains several sequence elements with the potential to bind transcription factors associated with various stress responses in plants, but as yet no specific transcription factors have been identified.
Several lines of evidence specifically point to complex I of the mitochondrial chain as a potentially important component for the retrograde signalling pathway in plants. Mutations in mitochondrial-encoded subunits of complex I in tobacco induce the expression of antioxidant enzymes throughout the cell, but importantly do not seem to alter the redox state of reduced compounds such as glutathione or ascorbate (Dutilleul et al., 2003). Likewise, addition of rotenone, an inhibitor of complex I, to Arabidopsis cell cultures induces the expression of AOX and NDs as might be expected (Clifton et al., 2005). In addition, the expression of many components of the mitochondrial import machinery and respiratory chain assembly pathway are also induced by rotenone (Lister et al., 2004). Thus in some manner, the impairment of complex I signals changes in mitochondrial biogenesis and alters cell redox balance in Arabidopsis. Additional evidence for the role of complex I in retrograde signalling in Arabidopsis comes from a study where mutants with reduced cold stress response were analysed. One mutant, frostbite1, was identified as an 18 kDa Fe-S protein of complex I (Lee et al., 2002). These plants have elevated levels of ROS but the 18 kDa complex I subunit itself is not induced by cold or other stresses.
Although the means of retrograde signalling is not known, it is tempting to propose the involvement of ROS or alteration of ADT/ATP and NAD(P)H/NAD(P)+ levels in the cell or the mitochondrion (Figure 5). The induction of alternative NDs allows the oxidation of NADH by mitochondria to continue, but with less ATP produced. This has the potential to alter ADP/ATP ratios that could trigger a signalling cascade. Alternatively, inhibition of complex I may lead to increases in matrix NAD(P)H/NAD(P)+, since the NDs have lower affinity for NAD(P)H. Another complication is the demonstration that the activity of the terminal enzyme in ascorbic acid synthesis, galactono-1,4-lactone dehydrogenase, is dependent on complex I activity (Millar et al., 2003). Thus any perturbation of complex I function could affect the synthesis of ascorbate and this would likely have a range of diverse affects. Mutants of ascorbic acid synthesis in Arabidopsis display altered expression of defence genes and increased synthesis of abscisic acid and associated signalling pathways (Pastori et al., 2003).
Coordination of protein synthesis in different organelles may also be achieved by common responses to external stimulae. An obvious example of this is the regulation of the expression of some respiratory genes by light. The most widely studied mitochondrial proteins that respond to light are the different subunits of the glycine decarboxylase complex. Despite its localization within the mitochondria, glycine decarboxylase is a photosynthetic enzyme and its expression, therefore, is controlled in much the same way as other photosynthetic enzymes. Glycine decarboxylase is present at low but detectable levels in all plant tissues (as it is in animal tissues) and is involved in the metabolic interconversion of one, two, and three carbon intermediates. This contrasts with the situation in green leaf tissues where it is the predominant enzyme in the matrix of mitochondria comprising nearly half of the total soluble protein in these organelles (Oliver et al., 1990). The light dependent expression of the genes for glycine decarboxylase shows strong control at the level of mRNA. During the transition from etiolated to green tissue in Arabidopsis, there is a lag of several hours before the level of mRNA for the H-protein changes and then the levels of mRNAs for the H-protein and the small subunit of Rubisco increase in parallel (Srinivasan and Oliver, 1995). Run-on transcription (Srinivasan et al., 1992) and promoter::GUS fusion expression experiments (Srinivasan and Oliver, 1995) showed that most of this increase in mRNA is controlled at the transcriptional level. The light dependent expression of the glycine decarboxylase genes appear to involve many of the same phytochrome, blue light, and chloroplast to nucleus communication events that are important for the expression of other photosynthetic genes. Analyses of the promoters for the H-protein (Srinivasan and Oliver, 1995) showed, in addition to other light responsive elements, a putative GT box. Deletion of the DNA region containing this element caused loss of both light-dependence and tissue specificity of H-protein expression. At least two proteins have been shown to bind to this regions of DNA (Oliver and Raman, 1996) including an unusual MYB-type factor whose binding has been partially characterized (Abbaraju et al., 2002). Similar elements have been identified in the promoter region of the T-protein (Vauclare et al., 1998). The L-protein makes an interesting exception to this rule because this protein is also important in the pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, and branched α-keto acid dehydrogenase complexes in plant mitochondria (Bourguignon et al., 1996). In order to fill this multiple role the regulation of this protein is more complex and in Arabidopsis there are two highly similar genes for the L-protein (Lutziger and Oliver, 2001). Although the genes are interchangeable enzymatically, demonstrated by disrupting one of the genes with a T-DNA insertion, one is expressed at low levels in all tissues while the other is expressed at high levels in green tissue in the light.
FUTURE MITOCHONDRIAL RESEARCH IN ARABIDOPSIS
With the increased data through genomic analysis and the increasing knowledge of the mitochondrial proteome, the development of new methods for the analysis and dissemination of this information is essential. Almost all the current generation of targeting prediction tools were developed before the recent influx of new experimental data from proteomics and high- throughput localisation by GFP tagging. It would be timely to retrain some of the more successful tools with the much larger and more representative data sets now available.
As plant mitochondrial research is advanced through application of new technologies such as proteomics, deep sequencing of mRNA and smRNA populations and advanced fluorescence imaging techniques, a series of biology questions can be asked of these data streams to advance our understanding of mitochondria – both in terms of their history and origins, and in terms of their regulation today.
Much of our current understanding of the mitochondrial proteome is qualitative, based on presence or absence. As this analysis becomes deeper we will continue to learn about new low-abundance components involved in specific features of plant mitochondria, such as genome structure, maintenance and inheritance, organelle transcription, post-transcriptional processing and translation. Determining the link between transcript abundances (inferred from deep sequencing, microarray data or EST pool sizes) and protein abundances can provide examples of post-transcriptional regulation of nuclear-encoded mitochondrial gene expression. Linking functional categorisation and protein abundances will also allow us to map biochemical pathways and model fluxes of the whole mitochondrial metabolic network. The development of large arrays of mutant lines with gain or loss of function phenotypes will aid in the understanding of these pathways. It should also be noted that plant mitochondria are not static entities. The overall proteome is a dynamic set of proteins that change over time, between different cells and tissues, and the genes primarily responsible for their form and function may be differentially expressed, both in time and space in plants. Analysing these dynamics will allow us to understand the different roles played by plant mitochondria in different environments, in different cell architectures, and in different metabolic backgrounds –such as leaves vs roots vs flowers, monocot vs dicot, oil vs starch accumulators, or C3 vs C4 plant species. Interspecies comparisons of mitochondrial proteomes will also give insights into the evolutionarily processes that have yielded species specific mitochondrial functions in plants.
A noticeable gap in our knowledge on retrograde signalling from mitochondria to the nucleus in Arabidopsis is the unambiguous identification of any of the key molecules or players involved in this process. As outlined above although various biochemical and molecular approaches have established the existence of several pathways it remains to be determined how these pathways interact with mainstream stress signalling pathways in plants cells and how and if mitochondrial retrograde signalling interacts with chloroplast retrograde signalling. Given the variety of mutants now available and the relative ease of mapping mutations it is expected that the identity of the components involved in both anterograde and retrograde regulation of genes encoding mitochondrial components will be identified in the near future. These are the footprints of the master control switches that define the biogenesis of this organelle.
Footnotes
Citation: Millar A.H., Small I.D., Day D.A., and Whelan J. (2008) Mitochondrial Biogenesis and Function in Arabidopsis. The Arabidopsis Book 6:e0111. doi:10.1199/tab.0111
elocation-id: e0111
This chapter was updated on July 9, 2008. The original version has been archived and is available in PDF format at http://dx.doi.org/10.1199/tab.0105.
REFERENCES
- Abbaraju H. R., Behal R. H., Oliver D. J. Identification of an unusual MYB-type factor that binds to the promoter of the H-proteins of glycine decarboxylase. J. Plant. Biochem. Biotechnol. 2002;116(1):65–71. [Google Scholar]
- Abdelnoor R. V., Yule R., Elo A., Christensen A. C., Meyer-Gauen G., Mackenzie S. A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA. 2003;1006(1):5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z., Adamska I., Nakabayashi K., Ostersetzer O., Haussuhl K., Manuell A., Zheng B., Vallon O., Rodermel S. R., Shinozaki K., Clarke A. K. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001;1256(1):1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. L., Daley D. O., Whelan J., Palmer J. D. Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell. 2002;146(1):931–943. doi: 10.1105/tpc.010483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi H., Nakamura A., Yokozeki-Misono Y., Inagaki A., Takahashi H., Mori K., Fujimura T. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 2004;1086(1):1449–1457. doi: 10.1007/s00122-004-1591-2. [DOI] [PubMed] [Google Scholar]
- Amirsadeghi S., Robson C. A., McDonald A. E., Vanlerberghe G. C. Changes in plant mitochondrial electron transport alter cellular levels of reactive oxygen species and susceptibility to cell death signaling molecules. Plant Cell Physiol. 2006;476(1):1509–19. doi: 10.1093/pcp/pcl016. [DOI] [PubMed] [Google Scholar]
- Andres C., Lurin C., Small I. D. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 2007;1296(1):14–22. [Google Scholar]
- Arimura S., Tsutsumi N. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA. 2002;996(1):5727–5731. doi: 10.1073/pnas.082663299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S., Aida G. P., Fujimoto M., Nakazono M., Tsutsumi N. Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 2004a;456(1):236–242. doi: 10.1093/pcp/pch024. [DOI] [PubMed] [Google Scholar]
- Arimura S., Yamamoto J., Aida G. P., Nakazono M., Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA. 2004b;1016(1):7805–7808. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L. C., Komiya T., Bergman B. E., Mihara K., Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem. 1997;2726(1):6510–6518. doi: 10.1074/jbc.272.10.6510. [DOI] [PubMed] [Google Scholar]
- Arnal N., Alban C., Quadrado M., Grandjean O., Mireau H. The Arabidopsis Bio2 protein requires mitochondrial targeting for activity. Plant Mol. Biol. 2006;626(1):471–479. doi: 10.1007/s11103-006-9034-x. [DOI] [PubMed] [Google Scholar]
- Aronsson H., Boij P., Patel R., Wardle A., Topel M., Jarvis P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007;526(1):53–68. doi: 10.1111/j.1365-313X.2007.03207.x. [DOI] [PubMed] [Google Scholar]
- Arpagaus S., Rawyler A., Braendle R. Occurrence and characteristics of the mitochondrial permeability transition in plants. J. Biol. Chem. 2002;2776(1):1780–1787. doi: 10.1074/jbc.M109416200. [DOI] [PubMed] [Google Scholar]
- Attallah C. V., Welchen E., Pujol C., Bonnard G., Gonzalez D. H. Characterization of Arabidopsis thaliana genes encoding functional homologues of the yeast metal chaperone Cox19p, involved in cytochrome c oxidase biogenesis. Plant Mol. Biol. 2007;656(1):343–355. doi: 10.1007/s11103-007-9224-1. [DOI] [PubMed] [Google Scholar]
- Baba K., Schmidt J., Espinosa-Ruiz A., Villarejo A., Shiina T., Gardestrom P., Sane A. P., Bhalerao R. P. Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J. 2004;386(1):38–48. doi: 10.1111/j.1365-313X.2004.02022.x. [DOI] [PubMed] [Google Scholar]
- Babiychuk E., Muller F., Eubel H., Braun H. P., Frentzen M., Kushnir S. Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J. 2003;336(1):899–909. doi: 10.1046/j.1365-313x.2003.01680.x. [DOI] [PubMed] [Google Scholar]
- Baldet P., Alban C., Douce R. Biotin synthesis in higher plants: purification and characterization of bioB gene product equivalent from Arabidopsis thaliana overexpressed in Escherichia coli and its subcellular localization in pea leaf cells. FEBS Lett. 1997;4196(1):206–210. doi: 10.1016/s0014-5793(97)01458-0. [DOI] [PubMed] [Google Scholar]
- Balk J., Leaver C. J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell. 2001;136(1):1803–1818. doi: 10.1105/TPC.010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Chew S. K., Leaver C. J., McCabe P. F. The intermembrane space of plant mitochondria contains a DNase activity that may be involved in programmed cell death. Plant J. 2003;346(1):573–583. doi: 10.1046/j.1365-313x.2003.01748.x. [DOI] [PubMed] [Google Scholar]
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;186(1):298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Bartoli C. G., Pastori G. M., Foyer C. H. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complex III and IV. Plant Physiol. 2000;1236(1):335–343. doi: 10.1104/pp.123.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M., Dietrich C., Nowak K., Sierralta W. D., Papenbrock J. Intracellular localization of Arabidopsis sulfurtransferases. Plant Physiol. 2004;1356(1):916–926. doi: 10.1104/pp.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H., Kolukisaoglu U. Genetic manipulation of glycine decarboxylation. J. Exp. Bot. 2003;546(1):1523–1535. doi: 10.1093/jxb/erg171. [DOI] [PubMed] [Google Scholar]
- Behal R. H., Oliver D. J. Biochemical and molecular characterization of fumarase from plants: purification and characterization of the enzyme—cloning, sequencing, and expression of the gene. Arch. Biochem. Biophys. 1997;3486(1):65–74. doi: 10.1006/abbi.1997.0359. [DOI] [PubMed] [Google Scholar]
- Behal R. H., Oliver D. J. NAD(+)-dependent isocitrate dehydrogenase from Arabidopsis thaliana. Characterization of two closely related subunits. Plant Mol. Biol. 1998;366(1):691–698. doi: 10.1023/a:1005923410940. [DOI] [PubMed] [Google Scholar]
- Bentolila S., Alfonso A. A., Hanson M. R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA. 2002;996(1):10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S., Chateigner-Boutin A. L., Hanson M. R. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol. 2005;1396(1):2006–2016. doi: 10.1104/pp.105.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M., Rogers R., Muralla R., Meinke D. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 2005;446(1):866–878. doi: 10.1111/j.1365-313X.2005.02580.x. [DOI] [PubMed] [Google Scholar]
- Bhushan S., Lefebvre B., Stahl A., Wright S. J., Bruce B. D., Boutry M., Glaser E. Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep. 2003;46(1):1073–1077. doi: 10.1038/sj.embor.7400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S., Brennicke A. Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2003;3586(1):181–188. doi: 10.1098/rstb.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;96(1):42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;86(1):26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Bouche N., Fait A., Bouchez D., Moller S. G., Fromm H. Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sc.i USA. 2003;1006(1):6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon J., Merand V., Rawsthorne S., Forest E., Douce R. Glycine decarboxylase and pyruvate dehydrogenase complexes share the same dihydrolipoamide dehydrogenase in pea leaf mitochondria: evidence from mass spectrometry and primary-structure analysis. Biochem. J. 1996;3136(1):229–234. doi: 10.1042/bj3130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry M., Nagy F., Poulsen C., Aoyagi K., Chua N. H. Targeting of bacterial chloramphenicol acetyltransferase to mitochondria in transgenic plants. Nature. 1987;3286(1):340–342. doi: 10.1038/328340a0. [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Fricker M., Hawes C. A greener world: the revolution in plant bioimaging. Nat. Rev. Mol. Cell Biol. 2002;36(1):520–530. doi: 10.1038/nrm861. [DOI] [PubMed] [Google Scholar]
- Bras M., Queenan B., Susin S. A. Programmed cell death via mitochondria: different modes of dying. Biochemistry. 2005;706(1):231–239. doi: 10.1007/s10541-005-0105-4. [DOI] [PubMed] [Google Scholar]
- Braun H. P., Jansch L., Kurft V., Schmitz U. K. The ‘Hinge’ protein of cytochrome c reductase from potato lacks the acidic domain and has no cleavable presequence. FEBS Lett. 1994;3476(1):90–94. doi: 10.1016/0014-5793(94)00515-x. [DOI] [PubMed] [Google Scholar]
- Brennicke A., Zabaleta E., Dombrowski S., Hoffmann M., Binder S. Transcription signals of mitochondrial and nuclear genes for mitochondrial proteins in dicot plants. J. Hered. 1999;906(1):345–350. doi: 10.1093/jhered/90.3.345. [DOI] [PubMed] [Google Scholar]
- Brown G. G., Formanova N., Jin H., Wargachuk R., Dendy C., Patil P., Laforest M., Zhang J., Cheung W. Y., Landry B. S. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 2003;356(1):262–272. doi: 10.1046/j.1365-313x.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- Burger G., Lang B. F., Braun H. P., Marx S. The enigmatic mitochondrial ORF ymf39 codes for ATP synthase chain b. Nucleic Acids Res. 2003;316(1):2353–2360. doi: 10.1093/nar/gkg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Piehler J., Fromm H. Plant succinic semi-aldehyde dehydrogenase: dissection of nucleotide binding by surface plasmon resonance and fluorescence spectroscopy. Biochemistry. 2000;396(1):10110–10117. doi: 10.1021/bi000589e. [DOI] [PubMed] [Google Scholar]
- Busch K. B., Fromm H. Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiol. 1999;1216(1):589–597. doi: 10.1104/pp.121.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon A., Stern D. B. Plastid transcription: a menage a trois. Trends Plant Sci. 2001;66(1):45–46. doi: 10.1016/s1360-1385(00)01858-6. [DOI] [PubMed] [Google Scholar]
- Caiveau O., Fortune D., Cantrel C., Zachowski A., Moreau F. Consequences of omega -6-oleate desaturase deficiency on lipid dynamics and functional properties of mitochondrial membranes of Arabidopsis thaliana. J. Biol. Chem. 2001;2766(1):5788–5794. doi: 10.1074/jbc.M006231200. [DOI] [PubMed] [Google Scholar]
- Carlsson J., Leino M., Glimelius K. Mitochondrial genotypes with variable parts of Arabidopsis thaliana DNA affect development in Brassica napus lines. Theor. Appl. Genet. 2007;1156(1):627–641. doi: 10.1007/s00122-007-0593-2. [DOI] [PubMed] [Google Scholar]
- Casolo V., Petrussa E., Krajnakova J., Macri F., Vianello A. Involvement of the mitochondrial K+ATP channel in H2O2-or NO-induced programmed death of soybean suspension cell cultures. J. Exp. Bot. 2005;566(1):997–1006. doi: 10.1093/jxb/eri093. [DOI] [PubMed] [Google Scholar]
- Catoni E., Schwab R., Hilpert M., Desimone M., Schwacke R., Flugge U. I., Schumacher K., Frommer W. B. Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett. 2003a;5346(1):87–92. doi: 10.1016/s0014-5793(02)03782-1. [DOI] [PubMed] [Google Scholar]
- Catoni E., Desimone M., Hilpert M., Wipf D., Kunze R., Schneider A., Flugge U. I., Schumacher K., Frommer W. B. Expression pattern of a nuclear encoded mitochondrial arginine- ornithine translocator gene from Arabidopsis. BMC Plant Biol. 2003b;36(1):1. doi: 10.1186/1471-2229-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabregas S. M., Luche D. D., Van Sluys M. A., Menck C. F., Silva-Filho M. C. Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J. Cell Sci. 2003;1166(1):285–291. doi: 10.1242/jcs.00228. [DOI] [PubMed] [Google Scholar]
- Chabregas S. M., Luche D. D., Farias L. P., Ribeiro A. F., van Sluys M. A., Menck C. F., Silva-Filho M. C. Dual targeting properties of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Mol. Biol. 2001;466(1):639–650. doi: 10.1023/a:1011628510711. [DOI] [PubMed] [Google Scholar]
- Chan N. C., Likic V. A., Waller R. F., Mulhern T. D., Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J. Mol. Biol. 2006;3586(1):1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Chase C. D. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;236(1):81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A. L., Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;356(1):e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Wurtele E. S., Nikolau B. J. Metabolic and environmental regulation of 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis. Plant Physiol. 2002;1296(1):625–637. doi: 10.1104/pp.001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Sanchez-Fernandez R., Lyver E. R., Dancis A., Rea P. A. Functional characterization of AtATM1, AtATM2, and AtATM3, a subfamily of Arabidopsis half-molecule ATP-binding cassette transporters implicated in iron homeostasis. J Biol. Chem. 2007;2826(1):21561–21571. doi: 10.1074/jbc.M702383200. [DOI] [PubMed] [Google Scholar]
- Chew O., Whelan J. Dual targeting ability of targeting signals is dependent on the nature of the mature protein. Func. Plant Biol. 2003;306(1):805–812. doi: 10.1071/FP03077. [DOI] [PubMed] [Google Scholar]
- Chew O., Whelan J., Millar A. H. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003a;2786(1):46869–46877. doi: 10.1074/jbc.M307525200. [DOI] [PubMed] [Google Scholar]
- Chew O., Rudhe C., Glaser E., Whelan J. Characterisation of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol. Biol. 2003b;536(1):341–356. doi: 10.1023/b:plan.0000006939.87660.4f. [DOI] [PubMed] [Google Scholar]
- Chew O., Lister R., Qbadou S., Heazlewood J. L., Soll J., Schleiff E., Millar A. H., Whelan J. A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 2004;5576(1):109–114. doi: 10.1016/s0014-5793(03)01457-1. [DOI] [PubMed] [Google Scholar]
- Christensen A. C., Lyznik A., Mohammed S., Elowsky C. G., Elo A., Yule R., Mackenzie S. A. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell. 2005;176(1):2805–2816. doi: 10.1105/tpc.105.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M. G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;2416(1):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Clausen C., Ilkavets I., Thomson R., Philippar K., Vojta A., Mohlmann T., Neuhaus E., Fulgosi H., Soll J. Intracellular localization of VDAC proteins in plants. Planta. 2004;2206(1):30–37. doi: 10.1007/s00425-004-1325-3. [DOI] [PubMed] [Google Scholar]
- Clifton R., Millar A. H., Whelan J. Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta. 2006;17576(1):730–741. doi: 10.1016/j.bbabio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Clifton R., Lister R., Parker K. L., Sappl P. G., Elhafez D., Millar A. H., Day D. A., Whelan J. Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 2005;586(1):193–212. doi: 10.1007/s11103-005-5514-7. [DOI] [PubMed] [Google Scholar]
- Considine M. J., Holtzapffel R. C., Day D. A., Whelan J., Millar A. H. Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 2002;1296(1):949–953. doi: 10.1104/pp.004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine M. J., Goodman M., Echtay K. S., Laloi M., Whelan J., Brand M. D., Sweetlove L. J. Superoxide stimulates a proton leak in potato mitochondria that is related to the activity of uncoupling protein. J. Biol. Chem. 2003;2786(1):22298–22302. doi: 10.1074/jbc.M301075200. [DOI] [PubMed] [Google Scholar]
- Contran N., Cerana R., Crosti P., Malerba M. Cyclosporin A inhibits programmed cell death and cytochrome c release induced by fusicoccin in sycamore cells. Protoplasma. 2007;2316(1):193–199. doi: 10.1007/s00709-007-0250-2. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Fearnley I. M., Walker J. E. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 2005;5796(1):4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- Cummings M. P., Myers D. S. Simple statistical models predict C-to-U edited sites in plant mitochondrial RNA. BMC Bioinformatics. 2004;56(1):132. doi: 10.1186/1471-2105-5-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunillera N., Boronat A., Ferrer A. The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J. Biol. Chem. 1997;2726(1):15381–15388. doi: 10.1074/jbc.272.24.15381. [DOI] [PubMed] [Google Scholar]
- Curi G. C., Welchen E., Chan R. L., Gonzalez D. H. Nuclear and mitochondrial genes encoding cytochrome c oxidase subunits respond differently to the same metabolic factors. Plant Physiol. Biochem. 2003;416(1):689–693. [Google Scholar]
- Daschner K., Couee I., Binder S. The mitochondrial isovaleryl-coenzyme a dehydrogenase of arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol. 2001;1266(1):601–612. doi: 10.1104/pp.126.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Vance J. E. Import of lipids into mitochondria. Prog. Lipid Res. 1997;366(1):103–130. doi: 10.1016/s0163-7827(97)00006-4. [DOI] [PubMed] [Google Scholar]
- Day D. A., Neuburger M., Douce R. Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust. J. Plant Physiol. 1985;126(1):219–228. [Google Scholar]
- Delage L., Giege P., Sakamoto M., Marechal-Drouard L. Four paralogues of RPL12 are differentially associated to ribosome in plant mitochondria. Biochimie. 2007;896(1):658–668. doi: 10.1016/j.biochi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Delannoy E., Stanley W. A., Bond C. S., Small I. D. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;356(1):1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- Desloire S., Gherbi H., Laloui W., Marhadour S., Clouet V., Cattolico L., Falentin C., Giancola S., Renard M., Budar F., Small I., Caboche M., Delourme R., Bendahmane A. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 2003;46(1):588–594. doi: 10.1038/sj.embor.embor848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessi P., Pavel P. F., Wallberg F., Rudhe C., Brack S., Whelan J., Glaser E. Investigations on the in vitro ability of mitochondrial precursor proteins synthesised in wheat germ transcription-translation extract. Plant Mol. Biol. 2003;526(1):259–271. doi: 10.1023/a:1023993107220. [DOI] [PubMed] [Google Scholar]
- Deuschle K., Funck D., Hellmann H., Daschner K., Binder S., Frommer W. B. A nuclear gene encoding mitochondrial Delta-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 2001;276(1):345–356. doi: 10.1046/j.1365-313x.2001.01101.x. [DOI] [PubMed] [Google Scholar]
- Diebold R., Schuster J., Daschner K., Binder S. The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol. 2002;1296(1):540–550. doi: 10.1104/pp.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich A., Small I., Cosset A., Weil J. H., Marechal-Drouard L. Editing and import: strategies for providing plant mitochondria with a complete set of functional transfer RNAs. Biochimie. 1996;786(1):518–529. doi: 10.1016/0300-9084(96)84758-4. [DOI] [PubMed] [Google Scholar]
- Djajanegara I., Finnegan P. M., Mathieu C., McCabe T., Whelan J., Day D. A. Regulation of alternative oxidase gene expression in soybean. Plant Mol. Biol. 2002;506(1):735–742. doi: 10.1023/a:1019942720636. [DOI] [PubMed] [Google Scholar]
- Dojcinovic D., Krosting J., Harris A. J., Wagner D. J., Rhoads D. M. Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol. Biol. 2005;586(1):159–175. doi: 10.1007/s11103-005-5390-1. [DOI] [PubMed] [Google Scholar]
- Douce R., Christensen E. L., Bonner W. D., Jr. Preparation of intact plant mitochondria. Biochim. Biophys. Acta. 1972;2756(1):148–160. doi: 10.1016/0005-2728(72)90035-7. [DOI] [PubMed] [Google Scholar]
- Drea S. C., Mould R. M., Hibberd J. M., Gray J. C., Kavanagh T. A. Tissue-specific and developmental-specific expression of an Arabidopsis thaliana gene encoding the lipoamide dehydrogenase component of the plastid pyruvate dehydrogenase complex. Plant Mol. Biol. 2001;466(1):705–715. doi: 10.1023/a:1011612921144. [DOI] [PubMed] [Google Scholar]
- Duchene A. M., Marechal-Drouard L. The chloroplast-derived trnW and trnM-e genes are not expressed in Arabidopsis mitochondria. Biochem. Biophys. Res. Commun. 2001;2856(1):1213–1216. doi: 10.1006/bbrc.2001.5303. [DOI] [PubMed] [Google Scholar]
- Duchene A. M., Giritch A., Hoffmann B., Cognat V., Lancelin D., Peeters N. M., Zaepfel M., Marechal-Drouard L., Small I. D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2005;1026(1):16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley T. P., Watson R., Griffin J. L., Dupree P., Lilley K. S. Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell Proteomics. 2004;36(1):1128–1134. doi: 10.1074/mcp.T400009-MCP200. [DOI] [PubMed] [Google Scholar]
- Dunkley T. P., Hester S., Shadforth I. P., Runions J., Weimar T., Hanton S. L., Griffin J. L., Bessant C., Brandizzi F., Hawes C., Watson R. B., Dupree P., Lilley K. S. Mapping the Arabidopsis organelle proteome. Proc Natl. Acad. Sci. USA. 2006;1036(1):6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C., Garmier M., Noctor G., Mathieu C., Chetrit P., Foyer C. H., de Paepe R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;156(1):1212–1226. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtay K. S., Murphy M. P., Smith R. A., Talbot D. A., Brand M. D. Superoxide activates mitochondrial uncoupling protein 2 from the matrix side. Studies using targeted antioxidants. J. Biol. Chem. 2002;2776(1):47129–47135. doi: 10.1074/jbc.M208262200. [DOI] [PubMed] [Google Scholar]
- Elhafez D., Murcha M. W., Clifton R., Soole K. L., Day D. A., Whelan J. Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant Cell Physiol. 2006;476(1):43–54. doi: 10.1093/pcp/pci221. [DOI] [PubMed] [Google Scholar]
- Elo A., Lyznik A., Gonzalez D. O., Kachman S. D., Mackenzie S. A. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell. 2003;156(1):1619–1631. doi: 10.1105/tpc.010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel C., von Groll U., Muller M., Borner T., Weihe A. Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta. 2006;2236(1):998–1009. doi: 10.1007/s00425-005-0159-y. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N- terminal amino acid sequence. J. Mol. Biol. 2000;3006(1):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Esser K., Tursun B., Ingenhoven M., Michaelis G., Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 2002;3236(1):835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- Eubel H., Jansch L., Braun H. P. New Insights into the Respiratory Chain of Plant Mitochondria. Supercomplexes and a Unique Composition of Complex II. Plant Physiol. 2003;1336(1):274–286. doi: 10.1104/pp.103.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H., Lee C. P., Kuo J., Meyer E. H., Taylor N. L., Millar A. H. Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J. 2007;526(1):583–594. doi: 10.1111/j.1365-313X.2007.03253.x. [DOI] [PubMed] [Google Scholar]
- Ewald R., Kolukisaoglu U., Bauwe U., Mikkat S., Bauwe H. Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol. 2007;1456(1):41–48. doi: 10.1104/pp.107.104000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon de Longevialle A., Meyer E. H., Andres C., Taylor N. L., Lurin C., Millar A. H., Small I. D. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell. 2007;196(1):3256–3265. doi: 10.1105/tpc.107.054841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Arimura S., Hirano H. Y., Sakamoto W., Tsutsumi N. Isolation of mutants with aberrant mitochondrial morphology from Arabidopsis thaliana. Genes Genet. Syst. 2004;796(1):301–305. doi: 10.1266/ggs.79.301. [DOI] [PubMed] [Google Scholar]
- Figueroa P., Leon G., Elorza A., Holuigue L., Jordana X. Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol. Biol. 2001;466(1):241–250. doi: 10.1023/a:1010612506070. [DOI] [PubMed] [Google Scholar]
- Figueroa P., Leon G., Elorza A., Holuigue L., Araya A., Jordana X. The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol. Biol. 2002;506(1):725–734. doi: 10.1023/a:1019926301981. [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L. J., Dietz K. J. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 2005;2806(1):12168–12180. doi: 10.1074/jbc.M413189200. [DOI] [PubMed] [Google Scholar]
- Fiorani F., Umbach A. L., Siedow J. N. The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiol. 2005;1396(1):1795–1805. doi: 10.1104/pp.105.070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J., Weber B., Thuss S., Wildum S., Binder S. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007;356(1):3676–3692. doi: 10.1093/nar/gkm270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes F., Castilho R. F., Catisti R., Carnieri E. G., Vercesi A. E. Ca2+ induces a cyclosporin A-insensitive permeability transition pore in isolated potato tuber mitochondria mediated by reactive oxygen species. J. Bioenerg Biomembr. 2001;336(1):43–51. doi: 10.1023/a:1005672623709. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Sato T., Ito M., Watanabe A. Isolation and characterization of cDNA clones for the E1b and E2 subunits of the branched-chain a-ketoacid dehydrogenase complex in Arabidopsis. J. Biol. Chem. 2000;2756(1):6007–6013. doi: 10.1074/jbc.275.8.6007. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Ito M., Nishida I., Watanabe A. Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett. 2001;4996(1):161–165. doi: 10.1016/s0014-5793(01)02536-4. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Ito M., Itoh T., Nishida I., Watanabe A. Activation of the promoters of Arabidopsis genes for the branched-chain alpha-keto acid dehydrogenase complex in transgenic tobacco BY-2 cells under sugar starvation. Plant Cell Physiol. 2002;436(1):275–280. doi: 10.1093/pcp/pcf032. [DOI] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T. S., Laloi C., Minkov I. N., Shulaev V., Apel K., Inze D., Mittler R., Van Breusegem F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;1416(1):436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D., Gualberto J. M. Gene Expression in Higher Plant Mitochondria. In: Day D. A., Millar A. H., Whelan J., editors. Advances in Photosynthesis and Respiration. 1. Vol. 6. Dordrecht; Kluwer: 2004. pp. 55–82. [Google Scholar]
- Geddy R., Brown G. G. Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genomics. 2007;86(1):130. doi: 10.1186/1471-2164-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler D. A., Broselid C., Hederstedt L., Rasmusson A. G. Ca2+-binding and Ca2+-independent respiratory NADH and NADPH dehydrogenases of Arabidopsis thaliana. J. Biol. Chem. 2007;2826(1):28455–28464. doi: 10.1074/jbc.M704674200. [DOI] [PubMed] [Google Scholar]
- Gerhardt B. Fatty acid degradation in plants. Prog. Lipid. Res. 1992;316(1):417–446. doi: 10.1016/0163-7827(92)90004-3. [DOI] [PubMed] [Google Scholar]
- Giege P., Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA. 1999;966(1):15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege P., Heazlewood J. L., Roessner-Tunali U., Millar A. H., Fernie A. R., Leaver C. J., Sweetlove L. J. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;156(1):2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman J. D., Bentolila S., Hanson M. R. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 2007;496(1):217–227. doi: 10.1111/j.1365-313X.2006.02953.x. [DOI] [PubMed] [Google Scholar]
- Giraud E., Ho L. H., Clifton R., Carroll A., Estavillo G., Tan Y. F., Howell K. A., Ivanova A., Pogson B. J., Millar A. H., Whelan J. The absence of Alternative Oxidase 1a in Arabidopsis thaliana results in acute sensitivity to combined light and drought stress. Plant Physiol. 2008;PMID6(1):18424626. doi: 10.1104/pp.107.115121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser E., Dessi P. Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J. Bioenerg. Biomembr. 1999;316(1):259–274. doi: 10.1023/a:1005475930477. [DOI] [PubMed] [Google Scholar]
- Glaser E., Whelan J. Import of nuclear-encoded mitochondrial proteins. Plant Mitochondria. Ann. Plant. Rev. 2007;316(1):97–128. In. [Google Scholar]
- Gomez-Casati D. F., Busi M. V., Gonzalez-Schain N., Mouras A., Zabaleta E. J., Araya A. A mitochondrial dysfunction induces the expression of nuclear-encoded complex I genes in engineered male sterile Arabidopsis thaliana. FEBS Lett. 2002;5326(1):70–74. doi: 10.1016/s0014-5793(02)03631-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez D. H., Welchen E., Attallah C. V., Comelli R. N., Mufarrege E. F. Transcriptional coordination of the biogenesis of the oxidative phosphorylation machinery in plants. Plant J. 2007;516(1):105–116. doi: 10.1111/j.1365-313X.2007.03121.x. [DOI] [PubMed] [Google Scholar]
- Graham I. A., Eastmond P. J. Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res. 2002;416(1):156–181. doi: 10.1016/s0163-7827(01)00022-4. [DOI] [PubMed] [Google Scholar]
- Green D. R., Reed J. C. Mitochondria and apoptosis. Science. 1998;2816(1):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Guan Y., Rawsthorne S., Scofield G., Shaw P., Doonan J. Cloning and characterization of a dihydrolipoamide acetyltransferase (E2) subunit of the pyruvate dehydrogenase complex from Arabidopsis thaliana. J. Biol. Chem. 1995;2706(1):5412–5417. doi: 10.1074/jbc.270.10.5412. [DOI] [PubMed] [Google Scholar]
- Haferkamp I., Hackstein J. H., Voncken F. G., Schmit G., Tjaden J. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 2002;2696(1):3172–3181. doi: 10.1046/j.1432-1033.2002.02991.x. [DOI] [PubMed] [Google Scholar]
- Halestrap A. P., Davidson A. M. Inhibition of Ca2_-induced largeamplitude swelling of liver and heart mitochondria by cyclosporin is probably caused by the inhibitor binding to mitochondrial-matrix peptidyl- prolyl cis-trans isomerase and preventing it interacting with the adenine nucleotide translocase. Biochem. J. 1990;2686(1):153–160. doi: 10.1042/bj2680153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P., Saint-Georges Y., de Pinto B., Lachacinski N., Altamura N., Dujardin G. Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol. Microbiol. 2004;516(1):307–317. doi: 10.1046/j.1365-2958.2003.03810.x. [DOI] [PubMed] [Google Scholar]
- Hartman N. T., Sicilia F., Lilley K. S., Dupree P. Proteomic complex detection using sedimentation. Anal. Chem. 2007;796(1):2078–2083. doi: 10.1021/ac061959t. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y., Saito K. Evidence for the existence of rhodanese (thiosulfate:cyanide sulfurtransferase) in plants: preliminary characterization of two rhodanese cDNAs from Arabidopsis thaliana. FEBS Lett. 2000;4706(1):147–150. doi: 10.1016/s0014-5793(00)01311-9. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y., Maruyama A., Schmidt A., Noji M., Ishizawa K., Saito K. beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiol. 2000;1236(1):1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C., Saint-Jore C., Martin B., Zheng H. Q. ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP!. Trends Plant Sci. 2001;66(1):245–246. doi: 10.1016/s1360-1385(01)01980-x. [DOI] [PubMed] [Google Scholar]
- Heazlewood J. L., Howell K. A., Millar A. H. Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta. 2003a;16046(1):159–169. doi: 10.1016/s0005-2728(03)00045-8. [DOI] [PubMed] [Google Scholar]
- Heazlewood J. L., Whelan J., Millar A. H. The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 2003b;5406(1):201–205. doi: 10.1016/s0014-5793(03)00264-3. [DOI] [PubMed] [Google Scholar]
- Heazlewood J. L., Tonti-Filippini J., Verboom R. E., Millar A. H. Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 2005;1396(1):598–609. doi: 10.1104/pp.105.065532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J. L., Verboom R. E., Tonti-Filippini J., Small I., Millar A. H. SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;356(1):D213–218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J. L., Tonti-Filippini J. S., Gout A., Day D. A., Whelan J. M., Millar A. H. Experimental Analysis of the Arabidopsis Mitochondrial Proteome Highlights Signalling and Regulatory Components, Provides Assessment of Targeting Prediction Programs and Points to Plant Specific Mitochondrial Proteins. Plant Cell. 2004;166(1):241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B., Borner T., Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;2776(1):809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- Hell K. The Erv1-Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim. Biophys. Acta. 2008;17836(1):601–609. doi: 10.1016/j.bbamcr.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Hill T. A., Broadhvest J., Kuzoff R. K., Gasser C. S. Arabidopsis SHORT INTEGUMENTS 2 is a mitochondrial DAR GT-Pase. Genetics. 2006;1746(1):707–718. doi: 10.1534/genetics.106.060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P., Small I., Kuiper M. T. European consortia building integrated resources for Arabidopsis functional genomics. Curr. Opin. Plant Biol. 2003;66(1):426–429. doi: 10.1016/s1369-5266(03)00086-4. [DOI] [PubMed] [Google Scholar]
- Ho L. H., Giraud E., Lister R., Thirkettle-Watts D., Low J., Clifton R., Howell K. A., Carrie C., Donald T., Whelan J. Characterization of the regulatory and expression context of an alternative oxidase gene provides insights into cyanide-insensitive respiration during growth and development. Plant Physiol. 2007;1436(1):1519–1533. doi: 10.1104/pp.106.091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund A., Donnes P., Blum T., Adolph H. W., Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;226(1):1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Holec S., Lange H., Kuhn K., Alioua M., Borner T., Gagliardi D. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell Biol. 2006;266(1):2869–2876. doi: 10.1128/MCB.26.7.2869-2876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos M. E., Palmieri L., Wertin T., Arrigoni R., Polacco J. C., Palmieri F. Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 2003;336(1):1027–1035. doi: 10.1046/j.1365-313x.2003.01685.x. [DOI] [PubMed] [Google Scholar]
- Hua S., Sun Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 2001;176(1):721–728. doi: 10.1093/bioinformatics/17.8.721. [DOI] [PubMed] [Google Scholar]
- Hugosson M., Andreu D., Boman H. G., Glaser E. Antibacterial peptides and mitochondrial presequences affect mitochondrial coupling, respiration and protein import. Eur J Biochem. 1994;2236(1):1027–1033. doi: 10.1111/j.1432-1033.1994.tb19081.x. [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Larson T. R., Schauer N., Fernie A. R., Graham I. A., Leaver C. J. The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell. 2005;176(1):2587–2600. doi: 10.1105/tpc.105.035162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Schauer N., Larson T. R., Graham I. A., Fernie A. R., Leaver C. J. The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 2006;476(1):751–760. doi: 10.1111/j.1365-313X.2006.02826.x. [DOI] [PubMed] [Google Scholar]
- Ito J., Heazlewood J. L., Millar A. H. Analysis of the soluble ATP-binding proteome of plant mitochondria identifies new proteins and nucleotide triphosphate interactions within the matrix. J. Proteome Res. 2006;56(1):3459–3469. doi: 10.1021/pr060403j. [DOI] [PubMed] [Google Scholar]
- Jin J. B., Bae H., Kim S. J., Jin Y. H., Goh C. H., Kim D. H., Lee Y. J., Tse Y. C., Jiang L., Hwang I. The Arabidopsis dynamin-like proteins ADL1C and ADL1E play a critical role in mitochondrial morphogenesis. Plant Cell. 2003;156(1):2357–2369. doi: 10.1105/tpc.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A., Bhushan S., Stahl A., Hallberg B. M., Frohn A., Glaser E., Eneqvist T. The closed structure of presequence protease PreP forms a unique 10,000 Angstroms3 chamber for proteolysis. EMBO J. 2006;256(1):1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost R., Berkowitz O., Wirtz M., Hopkins L., Hawkesford M. J., Hell R. Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene. 2000;2536(1):237–247. doi: 10.1016/s0378-1119(00)00261-4. [DOI] [PubMed] [Google Scholar]
- Jouhet J., Marechal E., Baldan B., Bligny R., Joyard J., Block M. A. Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 2004;1676(1):863–874. doi: 10.1083/jcb.200407022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg O., Canback B., Kurland C. G., Andersson S. G. The dual origin of the yeast mitochondrial proteome. Yeast. 2000;176(1):170–187. doi: 10.1002/1097-0061(20000930)17:3<170::AID-YEA25>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K., Sakurai I., Wada H. Identification of an Arabidopsis thaliana gene for cardiolipin synthase located in mitochondria. FEBS Lett. 2004;5776(1):193–198. doi: 10.1016/j.febslet.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M., Ohori Y., Uchimiya H. Dissection of Arabidopsis bax inhibitor-1 suppressing bax-, hydrogen peroxide-, and salicylic acidinduced cell death. Plant Cell. 2004;166(1):21–32. doi: 10.1105/tpc.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Yamada M., Saito Y., Jin L., Ogawa T., Kim K. M., Yu L. H., Tone Y., Hirata A., Umeda M., Uchimiya H. A novel Arabidopsis gene causes Bax-like lethality in Saccharomyces cerevisiae. J. Biol. Chem. 2005;2806(1):39468–39473. doi: 10.1074/jbc.M509632200. [DOI] [PubMed] [Google Scholar]
- Khazi F. R., Edmondson A. C., Nielsen B. L. An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol. Genet. Genomics. 2003;246(1):24. doi: 10.1007/s00438-003-0859-6. [DOI] [PubMed] [Google Scholar]
- Kim D. Y., Bovet L., Kushnir S., Noh E. W., Martinoia E., Lee Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006a;1406(1):922–932. doi: 10.1104/pp.105.074146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Lim J. H., Ahn C. S., Park K., Kim G. T., Kim W. T., Pai H. S. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell. 2006b;186(1):2341–2355. doi: 10.1105/tpc.106.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T., Yoshiba Y., Yamaguchi-Shinozaki K., Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996;86(1):1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. R., Klein P. E., Mullet J. E., Minx P., Rooney W. L., Schertz K. F. Fertility restorer locus Rf1 of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet. 2005;1116(1):994–1012. doi: 10.1007/s00122-005-2011-y. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Suzuki M., Tang J., Nagata N., Ohyama K., Seki H., Kiuchi R., Kaneko Y., Nakazawa M., Matsui M., Matsumoto S., Yoshida S., Muranaka T. Lovastatin insensitive 1, a Novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in Arabidopsis. Plant Cell Physiol. 2007;486(1):322–331. doi: 10.1093/pcp/pcm005. [DOI] [PubMed] [Google Scholar]
- Kocabek T., Repkova J., Dudova M., Hoyerova K., Vrba L. Isolation and characterization of a novel semi-lethal Arabidopsis thaliana mutant of gene for pentatricopeptide (PPR) repeat-containing protein. Genetica. 2006;1286(1):395–407. doi: 10.1007/s10709-006-7518-x. [DOI] [PubMed] [Google Scholar]
- Koizuka N., Imai R., Fujimoto H., Hayakawa T., Kimura Y., Kohno-Murase J., Sakai T., Kawasaki S., Imamura J. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 2003;346(1):407–415. doi: 10.1046/j.1365-313x.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- Komori T., Ohta S., Murai N., Takakura Y., Kuraya Y., Suzuki S., Hiei Y., Imaseki H., Nitta N. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 2004;376(1):315–325. doi: 10.1046/j.1365-313x.2003.01961.x. [DOI] [PubMed] [Google Scholar]
- Koppen M., Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 2007;426(1):221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Kosugi S., Suzuka I., Ohashi Y. Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J. 1995;76(1):877–886. doi: 10.1046/j.1365-313x.1995.07060877.x. [DOI] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;4336(1):326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T. C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;3166(1):715–719. [PubMed] [Google Scholar]
- Koyama H., Kawamura A., Kihara T., Hara T., Takita E., Shibata D. Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol. 2000;416(1):1030–1037. doi: 10.1093/pcp/pcd029. [DOI] [PubMed] [Google Scholar]
- Krause K., Kilbienski I., Mulisch M., Rodiger A., Schafer A., Krupinska K. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005;5796(1):3707–3712. doi: 10.1016/j.febslet.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Kruft V., Eubel H., Jansch L., Werhahn W., Braun H. P. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 2001;1276(1):1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kuhn K., Bohne A. V., Liere K., Weihe A., Borner T. Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes. Plant Cell. 2007;196(1):959–971. doi: 10.1105/tpc.106.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Cheung K. H., Miller P., Gerstein M., Roeder G. S., Snyder M. Subcellular localization of the yeast proteome. Genes Dev. 2002;166(1):707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir S., Babiychuk E., Storozhenko S., Davey M. W., Papenbrock J., De Rycke R., Engler G., Stephan U. W., Lange H., Kispal G., Lill R., Van Montagu M. A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell. 2001;136(1):89–100. doi: 10.1105/tpc.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cognata U., Landschutze V., Willmitzer L., Muller-Rober B. Structure and expression of mitochondrial citrate synthases from higher plants. Plant Cell Physiol. 1996;376(1):1022–1029. doi: 10.1093/oxfordjournals.pcp.a029033. [DOI] [PubMed] [Google Scholar]
- Laloi M. Plant mitochondrial carriers: an overview. Cell Mol. Life Sci. 1999;566(1):918–944. doi: 10.1007/s000180050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E., Kato N., Lawton M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature. 2001;4116(1):848–853. doi: 10.1038/35081184. [DOI] [PubMed] [Google Scholar]
- Landschutze V., Muller-Rober B., Willmitzer L. Mitochondrial citrate synthase from potato: predominant expression in mature leaves and young flower buds. Planta. 1995;1966(1):756–764. [PubMed] [Google Scholar]
- Larkin R. M., Alonso J. M., Ecker J. R., Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;2996(1):902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- Lee B. H., Lee H., Xiong L., Zhu J. K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell. 2002;146(1):1235–1251. doi: 10.1105/tpc.010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Sedman J., Neupert W., Stuart R. A. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J. Biol. Chem. 1999;2746(1):20937–20942. doi: 10.1074/jbc.274.30.20937. [DOI] [PubMed] [Google Scholar]
- Leino M., Landgren M., Glimelius K. Alloplasmic effects on mitochondrial transcriptional activity and RNA turnover result in accumulated transcripts of Arabidopsis orfs in cytoplasmic male-sterile Brassica napus. Plant J. 2005;426(1):469–480. doi: 10.1111/j.1365-313X.2005.02389.x. [DOI] [PubMed] [Google Scholar]
- Leino M., Sohlberg J., Sundström J. F., Glimelius K. Mitochondrial regulation of flower development. Mitochondrion. 2008;86(1):74–86. doi: 10.1016/j.mito.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Leister D. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene. 2005;3546(1):110–116. doi: 10.1016/j.gene.2005.03.039. [DOI] [PubMed] [Google Scholar]
- Lemaitre T., Hodges M. Expression analysis of Arabidopsis thaliana NAD-dependent isocitrate dehydrogenase genes shows the presence of a functional subunit that is mainly expressed in the pollen and absent from vegetative organs. Plant Cell Physiol. 2006;476(1):634–643. doi: 10.1093/pcp/pcj030. [DOI] [PubMed] [Google Scholar]
- Lemaitre T., Urbanczyk-Wochniak E., Flesch V., Bismuth E., Fernie A. R., Hodges M. NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol. 2007;1446(1):1546–1558. doi: 10.1104/pp.107.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. Y., Luo X., Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;4126(1):95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Liao X., Butow R. A. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;726(1):61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Lister R., Whelan J. Mitochondrial protein import: convergent solutions for receptor structure. Curr. Biol. 2006;166(1):R197–199. doi: 10.1016/j.cub.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Lister R., Murcha M. W., Whelan J. The Mitochondrial Protein Import Machinery of Plants (MPIMP) database. Nucleic Acids Res. 2003;316(1):325–327. doi: 10.1093/nar/gkg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Mowday B., Whelan J., Millar A. H. Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J. 2002;306(1):555–566. doi: 10.1046/j.1365-313x.2002.01316.x. [DOI] [PubMed] [Google Scholar]
- Lister R., Hulett J. M., Lithgow T., Whelan J. Protein import into mitochondria: origins and functions today (review). Mol. Membr. Biol. 2005;226(1):87–100. doi: 10.1080/09687860500041247. [DOI] [PubMed] [Google Scholar]
- Lister R., Carrie C., Duncan O., Ho L. H., Howell K. A., Murcha M. W., Whelan J. Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell. 2007;196(1):3739–3759. doi: 10.1105/tpc.107.050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Chew O., Lee M-N., Heazlewood J. L., Clifton R., Parker K. L., Millar A. H., Whelan J. A Transcriptomic and Proteomic Characterisation of the Arabidopsis Mitochondrial Protein Import Apparatus and its Response to Mitochondrial Stresses. Plant Physiol. 2004;1346(1):777–789. doi: 10.1104/pp.103.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D. C., Leaver C. J. Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot. 2000;516(1):865–871. [PubMed] [Google Scholar]
- Logan D. C., Scott I., Tobin A. K. The genetic control of plant mitochondrial morphology and dynamics. Plant J. 2003;366(1):500–509. doi: 10.1046/j.1365-313x.2003.01894.x. [DOI] [PubMed] [Google Scholar]
- Logan D. C., Scott I., Tobin A. K. ADL2a, like ADL2b, is involved in the control of higher plant mitochondrial morphology. J. Exp. Bot. 2004;556(1):783–785. doi: 10.1093/jxb/erh073. [DOI] [PubMed] [Google Scholar]
- Lohrmann J., Sweere U., Zabaleta E., Baurle I., Keitel C., Kozma-Bognar L., Brennicke A., Schafer E., Kudla J., Harter K. The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Genet. Genomics. 2001;2656(1):2–13. doi: 10.1007/s004380000400. [DOI] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Fauron C. M. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984;126(1):9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. L., de Noyer S. B., Hobbs D. H., Kang J., Wen Y., Krachtus D., Hills M. J. Expression pattern of diacylglycerol acyl-transferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Mol. Biol. 2003;526(1):31–41. doi: 10.1023/a:1023935605864. [DOI] [PubMed] [Google Scholar]
- Luethy M. H., Miernyk J. A., Randall D. D. The nucleotide and deduced amino acid sequences of a cDNA encoding the E1 beta-subunit of the Arabidopsis thaliana mitochondrial pyruvate dehydrogenase complex. Biochim. Biophys. Acta. 1994;11876(1):95–98. doi: 10.1016/0005-2728(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Luethy M. H., Miernyk J. A., Randall D. D. The mitochondrial pyruvate dehydrogenase complex: nucleotide and deduced amino-acid sequences of a cDNA encoding the Arabidopsis thaliana E1 alpha-subunit. Gene. 1995;1646(1):251–254. doi: 10.1016/0378-1119(95)00465-i. [DOI] [PubMed] [Google Scholar]
- Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Le Ret M., Martin-Magniette M. L., Mireau H., Peeters N., Renou J. P., Szurek B., Taconnat L., Small I. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;166(1):2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutziger I., Oliver D. J. Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol. 2001;1276(1):615–623. [PMC free article] [PubMed] [Google Scholar]
- Macasev D., Newbigin E., Whelan J., Lithgow T. How do plant mitochondria avoid importing chloroplast proteins? Components of the import apparatus Tom20 and Tom22 from Arabidopsis differ from their fungal counterparts. Plant Physiol. 2000;1236(1):811–816. doi: 10.1104/pp.123.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macasev D., Whelan J., Newbigin E., Silva-Filho M. C., Mulhern T. D., Lithgow T. Tom22′, an 8-kDa trans-site receptor in plants and protozoans, is a conserved feature of the TOM complex that appeared early in the evolution of eukaryotes. Mol. Biol. Evol. 2004;216(1):1557–1564. doi: 10.1093/molbev/msh166. [DOI] [PubMed] [Google Scholar]
- Mackenzie S., McIntosh L. Higher plant mitochondria. Plant Cell. 1999;116(1):571–585. doi: 10.1105/tpc.11.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia I. G., Benedetti C. E., Leite A., Turcinelli S. R., Vercesi A. E., Arruda P. AtPUMP: an Arabidopsis gene encoding a plant uncoupling mitochondrial protein. FEBS Lett. 1998;4296(1):403–406. doi: 10.1016/s0014-5793(98)00634-6. [DOI] [PubMed] [Google Scholar]
- Mannella C. A. The ‘ins’ and ‘outs’ of mitochondrial membrane channels. Trends Biochem. Sci. 1992;176(1):315–320. doi: 10.1016/0968-0004(92)90444-e. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Tedeschi H. Importance of the mitochondrial outer membrane channel as a model biological channel. J. Bioenerg. Biomembr. 1987;196(1):305–308. doi: 10.1007/BF00768533. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Pfeiffer D. R., Bradshaw P. C., Moraru, Slepchenko B., Loew L. M., Hsieh C. E., Buttle K., Marko M. Topology of the mitochondrial inner membrane: dynamics and bioenergetic implications. IUBMB Life. 2001;526(1):93–100. doi: 10.1080/15216540152845885. [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 2004;386(1):487–498. doi: 10.1111/j.1365-313X.2004.02063.x. [DOI] [PubMed] [Google Scholar]
- Marienfeld J., Unseld M., Brennicke A. The mitochondrial genome of Arabidopsis is composed of both native and immigrant information. Trends Plant Sci. 1999;46(1):495–502. doi: 10.1016/s1360-1385(99)01502-2. [DOI] [PubMed] [Google Scholar]
- Marillia E. F., Micallef B. J., Micallef M., Weninger A., Pedersen K. K., Zou J., Taylor D. C. Biochemical and physiological studies of Arabidopsis thaliana transgenic lines with repressed expression of the mitochondrial pyruvate dehydrogenase kinase. J. Exp. Bot. 2003;546(1):259–270. doi: 10.1093/jxb/erg020. [DOI] [PubMed] [Google Scholar]
- Matthes A., Schmidt-Gattung S., Kohler D., Forner J., Wildum S., Raabe M., Urlaub H., Binder S. Two DEAD-box proteins may be part of RNA-dependent high molecular weight protein complexes in Arabidopsis thaliana mitochondria. Plant Physiol. 2007;1456(1):1637–46. doi: 10.1104/pp.107.108076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell D. P., Wang Y., McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA. 1999;966(1):8271–6. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C. R., Hsu M., Painter J. E., Gagne J. M., Karlsberg S. D., Salome P. A. Integrated temporal regulation of the photorespiratory pathway. Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol. 2000;1236(1):381–392. doi: 10.1104/pp.123.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G. I., Ralph S. A. Dynamin: the endosymbiosis ring of power. Proc. Natl. Acad. Sci. USA. 2003;1006(1):3557–3559. doi: 10.1073/pnas.0831049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz R. I., Day D. A. Purification and characterization of a 43-kDa rotenone-insensitive NADH dehydrogenase from plant mitochondria. J. Biol. Chem. 1996;2716(1):23117–23120. doi: 10.1074/jbc.271.38.23117. [DOI] [PubMed] [Google Scholar]
- Meyer E. H., Heazlewood J. L., Millar A. H. Mitochondrial acyl carrier proteins in Arabidopsis thaliana are predominantly soluble matrix proteins and none can be confirmed as subunits of respiratory Complex I. Plant Mol. Biol. 2007;646(1):319–327. doi: 10.1007/s11103-007-9156-9. [DOI] [PubMed] [Google Scholar]
- Meyer E. H., Taylor N. L., Millar A. H. Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J. Proteome Res. 2008;76(1):786–794. doi: 10.1021/pr700595p. [DOI] [PubMed] [Google Scholar]
- Michalecka A. M., Svensson A. S., Johansson F. I., Agius S. C., Johanson U., Brennicke A., Binder S., Rasmusson A. G. Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary origin and show distinct responses to light. Plant Physiol. 2003;1336(1):642–652. doi: 10.1104/pp.103.024208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. H., Heazlewood J. L. Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 2003;1316(1):443–453. doi: 10.1104/pp.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. H., Liddell A., Leaver C. J. Isolation and sub-fractionation of mitochondria from plants. Methods Cell Biol. 2001a;656(1):53–74. doi: 10.1016/s0091-679x(01)65004-0. [DOI] [PubMed] [Google Scholar]
- Millar A. H., Whelan J., Small I. Recent surprises in protein targeting to mitochondria and plastids. Curr. Opin. Plant Biol. 2006;96(1):610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Millar A. H., Sweetlove L. J., Giege P., Leaver C. J. Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 2001b;1276(1):1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Millar A. H., Eubel H., Jansch L., Kruft V., Heazlewood J. L., Braun H. P. Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol. Biol. 2004;566(1):77–90. doi: 10.1007/s11103-004-2316-2. [DOI] [PubMed] [Google Scholar]
- Millar A. H., Mittova V., Kiddle G., Heazlewood J. L., Bartoli C. G., Theodoulou F. L., Foyer C. H. Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol. 2003;1336(1):443–447. doi: 10.1104/pp.103.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller I. M. PLANT MITOCHONDRIA AND OXIDATIVE STRESS: Electron Transport, NADPH Turnover, and Metabolism of Reactive Oxygen Species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;526(1):561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- Mollier P., Hoffmann B., Debast C., Small I. The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. 2002;406(1):405–409. doi: 10.1007/s00294-002-0271-5. [DOI] [PubMed] [Google Scholar]
- Moore C. S., Cook-Johnson R., Rudhe C., Whelan J., Day D. A., Wiskich J. T., Soole K. L. Identification of AtNDI1, an internal non-phosphorylating NAD(P)H dehydrogenase in Arabidopsis thaliana mitochondria. Plant Physiol. 2003;1336(1):1968–1978. doi: 10.1104/pp.103.029363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. J., Lehmann M., Schwarzländer M., Baxter C. J., Sienkiewicz-Porzucek A., Williams T. C. R., Schauer N., Fernie A. R., Fricker M. D., Ratcliffe R. G., Sweetlove L. J., Finkemeier I. Decrease in manganese superoxide dismutase leads to reduced root growth and affects TCA cycle flux and mitochondrial redox homeostasis. Plant Phyisol. 2008;1476(1):101–114. doi: 10.1104/pp.107.113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Yang C. S., Wei B., Weiner H. Precursor protein is readily degraded in mitochondrial matrix space if the leader is not processed by mitochondrial processing peptidase. J. Biol. Chem. 2007;2826(1):37266–37275. doi: 10.1074/jbc.M706594200. [DOI] [PubMed] [Google Scholar]
- Muller F., Frentzen M. Phosphatidylglycerophosphate synthases from Arabidopsis thaliana. FEBS Lett. 2001;5096(1):298–302. doi: 10.1016/s0014-5793(01)03163-5. [DOI] [PubMed] [Google Scholar]
- Mulligan R. M., Chang K. L., Chou C. C. Computational analysis of RNA editing sites in plant mitochondrial genomes reveals similar information content and a sporadic distribution of editing sites. Mol. Biol. Evol. 2007;246(1):1971–1981. doi: 10.1093/molbev/msm125. [DOI] [PubMed] [Google Scholar]
- Muralla R., Chen E., Sweeney C., Gray J. A., Dickerman A., Nikolau B. J., Meinke D. A Bifunctional Locus (BIO3-BIO1) Required for Biotin Biosynthesis in Arabidopsis. Plant Physiol. 2008;1466(1):60–73. doi: 10.1104/pp.107.107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha M. W., Lister R., Ho A. Y., Whelan J. Identification, expression, and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol. 2003;1316(1):1737–1747. doi: 10.1104/pp.102.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha M. W., Elhafez D., Millar A. H., Whelan J. The C-terminal region of TIM17 links the outer and inner mitochondrial membranes in Arabidopsis and is essential for protein import. J. Biol. Chem. 2005;2806(1):16476–16483. doi: 10.1074/jbc.M413299200. [DOI] [PubMed] [Google Scholar]
- Murcha M. W., Elhafez D., Lister R., Tonti-Filippini J., Baumgartner M., Philippar K., Carrie C., Mokranjac D., Soll J., Whelan J. Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 2007;1436(1):199–212. doi: 10.1104/pp.106.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R., Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;3486(1):85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Sakurai N. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2006;476(1):772–783. doi: 10.1093/pcp/pcj051. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Shimizu S., Watanabe T., Yamaguchi O., Otsu K., Yamagata H., Inohara H., Kubo T., Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;4346(1):652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;246(1):34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yamaguchi Y., Sano H. Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic activities. Eur. J. Biochem. 2000;2676(1):5621–5630. doi: 10.1046/j.1432-1327.2000.01633.x. [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Herve C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007;656(1):23–33. doi: 10.1007/s00239-006-0174-z. [DOI] [PubMed] [Google Scholar]
- Nerlich A., von Orlow M., Rontein D., Hanson A. D., Dormann P. Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol. 2007;1446(1):904–914. doi: 10.1104/pp.107.095414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuburger M. Preparation of plant mitochondria, criterea for assessment of mitochondrial integerity,and purity, survival in vitro. In: Douce R., Day D. A., editors. Higher Plant Cell Respiration. 1. Vol. 6. Berlin: Springer-Verlag; 1985. pp. 7–24. [Google Scholar]
- Neuburger M., Journet E., Bligny R., Carde J., Douce R. Purification of plant mitochondria by isopycnic centrifugation in density gradients of percoll. Archives of Biochemistry and Biophysics. 1982;2176(1):312–323. doi: 10.1016/0003-9861(82)90507-0. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;766(1):723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Newmeyer D. D., Ferguson-Miller S. Mitochondria. Releasing power for life and unleashing the machineries of death. Cell. 2003;1126(1):481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Noji M., Inoue K., Kimura N., Gouda A., Saito K. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J. Biol. Chem. 1998;2736(1):32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- Ohtsu K., Nakazono M., Tsutsumi N., Hirai A. Characterization and expression of the genes for cytochrome c oxidase subunit VIb (COX6b) from rice and Arabidopsis thaliana. Gene. 2001;2646(1):233–239. doi: 10.1016/s0378-1119(01)00334-1. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;396(1):503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA. 2007;1046(1):8178–8183. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. J., Raman R. Glycine decarboxylase: protein chemistry and molecular biology of the major protein in leaf mitochondria. J. Bioenerg. Biomem. 1996;276(1):407–414. doi: 10.1007/BF02110003. [DOI] [PubMed] [Google Scholar]
- Oliver D. J., Neuburger M., Bourguignon J., Douce R. Interaction between the component enzymes of the glycine decarboxylase multienzyme complex. Plant Physiol. 1990;946(1):833–839. doi: 10.1104/pp.94.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. G., Rasmussen A. V., von Wettstein-Knowles P., Henriksen A. Structure of the mitochondrial beta-ketoacyl-[acyl carrier protein] synthase from Arabidopsis and its role in fatty acid synthesis. FEBS Lett. 2004;5776(1):170–4. doi: 10.1016/j.febslet.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Picault N., Arrigoni R., Besin E., Palmieri F., Hodges M. Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem. J. 2008;4106(1):621–629. doi: 10.1042/BJ20070867. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Todd C. D., Arrigoni R., Hoyos M. E., Santoro A., Polacco J. C., Palmieri F. Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim. Biophys. Acta. 2006a;17576(1):1277–1283. doi: 10.1016/j.bbabio.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M. I., Studart-Guimaraes C., Fernie A. R., Palmieri F. Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 2006b;1426(1):855–865. doi: 10.1104/pp.106.086975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J., Schmidt A. Characterization of a sul-furtransferase from Arabidopsis thaliana. Eur. J. Biochem. 2000;2676(1):145–154. doi: 10.1046/j.1432-1327.2000.00980.x. [DOI] [PubMed] [Google Scholar]
- Parisi G., Perales M., Fornasari M. S., Colaneri A., Gonzalez-Schain N., Gomez-Casati D., Zimmermann S., Brennicke A., Araya A., Ferry J. G., Echave J., Zabaleta E. Gamma carbonic anhydrases in plant mitochondria. Plant Mol. Biol. 2004;556(1):193–207. doi: 10.1007/s11103-004-0149-7. [DOI] [PubMed] [Google Scholar]
- Pastori G. M., Kiddle G., Antoniw J., Bernard S., Veljovic-Jovanovic S., Verrier P. J., Noctor G., Foyer C. H. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell. 2003;156(1):939–951. doi: 10.1105/tpc.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N., Small I. Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta. 2001;15416(1):54–63. doi: 10.1016/s0167-4889(01)00146-x. [DOI] [PubMed] [Google Scholar]
- Peiffer W. E., Ingle R. T., Ferguson-Miller S. Structurally unique plant cytochrome c oxidase isolated from wheat germ, a rich source of plant mitochondrial enzymes. Biochemistry. 1990;296(1):8696–8701. doi: 10.1021/bi00489a027. [DOI] [PubMed] [Google Scholar]
- Perales M., Eubel H., Heinemeyer J., Colaneri A., Zabaleta E., Braun H. P. Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 2005;3506(1):263–277. doi: 10.1016/j.jmb.2005.04.062. [DOI] [PubMed] [Google Scholar]
- Perrin R., Lange H., Grienenberger J. M., Gagliardi D. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 2004a;326(1):5174–5182. doi: 10.1093/nar/gkh852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R., Meyer E. H., Zaepfel M., Kim Y. J., Mache R., Grienenberger J. M., Gualberto J. M., Gagliardi D. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 2004b;2796(1):25440–25446. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- Perry A. J., Hulett J. M., Likic V. A., Lithgow T., Gooley P. R. Convergent evolution of receptors for protein import into mitochondria. Curr. Biol. 2006;166(1):221–229. doi: 10.1016/j.cub.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Geissler A. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2001;26(1):339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- Picault N., Hodges M., Palmieri L., Palmieri F. The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 2004;96(1):138–146. doi: 10.1016/j.tplants.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Picault N., Palmieri L., Pisano I., Hodges M., Palmieri F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 2002;2776(1):24204–24211. doi: 10.1074/jbc.M202702200. [DOI] [PubMed] [Google Scholar]
- Picciocchi A., Douce R., Alban C. Biochemical characterization of the Arabidopsis biotin synthase reaction. The importance of mitochondria in biotin synthesis. Plant Physiol. 2001;1276(1):1224–1233. [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A., Douce R., Alban C. The plant biotin synthase reaction. Identification and characterization of essential mitochondrial accessory protein components. J. Biol. Chem. 2003;2786(1):24966–24975. doi: 10.1074/jbc.M302154200. [DOI] [PubMed] [Google Scholar]
- Portereiko M. F., Sandaklie-Nikolova L., Lloyd A., Dever C. A., Otsuga D., Drews G. N. NUCLEAR FUSION DEFEC-TIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 2006;1416(1):957–965. doi: 10.1104/pp.106.079319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S., Becker T., Mirus O., Tews I., Soll J., Schleiff E. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006;256(1):1836–1847. doi: 10.1038/sj.emboj.7601091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S., Becker T., Bionda T., Reger K., Ruprecht M., Soll J., Schleiff E. Toc64—a preprotein-receptor at the outer membrane with bipartide function. J. Mol. Biol. 2007;3676(1):1330–1346. doi: 10.1016/j.jmb.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Rasmusson A. G., Heiser V., Zabaleta E., Brennicke A., Grohmann L. Physiological, biochemical and molecular aspects of mitochondrial complex I in plants. Biochim. Biophys. Acta. 1998;13646(1):101–111. doi: 10.1016/s0005-2728(98)00021-8. [DOI] [PubMed] [Google Scholar]
- Ravanel S., Cherest H., Jabrin S., Grunwald D., Surdin-Kerjan Y., Douce R., Rebeille F. Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2001;986(1):15360–15365. doi: 10.1073/pnas.261585098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J. C. Mechanisms of apoptosis. Am. J. Pathol. 2000;1576(1):1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial D. V., Lombardo V. A., Ceccarelli E. A., Ottado J. The import of ferredoxin-NADP+ reductase precursor into chloroplasts is modulated by the region between the transit peptide and the mature core of the protein. Eur. J. Biochem. 2002;2696(1):5431–5439. doi: 10.1046/j.1432-1033.2002.03233.x. [DOI] [PubMed] [Google Scholar]
- Richly E., Chinnery P. F., Leister D. Evolutionary diversification of mitochondrial proteomes: implications for human disease. Trends Genet. 2003;186(1):356–362. doi: 10.1016/S0168-9525(03)00137-9. [DOI] [PubMed] [Google Scholar]
- Rikhvanov E. G., Gamburg K. Z., Varakina N. N., Rusaleva T. M., Fedoseeva I. V., Tauson E. L., Stupnikova I. V., Stepanov A. V., Borovskii G. B., Voinikov V. K. Nuclear-mitochondrial cross-talk during heat shock in Arabidopsis cell culture. Plant J. 2007;526(1):763–778. doi: 10.1111/j.1365-313X.2007.03275.x. [DOI] [PubMed] [Google Scholar]
- Robson C. A., Vanlerberghe G. C. Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria dependent- and -independent pathways of programmed cell death. Plant Physiol. 2002;1296(1):1908–1920. doi: 10.1104/pp.004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Roldan V., Garcia-Heredia J. M., Navarro J. A., Hervas M., De la Cerda B., Molina-Heredia F. P., De la Rosa M. A. A comparative kinetic analysis of the reactivity of plant, horse, and human respiratory cytochrome c towards cytochrome c oxidase. Biochem. Biophys. Res. Commun. 2006;3466(1):1108–1113. doi: 10.1016/j.bbrc.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Rosenbaum Hofmann N., Theg S. M. Toc64 is not required for import of proteins into chloroplasts in the moss Physcomitrella patens. Plant J. 2005;436(1):675–687. doi: 10.1111/j.1365-313X.2005.02483.x. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P., Coelho P. S., Roemer T., Agarwal S., Kumar A., Jansen R., Cheung K. H., Sheehan A., Symoniatis D., Umansky L., Heidtman M., Nelson F. K., Iwasaki H., Hager K., Gerstein M., Miller P., Roeder G. S., Snyder M. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;4026(1):413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Rudhe C., Chew O., Whelan J., Glaser E. A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J. 2002;306(1):213–220. doi: 10.1046/j.1365-313x.2002.01280.x. [DOI] [PubMed] [Google Scholar]
- Sabar M., Gagliardi D., Balk J., Leaver C. J. ORFB is a subunit of F(1)F(O)-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 2003;46(1):1–6. doi: 10.1038/sj.embor.embor800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D., Prasad A. M., Sujatha T. P., Kumar V., Jain P. K., Bhat S. R., Srinivasan R. In silico analysis of the lateral organ junction (LOJ) gene and promoter of Arabidopsis thaliana. In Silico Biol. 2007;76(1):7–19. [PubMed] [Google Scholar]
- Saisho D., Nambara E., Naito S., Tsutsumi N., Hirai A., Nakazono M. Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol Biol. 1997;356(1):585–596. doi: 10.1023/a:1005818507743. [DOI] [PubMed] [Google Scholar]
- Saisho D., Nakazono M., Lee K., Tsutsumi N., Akita S., Hirai A. The gene for alternative oxidase-2 (AOX2) from Arabidopsis thaliana consists of five exons unlike other AOX genes and is transcribed at an early stage during germination. Genes Genet. Syst. 2001;766(1):89–97. doi: 10.1266/ggs.76.89. [DOI] [PubMed] [Google Scholar]
- Salone V., Rudinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;5816(1):4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Fischer K., Ketelsen B., Krupinska K., Krause K. Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics. 2007;2776(1):631–646. doi: 10.1007/s00438-007-0214-4. [DOI] [PubMed] [Google Scholar]
- Scott I., Tobin A. K., Logan D. C. BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 2006;576(1):1275–1280. doi: 10.1093/jxb/erj096. [DOI] [PubMed] [Google Scholar]
- Sheahan M. B., McCurdy D. W., Rose R. J. Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 2005;446(1):744–755. doi: 10.1111/j.1365-313X.2005.02561.x. [DOI] [PubMed] [Google Scholar]
- Shedge V., Arrieta-Montiel M., Christensen A. C., Mackenzie S. A. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;196(1):1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol. Life Sci. 2006;636(1):698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Narita M., Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;3396(1):483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- Shintani D. K., Ohlrogge J. B. The characterization of a mitochondrial acyl carrier protein isoform isolated from Arabidopsis thaliana. Plant Physiol. 1994;1046(1):1221–1229. doi: 10.1104/pp.104.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H. E., Schonfisch B., Perschil I., Chacinska A., Guiard B., Rehling P., Pfanner N., Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 2003;1006(1):13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Filho M. C. One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr. Opin. Plant Biol. 2003;66(1):1–7. doi: 10.1016/j.pbi.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Small I., Wintz H., Akashi K., Mireau H. Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol. Biol. 1998;386(1):265–277. [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;46(1):1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Small I. D., Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;256(1):46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Somerville C. R., Ogren W. L. Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem. J. 1982;2026(1):373–380. doi: 10.1042/bj2020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Oliver D. J. Light-dependent and tissue-specific expression of the H-protein of the glycine decarboxylase complex. Plant Physiol. 1995;1096(1):161–168. doi: 10.1104/pp.109.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Kraus C., Oliver D. J. Developmental Expression of the Glycine Decarboxylase Multienzyme Complex in Green Pea Leaves. In: Lambers H., Plas L. H. Wvd, editors. Molecular, Biochemical and Physiological Aspects of Plant Respiration. 1. Vol. 6. The Hague: SPB Academic Publishing; 1992. pp. 323–334. [Google Scholar]
- Stahl A., Nilsson S., Lundberg P., Bhushan S., Biverstahl H., Moberg P., Morisset M., Vener A., Maler L., Langel U., Glaser E. Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J. Mol. Biol. 2005;3496(1):847–860. doi: 10.1016/j.jmb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Stähl A., Moberg P., Ytterberg J., Panfilov O., Brockenhuus Von Lowenhielm H., Nilsson F., Glaser E. Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. J. Biol. Chem. 2002;2776(1):41931–41939. doi: 10.1074/jbc.M205500200. [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Guiard B., Kozjak-Pavlovic V., Pfanner N., Meisinger C. Alternative function for the mitochondrial SAM complex in biogenesis of alpha-helical TOM proteins. J. Cell Biol. 2007;1796(1):881–893. doi: 10.1083/jcb.200706043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A., Asami T., Alonso J., Ecker J. R., Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;4216(1):79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- Sunderhaus S., Dudkina N. V., Jansch L., Klodmann J., Heinemeyer J., Perales M., Zabaleta E., Boekema E. J., Braun H. P. Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J. Biol. Chem. 2006;2816(1):6482–6488. doi: 10.1074/jbc.M511542200. [DOI] [PubMed] [Google Scholar]
- Sunderland P. A., West C. E., Waterworth W. M., Bray C. M. Choice of a start codon in a single transcript determines DNA ligase 1 isoform production and intracellular targeting in Arabidopsis thaliana. Biochem Soc. Trans. 2004;326(1):614–616. doi: 10.1042/BST0320614. [DOI] [PubMed] [Google Scholar]
- Sunderland P. A., West C. E., Waterworth W. M., Bray C. M. An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J. 2006;476(1):356–367. doi: 10.1111/j.1365-313X.2006.02791.x. [DOI] [PubMed] [Google Scholar]
- Sweetlove L. J., Heazlewood J. L., Herald V., Holtzapffel R., Day D. A., Leaver C. J., Millar A. H. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002;326(1):891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Sweetlove L. J., Lytovchenko A., Morgan M., Nunes-Nesi A., Taylor N. L., Baxter C. J., Eickmeier I., Fernie A. R. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sc.i USA. 2006;1036(1):19587–19592. doi: 10.1073/pnas.0607751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabatake R., Hata S., Taniguchi M., Kouchi H., Sugiyama T., Izui K. Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopis. Plant Mol. Biol. 1999;406(1):479–486. doi: 10.1023/a:1006285009435. [DOI] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C. A., Innes R. W. A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 2006;476(1):75–84. doi: 10.1111/j.1365-313X.2006.02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;276(1):306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H. Lipoic acid-dependent oxidative catabolism of 2-oxo acids in mitochondria provides evidence of branched chain amino acid catabolism in Arabidopsis. Plant Physiol. 2003a;1346(1):838–848. doi: 10.1104/pp.103.035675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. Characterization of the human heart mitochondrial proteome. Nature Biotech. 2003b;216(1):281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;4086(1):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Thelen J. J., Miernyk J. A., Randall D. D. Pyruvate dehydrogenase kinase from Arabidopsis thaliana: a protein histidine kinase that phosphorylates serine residues. Biochem. J. 2000;3496(1):195–201. doi: 10.1042/0264-6021:3490195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen J. J., Muszynski M. G., David N. R., Luethy M. H., Elthon T. E., Miernyk J. A., Randall D. D. The dihydrolipoamide S-acetyltransferase subunit of the mitochondrial pyruvate dehydrogenase complex from maize contains a single lipoyl domain. J. Biol. Chem. 1999;2746(1):21769–21775. doi: 10.1074/jbc.274.31.21769. [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D., McCabe T. C., Clifton R., Moore C., Finnegan P. M., Day D. A., Whelan J. Analysis of the Alternative Oxidase Promoters from Soybean. Plant Physiol. 2003;1336(1):1158–1169. doi: 10.1104/pp.103.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley H. E., McDonald K., Jenkins G. I., Knight M. R., Leaver C. J. Ceramides induce programmed cell death in Arabidopsis in a calcium-dependent manner. Biol. Chem. 2005;3866(1):161–166. doi: 10.1515/BC.2005.020. [DOI] [PubMed] [Google Scholar]
- Tremousaygue D., Garnier L., Bardet C., Dabos P., Herve C., Lescure B. Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 2003;336(1):957–966. doi: 10.1046/j.1365-313x.2003.01682.x. [DOI] [PubMed] [Google Scholar]
- Truscott K. N., Brandner K., Pfanner N. Mechanisms of protein import into mitochondria. Curr. Biol. 2003;136(1):R326–337. doi: 10.1016/s0960-9822(03)00239-2. [DOI] [PubMed] [Google Scholar]
- Umbach A. L., Fiorani F., Siedow J. N. Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 2005;1396(1):1806–20. doi: 10.1104/pp.105.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger E. A., Hand J. M., Cashmore A. R., Vasconcelos A. C. Isolation of a cDNA encoding mitochondrial citrate synthase from Arabidopsis thaliana. Plant Mol. Biol. 1989;136(1):411–418. doi: 10.1007/BF00015553. [DOI] [PubMed] [Google Scholar]
- Unseld M., Marienfeld J. R., Brandt P., Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997;156(1):57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- Van Cauwenberghe O. R., Makhmoudova A., McLean M. D., Clark S. M., Shelp B. J. Plant pyruvate-dependent gamma-aminobutyrate transaminase: identification of the Arabidopsis cDNA and its expression in Escherichia coli. Can. J. Bot. 2002;806(1):933–941. [Google Scholar]
- van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J., Moczko M., Wagner R., Meijer M., Guiard B., Honlinger A., Pfanner N. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;4016(1):485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G. C., McLntosh L. Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol. 1996;1116(1):589–595. doi: 10.1104/pp.111.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G. C., Robson C. A., Yip J. Y. Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol. 2002;1296(1):1829–1842. doi: 10.1104/pp.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclare P., Marcherel D., Douce R., Bourguignon J. The gene encoding T protein of the glycine decarboxylase complex involved in the mitochondrial step of the photorespiratory pathway in plants exhibits features of light-induced genes. Plant Mol. Biol. 1998;376(1):309–318. doi: 10.1023/a:1005954200042. [DOI] [PubMed] [Google Scholar]
- Verbruggen N., Hua X. J., May M., Van Montagu M. Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc. Natl. Acad. Sci. USA. 1996;936(1):8787–8791. doi: 10.1073/pnas.93.16.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercesi A. E., Borecky J., Maia Ide G., Arruda P., Cuccovia I. M., Chaimovich H. Plant uncoupling mitochondrial proteins. Annu. Rev. Plant Biol. 2006;576(1):383–404. doi: 10.1146/annurev.arplant.57.032905.105335. [DOI] [PubMed] [Google Scholar]
- Vermel M., Guermann B., Delage L., Grienenberger J. M., Marechal-Drouard L., Gualberto J. M. A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA. 2002;996(1):5866–5871. doi: 10.1073/pnas.092019599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Yasuno R., Jordan S. W., Cronan J. E., Jr., Wada H. Lipoic acid metabolism in Arabidopsis thaliana: cloning and characterization of a cDNA encoding lipoyltransferase. Plant Cell Physiol. 2001;426(1):650–656. doi: 10.1093/pcp/pce081. [DOI] [PubMed] [Google Scholar]
- Wall M. K., Mitchenall L. A., Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA. 2004;1016(1):7821–7826. doi: 10.1073/pnas.0400836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zou Y., Li X., Zhang Q., Chen L., Wu H., Su D., Chen Y., Guo J., Luo D., Long Y., Zhong Y., Liu Y. G. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;186(1):676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Nakazono M., Tsutsumi N., Hirai A. AtUCP2: a novel isoform of the mitochondrial uncoupling protein of Arabidopsis thaliana. Plant Cell Physiol. 1999;406(1):1160–1166. doi: 10.1093/oxfordjournals.pcp.a029501. [DOI] [PubMed] [Google Scholar]
- Welchen E., Gonzalez D. H. Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol. 2005;1396(1):88–100. doi: 10.1104/pp.105.065920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E., Gonzalez D. H. Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiol. 2006;1416(1):540–545. doi: 10.1104/pp.105.075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E., Chan R. L., Gonzalez D. H. Metabolic regulation of genes encoding cytochrome c and cytochrome c oxidase subunit Vb in Arabidopsis. Plant Cell Environ. 2002;256(1):1605–1615. [Google Scholar]
- Werhahn W., Braun H. P. Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis. 2002;236(1):640–646. doi: 10.1002/1522-2683(200202)23:4<640::AID-ELPS640>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Werhahn W., Jansch L., Braun H-P. Identification of novel subunits of the TOM complex from Arabidopsis thaliana. Plant Physiol. Biochem. 2003;416(1):407–416. [Google Scholar]
- Werhahn W., Niemeyer A., Jansch L., Kruft V. V., Schmitz U. K., Braun H. P. Purification and Characterization of the Preprotein Translocase of the Outer Mitochondrial Membrane from Arabidopsis. Identification of Multiple Forms of TOM20. Plant Physiol. 2001;1256(1):943–954. doi: 10.1104/pp.125.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J., Schleiff E. Protein targeting and import. In: Day D. A., Millar A. H., Whelan J., editors. Advances in Photosynthesis and Respiration. Dordrecht: Kluwer; 2004. [Google Scholar]
- Winger A. M., Millar A. H., Day D. A. Sensitivity of plant mitochondrial terminal oxidases to the lipid peroxidation product 4-hydroxy-2-nonenal (HNE). Biochem J. 2005;3876(1):865–870. doi: 10.1042/BJ20042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger A. M., Taylor N. L., Heazlewood J. L., Day D. A., Millar A. H. The Cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal covalently modifies a selective range of proteins linked to respiratory function in plant mitochondria. J. Biol. Chem. 2007;2826(1):37436–37447. doi: 10.1074/jbc.M702385200. [DOI] [PubMed] [Google Scholar]
- Wirtz M., Berkowitz O., Droux M., Hell R. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur. J. Biochem. 2001;2686(1):686–693. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- Woodson J. D., Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008;96(1):383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Yatsukawa Y. I., Esaki M., Hobbs A. E., Jensen R. E., Endo T. TOM20 and TOM22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 2008;2836(1):3799–3807. doi: 10.1074/jbc.M708339200. [DOI] [PubMed] [Google Scholar]
- Yao N., Eisfelder B. J., Marvin J., Greenberg J. T. The mitochondrion—an organelle commonly involved in programmed cell death in Arabidopsis thaliana. Plant J. 2004;406(1):596–610. doi: 10.1111/j.1365-313X.2004.02239.x. [DOI] [PubMed] [Google Scholar]
- Yasuno R., Wada H. Biosynthesis of lipoic acid in Arabidopsis: cloning and characterization of the cDNA for lipoic acid synthase. Plant Physiol. 1998;1186(1):935–943. doi: 10.1104/pp.118.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R., Wada H. The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett. 2002;5176(1):110–114. doi: 10.1016/s0014-5793(02)02589-9. [DOI] [PubMed] [Google Scholar]
- Yasuno R., von Wettstein-Knowles P., Wada H. Identification and molecular characterization of the beta-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase. J. Biol. Chem. 2004;2796(1):8242–8251. doi: 10.1074/jbc.M308894200. [DOI] [PubMed] [Google Scholar]
- Yu J., Nickels R., McIntosh L. A genome approach to mitochondrial-nuclear communication in Arabidopsis. Plant Physiol. Biochem. 2001;396(1):345–353. [Google Scholar]
- Zabaleta E., Heiser V., Grohmann L., Brennicke A. Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J. 1998;156(1):49–59. doi: 10.1046/j.1365-313x.1998.00177.x. [DOI] [PubMed] [Google Scholar]
- Zaegel V., Guermann B., Le Ret M., Andres C., Meyer D., Erhardt M., Canaday J., Gualberto J. M., Imbault P. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006;186(1):3548–3563. doi: 10.1105/tpc.106.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkovic J., Anderson S. L., Rhoads D. M. A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol. Biol. 2005;576(1):871–888. doi: 10.1007/s11103-005-3249-0. [DOI] [PubMed] [Google Scholar]
- Zhou J., Weiner H. The N-terminal portion of mature aldehyde dehydrogenase affects protein folding and assembly. Protein Science. 2001;106(1):1490–1497. doi: 10.1110/ps.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. F., Suzuki K., Saito T., Okada K., Tanaka K., Nakagawa T., Matsuda H., Kawamukai M. Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Mol. Biol. 1997;356(1):331–341. doi: 10.1023/a:1005898805326. [DOI] [PubMed] [Google Scholar]
- Zou J., Qi Q., Katavic V., Marillia E. F., Taylor D. C. Effects of antisense repression of an Arabidopsis thaliana pyruvate dehydrogenase kinase cDNA on plant development. Plant Mol. Biol. 1999;416(1):837–849. doi: 10.1023/a:1006393726018. [DOI] [PubMed] [Google Scholar]


