Abstract
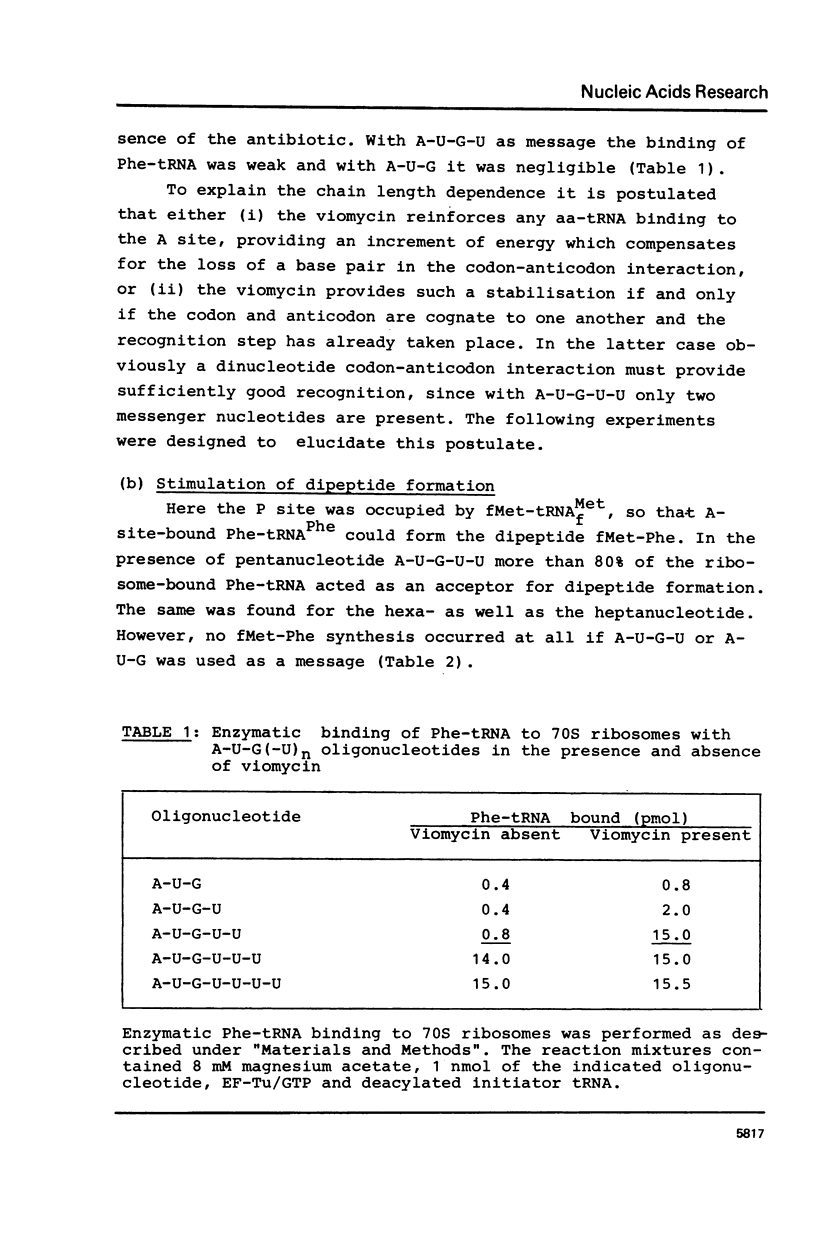
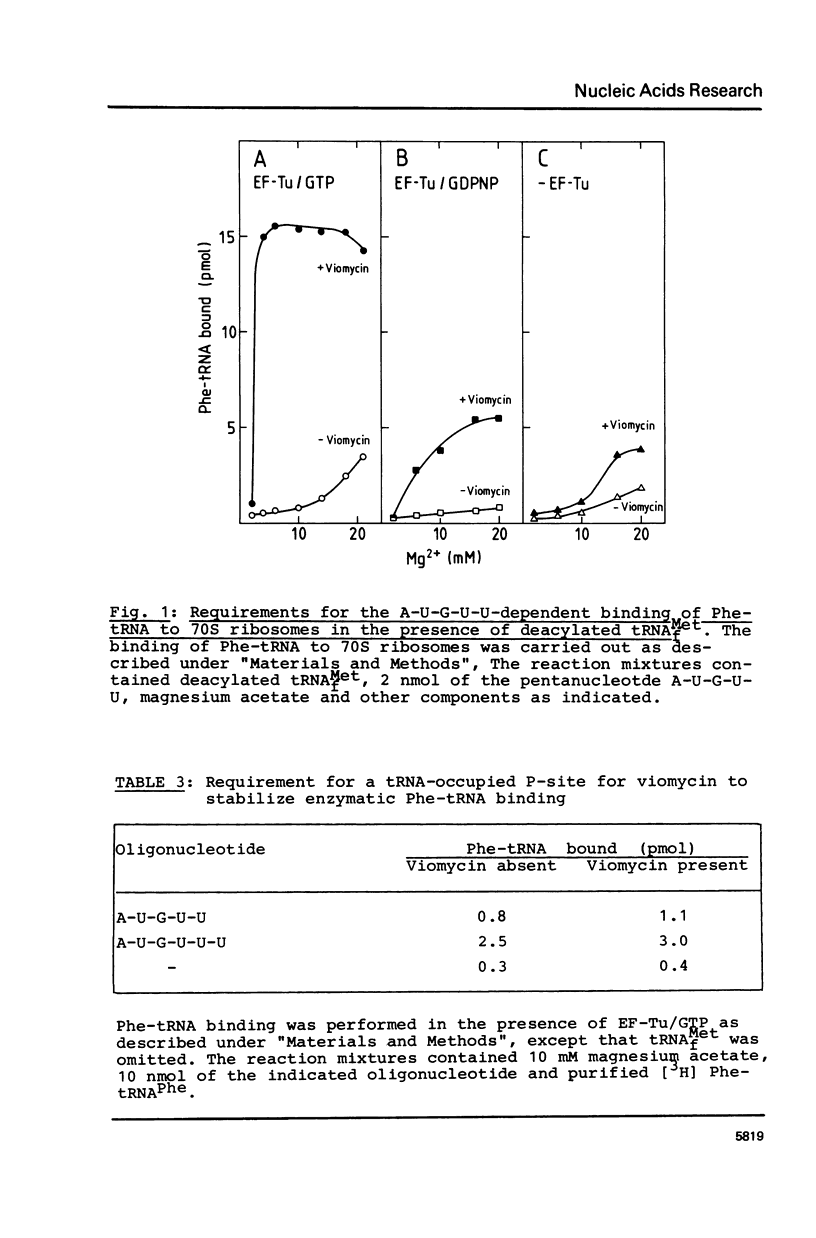
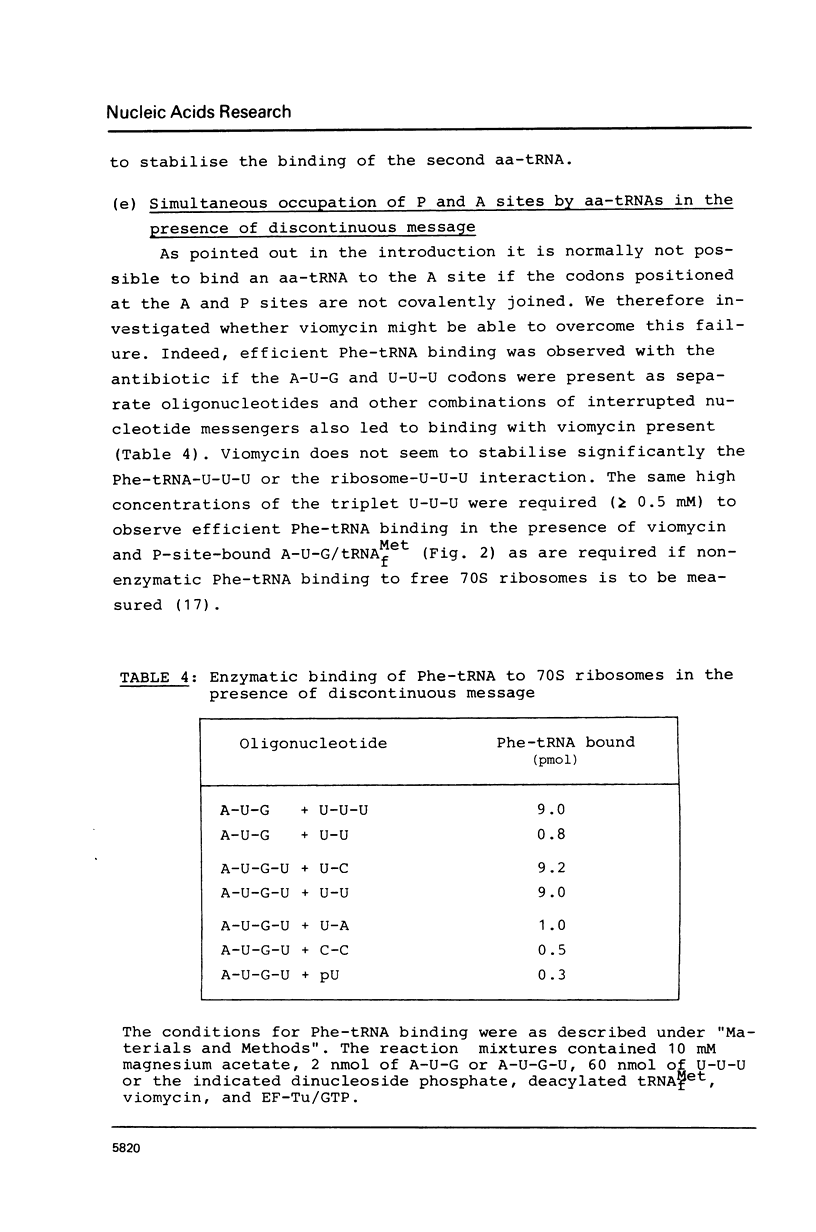
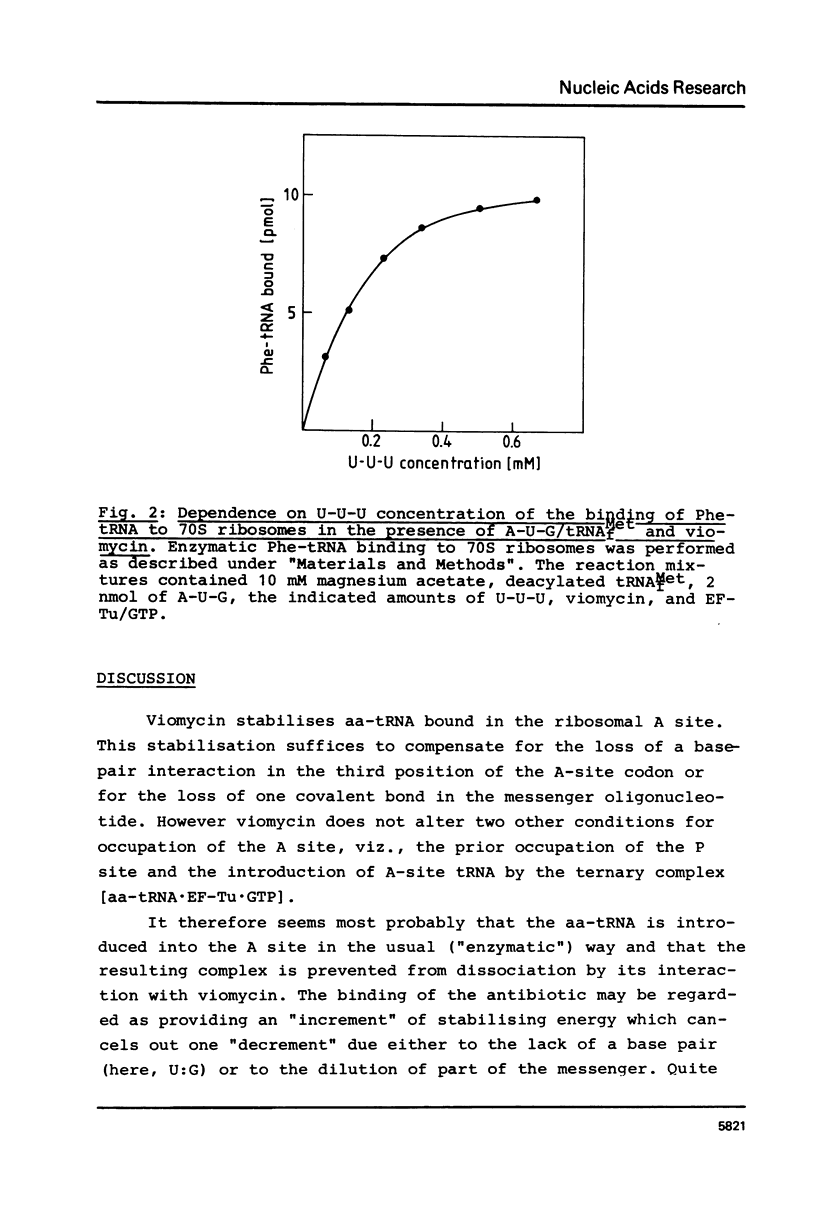
The requirements for the decoding process at the ribosomal A site have been investigated in the presence of viomycin. For these studies natural mRNA was replaced either by the synthetic oligonucleotide A-U-G(-U)n, with 0 less than or equal to n less than or equal to 4, or by a physical mixture of the oligonucleotides A-U-G and various oligo(U) sequences. Thus the effect of the "removal" of selected covalent bonds from the sequence A-U-G(U)n could be studied. When the ribosomal P site contains tRNAMetf, then normally the full hexanucleotide "messenger" A-U-G-U-U-U is needed for the EF-Tu-mediated binding of Phe-tRNA into the A site. However in presence of viomycin the pentanucleotide A-U-G-U-U suffices for this. It is also possible in the presence of viomycin to replace A-U-G-U and U-U. In all the above systems the binding of Phe-tRNA required the presence of EF-Tu and GTP. The results suggest that viomycin reinforces interactions between aa-tRNA and the A site after the codon-anticodon recognition step.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K. I., Kawakita M., Kaziro Y. Studies on polypeptide elongation factors from Escherichia coli. II. Purification of factors Tu-guanosine diphosphate, Ts, and Tu-Ts, and crystallization of Tu-guanosine diphosphate and Tu-Ts. J Biol Chem. 1972 Nov 10;247(21):7029–7037. [PubMed] [Google Scholar]
- De Groot N., Panet A., Lapidot Y. The binding of purified Phe-tRNA and peptidyl-tRNA Phe to Escherichia coli ribosomes. Eur J Biochem. 1971 Dec 10;23(3):523–527. doi: 10.1111/j.1432-1033.1971.tb01649.x. [DOI] [PubMed] [Google Scholar]
- Erbe R. W., Nau M. M., Leder P. Translation and translocation of defined RNA messengers. J Mol Biol. 1969 Feb 14;39(3):441–460. doi: 10.1016/0022-2836(69)90137-5. [DOI] [PubMed] [Google Scholar]
- Gassen H. G., Schetters H., Matthaei H. Codon-anticodon interaction studied with oligonucleotides containing 3 -deazauridine, 4 -deoxyuridine or 3 -deaza- 4 -deoxyuridine. II. Ribosome binding of oligonucleotides and phenylalanyl-tRNA. Biochim Biophys Acta. 1972 Jul 31;272(4):560–567. doi: 10.1016/0005-2787(72)90511-4. [DOI] [PubMed] [Google Scholar]
- Hershey J. W., Thach R. E. Role of guanosine 5'-triphosphate in the initiation of Peptide synthesis, I. Synthesis of formylmethionyl-puromycin. Proc Natl Acad Sci U S A. 1967 Mar;57(3):759–766. doi: 10.1073/pnas.57.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou Y. F., Tanaka N. Dual actions of viomycin on the ribosomal functions. Biochem Biophys Res Commun. 1976 Jul 26;71(2):477–483. doi: 10.1016/0006-291x(76)90812-3. [DOI] [PubMed] [Google Scholar]
- Lührmann R., Eckhardt H., Stöffler G. Codon-anticodon interaction at the ribosomal peptidyl-site. Nature. 1979 Aug 2;280(5721):423–425. doi: 10.1038/280423a0. [DOI] [PubMed] [Google Scholar]
- Misumi M., Tanaka N. Mechanism of inhibition of translocation by kanamycin and viomycin: a comparative study with fusidic acid. Biochem Biophys Res Commun. 1980 Jan 29;92(2):647–654. doi: 10.1016/0006-291x(80)90382-4. [DOI] [PubMed] [Google Scholar]
- Misumi M., Tanaka N., Shiba T. Binding of [14C]tuberactinomycin O, an antibiotic closely related to viomycin, to the bacterial ribosome. Biochem Biophys Res Commun. 1978 Jun 14;82(3):971–976. doi: 10.1016/0006-291x(78)90878-1. [DOI] [PubMed] [Google Scholar]
- Modolell J., Vázquez The inhibition of ribosomal translocation by viomycin. Eur J Biochem. 1977 Dec;81(3):491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- Mohr S. C., Thach R. E. Application of ribonuclease T1 to the synthesis of oligoribonucleotides of defined base sequence. J Biol Chem. 1969 Dec 25;244(24):6566–6576. [PubMed] [Google Scholar]
- NIRENBERG M., LEDER P. RNA CODEWORDS AND PROTEIN SYNTHESIS. THE EFFECT OF TRINUCLEOTIDES UPON THE BINDING OF SRNA TO RIBOSOMES. Science. 1964 Sep 25;145(3639):1399–1407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- Noll M., Hapke B., Schreier M. H., Noll H. Structural dynamics of bacterial ribosomes. I. Characterization of vacant couples and their relation to complexed ribosomes. J Mol Biol. 1973 Apr 5;75(2):281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- Springer M., Grunberg-Manago M. Characteristics of N-Ac-Phe-tRNA binding and its correlation with internal aminoacyl-tRNA recognition. Biochem Biophys Res Commun. 1972 Apr 28;47(2):477–484. doi: 10.1016/0006-291x(72)90739-5. [DOI] [PubMed] [Google Scholar]
- Turnowsky F., Högenauer G. Colicin E 3, an inactivating agent of the ribosomal A-site. Biochem Biophys Res Commun. 1973 Dec 19;55(4):1246–1254. doi: 10.1016/s0006-291x(73)80028-2. [DOI] [PubMed] [Google Scholar]
- Yamada T., Bierhaus K. H. Viomycin favours the formation of 70S ribosome couples. Mol Gen Genet. 1978 May 31;161(3):261–265. doi: 10.1007/BF00330999. [DOI] [PubMed] [Google Scholar]



