Abstract
Characterization of the stimulated release of neuropeptides from brain slices and individual cultured neurons requires efficient collection of the releasate from relatively large volumes of physiological saline. Here several collection approaches are optimized using particle-embedded monolithic capillaries (PEMCs) with poly(stearyl methacrylate-co-ethylene glycol dimethacrylate) monolith acting as a “glue.” Two distinct extraction particles, with either pyrrolidone (PY) or ethylenediamine (EDA) as the functional group on polystyrene backbone, have been embedded into capillaries having an inner diameter of 250 μm. The capillaries act as collection devices for sampling neuropeptide release; the collection protocols are described and the extraction efficiency of the probes characterized. Specifically, the binding of angiotensin II from a peptide mixture onto the PY and EDA columns was 16 and 28 pmol, respectively, in a volume of 20 μL of saline. The peptides released from these columns have been characterized via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry with low femtomole detection limits. By positioning the PEMC columns in close proximity to individual neurons and using 50 mM of KCl as the secretagogue, peptides released from individual identified cultured neurons isolated from Aplysia californica were collected and characterized.
Keywords: Particle-embedded monolithic capillary, neuropeptide, single neuron, release, MALDI MS
Introduction
Neuropeptides are among the most structurally and chemically diverse cell-to-cell signaling molecules in the nervous system. They are involved in a wide range of physiological processes such as neuromodulation, neurotransmission, cell outgrowth, cell survival, and hormonal signaling between organs.1,2 Investigations to monitor the activity-dependent release of neuropeptides in a chemically, spatially and temporally resolved manner aid in understanding neuropeptide function as well as neuronal network physiology. Although these studies can involve measuring peptides released from relatively larger nervous system areas such as discrete brain regions, ganglia or neuron clusters, we are interested in probing activity-dependent release from individual neurons. Why? Because even adjacent neurons have appreciably different biochemical profiles and thus, the response of a network to the activity of specific cells within that network varies significantly.3,4
The ability to collect and characterize cell-to-cell signaling peptides released from targeted neurons facilitates a number of neuroscience-related analyses. Neuropeptides can be active at low levels and across a broad dynamic range. They are diluted upon cellular release by orders of magnitude into a chemically complex extracellular environment containing high levels of inorganic salts, which can interfere with peptide collection and subsequent detection. Even with these challenges, several methods have been used successfully to characterize single-neuron peptide release. These include radioactive labeling using radioimmunoassays5 or pulse/chase experiments to investigate radiolabeled neuropeptides via their in situ synthesis.6,7 Also effective are electrochemical techniques, which typically focus on small molecules and use micro- or nanoelectrodes and electrode arrays to provide high spatial resolution and low mass detection limits.8–10 By monitoring the oxidation of tryptophan and tyrosine residues with an amperometric method, peptides released from single melanotrophs of rat pituitary have been characterized.11 Although such methods provide high selectivity and sensitivity and thus permit direct sampling from relatively large volumes of extracellular media around single cells, they can require peptide selection and ancillary analytical approaches to validate the identity of the peptides.
Mass spectrometry (MS) has been applied to vast variety of peptide measurement tasks, due to its ability to identify multiple analytes simultaneously.12,13 Among them, matrix-assisted laser desorption/ionization (MALDI)-time-of-flight (TOF) MS is especially well suited for single-cell peptide investigations. It allows measurements of small sample sizes while providing high sensitivity, and sample preparation requirements are relatively simple.14 When using MALDI, appropriate sample pretreatment, including desalting and concentrating, enhances MS detection in terms of signal intensity, peak abundance and reproducibility. Commercially available pipette tips packed with extraction media have been used for desalting biological samples such as releasates from rat brain slices before MALDI detection.15 However, pipette tips are not at the ideal size scale for many individual cell manipulations or for subcellular sampling. When detecting peptides released from single neurons, a collection probe capable of being placed close to the secretion site to efficiently collect releasates from high ionic strength extracellular media is desired. In earlier work, releasates from clusters of Aplysia californica bag cell neurons16 and from individual neurons17 were collected using capillaries followed by additional extraction and characterization via MS. Recently, single solid phase extraction beads were placed directly on an A. californica bag cell neuron and peptide releasates collected at a high spatial resolution, and where secretion profiles were found to be different in processes versus cell bodies.18,19 This technique however is manually intensive and achieving reproducibility has been difficult, primarily due to issues in placing and retrieving beads.
An alternative strategy relies upon polymer monolithic materials to achieve peptide separations and extractions.20–22 One advantage of these materials is the ease with which the chemical properties of the monolith can be optimized for specific analyte classes, either by selecting different monomers/cross-linkers or further modifying the monolith with the intended functional group after polymerization.23–26, This results in the availability of multiple surface chemistries and improved performance in extraction or separation of specific analytes. In addition to modifications made directly to the monolith itself, hybrid materials have also been fabricated where particles of different surface properties and size, varying from nanometer to micrometer, are embedded into the monolith. The variable surface chemistries of the embedded particles provide the hybrid material with dual or even multiple retention mechanisms or capturing abilities targeted to particular analytes.27–35 Moreover, incorporation of the particles into the monolith helps to increase the stationary phase to mobile phase ratio, leading to increased binding capacity.36
Here we have adapted hybrid materials for collecting peptides from single-neuron releasates using particle-embedded monolithic capillaries (PEMCs). Two distinct extraction particles, with either pyrrolidone (PY) or ethylenediamine (EDA) as the functional group, were embedded into individual poly(stearyl methacrylate-co-ethylene glycol dimethacrylate) (SMA-co-EDMA) monoliths inside capillaries with a 250 μm inner diameter (i.d.). The PEMC columns were efficient in extracting peptides from high ionic strength samples such as artificial sea water with detection limits down to the low femtomole regime with MALDI MS detection. The successful collection of releasates from single neurons isolated from A. californica with cell sizes ranging from 40 to 200 μm demonstrates the effectiveness of this approach.
Experimental Section
Materials and Reagents
Stearyl methacrylate (SMA), 2,2′-azobis(2-methylpropionitrile) (AIBN), 3-(trimethoxysilyl)propyl methacrylate 98% (TMSPM), iso-amyl alcohol (IAA) and 1,4-butanediol (BD) were purchased from Sigma-Aldrich (St. Louis, MO). Ethylene glycol dimethacrylate 98% (EDMA), methanol, acetonitrile (ACN) and formic acid (FA) were obtained from Fisher Scientific (Fairlawn, NJ).
Artificial sea water (ASW) consisting of 460 mM NaCl, 10 mM KCl, 10 mM CaCl2, 22 mM MgCl2, 26 mM MgSO4 and 10 mM HEPES, pH 7.8, was used as the sample matrix for extraction of the peptide standards. Seven peptide standards—bradykinin (1–7) (5.3 mM), angiotensin II (3.8 mM), angiotensin I (3.1 mM), substance P (3.0 mM), bombesin (2.5 mM), somatostatin (2.4 mM) and adrenocorticotropic hormone (18–39) (ACTH) (3.5 mM)—were obtained from Sigma-Aldrich and prepared in water as stock solutions at the concentrations indicated. They were mixed together and diluted 2000-fold to 1.2 to 2.7 μM in ASW for evaluating the binding capacity of the columns. Water used throughout the experiments was obtained from a Milli-Q water purification system (Millipore, Bedford, MA).
Fabrication of PEMC Columns
Fused silica capillaries (250 μm i.d. × 360 μm outer diameter (o.d.), Polymicro Technologies, Phoenix, AZ) were rinsed with 1.0 M aqueous sodium hydroxide for 2 h, 10 min with water, 2 h with 1.0 M hydrochloric acid, and again with water for 10 min. After drying under an N2 flow, each capillary was filled with a methanolic solution of TMSPM (50%) and allowed to react overnight at 40 °C with both capillary ends sealed. The capillaries were then rinsed with methanol after the inner wall modification. For preparing the PEMC extraction column, a piece of 5 cm-long capillary was used with the polyimide coating on one end peeled off to generate an ultraviolet (UV)-transparent window, 15 mm long.
The poly(SMA-co-EDMA) monoliths for embedding the particles were synthesized according to a previously published report37 with modification. A prepolymerization mixture was prepared as follows: IAA (836 mg), BD (167 mg), SMA (406 mg) and EDMA (136 mg) were mixed with AIBN at 1% (wt) and TMSPM at 5% (v:v). Particles of PY (Strata-X™) or EDA (Strata-X-AW™) (Phenomenex, Torrance, CA), 20 mg of each with an average size of 30 μm, were added into 100 μL of the prepolymerization mixture to make a slurry, which was then filled into the capillaries to cover the entire length of the UV-transparent windows. For polymerization, each capillary was exposed to 365-nm longwave UV radiation with a hand-held UV lamp (UVP, Upland, CA) at a distance of 3 cm for 30 min. The capillaries were then thoroughly rinsed with methanol to eliminate residual monomer and porogen. A poly(SMA-co-EDMA) monolithic column without particles embedded was also fabricated for comparison.
Scanning Electron Microscopy (SEM) Characterization
For each type of column, the structure of the polymer monolith with the particles embedded was characterized with a Philips XL30 field-emission environmental scanning electron microscope (FEI Company, Hillsboro, OR) using a secondary electron detector. The capillaries, with their cross-sections coated with Pd/Au using a Desk II turbo sputter coater (Denton Vacuum, Moorestown, NJ), were positioned on a 45° sample stage for further characterization.
Extraction Efficiency of PEMC Columns
The extraction efficiency of the PEMC columns was evaluated by extracting peptide standards prepared in ASW. Each column was connected to a syringe mounted on a KDS270 syringe pump (KD Scientific, Holliston, MA) through a Tygon® tube, 250 μm i.d. (Cole-Parmer, Vernon Hills, IL) with the particle-embedded end positioned in the sample solution. The columns were first conditioned with methanol and equilibrated with water (PY column) or water acidified with HCl to pH 2.5 (EDA column). Then the sample solution was loaded onto the columns at 0.25 μL/min by setting the syringe pump to withdraw mode. After extraction, the columns were washed with 5 μL of water (PY column) or 25 mM ammonium acetate (EDA column). Next the analytes were eluted with 3 μL of 90% methanol containing 2% FA, and further analyzed by MALDI-TOF MS/capillary liquid chromatography (CapLC)-UV/scintillation counting assays, as described below.
Radioactive assays
[3H]angiotensin II (Ang II [tyr-3,5-3H(N)], catalog no. ART 0700-250μCi, American Radiolabeled Chemicals Inc., St. Louis, MO), was diluted in ASW at a final concentration ranging, as appropriate, from 10 nM to 500 nM for binding assays. After extraction, the bound [3H]angiotensin II was eluted into 5 mL of Ultima Gold™ LLT scintillation solution (PerkinElmer, Waltham, MA) and subjected to further analysis by an LS 6000IC liquid scintillation counter (Beckman Coulter, Fullerton, CA).
CapLC-UV/electrospray ionization (ESI)-MS assays
The eluents from the PEMC columns were first dried using a SpeedVac concentrator (Thermo Scientific, San Jose, CA) and redissolved in 25 μL of a solution containing 99% H2O, 1% ACN and 0.05% trifluoroacetic acid (TFA). Liquid chromatographic separation was performed with a U3000 CapLC system (Dionex, Sunnyvale, CA) equipped with an Acclaim® PepMap RSLC C18 column (300 μm, 15 cm, 2 μm 100 Å) with an elution gradient of 0–80% solvent B (80% ACN with 0.05% TFA) and solvent A (100% H2O with 0.05% TFA) in 35 min. Detection was performed with a diode array detector set at 214 nm, coupled to an HCTultra™ ESI mass spectrometer (Bruker Daltonics, Billerica, MA) for peak assignment.
MALDI-TOF mass spectrometric analysis
MALDI-TOF MS was performed with an ultrafleXtreme™ MALDI TOF/TOF mass spectrometer equipped with a smartbeam™-II laser and flexControl software (Bruker Daltonics). Eluents (0.25 μL) were directly applied onto targets prespotted with matrix (AnchorChip, Bruker Daltonics). Mass spectra were acquired with external calibration in reflectron mode with typical mass accuracies within 100 ppm. Resulting spectra represent the accumulation of 2000 laser shots without baseline correction or data filtering; flexAnalysis software (Bruker Daltonics) was used to process the spectra.
Single-Neuron Release Assays
A. californica, weighing 100–150 g, were obtained from the University of Miami/NIH National Resource for Aplysia and kept in an aquarium containing aerated and filtered artificial sea water (Instant Ocean, Aquarium Systems Inc., Mentor, OH) maintained at 14 °C. Before dissection, animals were anesthetized by injection of isotonic MgCl2 into the body cavity at 30–50% of body weight.
The abdominal, pedal and buccal ganglia were surgically dissected, followed by treatment with 1% protease (type IX Bacterial, Sigma-Aldrich) prepared in ASW antibiotic solution (ASW containing penicillin G, gentamycin and streptomycin) 45–120 min at 34 °C to help remove connective tissues and reduce adherence between cells. After rinsing the ganglia with ASW to remove extra protease, bag cell neurons from the abdominal ganglia, pedal neurons from the pedal ganglia, and B1/B2 neurons from the buccal ganglia were isolated manually under the guidance of a Leica MZ 7.5 high-performance stereomicroscope (Leica Microsystems Inc., Bannockburn, IL) and placed in a petri dish containing 4 mL ASW. The neurons were cultured overnight before use.
The secretagogue KCl (50 mM) was prepared in ASW and used to stimulate the cultured neurons. NaCl concentration was reduced accordingly in order to maintain the osmolarity. For each experiment, a bare capillary for delivering the KCl and a preconditioned PEMC column were both positioned as close as possible to the targeted neuron (no direct contact). KCl was infused over the cell body at a flow rate of 0.25 μL/min and releasates were collected simultaneously at the same rate for 30 min. After collection, the PEMC columns were rinsed with 5 μL of water (PY column) or 25 mM ammonium acetate (EDA column) to reduce inorganic salts. The releasates were then eluted with 3 μL of 2% FA in 90% MeOH. An aliquot (0.25 μL) of each eluent was applied on a prespotted matrix target for further MALDI-TOF MS analysis. Releasates determined to be stimulation-dependent were distinguished from the chemical background by comparing the mass spectra from samples obtained before and during stimulation. Peptides were identified by their mass-to-charge ratio (m/z) and according to prior extensive MS-based (and tandem MS) studies on these specific cells.
Results and Discussion
Fabrication of the PEMC Columns
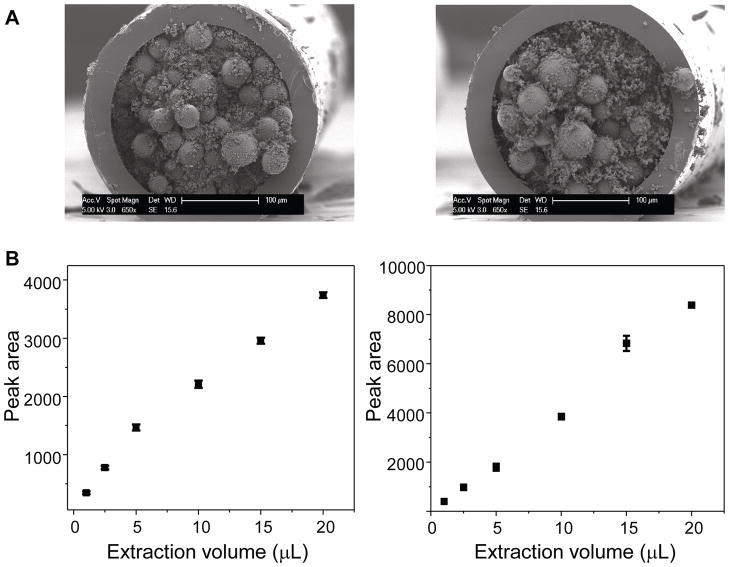
By selecting a capillary with a dimension comparable to the cells of interest and filling it with the hybrid extraction/collection material, a PEMC column can be tailored to accomplish specific measurement tasks. The column is fabricated either by first packing the capillary with particles, followed by infusion of a prepolymerization mixture and in-situ polymerization,36 or by making a mixture of particles and prepolymerization solution and filling the capillary, followed by in-situ polymerization.30,32 The latter approach was used here. A previously published polymerization protocol37 was used with modification, which generated monoliths with high permeability that facilitated particle embedding. Poly(SMA-co-EDMA) monolith was selected as the particle embedding material because it offers hydrophobic interactions. Besides hydrophobic interaction, PY particles provide π-π interactions, hydrogen bonding and di-polar interactions, while EDA particles offer π-π interactions and weak anion-exchanging abilities. The particles were directly added into the prepolymerization mixture, creating a slurry that was immediately filled into the pretreated capillaries. UV initiation was used in order to shorten the polymerization time and thereby avoid inhomogeneous distribution of the particles inside the monolith that might result from their aggregation during this process. Fabrication parameters were then fine-tuned in terms of particle concentration, polymerization time and UV irradiation distance, taking into account the permeability, mechanical strength and extraction efficiency of the PEMC columns. The resulting optimized prepolymerization mixture, with a particle concentration of 20 mg/100 μL, was used to fill the capillaries, followed by a 30 min polymerization with the columns exposed to UV light at a distance of 3 cm. An addition of 5% (v/v) TMSPM to the prepolymerization mixture helped to tighten the particle-embedded monolith to the capillary wall. Figure 1A shows SEM images of the cross-section of both the PY and EDA columns. No obvious structure change in the poly(SMA-co-EDMA) monolith was found, and the particles were homogeneously embedded in the monolith with high density, providing a large binding capacity for peptide collection.
Figure 1.
(A) SEM images of a cross-section of PY (left) and EDA (right) columns. (B) Binding of angiotensin II onto the PY (left) and EDA (right) PEMC columns. Angiotensin II was prepared at 1.9 μM in ASW containing six other peptide standards at 1.2 to 2.7 μM. Each data point represents average extraction results from three individual columns ± standard deviation.
Optimization of Peptide Collection and Performance Specifications
Before using the PEMC columns for single-neuron release assays, optimization of the collection conditions for the peptides prepared in ASW was required. In order to quantify peptide binding and collection, we used a radioactive assay with 3H isotope-labeled angiotensin II prepared in ASW at various concentrations. Using 5 μL of 3H-angiotensin II at 10 nM, an extraction flow rate that varied from 0.1 to 1.0 μL/min was investigated for both the PY and EDA columns. While the extraction efficiency decreased slightly with flow rates higher than 0.25 μL/min, the decrease was not critical given that the extraction efficiency at 1 μL/min was 68% (PY column) and 82% (EDA column) of that at 0.25 μL/min, respectively.
Modifying the sample matrix can increase extraction efficiency by acidifying, alkalifying, or adding ion-pair reagents. However, this is not possible when the peptides are directly collected from physiological saline because many such additives affect cell physiology. Therefore, although an acidified sample matrix could help to enhance the extraction efficiency, in our experiments, the EDA columns were evaluated using ASW as the sample matrix without further modification.
The peptide extraction was followed by a washing step in order to remove excess inorganic salts and unbound molecules, as this step dramatically improves MALDI-TOF MS results.18 An extraction test mixture containing seven peptide standards was prepared in ASW from 24 to 53 nM, followed by a 5 μL washing step of either water (PY column) or 25 mM ammonium acetate buffer (EDA column).38 The washing step removed most of the inorganic salts, which caused a large increase in the peptide MALDI-MS signals. The percentage of peptides removed during the washing process was assessed via 3H-angiotensin II; 4.6% and 2.7% of bound 3H-angiotensin II was lost from the PY and EDA columns, respectively. A methanolic solution combined with FA allowed efficient peptide elution for both PEMC columns. We found that 3 μL of elution solution was adequate to elute the bound peptides from both columns as reduced (or no detectable) peptide signals were found via MALDI MS when larger volumes of eluents were used.
Using these optimized conditions, the binding capacities of the PEMC columns were assessed for our peptide standard mixtures prepared in ASW. Given that we would expect to see multiple peptides in naturally occurring peptide releasates, binding curves were constructed using angiotensin II at 1.9 μM prepared in ASW with the other six peptide standards at various concentration levels (1.2 to 2.7 μM). When the sample volume was increased from 1 to 20 μL, the angiotensin II signal increased accordingly, as shown in Figure 1B. A loading volume of 20 μL did not saturate either column, and the amounts of angiotensin II binding to the PEMC columns were 16.5 and 28.2 pmol for the PY and EDA columns, respectively. Further increases in the extraction volumes led to long extraction times, which were not practical for actual sample assays and thus were not assessed.
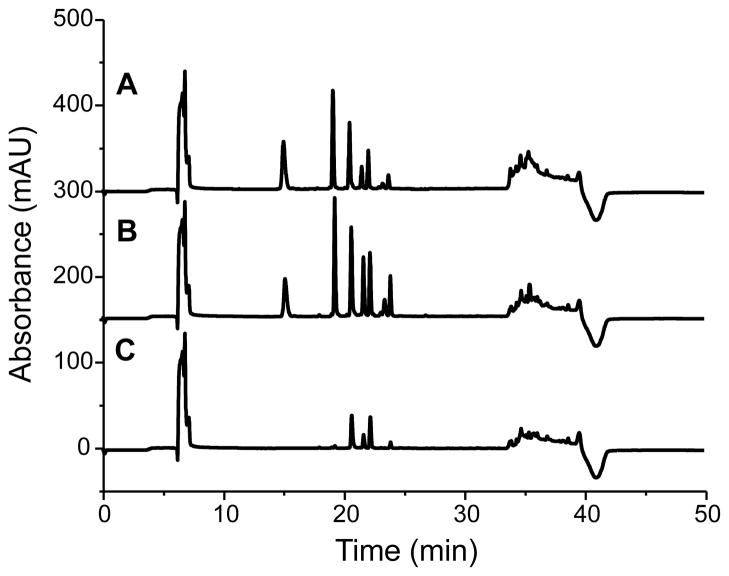
The ability of the PY and EDA PEMC columns to collect peptides from ASW was compared to the poly(SMA-co-EDMA) column (without embedded particles) for the same monolith length. The CapLC-UV was used to simultaneously monitor multiple analytes quantitatively. As seen in Figure 2, the peptide extraction efficiency of the PY and EDA columns was superior to that of the poly(SMA-co-EDMA) monolithic column; specifically, more peptides were detected with higher peak intensities. Hydrophilic peptides such as bradykinin (1–7) and bombesin, which were almost missed from the poly(SMA-co-EDMA) column, were clearly visible in the eluents from the PY and EDA columns under the selected extraction conditions. This suggests that in addition to a hydrophobic interaction, the π-π bonding, hydrogen bonding and ion-exchange properties of the PEMC columns enhance peptide collection from high ionic strength samples. A slightly higher extraction efficiency was achieved with the EDA column compared to the PY column. The increased ACTH (18–39) retention on the EDA column implied the presence of weak anion-exchanging interactions. Although the binding capacity of the PEMC columns was within the same orders of magnitude reported for various monolith materials in extracting different analytes,39–41 the PEMC columns showed better performance than the polymer monolith capillary in extracting peptides from cell culture media without modifying the sample.
Figure 2.
Comparison of the peptide collection efficiency from ASW by column: (A) PY, (B) EDA and (C) poly(SMA-co-EDMA) monolith. The eluents from the columns were dried and redissolved in loading solution for CapLC-UV characterization with the chromatograms recorded. Peaks from the left: bradykinin (1-7), angiotensin II, angiotensin I, bombesin, substance P, ACTH (18-39) and somatostatin.
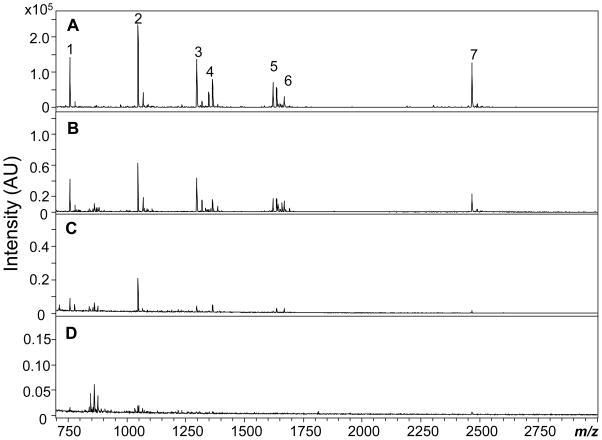
The detection limits of the PEMC column collection combined with MALDI-TOF MS were determined by extracting consecutively diluted peptide standards prepared in ASW. With a loading volume of 10 μL, peptides diluted to picomolar concentrations could still be detected. The MALDI MS spectra obtained by extraction of peptide standards with the PY column are shown in Figure 3. When the concentration of the peptide standards was lowered to 0.24 to 0.53 nM, most of the peptides were still detectable with a signal-to-noise ratio larger than 3 (Figure 3C). Angiotensin II at 0.38 nM showed the highest intensity. When the peptide concentration was further decreased to 50–110 pM, the peptide intensities also decreased so that some peptides were no longer detectable (Figure 3D); for example, 110 pM bradykinin (1–7), 80 pM angiotensin II and 60 pM ACTH (18–39) were visible, corresponding to 1.0, 0.8 and 0.6 fmol, respectively. Similar results were also obtained with the EDA column. Compared to direct MALDI MS detection of the peptides prepared in ASW, PEMC columns improved the detection limits by three orders of magnitude.
Figure 3.
Limits of detection by a PY column with extraction peptide standards consecutively diluted in ASW followed by MALDI-TOF MS detection. Peptide concentrations: (A) 24–53 nM; (B) 2.4–5.3 M; (C) 0.24–0.53 nM; and (D) 0.05–0.11 nM. Peak identities: 1, bradykinin (1–7); 2, angiotensin II; 3, angiotensin I; 4, substance P; 5, bombesin; 6, somatostatin; 7, ACTH (18–39).
Single-Neuron Release
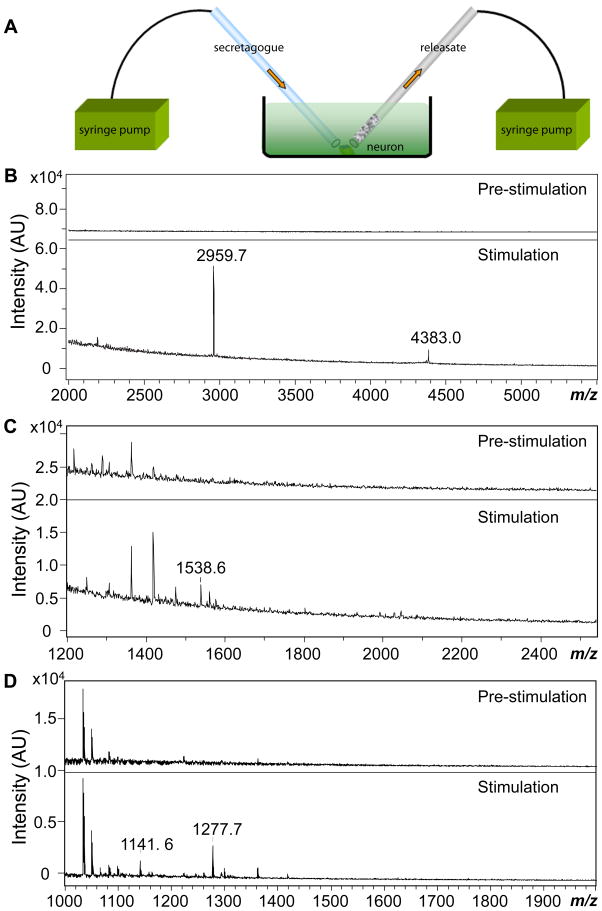
Neuropeptides are released and active at micromolar to picomolar levels; in the case of A. californica peptidergic neurons, femtomole amounts of neuropeptides are oftentimes bserved under stimulation.5,42,43 Therefore, the herein described protocol combining PEMC column collection and MALDI-TOF MS detection should be capable of detecting releasates even from single neurons. To demonstrate this, selected neurons isolated from the ganglia of A. californica were cultured overnight in ASW before conducting the release experiments. KCl at an elevated concentration (50 mM) in ASW was used as the secretagogue. The increase in the extracellular concentration of potassium ions alters the electrochemical gradient across neuronal membranes, leading to cell depolarization and subsequent exocytosis of neurotransmitters.18 This enabled us to validate single-neuron release experiments using three separate test cases: neurons from a homogeneous cluster, unknown neurons with known peptides, and individual identified neurons. In each case, we applied the secretagogue to the individual neurons with one capillary and collected releasates with a PEMC column, as shown in Figure 4A.
Figure 4.
(A) Schematic illustration of single-neuron releasate collection with a particle-embedded monolithic capillary. Both the stimulant-delivering capillary and the particle-embedded monolithic capillary ends are positioned near the target neuron. (B–D) MALDI MS spectra shown for neuronal releasates collected both pre-stimulation (showing few if any peaks) and collected during/after chemical stimulation of the neuron: (B) Bag cell neuron releasates with acidic peptide (AP) at m/z 2959.7 and egg-laying hormones (ELH) at m/z 4383.0 labeled; (C) individual unknown pedal neuron, with the peak at m/z 1538.6 being pedal peptide; (D) individual B1/B2 neuron release showing the expected small cardioactive peptides A (SCPA) at m/z 1277.7 and B (SCPB) at m/z 1141.6.
A.californica bag cell neurons have been extensively studied under stimulation with peptide profiles well-documented,16–19,39,44,45 making them good models for testing novel peptide collection protocols. We used PY columns to collect the extracellular media before and during KCl stimulation. As shown in Figure 4B, samples collected before KCl stimulation did not reveal known peptides. For samples collected immediately upon KCl introduction to the cell body of a single bag cell neuron for 30 min, α-bag cell peptide (m/z 1122.7) (not shown in figure), acidic peptide (m/z 2959.7), and egg-laying hormone (m/z 4383.0) were detected, similar to results shown in previous bag cell release studies.17,18 The collection was also performed with EDA columns, where similar releasate profiles were observed.
Pedal peptide, which can be released in a calcium- and stimulation-dependent manner,6,43 should be the predominant peptide found in pedal neurons.6,46 Under the same stimulation protocol used for bag cell neurons, pedal peptide (m/z 1538.6) was detected in a 30 min collection of releasates with the EDA column (Figure 4C). Several additional peaks ranging from m/z 1300 to 1480 (m/z 1306.5, 1362.5, 1418.6, 1474.6) with a mass difference of 56.0 were detected; they were not activity-dependent releasates as they appeared in the releasate before stimulation; therefore, these appear to be contamination in the media or matrix.
B1 and B2 neurons are large motor neurons located in A. californica buccal ganglia that produce large amounts of the small cardioactive peptides SCPA and SCPB.47–49 The release of these two peptides in a calcium-dependent fashion upon intracellular electrical stimulation was demonstrated by radioactively labeling in situ synthesized peptides followed by a detection approach combining high performance LC and liquid scintillation counting.7,50 They have also been characterized by on-plate microextraction methods followed by MALDI MS to assess entire neurotransmitter profiles.51 As the soma is approximately 200 μm in diameter, these are relatively fragile neurons but they survived the stimulation and collection conditions used here. Within the 30 min collection period, the characteristic small cardioactive peptides at m/z 1141.6 and 1277.7 were detected in the stimulation-dependent releasates, as shown in Figure 4D. Again, several unknown/contamination peaks found in the spectra, including m/z 1034.3, 1043.4, 1050.2, 1066.2, were determined to be matrix-related and not from neuron release.
Conclusions
Polymer monolithic capillaries embedded with extraction particles were fabricated as sample pretreatment tools for analyzing single-neuron release by MALDI-TOF MS. The extraction particles, together with the hydrophobic polymer monolith “glue”, provide a broad array of interactions to enable the collection of a wide range of neuropeptides. By directly positioning a collection capillary very close to individual cultured neurons, releasates resulting from KCl stimulation of specific identified neurons, individual neurons from a homogenous cluster and unknown neurons from a defined ganglion have been characterized. The approach is general and will facilitate a broad range of neuropeptide experiments. Lastly, by embedding unique particles or other types of extraction media having particular properties, the collection protocol can be easily tailored to target specific analytes in the releasates.
Acknowledgments
We are grateful to Scott Robinson from the Image Technology Group, Beckman Institute, UIUC for assistance in SEM, Susan Martinis from the Center of Biophysics and Computational Biology, UIUC. We also thank Callie Croushore for help with the radioactive assays and Elena Romanova for critical discussions. This material is based on work supported by Award No. DA018309 from the National Institute for Drug Abuse (NIDA) and Award No. NS031609 from the National Institute of Neurological Disorders and Stroke (NINDS). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIDA, NINDS or the National Institutes of Health.
References
- 1.Strand FL. MIT Press. Cambridge, MA: 1999. [Google Scholar]
- 2.Sandman CA. New York Academy of Sciences. New York: 1999. p. 897. [DOI] [PubMed] [Google Scholar]
- 3.Hummon AB, Sweedler JV, Corbin RW. TrAC, Trends Anal Chem. 2003;22:515–521. [Google Scholar]
- 4.Rubakhin SS, Sweedler JV. Anal Chem. 2008;80:7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilim FS, Cropper EC, Price DA, Kupfermann I, Weiss KR. J Neurosci. 1996;16:8105–8114. doi: 10.1523/JNEUROSCI.16-24-08105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall JD, Lloyd PE. J Neurobiol. 1991;22:583–589. doi: 10.1002/neu.480220604. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd PE, Schacher S, Kupfermann I, Weiss KR. Proc Natl Acad Sci U S A. 1986;83:9794–9798. doi: 10.1073/pnas.83.24.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Omiatek DM, Dong Y, Heien ML, Ewing AG. ACS Chem Neurosci. 2010;1:234–245. doi: 10.1021/cn900040e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Adams KL, Luber SJ, Eves DJ, Heien ML, Ewing AG. Anal Chem. 2008;80:1394–1400. doi: 10.1021/ac702409s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paras CD, Qian WJ, Lakey JR, Tan WH, Kennedy RT. Cell Biochem Biophys. 2000;33:227–240. doi: 10.1385/cbb:33:3:227. [DOI] [PubMed] [Google Scholar]
- 11.Paras CD, Kennedy RT. Anal Chem. 1995;67:3633–3637. doi: 10.1021/ac00116a003. [DOI] [PubMed] [Google Scholar]
- 12.Baggerman G, Verleyen P, Clynen E, Huybrechts J, De Loof A, Schoofs L. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;803:3–16. doi: 10.1016/j.jchromb.2003.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L. J Sep Sci. 2008;31:427–445. doi: 10.1002/jssc.200700450. [DOI] [PubMed] [Google Scholar]
- 14.Li LJ, Garden RW, Sweedler JV. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- 15.Hatcher NG, Atkins N, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Proc Natl Acad Sci U S A. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garden RW, Shippy SA, Li LJ, Moroz TP, Sweedler JV. Proc Natl Acad Sci U S A. 1998;95:3972–3977. doi: 10.1073/pnas.95.7.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubakhin SS, Page JS, Monroe BR, Sweedler JV. Electrophoresis. 2001;22:3752–3758. doi: 10.1002/1522-2683(200109)22:17<3752::AID-ELPS3752>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 18.Hatcher NG, Richmond TA, Rubakhin SS, Sweedler JV. Anal Chem. 2005;77:1580–1587. doi: 10.1021/ac0487909. [DOI] [PubMed] [Google Scholar]
- 19.Hatcher NG, Sweedler JV. J Neurophysiol. 2008;99:333–343. doi: 10.1152/jn.00968.2007. [DOI] [PubMed] [Google Scholar]
- 20.Potter OG, Hilder EF. J Sep Sci. 2008;31:1881–1906. doi: 10.1002/jssc.200800116. [DOI] [PubMed] [Google Scholar]
- 21.Peterson DS, Rohr T, Svec F, Frechet JMJ. Anal Chem. 2003;75:5328–5335. doi: 10.1021/ac034108j. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Lee ML. J Sep Sci. 2009;32:3369–3378. doi: 10.1002/jssc.200900478. [DOI] [PubMed] [Google Scholar]
- 23.Svec F. J Chromatogr. 2010;1217:902–924. doi: 10.1016/j.chroma.2009.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlakh EG, Tennikova TB. J Sep Sci. 2007;30:2801–2813. doi: 10.1002/jssc.200700284. [DOI] [PubMed] [Google Scholar]
- 25.Gu B, Li Y, Lee ML. Anal Chem. 2007;79:5848–5855. doi: 10.1021/ac0623585. [DOI] [PubMed] [Google Scholar]
- 26.Hutchinson JP, Zakaria P, Bowiet AR, Macka M, Avdalovic N, Haddad PR. Anal Chem. 2005;77:407–416. doi: 10.1021/ac048748d. [DOI] [PubMed] [Google Scholar]
- 27.Chirica GS, Remcho VT. Anal Chem. 2000;72:3605–10. doi: 10.1021/ac000179w. [DOI] [PubMed] [Google Scholar]
- 28.Dulay MT, Kulkarni RP, Zare RN. Anal Chem. 1998;70:5103–5107. doi: 10.1021/ac9806456. [DOI] [PubMed] [Google Scholar]
- 29.Erzengin M, Unlu N, Odabasi M. J Chromatogr A. 2011;1218:484–490. doi: 10.1016/j.chroma.2010.11.074. [DOI] [PubMed] [Google Scholar]
- 30.Kato M, Dulay MT, Bennett B, Chen JR, Zare RN. Electrophoresis. 2000;21:3145–3151. doi: 10.1002/1522-2683(20000901)21:15<3145::AID-ELPS3145>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 31.Krenkova J, Lacher NA, Svec F. Anal Chem. 2010;82:8335–8341. doi: 10.1021/ac1018815. [DOI] [PubMed] [Google Scholar]
- 32.Rainer M, Sonderegger H, Bakry R, Huck CW, Morandell S, Huber LA, Gjerde DT, Bonn GK. Proteomics. 2008;8:4593–4602. doi: 10.1002/pmic.200800448. [DOI] [PubMed] [Google Scholar]
- 33.Ratnayake CK, Oh CS, Henry MP. J High Resolut Chromatogr. 2000;23:81–88. [Google Scholar]
- 34.Tang Q, Lee ML. In: Monolithic materials: preparation, properties, and applications. Svec F, Tennikova TB, Deyl Z, editors. Elsevier; Amsterdam: 2003. pp. 197–211. [Google Scholar]
- 35.Xu Y, Cao Q, Svec F, Frechet JMJ. Anal Chem. 2010;82:3352–3358. doi: 10.1021/ac1002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jandera P, Urban J, Skerikova V, Langmaier P, Kubickova R, Planeta J. J Chromatogr. 2010;1217:22–33. doi: 10.1016/j.chroma.2009.09.041. [DOI] [PubMed] [Google Scholar]
- 37.Jiang ZJ, Smith NW, Ferguson PD, Taylor MR. J Biochem Biophys Methods. 2007;70:39–45. doi: 10.1016/j.jbbm.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Phenomenex product library. https://phenomenex.blob.core.windows.net/documents/b50e7828-7b9b-431f-9490-fd9e4498a660.pdf.
- 39.Iannacone JM, Ren SF, Hatcher NG, Sweedler JV. Anal Chem. 2009;81:5433–5438. doi: 10.1021/ac9005843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan Y, Feng YQ, Da SL, Shi ZG. Anal Chim Acta. 2004;523:251–258. [Google Scholar]
- 41.Namera A, Nakamoto A, Saito T, Miyazaki S. J Sep Sci. 2011;34:901–924. doi: 10.1002/jssc.201000795. [DOI] [PubMed] [Google Scholar]
- 42.Kennedy RT, Watson CJ, Haskins WE, Powell DH, Strecker RE. Curr Opin Chem Biol. 2002;6:659–665. doi: 10.1016/s1367-5931(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 43.Vilim FS, Price DA, Lesser W, Kupfermann I, Weiss KR. J Neurosci. 1996;16:8092–8104. doi: 10.1523/JNEUROSCI.16-24-08092.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newcomb RW, Scheller RH. Brain Res. 1990;521:229–237. doi: 10.1016/0006-8993(90)91547-t. [DOI] [PubMed] [Google Scholar]
- 45.Loechner KJ, Azhderian EM, Dreyer R, Kaczmarek LK. J Neurophysiol. 1990;63:738–744. doi: 10.1152/jn.1990.63.4.738. [DOI] [PubMed] [Google Scholar]
- 46.Lloyd PE, Connolly CM. J Neurosci. 1989;9:312–317. doi: 10.1523/JNEUROSCI.09-01-00312.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lloyd PEKI, Weiss KR. Soc Neurosci Abstr. 1984;10:153. [Google Scholar]
- 48.Morris HR, Panico M, Karplus A, Lloyd PE, Riniker B. Nature. 1982;300:643–645. doi: 10.1038/300643a0. [DOI] [PubMed] [Google Scholar]
- 49.Lloyd PE, Mahon AC, Kupfermann I, Cohen JL, Scheller RH, Weiss KR. J Neurosci. 1985;5:1851–1861. doi: 10.1523/JNEUROSCI.05-07-01851.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whim MD, Lloyd PE. J Neurosci. 1992;12:3545–3553. doi: 10.1523/JNEUROSCI.12-09-03545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li LJ, Romanova EV, Rubakhin SS, Alexeeva V, Weiss KR, Vilim FS, Sweedler JV. Anal Chem. 2000;72:3867–3874. doi: 10.1021/ac000260z. [DOI] [PubMed] [Google Scholar]