Abstract
Pro-inflammatory cytokines, such as tumour necrosis factor-α (TNF-α), are increased in serum and CSF in Alzheimer's disease (AD). We investigated the effect of TNF-α on gene and protein expression levels of Aβ degrading enzymes (ACE, ECE-1, ECE- 2, IDE and NEP) in vitro. Differentiated (DC) and non-differentiated (NDC) neuroblastoma cells (SH-SY5Y) were exposed to TNF-α for 15 minutes and 3 hours and protein and gene expression levels measured using western blotting or sandwich ELISA (ECE-2), and real time-PCR (RT-PCR). Only ECE-2 protein levels decreased significantly in NDCs in a dose-dependent manner after 15 minutes of TNF-α exposure but reverted to basal levels after 3 hours. Basal NEP gene expression levels were higher in control DCs compared to NDCs but TNF-α treatment did not significantly alter the levels of expression of any of the Aβ degrading enzymes. In conclusion, apart from a transient reduction in ECE-2 protein levels, TNF-α had no impact in our in vitro experimental system on transcription or translation of any of our selected mediators of Aβ degradation.
Keywords: Alzheimer's disease, inflammation, TNF-α, SH-SY5Y cells, RT-PCR, western blot
Introduction
Alzheimer's disease (AD) is a chronic neurode-generative disorder characterized clinically by progressive loss of memory and cognitive function and pathologically by the progressive accumulation of insoluble amyloid-β (Aβ) and hyper-phosphorylated tau protein in the brain.
The accumulation of Aβ reflects an imbalance between its production and removal. Aβ is eliminated from the brain by a range of processes including microglial phagocytosis [1-3], receptor -mediated transport across the blood-brain barrier [4-5], enzymatic degradation [6-8], and drainage along perivascular basement membranes to the cervical lymphatics and possibly CSF [9-11].
The role of the immune system in clearing Aβ from the brain has been the subject of a number of recent studies. Multiple cytokines are produced in response to Aβ activation of microglia and can be detected at increased levels in the serum, CSF and brain in AD [12-14]. Although neurons were generally believed to be passive bystanders in neuroinflammation, there is now evidence that neurons contribute to the production of cytokines such as IL-1β, IL-6 and TNF-α [15, 16] which may aggravate local inflammation and neurodegeneration in AD brains [13].
TNF-α is an important pro-inflammatory cytokine, which is elevated in the serum and CSF in AD [17, 18] and has been implicated in the development of the disease [19]. One way which TNF-α might influence the development of AD is through its effects on the production of Aβ-degrading enzymes, including neprilysin (NEP), endothelin-converting enzymes (ECEs), insulin-degrading enzyme (IDE) and angiotensin-converting enzyme (ACE). TNF-α was shown to increase the expression of NEP in granulocytes [20] and ECE-1 in endothelial cells [21] and to down-regulate IDE in murine microglial cells [22] and ACE in both human endothelial cells [23] and macrophages [24]. However, little is known of the effects of TNF-α on Aβ-degrading enzymes in neurons.
This study investigated the effect of TNF-α on Aβ degrading enzymes NEP, IDE, ECE-1, ECE-2 and ACE in differentiated (DC) and non-differentiated neuroblastoma cells (NDC) (SH-SY5Y).
Materials and methods
Cell culture and TNF-α exposure
SH-SY5Y human neuroblastoma cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma) (2 mM glutamine (Sigma), 1% essential amino acids (Sigma), 15% fetal bovine calf serum (FCS) (Autogene Bioclear). Cells were differentiated by addition of 10 μM retinoic acid (RA) (Sigma-Aldrich, Dorset, UK) for 5 days and 50 ng/ml BDNF for 2 days in DMEM. Non-differentiated (NDC) and differentiated (DC) cells were incubated in serum-free medium for 12-14 h before exposure to 1 ng/ml, 10 ng/ml or 50 ng/ml TNF-α (R&D systems, Abingdon, UK) for 15 min and 3 h. All cells were discarded after 15 passages as the sensitivity of SH-SY5Y cells to TNF-α was shown to decrease with increased passages [25]. All experiments were repeated 3 times.
Real-time PCR (RT-PCR)
RT-PCR was performed as previously described [26] and according to manufacturer's instructions, by use of the ABI 7000 sequencing detection system (ABI Prism) with the following Assay-on-Demand Gene Expression Products (detectors) (TaqMan MGB probes, FAM dyelabeled): NEP (Hs00153510), IDE (Hs00610438), ECE-1 (Hs00154837), ECE-2 (Hs00206701), ACE (Hs01104605), TRAF-2 (Hs00184186), GAPDH (Hs99999905) and TaqMan Universal PCR Master Mix. All samples were analysed in triplicate. Gene expression for all targets was calibrated according to the level of GAPDH mRNA by the 2-DDCt method, and the fold difference in gene expression relative to that in untreated control cells calculated [27]. The mRNA levels were therefore expressed as exponential functions (the fold-change relative to untreated controls) and the values from repeat analyses were presented as the geometric means (with 95% confidence intervals).
Western blotting
Gel electrophoresis and western blotting were performed as previously described [26]. Blotted membranes were incubated for 1 h (ECE-1 for 3 h) with primary antibodies diluted as follows: NEP/ CD10 (polyclonal anti-rabbit, H-321: sc-9149, Santa Cruz Biotechnology, Inc, 1:250), IDE (monoclonal anti-mouse, AB2496, R&D systems, 1:1500) ECE-1 (biotinylated anti-goat, BAF1784, R&D Systems, 1:1000), ACE (monoclonal anti-mouse, MAB9291, R&D systems, 1:250), TRAF-2 (polyclonal anti-rabbit, sc876, Santa Cruz Biotechnology, Inc, 1:250) This was followed by incubation with peroxidase -conjugated anti-rabbit, anti-mouse or anti-goat IgG (1:10000) for 1 h. Specific labeling was detected by enhanced chemiluminescence (ECL, Amersham Biosciences, Little Chalfont, UK). The membranes were exposed to film (Kodak), developed, and the integrated band densities analyzed using Image J software (US National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997-2011). Values in TNF-α-treated cells were expressed as the fold change relative to untreated control cells ± the standard error of the mean (SEM).
ECE-2 sandwich ELISA
ECE-2 sandwich ELISA was performed as previously described [28]. Maxisorp ELISA plates (Nunc Maxisorp, Fisher Scientific, Loughborough, UK) were coated with ECE-2 antibody (goat anti-human, AF 1645, R&D Systems, 1:20) overnight at 4°C. Serial dilutions of recombinant human ECE-2 (R&D Systems) were used to produce a standard curve (final concentrations 27 to 432 μg/ml). Standards and sample homogenates (1 mg/ml) were incubated for 2 h, then incubated with rat-anti-human ECE-2 (1 μg/ml, R&S Systems) for 90 min. Peroxidase anti-rat conjugate (1:500dil, Cell Signaling Technology, Massachusetts, USA) was added for 1 h at room temperature, peroxidase substrate (R&D Systems) added for 20 min in the dark before adding stop solution (R&D Systems). The absorbance was read at 450 nm and concentrations were determined by interpolation against the standard curve. ECE-2 concentrations were expressed as the mean value (in ng/ml) ± the SEM.
Statistical analysis
Statistical tests were performed using Graph-Pad Prism v5 for Windows. Kruskal-Wallis test with Dunn's multiple comparison post test was used for analysis among groups. Mann-Whitney test was used for the comparison of protein and gene expression levels between treated and untreated cells. Differences with a p-value less than 0.05 were considered significant.
Results
Gene expression (RT-PCR)
All gene transcripts studied were detected in all cell samples. The results are summarized in Tables 1-3. Exposure of NDCs and DCs to TNF-α had no significant effect on ACE, IDE, NEP, ECE-1 and ECE-2 gene expression levels. TNF-receptor-associated-factor 2 (TRAF-2) gene expression, which we included as a control for the activation by TNF-α exposure of the TNF-α/TNF-R signaling pathway [29], was significantly increased in DCs (p = 0.018) but not in NDCs (p = 0.085) (Table 2). The increase was significant after 3 h exposure. We also noticed that gene expression of NEP and TRAF-2 was significantly higher in DCs than untreated NDCs that had not been exposed to TNF-α (p = 0.0003 and 0.032 respectively).
Table 1.
Gene expression of Aβ degrading enzymes in NDC and DC SH-SY5Y cells treated with TNF-α
| TNF-α | IDE | ACE | NEP | ECE-1 | ||||
|---|---|---|---|---|---|---|---|---|
| NDC | DC | NDC | DC | NDC | DC | NDC | DC | |
| Control | *1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| (0.78-1.29) | (0.76-1.31) | (0.71-0.41) | (0.89-1.12) | (0.74-1.36) | (0.63-1.58) | (0.77-1.3) | (0.77-1.3) | |
| 1 ng, | 1.12 | 1.13 | 0.83 | 1.16 | 1.13 | 1.03 | 1.07 | 1.34 |
| 15 min | (0.89-1.41) | (0.9-1.43) | (0.55-1.26) | (0.97-1.37) | (0.88-1.46) | (0.51-2.1) | (0.74-1.54) | (0.98-1.84) |
| 10 ng, | 1.25 | 0.97 | 0.88 | 1.26 | 1.04 | 1.08 | 1.12 | 1.24 |
| 15 min | (1.1-1.43) | (0.97-1.08) | (0.68-1.14) | (0.96-1.65) | (0.78-1.38) | (0.51-2.3) | (0.96-1.3) | (0.95-1.62) |
| 50 ng, | 1.39 | 0.92 | 0.94 | 0.82 | 1.19 | 0.92 | 1.13 | 1.12 |
| 15 min | (1.12-1.71) | (0.72-1.17) | (0.61-1.45) | (0.63-1.07) | (0.79-1.8) | (0.39-2.18) | (0.96-1.34) | (0.76-1.64) |
| 1 ng, | 1.14 | 1.28 | 0.94 | 1.31 | 1.36 | 1.44 | 1.06 | 1.31 |
| 3 h | (1.04-1.24) | (0.99-1.66) | (0.61-1.45) | (1.01-1.7) | (1.11-1.67) | (0.79-2.6) | (0.87-1.28) | (1.09-1.57) |
| 10 ng, | 1.28 | 1.02 | 0.80 | 1.49 | 1.49 | 1.58 | 1.21 | 1.44 |
| 3 h | (1.13-1.44) | (0.7-1.49) | (0.6-1.06) | (1.29-1.72) | (1.22-1.82) | (0.87-2.84) | (0.93-1.57) | (1.11-1.87) |
| 50 ng, | 1.15 | 0.95 | 0.98 | 1.56 | 1.54 | 1.64 | 1.40 | 1.53 |
| 3 h | (0.98-1.34) | (0.65-1.41) | (0.82-1.18) | (1.15-2.12) | (1.15-2.07) | (0.85-3.15) | (1.14-1.72) | (1.16-2.03) |
Numbers are the geometric means (with 95% confidence intervals) of the fold change in gene expression in each set of experiments.
Table 3.
ECE-2 gene and protein expression in NDC and DC SHSY5Y cells treated with TNF-α
| ECE-2 | ||||
|---|---|---|---|---|
| TNF-α | NDC | DC | ||
| gene | protein | gene | protein | |
| (fold change) | mean (ng/ml) (SEM) | (fold change) | mean (ng/ml) (SEM) | |
| Control | ‡1 (0.74-1.34) | 166.85 (12.05) | 1 (0.47-2.12) | 54.25 (7.19) |
| 1 ng, 15 min | 0.93 (0.63-1.38) | 116.70* (9.21) | 1.00 (0.41-2.48) | 73.73 (9.9) |
| 10 ng, 15 min | 0.95 (0.72-1.23) | 104.91** (8.9) | 1.04 (0.43-2.51) | 98.21 (11.41) |
| 50 ng, 15 min | 0.94 (0.68-1.29) | 80.16*** (8.9) | 1.13 (0.6-2.1) | 86.54 (7.55) |
| 1 ng, 3h | 0.75 (0.55-1.02) | 175.2 (10.02) | 1.06 (0.52-2.17) | 80.81 (8.0) |
| 10 ng, 3h | 0.88 (0.62-1.26) | 181.2 (13.79) | 1.16 (0.55-2.44) | 78.31 (6.12) |
| 50 ng, 3h | 0.94 (0.63-1.4) | 201.0 (10.9) | 1.47 (0.73-2.95) | 66.75 (12.11) |
Numbers showing gene expression are geometric means (with 95% confidence intervals) and those showing protein concentration are arithmetic means and the standard error of the mean (SEM).
p = 0.016
p = 0.002
p = 0.002 (Mann-Whitney test)
Table 2.
TRAF-2 gene and protein expression in NDC and Dc SH-SY5Y cells after TNF-α treatment
| TNF-α | TRAF-2 | |||
|---|---|---|---|---|
| NDC | DC | |||
| Gene | protein | gene | protein | |
| (fold change) | Mean % of total (SEM) | (fold change) | Mean % of total (SEM) | |
| Control | ‡1 | +12.39 | 1 | 13.48 |
| (0.75-1.34) | (0.09) | (0.82-1.22) | (0.06) | |
| 1 ng, | 1.03 | 13.39 | 1.11 | 12.44 |
| 15 min | (0.66-1.61) | (0.13) | (0.79-1.55) | (0.14) |
| 10 ng, | 1.01 | 14.31 | 1.09 | 13.33 |
| 15 min | (0.72-1.42) | (0.08) | (0.75-1.57) | (0.14) |
| 50 ng, | 1.08 | 14.57 | 0.88 | 13.70 |
| 15 min | (0.74-1.57) | (0.15) | (0.78-0.98) | (0.08) |
| 1 ng, 3h | 1.15 | 13.51 | 1.33 | 13.48 |
| (0.71-1.87) | (0.09) | (0.74-2.39) | (0.09) | |
| 10 ng, 3h | 1.49 | 14.44 | 1.71* | 15.86 |
| (1.06-2.08) | (0.10) | (1.08-2.72) | (0.15) | |
| 50 ng, 3h | 1.59 | 16.30 | 1.91** | 17.72 |
| (1.03-2.46) | (0.04) | (1.22-2.98) | (0.11) | |
Numbers are the geometric means (with 95% confidence intervals) of the fold change in gene expression and the + mean % of total protein as measured by Image J software with standard error of the mean (SEM) in each set of experiments.
p=0.032 (Mann-Whintney)
p=0.008 (Mann-Whitney)
Protein expression (western blotting and ECE-2 ELISA)
All proteins studied were readily detected in homogenates of non-differentiated and differentiated SH-SY5Ys by western blotting except for ECE-2, for which ELISA was used. Western blotting of ECE-1 showed 2 specific bands with the expected molecular weights (MW) of 110 kDA and 75 kDa representing the membrane-bound version ECE-1 and its smaller soluble splice variant. Only single bands of the expected MW were detected on probing for NEP, ACE and IDE. We observed no significant effect of time or dose of TNF-α on NEP, ACE, ECE-1 or IDE protein level.
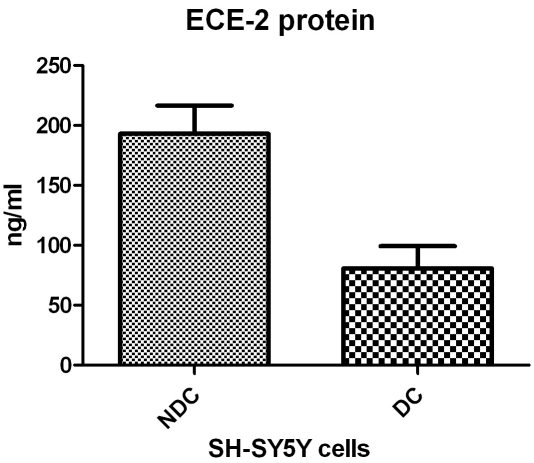
Although we were able to detect recombinant ECE-2 by western blotting, we were unable to detect ECE-2 in our cell homogenates and therefore determined its concentration by a more sensitive validated ELISA method [28]. We observed a significant effect of TNF-α on ECE-2 protein level in NDCs (p < 0.001) but not in DCs (p = 0.10) (Table 3). The increase in ECE-2 in NDCs was seen after 15 min and the concentration had returned to basal levels by 3 h (Table 3). In the absence of TNF-α, ECE-2 protein level was significantly lower in DCs than NDCs (p = 0.01, Mann-Whitney test) (Figure 1) but TRAF-2 protein expression was not significant increased in either cell line (Table 2).
Figure 1.
ECE-2 protein levels in NDCs and DCs in the absence of TNF-α p = 0.01 (Mann-Whitney test).
Discussion
Although the immune system may play a protective role in the early stages of AD, by mediating clearance of Aβ, uptake of glutamate and production of cytokines such as transforming growth factor-β1 [30, 31], but with disease progression inflammation seems to promotes neurodegeneration, partly as a result of excessive secretion of cytokines such as interleukin-1β and TNF-α [14, 3]. TNF-α production may contribute to Aβ accumulation through reduced microglial expression of SRA (scavenger receptor A) and CD36 (scavenger receptor B) both of which participate in Aβ phagocytosis [3], and up -regulation of BACE expression, demonstrated by Yamamoto et al (2007) [32] in Swedish mutant APP transgenic mice. In vitro, Liao et al, 2004 [33] reported that TNF-α stimulated γ-secretase activity, increasing the production of Aβ and the intracellular domain of APP (AICD).
We have now shown that TNF-α also causes a short-lived decline in ECE-2 in SY-SY5Y human neuroblastoma cells. The decrease has potential implications for AD. ECE-2 is capable of degrading Aβ; ECE-2 homozygous knock-out mice have significantly elevated levels of Aβ1-40 and Aβ1-42 in the brain and show impairment of learning and memory [34, 35]. However, ECE-2 is present in elevated concentration in the brain in AD and may contribute to the reduction in cerebral blood flow in this disease [28, 36] through increased production of the potent vasoconstrictor endothelin-1 [37, 28, 36]. We previously showed that exposure of SH-SY5Y cells to Aβ caused a marked rise in ECE-2 expression [28]. TNF-α may moderate this increase, although at the possible expense of reducing ECE-2-mediated degradation of Aβ.
At low concentrations, TNF-α was previously shown to promote SH-SY5Y cell survival by activating multiple survival pathways through upregulation of c-junc N-terminal kinase (JNK) and nuclear factor kappa-B (NF-κB) [38]. At higher concentrations it inhibited growth by activating apoptotic pathways [39]. A study on mouse embryonic fibroblasts found that the duration and dose of TNF-α activation had little effect on the duration of the initial NF-κB response: some NF-κB-regulated genes were expressed after only a very brief TNF-α stimulus, NF-κB itself responding very sensitively to a wide range of TNF-α concentrations [40]. We observed a significant decrease in ECE-2 protein levels in NDCs after 15 min exposure to TNF-α but the effect was transient and levels returned to normal levels after longer incubation. Our results suggest that the increased levels of ECE-2 seen in AD are not mediated through a TNF-α mechanism.
TRAF-2, an important protein of the TNF-α/TNF-R signaling pathway, was included in this study as a positive control for the activation of the TNF-α/TNF-R signaling pathway. We previously reported that TRAF-2 is present in neuritic plaques and neurofibrillary tangles in AD [26]. In this study TRAF-2 gene expression was significantly increased after 3 h exposure to TNF-α but only in differentiated SH-SY5Y cells. The differentiation process on its own activates various intracellular pathways mediating cellular stress and inhibiting cell growth. These activated pathways could enhance or counteract the effect of exogenous TNF-α and this could explain why gene expression levels of TRAF-2 vary between differentiated and non-differentiated cell samples with the same TNF-α treatment.
Surprisingly TNF-α had no effect on TRAF-2 protein levels. Previously TRAF-2 protein levels were shown to increase significantly in HUVECs within the first and third hour of TNF-α exposure [29]. TRAF-2 protein levels in that study were measured at 15-minute intervals and its expression was shown to oscillate within and over several hours. We only used 2 time points in our experiments and it is possible that we missed an intervening increase in the protein.
Differentiation of SH-SY5Y cells significantly increased NEP and TRAF-2 gene expression and significantly decreased ECE-2 protein level. The influence of retinoic acid on NEP expression was previously shown to be cell-line dependent [41]. There was no correlation between either TRAF-2 or ECE-2 mRNA and the corresponding protein level. Several other studies have found little or no correlation between the level of mRNA and that of its encoded protein [42]. This is thought to be due, at least partly, to post-transcriptional regulation by non-coding RNAs, such as miRNAs, small nucleolarRNAs (snoRNAs) and small nuclear RNAs (snRNAs) [42]. The significant impact of post-transcriptional mechanisms in regulating protein translation highlights the need to study both mRNA and protein levels to understand the impact of extracellular stimuli on individual protein production.
Acknowledgments
Salary (DC) and infrastructural support for this study was provided by BRACE (Bristol Research into Alzheimer's and Care of the Elderly). Some of the equipment was funded by grants from Alzheimer's Research UK and BRACE.
References
- 1.Paresce DM, Chung H, Maxfield FR. Slow Degradation of Aggregates of the Alzheimer's Disease Amyloid β-Protein by Microglial Cells. Journal of Biological Chemistry. 1997;272:29390–29397. doi: 10.1074/jbc.272.46.29390. [DOI] [PubMed] [Google Scholar]
- 2.Qiu WQ, Ye Z, Kholodenko D, Seubert P, Selkoe DJ. Degradation of Amyloid β-Protein by a Metalloprotease Secreted by Microglia and Other Neural and Non-neural Cells. Journal of Biological Chemistry. 1997;272:6641–6646. doi: 10.1074/jbc.272.10.6641. [DOI] [PubMed] [Google Scholar]
- 3.Hickman SE, Allison EK, El Khoury J. Microglial Dysfunction and Defective β-Amyloid Clearance Pathways in Aging Alzheimer's Disease Mice. J Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, Zlokovic BV. Clearance of Alzheimer's amyloid-β1-40 peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. The Journal of Clinical Investigation. 2000;106:1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhnke D, Jedlitschky G, Grube M, Krohn M, Jucker M, Mosyagin I, Cascorbi I, Walker LC, Kroemer HK, Warzok RW, Vogelgesang S. MDR1-P-Glycoprotein (ABCB1) Mediates Transport of Alzheimer's Amyloid-β Peptides-Implications for the Mechanisms of Aβ Clearance at the Blood-Brain Barrier. Brain Pathology. 2007;17:347–353. doi: 10.1111/j.1750-3639.2007.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, Frosch MP, Selkoe DJ. Enhanced Proteolysis of β-Amyloid in APP Transgenic Mice Prevents Plaque Formation, Secondary Pathology, and Premature Death. Neuron. 2003;40:1087–1093. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- 7.Deng-Shun Wang DWD, James S, Malter β-Amyloid Degradation and Alzheimer's Disease. Journal of Biomedicine and Biotechnology. 2006 doi: 10.1155/JBB/2006/58406. Article ID 58406 doi: 10.1155/JBB/2006/58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, Love S. Abeta-degrading enzymes in Alzheimer's disease. Brain Pathology. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weller RO, Massey A, Newman TA, Hutchings M, Kuo YM, Roher AE. Cerebral Amyloid Angiopathy Amyloid β Accumulates in Putative Interstitial Fluid Drainage Pathways in Alzheimer's Disease. Am J Pathol. 1998;153:725–733. doi: 10.1016/s0002-9440(10)65616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller RO, Massey A, Newman TA, Hutchings M, Kuo YM, Roher AE. Cerebral amyloid angiopathy: amyloid β accumulates in putative interstitial fluid drainage pathways in Alzheimer's disease. Am J Pathol. 1998;153:725–33. doi: 10.1016/s0002-9440(10)65616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preston SD, Steart PV, Wilkinson A, Nicoll JAR, Weller RO. Capillary and arterial cerebral amyloid angiopathy in Alzheimer's disease: defining the perivascular route for the elimination of amyloid β from the human brain. Neuro-pathology and Applied Neurobiology. 2003;29:106–117. doi: 10.1046/j.1365-2990.2003.00424.x. [DOI] [PubMed] [Google Scholar]
- 12.McGeer P, McGeer E. Local neuroinflammation and the progression of Alzheimer's disease. J Neurovirol. 2002;8:529–538. doi: 10.1080/13550280290100969. [DOI] [PubMed] [Google Scholar]
- 13.Heneka MT, O'Banion MK. Inflammatory processes in Alzheimer's disease. Journal of Neuroimmunology. 2007;184:69–91. doi: 10.1016/j.jneuroim.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Boche D, Nicoll JA. SYMPOSIUM: Clearance of Aβ from the Brain in Alzheimer’ Disease: The Role of the Immune System in Clearance of Aβ from the Brain. Brain Pathology. 2008;18:267–278. doi: 10.1111/j.1750-3639.2008.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong C, Qin Z, Betz AL, Liu XH, Yang GY. Cellular localization of tumor necrosis factor alpha following focal cerebral ischemia in mice. Brain Research. 1998;801:1–8. doi: 10.1016/s0006-8993(98)00489-2. [DOI] [PubMed] [Google Scholar]
- 16.Orzylowska O, Oderfeld-Nowak B, Zaremba M, Januszewski S, Mossakowski M. Prolonged and concomitant induction of astroglial immunoreactivity of interleukin-1β and interleukin-6 in the rat hippocampus after transient global ischemia. Neuroscience Letters. 1999;263:72–76. doi: 10.1016/s0304-3940(99)00043-9. [DOI] [PubMed] [Google Scholar]
- 17.Chao C, Hu S, Frey W, 2nd, Ala T, Tourtellotte W, Peterson P. Transforming growth factor beta in Alzheimer's disease. Clin Diagn Lab Immunol. 1994;1:109–110. doi: 10.1128/cdli.1.1.109-110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarkowski E, Blennow K, Wallin A, Tarkowski A. Intracerebral production of tumor necrosis factor-α, a local neuroprotective agent, in Alzheimer disease and vascular dementia. J Clin Immunol. 1999;19:223–230. doi: 10.1023/a:1020568013953. [DOI] [PubMed] [Google Scholar]
- 19.Perry RT, Collins JS, Wiener H, Acton R, Go RCP. The role of TNF and its receptors in Alzheimer's disease. Neurobiology of Aging. 2001;22:873–883. doi: 10.1016/s0197-4580(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 20.Stefano GB, Paemen LR, Hughes TK., Jr Autoimmunoregulation: differential modulation of CD10/neutral endopeptidase 24.11 by tumor necrosis factor and neuropeptides. Journal of Neuroimmunology. 1992;41:9–14. doi: 10.1016/0165-5728(92)90189-r. [DOI] [PubMed] [Google Scholar]
- 21.Zhao RZ, Chen X, Yao Q, Chen C. TNF-α induces interleukin-8 and endothelin-1 expression in human endothelial cells with different redox pathways. Biochemical and Biophysical Research Communications. 2005;327:985–992. doi: 10.1016/j.bbrc.2004.12.109. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M, Kiyota T, Walsh SM, Liu J, Kipnis J, Ikezu T. Cytokine-Mediated Inhibition of Fibrillar Amyloid-β Peptide Degradation by Human Mononuclear Phagocytes. The Journal of Immunology. 2008;181:3877–3886. doi: 10.4049/jimmunol.181.6.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saijonmaa O, Nyman T, Fyhrquist F. Down-regulation of Angiotensin-Converting Enzyme by Tumor Necrosis Factor-α and Interleukin-1β in Cultured Human Endothelial Cells. Journal of Vascular Research. 2001;38:370–378. doi: 10.1159/000051068. [DOI] [PubMed] [Google Scholar]
- 24.Viinikainen A, Nyman T, Fyhrquist F, Saijonmaa O. Downregulation of angiotensin converting enzyme by TNF-α in differentiating human macrophages. Cytokine. 2002;18:304–310. doi: 10.1006/cyto.2002.1047. [DOI] [PubMed] [Google Scholar]
- 25.Kenchappa P, Yadav A, Singh G, Nandana S, Banerjee K. Rescue of TNFα-inhibited neuronal cells by IGF-1 involves Akt and c-Jun N-terminal kinases. Journal of Neuroscience Research. 2004;76:466–474. doi: 10.1002/jnr.20081. [DOI] [PubMed] [Google Scholar]
- 26.Culpan D, Cram D, Chalmers K, Cornish A, Palmer L, Palmer J, Hughes A, Passmore P, Craigs D, Wilcock GK, Kehoe PG, Love S. TNFR-associated factor-2 (TRAF-2) in Alzheimer's disease. Neurobiology of Aging. 2009;30:1052–1060. doi: 10.1016/j.neurobiolaging.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Palmer JC, Baig S, Kehoe PG, Love S. Endothelin-Converting Enzyme-2 Is Increased in Alzheimer's Disease and Up-Regulated by Aβ. Am J Pathol. 2009;175:262–270. doi: 10.2353/ajpath.2009.081054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang SK, Wang YC, Chao CC, Chuang YJ, Lan CY, Chen BS. Dynamic cross-talk analysis among TNF-R, TLR-4 and IL-1R signalings in TNFα-induced inflammatory responses. BMC Medical Genomics. 2010;3:19. doi: 10.1186/1755-8794-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streit W. Microglia and Alzheimer's disease pathogenesis. J Neurosci Res. 2004;77:1–8. doi: 10.1002/jnr.20093. [DOI] [PubMed] [Google Scholar]
- 31.Krause DL, Müller N. Neuroinflammation, Microglia and Implications for Anti-Inflammatory Treatment in Alzheimer's Disease. International Journal of Alzheimer's Disease. 2010 doi: 10.4061/2010/732806. Article ID 732806. doi: 10.4061/2010/732806 vol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto M, Kiyota T, Horiba M, Buescher JL, Walsh SM, Gendelman HE, Ikezu T. Interferon-γ and Tumor Necrosis Factor-α Regulate Amyloid-β Plaque Deposition and β-Secretase Expression in Swedish Mutant APP Transgenic Mice. Am J Pathol. 2007;170:680–692. doi: 10.2353/ajpath.2007.060378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao YF, Wang BJ, Cheng HT, Kuo LH, Wolfe MS. Tumor Necrosis Factor-α, Interleukin-1β, and Interferon-γ Stimulate γ-Secretase-mediated Cleavage of Amyloid Precursor Protein through a JNK-dependent MAPK Pathway. Journal of Biological Chemistry. 2004;279:49523–49532. doi: 10.1074/jbc.M402034200. [DOI] [PubMed] [Google Scholar]
- 34.Eckman EA, Watson M, Marlow L, Sambamurti K, Eckman CB. Alzheimer's Disease β-Amyloid Peptide Is Increased in Mice Deficient in Endothelin-converting Enzyme. Journal of Biological Chemistry. 2003;278:2081–2084. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguiz RM, Gadnidze K, Ragnauth A, Dorr N, Yanagisawa M, Wetsel WC, Devi LA. Animals lacking endothelin-converting enzyme-2 are deficient in learning and memory. Genes, Brain and Behavior. 2008;7:418–426. doi: 10.1111/j.1601-183X.2007.00365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer J, Love S. Endothelin receptor antagonists: Potential in Alzheimer's disease. Pharmacological Research. 2011;63:525–531. doi: 10.1016/j.phrs.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 37.Emoto N, Yanagisawa M. Endothelin-converting Enzyme-2 Is a Membrane-bound, Phosphorami-don-sensitive Metalloprotease with Acidic pH Optimum. Journal of Biological Chemistry. 1995;270:15262–15268. doi: 10.1074/jbc.270.25.15262. [DOI] [PubMed] [Google Scholar]
- 38.Cheng B, Christakos S, Mattson MP. Tumor necrosis factors protect neurons against meta-bolic-excitotoxic insults and promote maintenance of calcium homeostasis. Neuron. 1994;12:139–153. doi: 10.1016/0896-6273(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 39.Berk B, Abe J, Min W, Surapisitchat J, Yan C. Endothelial atheroprotective and anti-inflammatory mechanisms. Ann N Y Acad Sci. 2001;947:93–109. doi: 10.1111/j.1749-6632.2001.tb03932.x. [DOI] [PubMed] [Google Scholar]
- 40.Cheong R, Bergmann A, Werner S, Regal J, Hoffmann A, Levchenko A. Transient IκB kinase activity mediates temporal NF-κB dynamics in response to a wide range of tumor necrosis factor-alpha doses. J Biol Chem. 2006;281:2945–2950. doi: 10.1074/jbc.M510085200. [DOI] [PubMed] [Google Scholar]
- 41.Erhuma M, Köbel M, Mustafa T, Wulfänger J, Dralle H, Hoang-Vu C, Langner J, Seliger B, Kehlen A. Expression of neutral endopeptidase (NEP/CD10) on pancreatic tumor cell lines, pancreatitis and pancreatic tumor tissues. International Journal of Cancer. 2007;120:2393–2400. doi: 10.1002/ijc.22252. [DOI] [PubMed] [Google Scholar]
- 42.Nelson PT, Keller JN. RNA in Brain Disease: No Longer Just “The Messenger in the Middle". J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]