Abstract
Saturated branched chain fatty acids (BCFA) are present as complex mixtures in numerous biological samples. The traditional method for structure elucidation, electron ionization (EI) mass spectrometry, sometimes does not unambiguously enable assignment of branching in isomeric BCFA. Zirrolli and Murphy (Zirrolli , J. A. , and R. A. Murphy. 1993. Low-energy tandem mass spectrometry of the molecular ion derived from fatty acid methyl esters: a novel method for analysis of branched-chain fatty acids. J. Am. Soc. Mass Spectrom. 4: 223–229.) showed that the molecular ions of four BCFA methyl ester (BCFAME) yield highly characteristic fragments upon collisional dissociation using a triple quadrupole instrument. Here, we confirm and extend these results by analysis using a tabletop 3-D ion trap for activated molecular ion EI-MS/MS to 30 BCFAME. iso-BCFAME produces a prominent ion (30-100% of base peak) for [M-43] (M-C3H7), corresponding to the terminal isopropyl moiety in the original iso-BCFAME. Anteiso-FAME yield prominent ions (20-100% of base peak) corresponding to losses on both side of the methyl branch, [M-29] and [M-57], and tend to produce more prominent m/z 115 peaks corresponding to a cyclization product around the ester. Dimethyl and tetramethyl FAME, with branches separated by at least one methylene group, yield fragment on both sides of the sites of methyl branches that are more than 6 C away from the carboxyl carbon. EI-MS/MS yields uniquely specific ions that enable highly confident structural identification and quantification of BCFAME.
Keywords: branched chain fatty acid, saturated fatty acid, structural characterization, electron ionization tandem mass spectrometry
Methyl branched chain fatty acids (BCFA) are prevalent in a wide range of biological samples. They are especially rich in the membranes of some bacteria (1) and in animal skin and secretions, for instance, in sebum (2), cerumen (3), and meibomian gland (4) of the human eyelid and in the Harderian gland of rodents (5). They are also produced by rumen bacteria and are a substantial constituent of rumen tissue and milk. We recently showed that BCFA constitute 2% of fatty acids in the United States milk supply (6). The most prevalent monomethyl BCFA have branching on the n-2 or n-3 carbons, referred to as iso and anteiso, respectively. Polymethyl BCFA arising in prenol lipids are also common, specifically phytanic and pristanic acids.
The routine method of fatty acid analysis is to convert them from their nature lipid class into fatty acid methyl esters (FAME), which have superb characteristics on capillary gas chromatograph columns with respect to peak shape and baseline separation. The usual approach for identifying branching in iso or anteiso branched chain FAME (BCFAME) is to examine the electron ionization (EI) mass spectrum for losses corresponding to branching at the end of the molecule. Although this approach is satisfactory in many cases, peak intensities are often low and sometimes do not enable unambiguous assignment. For this reason, specialized esters that localize charge and enhance structure-specific charge-remote fragmentation, such as dimethyloxazoline (DMOX) and picolinyl esters, are typically prepared for EI-MS analysis (7, 8). However, preparation of DMOX, picolinyl, or other esters requires derivatization chemistry beyond routine methylation. Specialized esters have chromatographic characteristics that differ from FAME, requiring customized chromatography; peaks change retention times, sometimes inverting compared with FAME; establishing correspondence between FAME and other fatty esters can be challenging. High energy collisionally activated dissociation (CAD) in a tandem time-of-flight (TOF-TOF) mass spectrometer initiated by MALDI has been applied to locate branching in a single BCFAME recently (9); however, this method has not been further developed. Finally, all methods thus examined tend to yield abundant nondiagnostic product ions. This distributes the signal among several ions, which effectively reduces the sensitivity for selected ion monitoring and quantitative analysis. Methods for direct analysis of FAME that yield a few intense peaks are therefore preferred.
Zirroli and Murphy presented a method for direct analysis of a limited number of BCFAME by tandem mass spectrometry of the molecular ion using a triple quadrupole mass spectrometer (10). BCFAME were ionized by 70 eV electrons and the molecular ion isolated. CAD from 1-200 eV yield a striking, novel mass spectrum distinct from the MS-1 EI spectrum, devoid of the McLafferty rearrangement (11, 12) peak at m/z 74 that is usually at high abundance and the base peak in many FAME spectra. CAD spectra of the molecular ion yielded a novel series of peaks with intensities depending on CAD energy. CAD of BCFAME yielded intense peaks that unambiguously reveal the locus of branching for three monomethyl BCFAME [methyl 10-methyl-octadecanoate, methyl 13-methylpentadecanoate (anteiso-16:0), and methyl 14-methylpentadecanoate (iso-16:0)]. A series of fragments also were unique to the tetramethyl BCFAME methyl phytanate (3,7,11,15-tetramethyl 16:0).
We recently characterized the BCFA of human vernix caseosa, the waxy substance unique to humans that covers the fetus at birth, compared with BCFA in meconium from the same infants (13). In vernix, more than 20 monomethyl and dimethyl BCFA with 11 to 26 carbons were found at a total concentration of about 29% of fatty acids (13). Using vernix and lanolin, which contains some BCFA that are not present in vernix, we performed MS/MS on molecular ions of BCFAME using a tabletop internal ionization ion trap mass spectrometer. We report here spectra from BCFAME from these natural mixtures to compare and contrast spectra, and we present evidence that the technique applies to a broad range of BCFAME.
EXPERIMENTAL METHODS
Instrumentation
Data were generated on a tabletop Varian Star 3400CX gas chromatograph equipped with a 1078 split/splitless injector coupled to a Varian Saturn 2000 3D ion trap (Varian Inc., Walnut Creek, CA). BCFAME were separated on a BPX70 capillary column (60 m × 0.32 mm × 0.25 µm; SGE Inc., Austin, TX). GC conditions were as follows: injector temperature was 250°C in splitless mode with a purge at 0.85 min after injection; initial oven temperature was 60°C held for 1 min, then ramped to 170°C at 50°C/min and held for 6 min, then ramped to 200°C at 2.5°C/min and held for 3 min, then ramped up to 222°C at 10°C/min and held for 8.7 min, and then ramped to 255°C at 50°C/min and held for 1 min. Total run time was 36.8 min.
M+ ions for BCFAME were isolated for fragmentation in EIMS2 mode. The ionization mode was set to “EI auto mode” using the default parameters set by the Varian Saturn software V5.5.2. Ion preparation parameters were as follows: isolation window 3.0 amu; waveform type residence; excitation storage level was calculated using a q value of 0.215; excitation amplitude was set to 0.80 V. Segment set point parameters were: scan rate 1 s; count threshold 1; emission current 0.5 µA. All spectra were collected under the identical instrument settings, including collision energy (excitation amplitude) and mass isolation window. In our hands, these conditions provided suitable fragments intensities across all BCFAME without the need to customize parameters. The GC elution order for all BCFAME is iso, anteiso, normal under our conditions. Multiply branched FAME tend to elute prior to the iso isomer.
Samples
Very few BCFA standards are readily available. A limited number of pure BCFA are available commercially at high cost. Vernix caseosa from a previous study (13) and lanolin purchased from a local retail store were methylated using BF3/methanol and served as sources of BCFA. Methyl phytanate was obtained from Matreya, LLC (Bellefonte, PA). Identity of n-FAME and BCFAME are established by molecular weight and retention time. Saturated FAME appearing at retention times that did not correspond to normal, iso, or anteiso BCFA with internal branching were made by applying rules for interpretation developed for the knowns. Thus, internal and multiple branching spectra are labeled based on the spectra themselves, and as they were not validated by an independent structure-specific method, they should be considered tentative identifications. Their spectra are shown to illustrate fragmentation for internal and multiply branched FAME.
Spectra are presented in the text and in the supplementary data. For convenience, Table 1 provides figure numbers for all EI-MS/MS spectra. For simplicity, specific FAME are discussed according to their fatty acid designations (e.g., n-17:0) and the leading “methyl” [e.g, methyl(n-17:0)] is dropped.
TABLE 1.
FAME and BCFAME activated molecular ion EI-MS/MS spectra presented in this article according to figure number; those labeled with a roman numeral are in the online supplementary data.
| Acyl Chain Carbon | Normal (n) | Iso | Anteiso | Other (Assigned Location of Branch Point) |
| 12 | I-A | I-B | ||
| 13 | II-A | 4A | 4B | 6,9 (II-B) |
| 14 | III | 4C | 4D | |
| 15 | IV-A | 4E | 4F | 8,11 (IV-B) |
| 16 | V-A | V-B | 9 (V-C) | |
| 17 | 2A | 2B | 2C | 10,13 (2D) |
| 18 | VI-A | VI-B | 12,14 (VI-C) | |
| 19 | VII-A | VII-B | ||
| 20 | 3A | 3B | 3,7,11,15 (3C) | |
| 21 | VIII-A | VIII-B | VIII-C | |
| 22 | IX-A | IX-B | ||
| 24 | X-A | X-B | ||
| 25 | 6A | 6B | 6C | |
| 26 | 5A | 5B | ||
| 27 | 6D | |||
| 28 | 5C | 5D | ||
| 29 | 6E | |||
| 31 | 6F |
RESULTS AND DISCUSSION
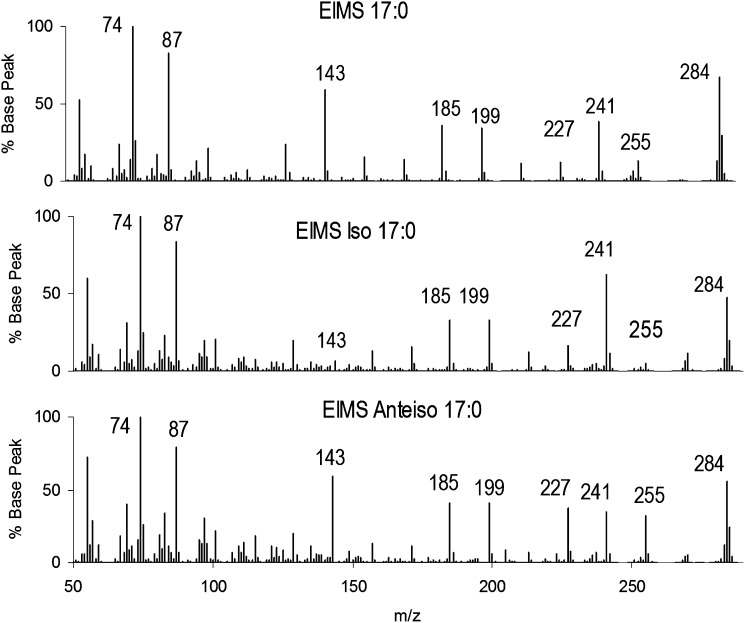
We first present the conventional single-stage EI-MS of isomeric saturated FAME to contrast with the MS/MS method. Fig. 1 illustrates the single-stage EIMS spectra of three isomeric 17:0. Careful inspection shows that the three spectra contain no unique ions; thus, peak intensities must suffice to distinguish unique isomers. The molecular ions at m/z 284 are present at 50-80% of the m/z 74 (McLafferty rearrangement) peak, with the accompanying m/z 87 peak at about 90%. iso-17:0 can be distinguished from n-17:0 and anteiso-17:0 by the intensity of the m/z 143 peak. n-17:0 and anteiso-17:0 spectra have minor intensity differences in m/z 227 and 255, but they are otherwise nearly indistinguishable.
Fig.1.
Electron ionization mass spectra (EIMS) for isomeric C17 FAME. (A) n-17:0, (B) iso-17:0, and (C) anteiso-17:0. Structural differences produce differences in ion intensities but no unique major ions.
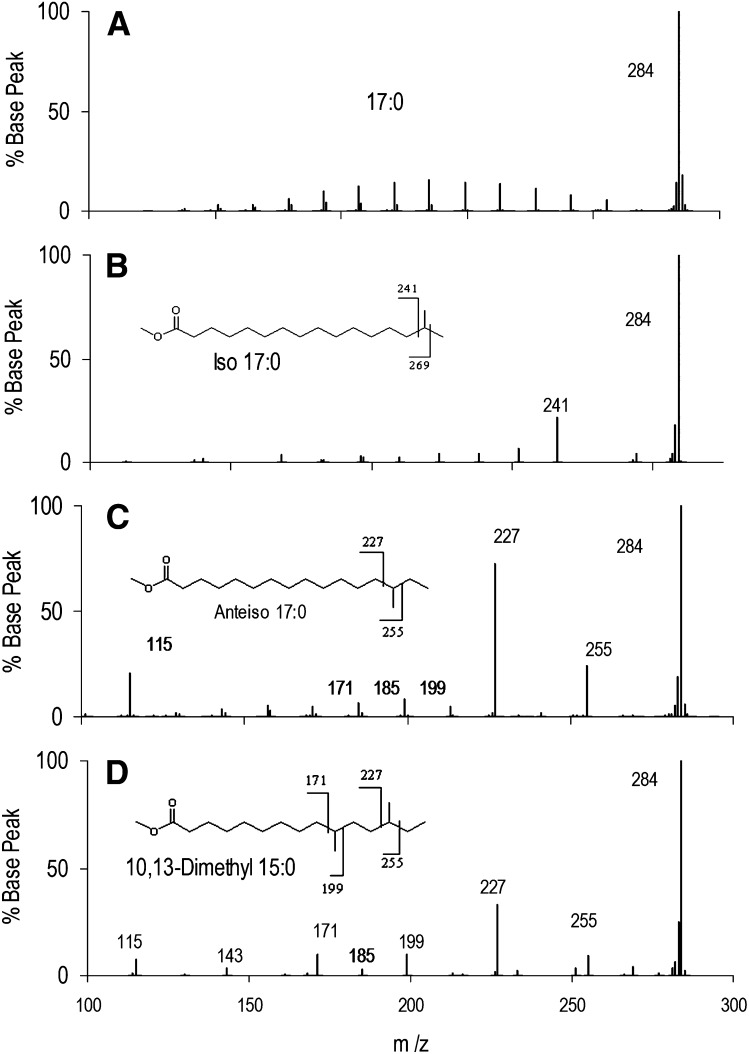
Fig. 2 presents the EI-MS/MS spectra of the same three isomeric 17:0 shown in Fig. 1, as well as a dimethyl isomer. Upon collisional dissociation of the m/z 284 peak, n-17:0 yields a series of ions of uniformly low intensity due to fragmentation between each C-C bond. In contrast, iso-17:0 yields a unique peak at m/z 241 and very weak fragment ions. The major fragment (m/z 241) arises by loss of the terminal isopropyl group [M-43] (M-C3H7). Anteiso-17:0 yields a peak at m/z 227, corresponding to loss of the terminal isobutyl group [M-57] (M-C4H9), and another at m/z 255 for loss of the terminal ethyl group [M-29] (M-C2H5). On the basis of these fragments, we posit that the dimethyl species is 10,13-dimethyl-15:0. It has fragments characteristic of branching at the anteiso position C13, analogous to anteiso-17:0. Interpreting peaks at m/z 171 and 199 in the same manner leads to the conclusion that fragments represent loss or retention of the neutral fragment containing the tertiary carbon at position 10, which are at greater intensity than those retaining or losing an addition 14 amu along the carbon chain (m/z 213 or m/z 185, or m/z 157, which is absent). This fragmentation is consistent with that observed for phytanic acid as shown below. Another notable ion is found at m/z 115 in the anteiso and 10,13-dimethyl-15:0 spectra but is absent in the iso spectrum. As shown below, this ion appears in most anteiso spectra, is absent or at low intensity in iso spectra, and is not observed in BCFAME with methyl branching at C3. Formation of FAME product ions has been extensively at low and high collision energies. Isotope labeling shows that it is often accompanied by H or ester group migration (14). The m/z 115 ion corresponds to alkyl radical loss with cleavage between C5 and C6 and no rearrangements under our low energy collision conditions. The structure of this common ion cannot be established by the data here, and the fragmentation most consistent with labeling studies makes no obvious prediction as to differences in fragment formation between iso and anteiso isomers (14). However, the appearance of m/z 115 can be rationalized based on cyclization and/or resonance stabilization of a product ion as either a direct cyclization (Scheme 1) or, perhaps less plausibly, as a resonance stabilized diradical cation (Scheme 2). The final m/z 115 product may be a resonance stabilized oxane cation.
Fig.2.
EI-MS/MS of isomeric C17 FAME. (A) n-17:0 yields exclusively a series of ions reflecting cleavage between C-C bonds; adjacent fragment ions appear as an envelope of intensities. (B) iso-17:0 yields a major ion at m/z 241 due to loss of the terminal isopropyl group [M-43] in the parent FAME as well as minor ions from C-C bond cleavage. (C) anteiso-17:0 yields two major ions reflecting bond breakage on both sides of the branch point in the parent FAME, at m/z 227 [M-57] and 255 [M-29]. (D) 10,13-dimethyl-15:0 yields ions reflecting bond breakage around the branch points. A branch at the 10 position is evident because of the negligible intensity of m/z 185 at the intermediate branch point.
Finally, there is no evidence for C-H bond breakage due to collisional activation of the molecular ion. The M-1 and M-2 H-loss peaks are evident in the first-stage EI spectrum of Fig. 1 and are retained in MS/MS due to imperfect isolation of the molecular ion. All EI-MS/MS spectra are consistent with this result.
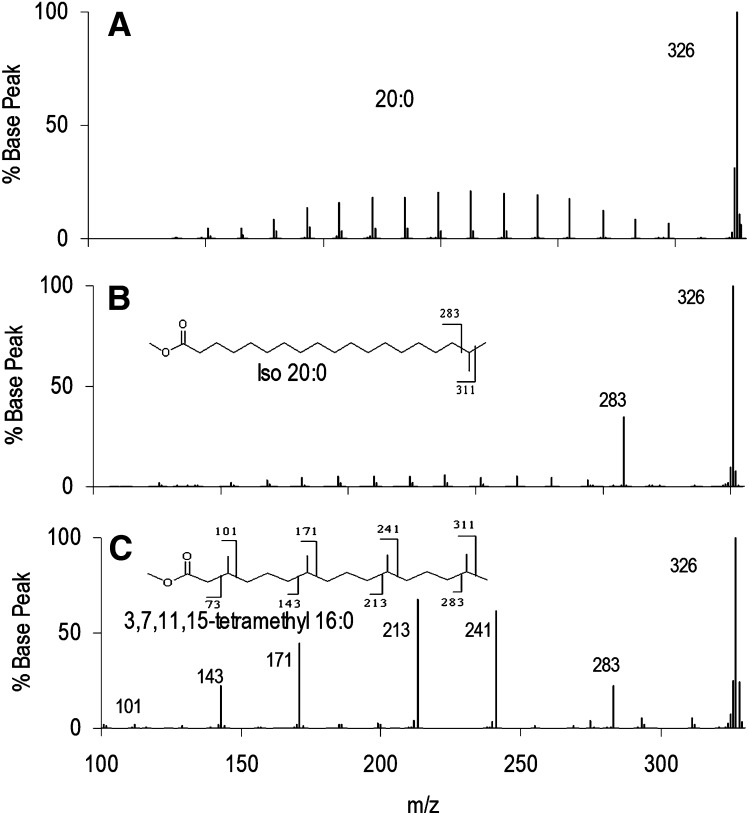
Fig. 3 presents a similar series of three isomeric C20, including one multiply branched FAME. Again, n-20:0 EI-MS/MS spectra yield an envelope of C-C bond breakage ions, and iso-20:0 presents a prominent fragment representing loss of the tertiary carbon. Phytanic acid (3,7,11,15-tetramethyl 16:0) produces an envelope of major peaks corresponding to fragmentation around the methyl branch points, with the exception of the branch at C3 (m/z 101), which gave very low signal. These results are similar to those presented previously for this molecule (10).
Fig.3.
EI-MS/MS of isomeric C20 FAME. (A) n-20:0, (B) iso-20:0, and (C) 3,7,11,15-tetramethyl-16:0 showing most intense fragments corresponding to cleavage at the branch points distal from the ester. The relative intensities decrease for branch points closer to the ester.
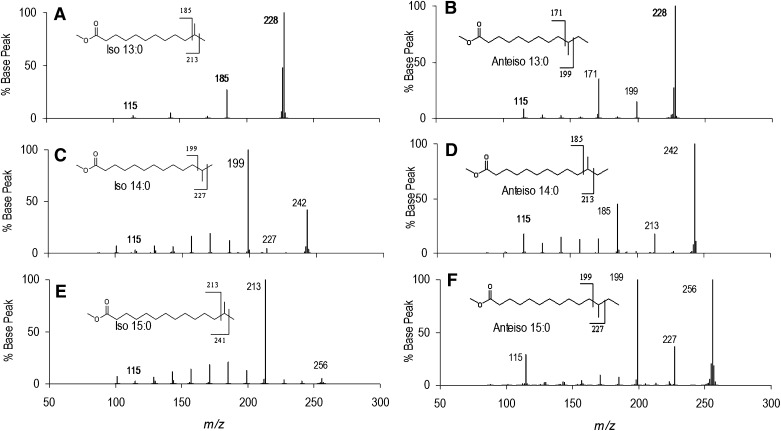
A series of iso and anteiso BCFAME spectra are presented in Fig. 4 for C13-C15. The iso BCFAME consistently yield strong peaks for the isopropyl loss fragments, although their intensities and the intensities of the molecular ions vary substantially. The anteiso BCFAME consistently yield strong signals for fragmentation on both sides of the branch point. Intensities are more consistent than for iso BCFAME, with the molecular ion as the base peak in all spectra and the ethyl fragment loss peak of lowest intensity. The m/z 115 peak is below 5% abundance in all the iso spectra, whereas it was 10-30% abundance in all the anteiso spectra. Although the mechanism for this intensity difference is unclear, it provides confirmatory diagnostic information for structure assignment.
Fig.4.
EI-MS/MS of a series of iso and anteiso C13-C15 BCFAME spectra. (A) iso-13:0, (B) anteiso-13:0, (C) iso-14:0, (D) anteiso-14:0, (E) iso-15:0, and (F) anteiso-15:0. The iso BCFAME yield peaks corresponding to the loss of the terminal isopropyl group. The anteiso BCFAME yield signals for fragmentation corresponding to loss of the terminal isobutyl and ethyl groups. m/z 115 is below 5% abundance in iso spectra, and above 10% abundance in anteiso spectra.
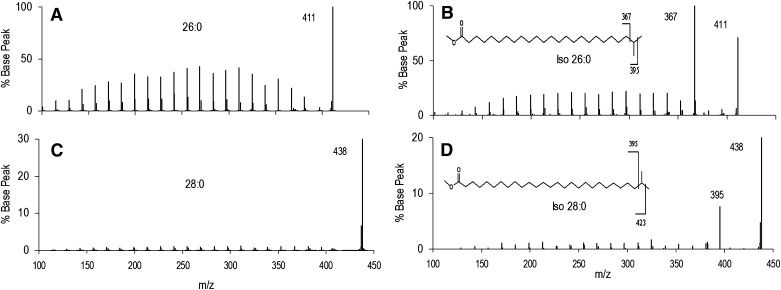
The spectra of straight chain and iso FAME are presented in Fig. 5 for very long chain (VLC) FAME (≥24 carbons). n-26:0 and iso-26:0 yield the analogous fragments observed for shorter chain FAME. n-28:0 and iso-28:0 also yield the expected fragments, although the abundances are much lower than for n-26:0 and iso-26:0 (note change in y axis scale). These data were obtained in separate analyses, and the differences in fragment yields are very likely to be due to differences in collision energy rather than the two-carbon difference in structure.
Fig.5.
EI-MS/MS spectra of very long chain iso FAME. (A, B) Mass spectra of n-26:0 and iso-26:0. (C, D) Mass spectra of 28:0 and iso-28:0. The loss of terminal isopropyl fragment is a major peak in both iso species.
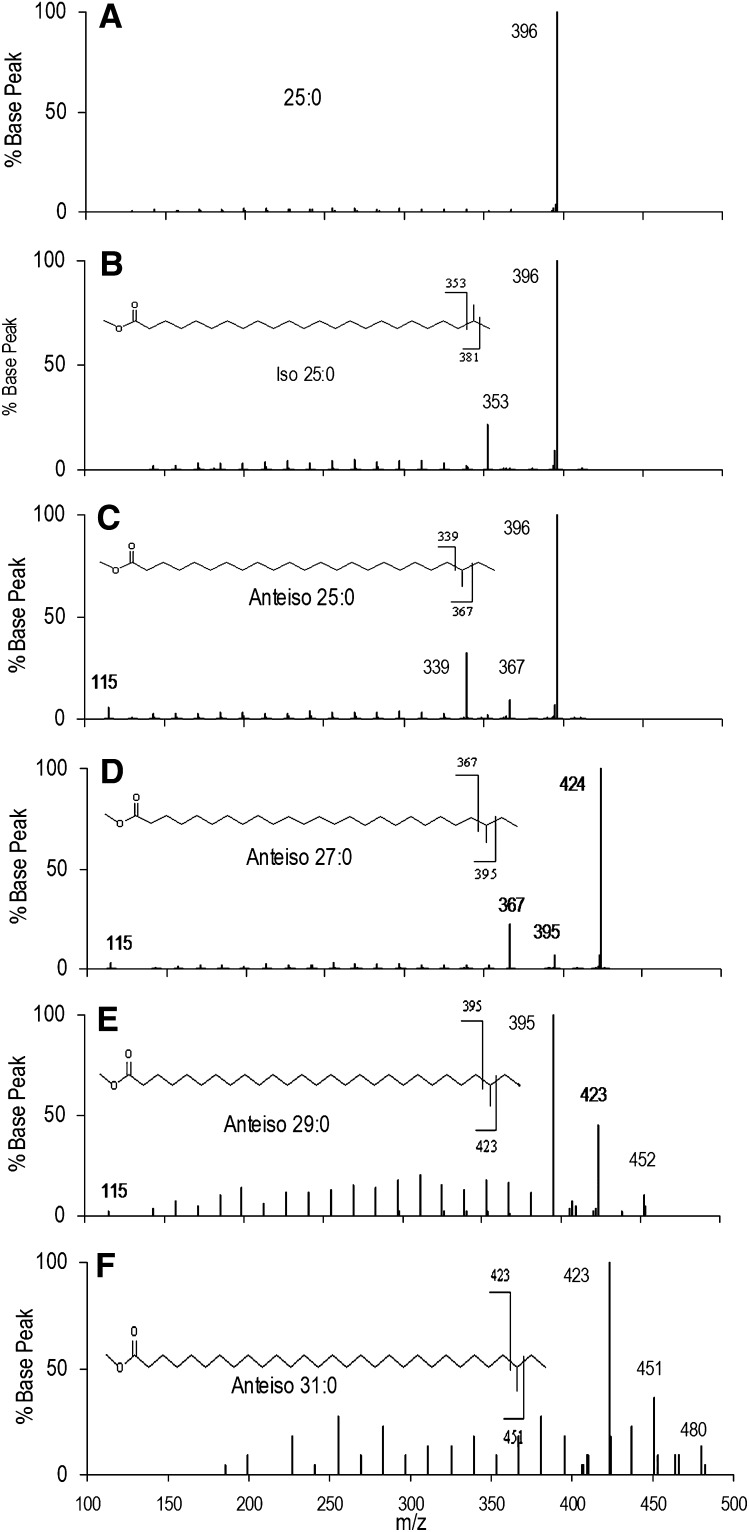
Several spectra of anteiso VLC-BCFAME are compared in Fig. 6. Fig. 6AndashC are spectra of n-25:0, iso-25:0, and anteiso-25:0, again all showing the expected fragments. Fig. 6DndashF are spectra for anteiso-27:0, anteiso-29:0, and anteiso-31:0. Fragmentation around the methyl branch point is found for all anteiso spectra. However, lower total concentrations of anteiso-29:0 and anteiso-31:0 led to lower signal-to-noise spectra in which the molecular ions are of low abundance. The anteiso-31:0 spectrum has no peak for m/z 115 and is the only anteiso spectrum in our dataset in which it does not appear, possibly due to the general low abundance of anteiso-31:0 and inefficient trapping at low masses. Figs. 5 and 6 show that the method yields useful results for large differences in fragmentation efficiency and signal-to-noise.
Fig.6.
EI-MS/MS spectra of very long chain anteiso FAME. (A) Mass spectrum of n-25:0, (B) iso-25:0, and (C) anteiso-25:0. The observed fragments are analogous to those observed for shorter chain BCFAME. (D, E, F) Mass spectra of anteiso-27:0, anteiso-29:0, and anteiso-31:0 show similar fragments to shorter chain anteiso BCFAME, although signal-to-noise is lower for these rarer FAME. The m/z 115 ion is observed for most anteiso VLC-BCFAME.
Taken together, these spectra demonstrate greater specificity in locating branch points in isomeric saturated FAME than is available in single-stage EIMS spectra. Moreover, the highly abundant ions unique to specific structures facilitate chromatographic resolution by selected ion plots as well as improve quantitative analysis.
Table 2 presents expected fragments for monomethyl branches at the iso, anteiso, and mid-chain positions for BCFAME from 12:0 to 26:0. Molecular weights of the methyl esters are located below the FAME designation. Branching at the iso position results in [M-43] as the main loss peak and is recorded two cells below the iso headings. Branching at the anteiso position results in [M-29] and [M-57] as the main loss peaks, both recorded under the anteiso headings. Branching at other positions are recorded under their respective headings as fragments that do or do not retain the branching carbon and methyl groups. Thus, fragments are 28 Da different in mass. Fragment intensities are expected to be very low for branches within four carbons of the carboxyl group. Finally, although it is recorded in the table, by itself m/z 115 is not a reliable diagnostic ion to assign branching position.
TABLE 2.
Diagnostic ions expected for branch points upon collisional dissociation of BCFAME-activated molecular ions
| Branch Point | ||||||||||||||||||||||||||||
| 12:0 | Iso | Anteiso | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||||||||||||||||
| 214 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||||||||||||||
| 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||||||||||||||
| 13:0 | Iso | Anteiso | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||||||||||||||
| 228 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||||||||||||||
| 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||||||||||||||
| 14:0 | Iso | Anteiso | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||||||||||||||
| 242 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||||||||||||
| 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||||||||||||
| 15:0 | Iso | Anteiso | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||||||||||||
| 256 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||||||||||||
| 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||||||||||||
| 16:0 | Iso | Anteiso | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||||||||||||
| 270 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||||||||||
| 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||||||||||
| 17:0 | Iso | Anteiso | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||||||||||
| 284 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||||||||||
| 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||||||||||
| 18:0 | Iso | Anteiso | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||||||||||
| 298 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||||||||
| 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||||||||
| 19:0 | Iso | Anteiso | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||||||||
| 312 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||||||||
| 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||||||||
| 20:0 | Iso | Anteiso | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||||||||
| 326 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||||||
| 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||||||
| 21:0 | Iso | Anteiso | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||||||
| 340 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||||||
| 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||||||
| 22:0 | Iso | Anteiso | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 | |||||||||
| 354 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||||
| 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||||
| 23:0 | Iso | Anteiso | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||||
| 368 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||||
| 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||||
| 24:0 | Iso | Anteiso | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||||
| 382 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||||
| 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||||
| 25:0 | Iso | Anteiso | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||||
| 396 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||||
| 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||||
| 26:0 | Iso | Anteiso | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||||
| 410 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||||
| 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||||
| 27:0 | Iso | Anteiso | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||||
| 424 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||||
| 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||||
| 28:0 | Iso | Anteiso | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |||
| 438 | 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||||
| 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||||
| 29:0 | Iso | Anteiso | 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | ||
| 452 | 423 | 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |||
| 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |||
| 30:0 | Iso | Anteiso | 26 | 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | |
| 466 | 437 | 423 | 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | ||
| 423 | 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | ||
| 31:0 | Iso | Anteiso | 27 | 26 | 25 | 24 | 23 | 22 | 21 | 20 | 19 | 18 | 17 | 16 | 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 |
| 480 | 451 | 437 | 423 | 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | |
| 437 | 423 | 409 | 395 | 381 | 367 | 353 | 339 | 325 | 311 | 297 | 283 | 269 | 255 | 241 | 227 | 213 | 199 | 185 | 171 | 157 | 143 | 129 | 115 | 101 | 87 | 73 | 59 | |
The molecular weights (molecular masses) are listed below the BCFAME designations (e.g., MW(Me12:0) = 214). For iso-BCFAME, the loss of methyl [M-15] (e.g., 12:0 m/z 199) is absent or weak and is not shown; the loss of isopropyl dominates [M-43] (e.g., 12:0 m/z 171). Bold numbers denote carbon number of the branch point.
These data document a convenient and reliable method for assignment of branching in methyl BCFAME. The strong and distinct peak intensities enable assignment with high confidence. Moreover, this approach concentrates signal in a small number of characteristic fragments rather than a distributing signal among many overlapping fragment ions characteristic of methods relying on specialized esters. Quantitative analysis via selected ion monitoring is more sensitive and precise with a small number of high-intensity ions. Moreover, the ability to analyze FAME directly obviates the need for special chemistry or chromatography for specialized esters. Implementation on inexpensive tabletop ion traps capable of MS/MS is a straightforward, very attractive method for BCFAME analysis.
Supplementary Material
Acknowledgments
The authors thank Robert Murphy for a helpful discussion.
Footnotes
Abbreviations:
- BCFA
- branched chain fatty acid
- BCFAME
- branched chain FAME
- CAD
- collisionally activated dissociation
- FAME
- fatty acid methyl ester
This work was supported by National Institutes of Health Grant R21 HD-064604. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and one table.
REFERENCES
- 1.Kaneda T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55: 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart M. E. 1992. Sebaceous gland lipids. Semin. Dermatol. 11: 100–105. [PubMed] [Google Scholar]
- 3.Harvey D. J. 1989. Identification of long-chain fatty acids and alcohols from human cerumen by the use of picolinyl and nicotinate esters. Biomed. Environ. Mass Spectrom. 18: 719–723. [DOI] [PubMed] [Google Scholar]
- 4.Harvey D. J., Tiffany J. M. 1984. Identification of meibomian gland lipids by gas chromatography-mass spectrometry: application to the meibomian lipids of the mouse. J. Chromatogr. 301: 173–187. [DOI] [PubMed] [Google Scholar]
- 5.Seyama Y., Ohashi K., Imamura T., Kasama T., Otsuka H. 1983. Branched chain fatty acids in phospholipids of guinea pig Harderian gland. J. Biochem. 94: 1231–1239. [DOI] [PubMed] [Google Scholar]
- 6.Ran-Ressler R. R., Sim D., O'Donnell-Megaro A. M., Bauman D. E., Barbano D. M., Brenna J. T. 2011. Branched chain fatty acid content of United States retail cow's milk and implications for dietary intake. Lipids. 46: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey D. J. 1982. Picolinyl esters as derivatives for the structural determination of long chain branched and unsaturated fatty acids. Biolog. Mass Spectrom. 9: 33–38. [Google Scholar]
- 8.Yu Q. T., Liu B. N., Zhang J. Y., Huang Z. H. 1988. Location of methyl branchings in fatty acids: fatty acids in uropygial secretion of Shanghai duck by GC-MS of 4,4-dimethyloxazoline derivatives. Lipids. 23: 804–810. [DOI] [PubMed] [Google Scholar]
- 9.Trimpin S., Clemmer D. E., McEwen C. N. 2007. Charge-remote fragmentation of lithiated fatty acids on a TOF-TOF instrument using matrix-ionization. J. Am. Soc. Mass Spectrom. 18: 1967–1972. [DOI] [PubMed] [Google Scholar]
- 10.Zirrolli J. A., Murphy R. A. 1993. Low-energy tandem mass spectrometry of the molecular ion derived from fatty acid methyl esters: a novel method for analysis of branched-chain fatty acids. J. Am. Soc. Mass Spectrom. 4: 223–229. [DOI] [PubMed] [Google Scholar]
- 11.Happ G. P., Stewart D. W. 1952. Rearrangement peaks in the mass spectra of certain aliphatic acids. J. Am. Chem. Soc. 74: 4404–4408. [Google Scholar]
- 12.McLafferty F. W. 1956. Mass spectrometric analysis. Broad applicability to chemical research. Anal. Chem. 28: 306–316. [Google Scholar]
- 13.Ran-Ressler R. R., Devapatla S., Lawrence P., Brenna J. T. 2008. Branched chain fatty acids are constituents of the normal healthy newborn gastrointestinal tract. Pediatr. Res. 64: 605–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidavsky I., Chorush R. A., Longevialle P., McLafferty F. W. 1994. Functional group migration in ionized long-chain compounds. J. Am. Chem. Soc. 116: 5865–5872. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.