Abstract
Lipid droplets are evolutionarily conserved organelles where cellular fat storage and mobilization are exquisitely regulated. Recent studies have defined lipid droplets in C. elegans and explored how they are regulated by genetic and dietary factors. C. elegans offers unique opportunities to visualize lipid droplets at single-cell resolution in live animals. The development of novel microscopy techniques and protein markers for lipid droplets will accelerate studies on how nutritional states and subcellular organization are linked in vivo. Together with powerful tools for genetic and biochemical analysis of metabolic pathways, alteration in lipid droplet abundance, size, and distribution in C. elegans can be readily connected to whole-animal energy homeostasis, behavior, and life span. Therefore, further studies on lipid droplets in C. elegans promise to yield valuable insights that complement our knowledge gained from yeast, Drosophila, and mammalian systems on cellular and organismal fat storage.
Lipid droplets are ubiquitous fat storage organelles that are conserved in yeast, C. elegans, Drosophila, and mammals (1–3). They are also sites of regulated release of stored fat by lipases during cell growth and fasting (4). Therefore, lipid droplets are central to energy balance at cellular and organismal levels. Key structural and biochemical features of lipid droplets have been defined, and it is generally accepted that neutral lipids such as triglycerides (TAG) and cholesterol esters (CE) are stored in the interior of the organelle that is delimited by a phospholipid monolayer (5–7). Current models suggest that fatty acid influx can be accommodated by neutral lipid synthesis and a concomitant change in lipid droplet size or number in a tissue-specific manner (1, 8). For example, differentiation of mammalian white adipose cells is characterized by a decrease in lipid droplet number and an increase in lipid droplet size until a dominant unilocular lipid droplet remains (9, 10). The mechanisms that govern lipid droplet size and number in white adipose cells and other cell types are not fully understood.
To study regulators of fat storage and lipid droplet size in metazoans, most studies have focused on the use of tissue culture cells and overexpressed lipid droplet-associated proteins or vital dyes as markers. These studies yielded important insights into how conserved proteins and signaling pathways are coupled to control cellular fat storage. Nevertheless, extension of observations made in tissue culture cells into whole-animal models is often time consuming and can lead to surprising results. In the last few years, C. elegans has emerged as an attractive model for studying fat storage in vivo (11, 12). Although C. elegans lacks dedicated adipose tissues, its intestine serves as the main site for fat storage. Yolk lipoproteins are synthesized in the intestine, exported into the body cavity (pseudocoelomic space), and taken up by developing oocytes (13–15). This is remarkably similar to yolk deposition in chicken, where yolk is synthesized in the liver and transported to the ovum via the bloodstream (16). Therefore, the C. elegans intestine may be viewed as a multifunctional organ that fulfills the roles of the liver and adipose tissues.
Inspection of the sequenced genome of C. elegans and functional studies through analysis of genetic mutants revealed extensive conservation of genes involved in fatty acid synthesis, elongation, desaturation, and degradation (17–23). Furthermore, fat storage in C. elegans appears to be controlled by conserved insulin, TGF-β, serotonin, and mammalian target of rapamycin (mTOR) signaling pathways (24–29). A number of reverse genetic techniques will accelerate functional dissection of signaling and protein interaction networks (30–32). The development of novel transgenic technologies has also enabled expression of native or heterologous proteins to near-physiological levels (33). This is critical for structure-function analysis of specific proteins in vivo and the accurate targeting of fluorescent protein markers to lipid droplets and other subcellular organelles. The use of C. elegans offers a unique advantage of imaging lipid droplets in live animals at single-cell resolution. Their intestinal cells support lipid droplet expansion to >10 µm in diameter, similar to lipid droplets found in mammalian adipocytes (34). In contrast, adherent tissue culture cells have limited depth, which restricts spherical expansion of lipid droplets. Finally, C. elegans offers ample opportunities to connect alteration in cellular fat storage to whole-animal physiology, behavior, and life span (18, 35, 36). This review will focus on the discovery of lipid droplets in C. elegans, the methods to detect lipid droplets, and the proteins that associate with lipid droplets in this model organism. Thorough understanding of these areas will no doubt provide answers to fundamental questions regarding how lipid droplets accommodate changes in the cellular demand and supply of fat in an evolutionarily conserved manner.
IDENTIFICATION OF LIPID DROPLETS IN C. ELEGANS
In the absence of a specialized fat storage tissue, C. elegans store their fat primarily in the intestine. It is remarkable that the entire intestine is composed of only 20 cells, arranged in rings of two or four cells that enclose the lumen, which spans almost the entire length of the animal (∼1 mm) (37, 38). Each intestinal cell is derived from a single embryonic blastomere; cell division ceases and organogenesis is complete before the hatching of a free-living animal (37, 38). Nevertheless, the intestinal cells increase in size that matches the growth of the animal as it progresses through four larval stages before maturing into a reproductive adult.
One of the defining features of lipid droplets is the phospholipid monolayer that encloses their neutral lipid core, as demonstrated by transmission and freeze-fracture electron microscopy (5–7). This sets lipid droplets apart from all other intracellular organelles that are bound by a phospholipid bilayer. Extensive ultrastructural studies have been performed in C. elegans that revealed numerous vesicular compartments of similar size in the intestinal cells (37–39). These compartments may be differentiated by their electron densities. Two putative fat and lipoprotein storage compartments have been proposed: electron-lucent lipid droplets and electron-opaque gut granules (37–40). Gut granules have also been detected using light microscopy and were subsequently defined as lysosome-related organelles (LRO) (41, 42). The appearance and relative abundance of these compartments are dependent on the developmental stages of the animal. However, membrane structures that surrounded these compartments were not studied in detail until recently.
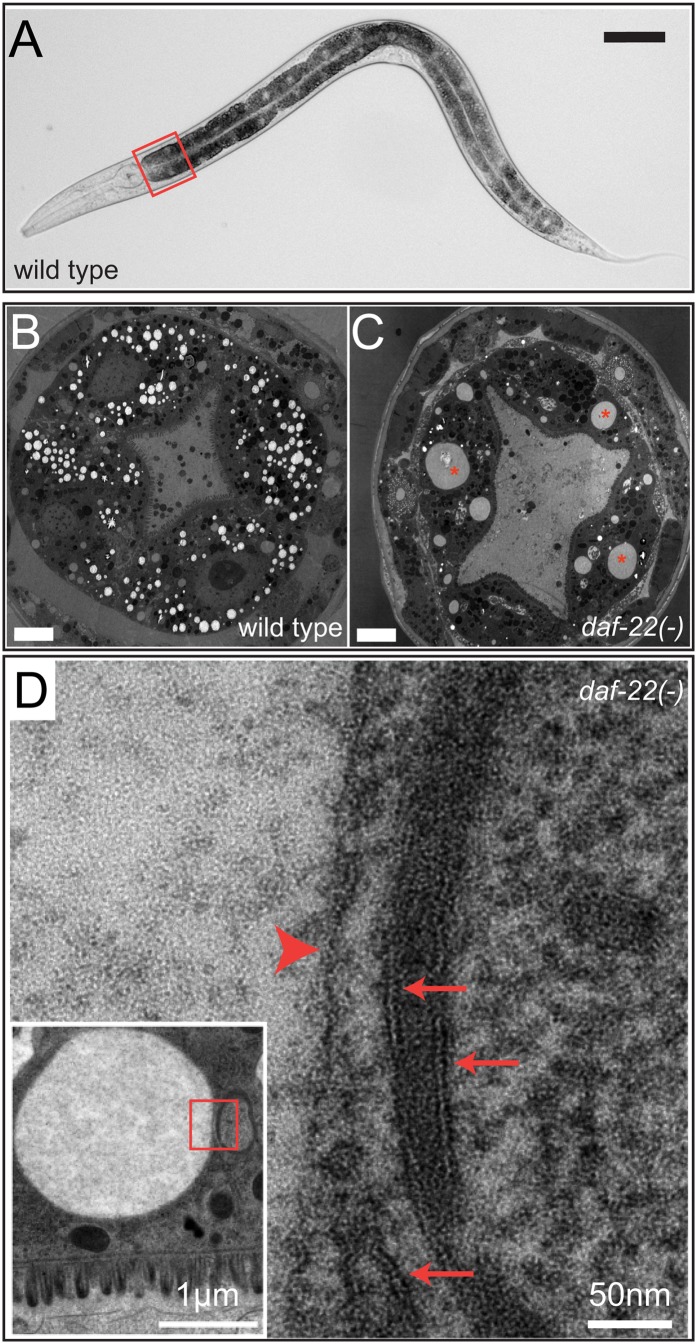
To positively identify lipid droplets using electron microscopy in C. elegans, Zhang et al. took advantage of a mutant that appeared to undergo selective expansion of fat storage compartments (34) (Fig. 1). The daf-22/thiolase gene encodes the terminal enzyme in a peroxisomal β-oxidation pathway. DAF-22/thiolase was originally identified as a key enzyme for processing the fatty acid moiety of the C. elegans dauer pheromone (43). Loss of DAF-22/thiolase function in C. elegans specifically increases the level of triglycerides that are accommodated by expanded fat storage compartments in the intestinal cells (34). As most other vesicular structures are <2 µm in diameter, electron-lucent, grossly expanded compartments (>10 µm in diameter) could be readily identified by size. Indeed, it was found that such compartments were delimited by a phospholipid monolayer, thus confirming the existence of lipid droplets in C. elegans (34). Additional support came from the targeting of the adipose triglyceride lipase (ATGL) ortholog ATGL-1 to the surface of expanded lipid droplets in daf-22 mutant animals (34). Since the initial report, lipid droplets have also been identified in wild-type animals by transmission electron microscopy (40). It was also determined that lipid droplets are distinct from LROs (40), a compartment that was previously proposed to store fat in C. elegans (44). Furthermore, expanded lipid droplets have been reported in mutant animals that lack ACS-3/acyl-CoA synthetase (45).
Fig. 1.
Lipid droplets in C. elegans intestinal cells. A: A wild-type larval stage L4 animal. The first intestinal segment is boxed in red. Scale bar, 100 µm. B, C: Electron micrographs showing cross-sections of the first intestinal segment in wild-type (B) and daf-22(−) (C) mutant animals. Red asterisks indicate enlarged lipid droplets. Scale bar, 5 µm. (D) Electron micrograph showing a phospholipid monolayer (red arrowhead) that surrounds an expanded lipid droplet (boxed area in inset) in contrast to phospholipid bilayers (red arrows) of nearby organelles. The lumen of the intestine is at the bottom of the inset. B–D: Reprint from Ref. 34.
VISUALIZATION OF FAT STORAGE IN C. ELEGANS
Attempts to visualize and quantitate fat storage long preceded the definitive identification of lipid droplets in C. elegans. Sudan Black has been widely adopted as a stain for intracellular fat since its first usage on bacteria (46). In C. elegans, animals that expressed a defective insulin/IGF receptor ortholog DAF-2 showed more intense Sudan Black staining than wild-type animals (29). Subsequent biochemical measurement confirmed that loss of insulin signaling elevated triglyceride levels in C. elegans, validating Sudan Black staining as a method for visualization of fat storage (47). Sudan Black staining was most prominent in intestinal cells, thus giving rise to the notion that the intestine is the major fat storage organ in C. elegans. In addition, Sudan Black staining was observed in the hypodermis and gonad, suggesting that fat is exported from the intestine to other tissues. Although Sudan Black staining was widely employed in other studies (27, 48–50), the fixation protocol was not readily adaptable for large-scale genetic and functional genomic screens. To overcome this, Nile Red and BODIPY-conjugated fatty acid were used as alternatives for fat staining in live animals (47). Nile Red was first introduced as a fluorescent vital stain for intracellular lipid droplets in mammalian cells (51), whereas BODIPY-conjugated fatty acid had been used to monitor fatty acid uptake in mammalian cells (52). It was assumed that worms would ingest Nile Red or BODIPY-conjugated fatty acid from E. coli that had been grown in the presence of these dyes on agar plates. This was followed by absorption of the dyes through the intestinal epithelium and their accumulation in fat storage compartments in intestinal cells. The precise mechanisms of absorption and turnover of these dyes are currently unknown.
Nile Red staining was initially used to conduct a whole-genome RNAi screen, which resulted in the discovery of hundreds of gene inactivations that altered the intensity and/or pattern of staining within intestinal cells (47). Many genes with conserved function in fat metabolism were uncovered, suggesting that Nile Red staining might serve as an indicator of fat storage in C. elegans. However, recent evidence suggests that vital staining with Nile Red and BODIPY-conjugated fatty acid also illuminates LROs, and that their accumulation in lipid droplets is highly dependent on genetic backgrounds (40, 44, 53–56). Therefore, future use of Nile Red as a vital dye for monitoring fat storage in C. elegans should always be validated with biochemical measurement of fat content. Further refinement may include analysis of the emission spectrum of Nile Red, which differs in lipid droplets versus lysosome related organelles (40, 45). This is because Nile Red in a neutral lipid environment (i.e., lipid droplets) will emit yellow-gold fluorescence instead of red fluorescence (51). Note that Nile Red, BODIPY, or Oil-Red-O staining on fixed C. elegans samples appears to faithfully highlight lipid droplets and reflect biochemical measurements of fat content (40, 54–57).
The continual demand for identification of fat storage compartments in live samples has led to the development and implementation of multiple label-free lipid imaging techniques (58–60). The vibrations of C-H bond in lipids allow sensitive, label-free detection of lipid droplets using coherent anti-Stokes Raman scattering (CARS) microscopy in mammalian cells (59). This method was subsequently used to visualize fat storage compartments in live C. elegans (54, 61, 62). By exploiting vibrational characteristics of C = C double bonds, CARS microscopy also allowed the detection of changes in relative abundance of unsaturated fatty acids in mutant C. elegans that lacked specific fatty acid desaturases (62, 63). More recently, stimulated Raman scattering (SRS) microscopy has been introduced (58). Compared with CARS microscopy, SRS microscopy was reported to be more sensitive and less prone to background noise from structures unrelated to lipids. It was demonstrated in C. elegans that SRS microscopy could be used to visualize intracellular fat storage compartments and measure organismal fat content quantitatively (64). Using SRS microscopy, 272 gene inactivations by RNAi have been surveyed, which yielded 9 potentially new regulators of fat content in C. elegans (64). Although vesicular structures detected by CARS or SRS microscopy in C. elegans are likely to be lipid droplets as in mammalian cells, simultaneous imaging of fluorescent protein markers would be a logical step to determine if these label-free imaging techniques detect all lipid droplets.
LIPID DROPLET-ASSOCIATED PROTEINS IN C. ELEGANS
A large number of lipid droplet-associated proteins have been identified from proteomics studies using mammalian and Drosophila cell and tissue extracts (65–68). In contrast, similar proteomics studies have not been carried out in C. elegans. This is in part because existing protocols for large-scale protein extraction and purification using frozen worm samples is incompatible with lipid droplet preservation (Dawn Brasaemle; personal communication). Intriguingly, the Perilipin and CIDE families of proteins, which have been widely used as reference markers for lipid droplets in other organisms, are absent in C. elegans. Therefore, few C. elegans proteins have been reported to be genuinely associated with lipid droplets. A strategy forward will involve selecting conserved proteins from mammalian or Drosophila lipid droplet proteomes and interrogate their localization in C. elegans. It may be prudent to concentrate on proteins that have a clear role in fat metabolism or regulation of lipid droplet size or number. One such candidate is ATGL-1. The C. elegans ATGL-1 lipase is crucial for the balance of fat utilization and preservation in the developmentally arrested dauer larval stage (69). Long term (>1 month) survival of dauers is dependent on tight regulation of ATGL-1 activity by the AMP kinase (69). In Drosophila, the ATGL ortholog Brummer, when fused to green fluorescent protein (GFP), has been shown to localize to the lipid droplet surface (70). On the basis of these results, it was satisfying to observe that the C. elegans ATGL-1::GFP fusion protein was also localized to the lipid droplet surface, both in wild-type and mutant animals with enlarged lipid droplets (40, 45). By coupling confocal microscopy with spectral analysis and linear unmixing procedures, it was further demonstrated that ATGL-1::GFP-marked lipid droplets are distinct from the autofluorescent lysosome related organelles (Fig. 2) (34, 40). Fat mobilization from lipid droplets is a highly regulated process that involves not only adipose triglyceride lipase but also its activator CGI58 and hormone-sensitive lipase (4). Therefore, the C. elegans orthologs of CGI58 and hormone-sensitive lipase are prime candidate markers for lipid droplets in future studies.
Fig. 2.
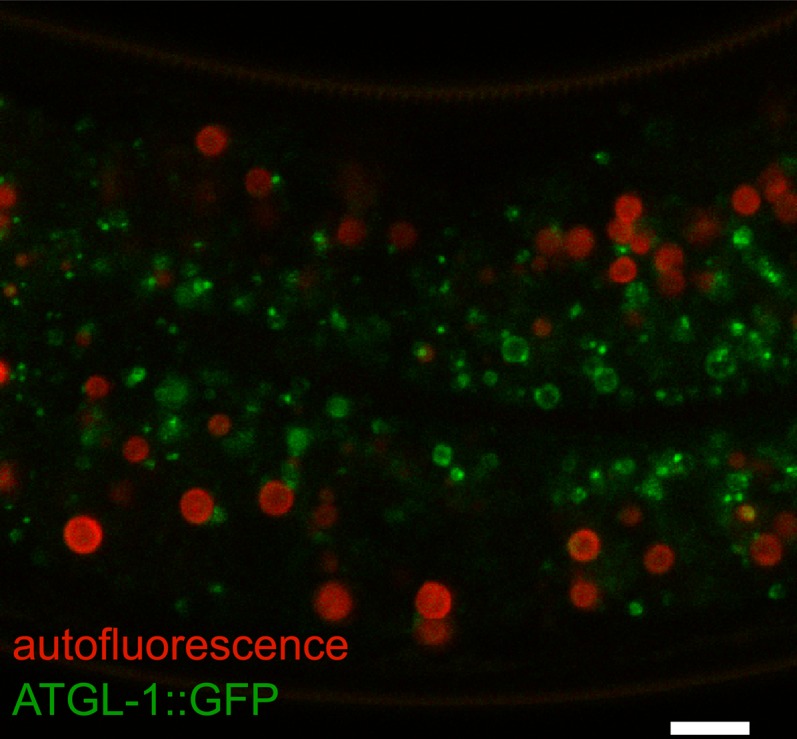
ATGL-1::GFP as a marker for lipid droplets in C. elegans intestinal cells. Confocal microscopy and linear unmixing distinguishes lipid droplets that are marked by ATGL-1::GFP fusion protein (in green) and autofluorescent LROs (in red). Scale bar, 10 µm.
The absence of Perilipin and CIDE families of proteins warrants further discussion. The Perilipin family members are found in mammals and invertebrates, including Drosophila and Dictyostelium (71). However, no Perilipin orthologs have been identified through sequence homology in C. elegans or other nematode species. As Dictyostelium is more primitive than nematodes, it is plausible that the Perilipin family was lost during evolution. Perilipin is known to play an important role in basal and hormone-stimulated lipolysis through its association with the lipid droplet surface and its stimulatory activity toward ATGL and hormone-sensitive lipase (HSL) (72–77). The absence of Perilipin in C. elegans gives rise to an intriguing possibility that additional Perilipin-independent regulatory mechanisms may exist for ATGL- and HSL-mediated lipolysis. Alternatively, proteins that share structural and functional similarity with Perilipin may have greatly diverged in the primary sequence that precludes their discovery through homology searches.
In contrast to the Perilipins, the CIDE family proteins are found only in vertebrates (78). All three family members, CIDEA, CIDEB, and CIDEC, have been shown to associate with lipid droplets, and they play important roles in regulating whole-body fat storage and energy balance in mice and human (79–86). Ectopic overexpression of CIDE proteins modulates lipid droplet size (82, 83). One may speculate that the CIDE proteins have evolved in vertebrates to add an additional level of regulation for fat storage in lipid droplets. It is also plausible that divergent proteins substitute for CIDE protein functions in C. elegans and other invertebrates.
PERSPECTIVES
With the definitive identification of lipid droplets as fat storage organelles in C. elegans, now is the time to harness the forward and reverse genetic, biochemical, and imaging tools available to study the cell biology of fat storage and mobilization in this well-established model organism. Because the diet of C. elegans can be easily manipulated by feeding with different E. coli strains, analysis of lipid species and other metabolites will shed light on how dietary factors affect fat storage in lipid droplets (34, 56, 87, 88). Cell biological studies can also be coupled with powerful analytical methods that allow simultaneous monitoring of fatty acid absorption, elongation, and de novo synthesis in C. elegans, which have yet to be employed in other metazoans (89). Although the intestinal cells are the major site of fat storage in C. elegans, with the advent of label-free imaging techniques and fluorescent protein markers for lipid droplets, it will be of great interest to determine whether the regulators of fat storage in intestinal cells function in a similar fashion in tissues such as the hypodermis and gonad. It is plausible that additional mechanisms may operate in these tissues to support rapid turnover of fat for energy and reproduction. For example, small lipid droplets provide a larger surface area-to-volume ratio that favors lipolysis. The size of lipid droplets in the hypodermis and gonad of C. elegans may, therefore, be tightly regulated.
The unique advantage of live animal imaging at single-cell resolution throughout the life cycle should see the use of C. elegans in addressing additional fundamental questions on lipid droplets. How is the interaction between lipid droplets and other subcellular organelles, such as the endoplasmic reticulum and mitochondria, maintained? What is the functional consequence of such interactions? Are the distribution and movement of lipid droplets regulated in different developmental stages and nutrient levels? Careful implementation of genetic, biochemical, and microscopy techniques should yield insights that can be readily extended from C. elegans to mammals.
Acknowledgments
The author thanks Ronald Cole for providing the image for Figure 2 and Karen Reue and Meng Wang for comments and suggestions on the manuscript.
Footnotes
Abbreviations:
- ATGL
- adipose triglyceride lipase
- GFP
- green fluorescent protein
- LRO
- lysosome-related organelle
This work was supported by the Stowers Institute for Medical Research and by March of Dimes Foundation Research Grant 5-FY07-662.
REFERENCES
- 1.Farese R. V., Jr, Walther T. C. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 139: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman J. M. 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50: 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin S., Parton R. G. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7: 373–378. [DOI] [PubMed] [Google Scholar]
- 4.Zechner R., Kienesberger P. C., Haemmerle G., Zimmermann R., Lass A. 2009. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 50: 3–21. [DOI] [PubMed] [Google Scholar]
- 5.Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36: 1211–1226. [PubMed] [Google Scholar]
- 6.Robenek H., Buers I., Hofnagel O., Robenek M. J., Troyer D., Severs N. J. 2009. Compartmentalization of proteins in lipid droplet biogenesis. Biochim. Biophys. Acta. 1791: 408–418. [DOI] [PubMed] [Google Scholar]
- 7.Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277: 44507–44512. [DOI] [PubMed] [Google Scholar]
- 8.Kuerschner L., Moessinger C., Thiele C. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 9: 338–352. [DOI] [PubMed] [Google Scholar]
- 9.Hausman G. J., Richardson R. L. 1983. Cellular and vascular development in immature rat adipose tissue. J. Lipid Res. 24: 522–532. [PubMed] [Google Scholar]
- 10.Slavin B. G. 1979. Fine structural studies on white adipocyte differentiation. Anat. Rec. 195: 63–72. [DOI] [PubMed] [Google Scholar]
- 11.Watts J. L. 2009. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol. Metab. 20: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullaney B. C., Ashrafi K. 2009. C. elegans fat storage and metabolic regulation. Biochim. Biophys. Acta. 1791: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant B., Hirsh D. 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 10: 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., Rose K. L., Hobert O., Greenstein D. 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. [DOI] [PubMed] [Google Scholar]
- 15.Kimble J., Sharrock W. J. 1983. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev. Biol. 96: 189–196. [DOI] [PubMed] [Google Scholar]
- 16.Schneider W. J. 1996. Vitellogenin receptors: oocyte-specific members of the low-density lipoprotein receptor supergene family. Int. Rev. Cytol. 166: 103–137. [DOI] [PubMed] [Google Scholar]
- 17.Mak H. Y., Nelson L. S., Basson M., Johnson C. D., Ruvkun G. 2006. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 38: 363–368. [DOI] [PubMed] [Google Scholar]
- 18.Kniazeva M., Crawford Q. T., Seiber M., Wang C. Y., Han M. 2004. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2: E257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kniazeva M., Sieber M., McCauley S., Zhang K., Watts J. L., Han M. 2003. Suppression of the ELO-2 FA elongation activity results in alterations of the fatty acid composition and multiple physiological defects, including abnormal ultradian rhythms, in Caenorhabditis elegans. Genetics. 163: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker A. K., Yang F., Jiang K., Ji J. Y., Watts J. L., Purushotham A., Boss O., Hirsch M. L., Ribich S., Smith J. J., et al. 2010. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 24: 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watts J. L., Browse J. 2002. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 99: 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F., Vought B. W., Satterlee J. S., Walker A. K., Sun Z.-Y. J., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., et al. 2006. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 442: 700–704. [DOI] [PubMed] [Google Scholar]
- 23.Van Gilst M. R., Hadjivassiliou H., Jolly A., Yamamoto K. R. 2005. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greer E. R., Perez C. L., Van Gilst M. R., Lee B. H., Ashrafi K. 2008. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 8: 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G. 2009. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23: 496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan S., Sadegh L., Elle I. C., Christensen A. G., Faergeman N. J., Ashrafi K. 2008. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 7: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sze J. Y., Victor M., Loer C., Shi Y., Ruvkun G. 2000. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 403: 560–564. [DOI] [PubMed] [Google Scholar]
- 28.Jones K. T., Greer E. R., Pearce D., Ashrafi K. 2009. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 277: 942–946. [DOI] [PubMed] [Google Scholar]
- 30.Frøkjaer-Jensen C., Davis M. W., Hollopeter G., Taylor J., Harris T. W., Nix P., Lofgren R., Prestgard-Duke M., Bastiani M., Moerman D. G., et al. 2010. Targeted gene deletions in C. elegans using transposon excision. Nat. Methods. 7: 451–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robert V., Bessereau J. L. 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26: 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S. P., Grunnet M., Jorgensen E. M. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S. O., Box A. C., Xu N., Le Men J., Yu J., Guo F., Trimble R., Mak H. Y. 2010. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 107: 4640–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brock T. J., Browse J., Watts J. L. 2006. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M. C., O'Rourke E. J., Ruvkun G. 2008. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 322: 957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung B., Hermann G. J., Priess J. R. 1999. Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216: 114–134. [DOI] [PubMed] [Google Scholar]
- 38.Altun Z. F., Hall D. H. 2009. Alimentary system: intestine. In WormAtlas. doi:10.3908/wormatlas.1.4. Accessed at http://www.wormatlas.org/hermaphrodite/intestine/Intframeset.html.
- 39.Albert P. S., Riddle D. L. 1988. Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 126: 270–293. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S. O., Trimble R., Guo F., Mak H. Y. 2010. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 11: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clokey G. V., Jacobson L. A. 1986. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 35: 79–94. [DOI] [PubMed] [Google Scholar]
- 42.Hermann G. J., Schroeder L. K., Hieb C. A., Kershner A. M., Rabbitts B. M., Fonarev P., Grant B. D., Priess J. R. 2005. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 16: 3273–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butcher R. A., Ragains J. R., Li W., Ruvkun G., Clardy J., Mak H. Y. 2009. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc. Natl. Acad. Sci. USA. 106: 1875–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder L. K., Kremer S., Kramer M. J., Currie E., Kwan E., Watts J. L., Lawrenson A. L., Hermann G. J. 2007. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol. Biol. Cell. 18: 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullaney B. C., Blind R. D., Lemieux G. A., Perez C. L., Elle I. C., Faergeman N. J., Van Gilst M. R., Ingraham H. A., Ashrafi K. 2010. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 12: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burdon K. L., Stokes J. C., Kimbrough C. E. 1942. Studies of the common aerobic spore-forming bacilli: I. Staining for fat with Sudan Black B-safranin. J. Bacteriol. 43: 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 421: 268–272. [DOI] [PubMed] [Google Scholar]
- 48.Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., Ruvkun G. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 389: 994–999. [DOI] [PubMed] [Google Scholar]
- 49.Ogg S., Ruvkun G. 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell. 2: 887–893. [DOI] [PubMed] [Google Scholar]
- 50.McKay R. M., McKay J. P., Avery L., Graff J. M. 2003. C elegans: a model for exploring the genetics of fat storage. Dev. Cell. 4: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greenspan P., Mayer E. P., Fowler S. D. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaffer J. E., Lodish H. F. 1994. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell. 79: 427–436. [DOI] [PubMed] [Google Scholar]
- 53.Märck C., Olsen L., Kurth C., Persson A., Storm N. J., Svensson E., Jansson J. O., Hellqvist M., Enejder A., Faergeman N. J., et al. 2009. Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 106: 18285–18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yen K., Le T. T., Bansal A., Narasimhan S. D., Cheng J. X., Tissenbaum H. A. 2010. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS ONE. 5 pii:e12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Rourke E. J., Soukas A. A., Carr C. E., Ruvkun G. 2009. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks K. K., Liang B., Watts J. L. 2009. The influence of bacterial diet on fat storage in C. elegans. PLoS ONE. 4: e7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klapper M., Ehmke M., Palgunow D., Böhme M., Matthäus C., Bergner G., Dietzek B., Popp J., Döring F. 2011. Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J. Lipid Res. 52: 1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Freudiger C. W., Min W., Saar B. G., Lu S., Holtom G. R., He C., Tsai J. C., Kang J. X., Xie X. S. 2008. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 322: 1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nan X., Cheng J. X., Xie X. S. 2003. Vibrational imaging of lipid droplets in live fibroblast cells with coherent anti-Stokes Raman scattering microscopy. J. Lipid Res. 44: 2202–2208. [DOI] [PubMed] [Google Scholar]
- 60.Evans C. L., Xie X. S. 2008. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu. Rev. Anal. Chem. (Palo Alto Calif.). 1: 883–909. [DOI] [PubMed] [Google Scholar]
- 61.Hellerer T., Axäng C., Brackmann C., Hillertz P., Pilon M., Enejder A. 2007. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. USA. 104: 14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le T. T., Duren H. M., Slipchenko M. N., Hu C. D., Cheng J. X. 2010. Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J. Lipid Res. 51: 672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rinia H. A., Burger K. N., Bonn M., Muller M. 2008. Quantitative label-free imaging of lipid composition and packing of individual cellular lipid droplets using multiplex CARS microscopy. Biophys. J. 95: 4908–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M. C., Min W., Freudiger C. W., Ruvkun G., Xie X. S. 2011. RNAi screening for fat regulatory genes with SRS microscopy. Nat. Methods. 8: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beller M., Riedel D., Jänsch L., Dieterich G., Wehland J., Jäckle H., Kühnlein R. P. 2006. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics. 5:1082–1094. [DOI] [PubMed] [Google Scholar]
- 66.Brasaemle D. L., Dolios G., Shapiro L., Wang R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 279: 46835–46842. [DOI] [PubMed] [Google Scholar]
- 67.Cermelli S, Guo Y., Gross S. P., Welte M. A. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16: 1783–1795. [DOI] [PubMed] [Google Scholar]
- 68.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279: 3787–3792. [DOI] [PubMed] [Google Scholar]
- 69.Narbonne P., Roy R. 2009. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 457: 210–214. [DOI] [PubMed] [Google Scholar]
- 70.GrÖnke S., Mildner A., Fellert S., Tennagels N., Petry S., Müller G., Jäckle H., Kühnlein R. P. 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1: 323–330. [DOI] [PubMed] [Google Scholar]
- 71.Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., Kimmel A. R. 2002. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277: 32253–32257. [DOI] [PubMed] [Google Scholar]
- 72.Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., Londos C. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266: 11341–11346. [PubMed] [Google Scholar]
- 73.Miyoshi H., Perfield J. W., 2nd, Souza S. C., Shen W. J., Zhang H. H., Stancheva Z. S., Kraemer F. B., Obin M. S., Greenberg A. S. 2007. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J. Biol. Chem. 282: 996–1002. [DOI] [PubMed] [Google Scholar]
- 74.Miyoshi H., Souza S. C., Zhang H. H., Strissel K. J., Christoffolete M. A., Kovsan J., Rudich A., Kraemer F. B., Bianco A. C., Obin M. S., et al. 2006. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J. Biol. Chem. 281: 15837–15844. [DOI] [PubMed] [Google Scholar]
- 75.Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., Londos C. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., et al. 2004. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3–L1 adipocytes. J. Biol. Chem. 279: 42062–42071. [DOI] [PubMed] [Google Scholar]
- 77.Yamaguchi T., Omatsu N., Matsushita S., Osumi T. 2004. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J. Biol. Chem. 279: 30490–30497. [DOI] [PubMed] [Google Scholar]
- 78.Wu C., Zhang Y., Sun Z., Li P. 2008. Molecular evolution of Cide family proteins: novel domain formation in early vertebrates and the subsequent divergence. BMC Evol. Biol. 8: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubio-Cabezas O, Puri V., Murano I., Saudek V., Semple R. K., Dash S., Hyden C. S., Bottomley W., Vigouroux C., Magré J., et al. (2009) Partial lipodystrophy and insulin resistant diabetes in a patient with a homozygous nonsense mutation in CIDEC. EMBO Mol Med. 1: 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keller P., Petrie J. T., De Rose P., Gerin I., Wright W. S., Chiang S. H., Nielsen A. R., Fischer C. P., Pedersen B. K., MacDougald O. A. 2008. Fat-specific protein 27 regulates storage of triacylglycerol. J. Biol. Chem. 283: 14355–14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J. Z., Ye J., Xue B., Qi J., Zhang J., Zhou Z., Li Q., Wen Z., Li P. 2007. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 56: 2523–2532. [DOI] [PubMed] [Google Scholar]
- 82.Puri V., Konda S., Ranjit S., Aouadi M., Chawla A., Chouinard M., Chakladar A., Czech M. P. 2007. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J. Biol. Chem. 282: 34213–34218. [DOI] [PubMed] [Google Scholar]
- 83.Puri V., Ranjit S., Konda S., Nicoloro S. M., Straubhaar J., Chawla A., Chouinard M., Lin C., Burkart A., Corvera S., et al. 2008. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc. Natl. Acad. Sci. USA. 105: 7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou Z., Yon Toh S., Chen Z., Guo K., Ng C. P., Ponniah S., Lin S. C., Hong W., Li P. 2003. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 35: 49–56. [DOI] [PubMed] [Google Scholar]
- 85.Nishino N., Tamori Y., Tateya S., Kawaguchi T., Shibakusa T., Mizunoya W., Inoue K., Kitazawa R., Kitazawa S., Matsuki Y., et al. 2008. FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J. Clin. Invest. 118: 2808–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Toh S. Y., Gong J., Du G., Li J. Z., Yang S., Ye J., Yao H., Zhang Y., Xue B., Li Q., et al. 2008. Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS ONE. 3: e2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geier F. M., Want E. J., Leroi A. M., Bundy J. G. 2011. Cross-platform comparison of Caenorhabditis elegans tissue extraction strategies for comprehensive metabolome coverage. Anal. Chem. 83: 3730–3736. [DOI] [PubMed] [Google Scholar]
- 88.Pungaliya C., Srinivasan J., Fox B. W., Malik R. U., Ludewig A. H., Sternberg P. W., Schroeder F. C. 2009. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 106: 7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perez C. L., Van Gilst M. R. 2008. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 8: 266–274. [DOI] [PubMed] [Google Scholar]