Abstract
The human bile acid pool composition is composed of both primary bile acids (cholic acid and chenodeoxycholic acid) and secondary bile acids (deoxycholic acid and lithocholic acid). Secondary bile acids are formed by the 7α-dehydroxylation of primary bile acids carried out by intestinal anaerobic bacteria. We have previously described a multistep biochemical pathway in Clostridium scindens that is responsible for bile acid 7α-dehydroxylation. We have identified a large (12 kb) bile acid inducible (bai) operon in this bacterium that encodes eight genes involved in bile acid 7α-dehydroxylation. However, the function of the baiF gene product in this operon has not been elucidated. In the current study, we cloned and expressed the baiF gene in E. coli and discovered it has bile acid CoA transferase activity. In addition, we discovered a second bai operon encoding three genes. The baiK gene in this operon was expressed in E. coli and found to encode a second bile acid CoA transferase. Both bile acid CoA transferases were determined to be members of the type III family by amino acid sequence comparisons. Both bile acid CoA transferases had broad substrate specificity, except the baiK gene product, which failed to use lithocholyl-CoA as a CoA donor. Primary bile acids are ligated to CoA via an ATP-dependent mechanism during the initial steps of 7α-dehydroxylation. The bile acid CoA transferases conserve the thioester bond energy, saving the cell ATP molecules during bile acid 7α-dehydroxylation. ATP-dependent CoA ligation is likely quickly supplanted by ATP-independent CoA transfer.
Keywords: secondary bile acids, deoxycholic acid, HPLC, protein expression
The human liver synthesizes two primary bile acids from cholesterol, chenodeoxycholic acid (CDCA; 3α, 7α-dihydro-5β-cholan-24-oic acid) and cholic acid (CA; 3α, 7α, 12α-trihydroxy-5β-cholan-24-oic acid). Bile acids are conjugated to either taurine or glycine before active secretion into bile and in this form are termed bile salts. Bile salts function to solubilize lipids and lipid soluble vitamins from the duodenum through the jejunum of the small intestine. Upon reaching the terminal ileum, bile salts are actively transported across the intestinal epithelium into the portal blood and returned to the liver. This process is termed enterohepatic circulation and is ∼95% efficient. However, roughly 400-600 mg of bile acids enter the large intestine every day, where they are metabolized by a diversity of microorganisms (1). Bile salts are rapidly deconjugated and converted to the secondary bile acids, deoxycholic acid (3α,12α-trihydroxy-5β-cholanoic acid) and lithocholic acid (3α-hydroxy-5β-cholan-24-oic acid) from CA and CDCA, respectively.
Decades of research strongly suggest that secondary bile acids are involved in human disease processes, including cancers of the colon, esophagus, and biliary track (2), as well as cholesterol gallstone disease in some patients with high levels of deoxycholic acid in bile (3). The level of deoxycholic acid in bile is believed to be controlled by two major factors: i) levels and activities of bile acid 7α-dehydroxylating gut bacteria (3) and ii) colonic transit time (4). Colonic pH may also be a minor factor. Therefore, an understanding of the genetics and enzymology of bile acid 7α-dehydroxylation is an important first step in finding ways to decrease secondary bile acids with potentially beneficial outcomes to the host.
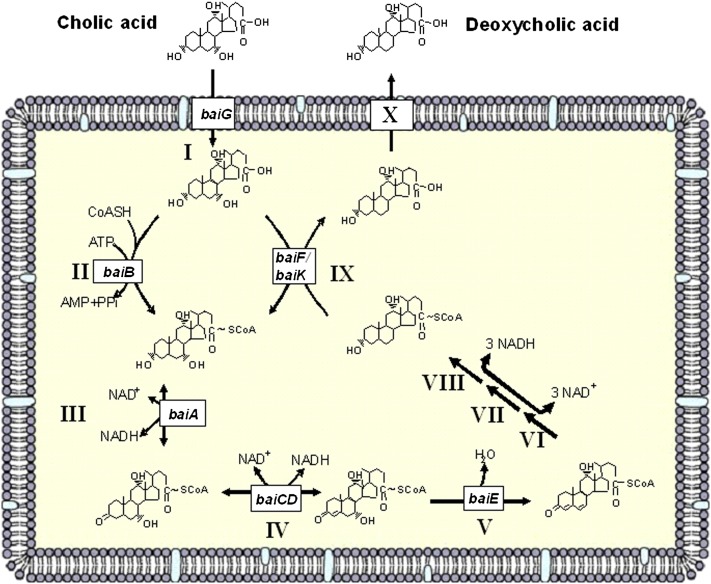
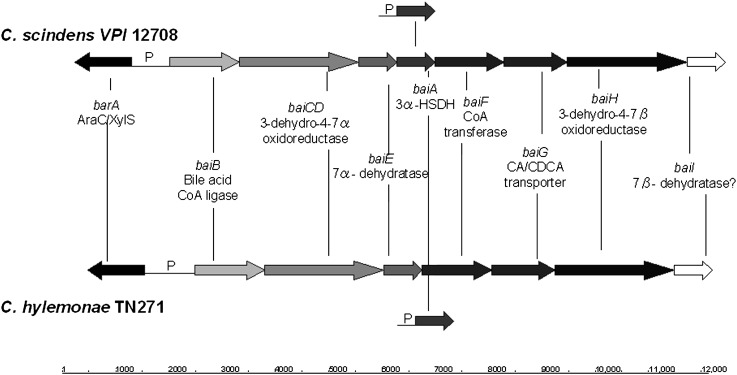
Several bacterial species in the genus Clostridium have been isolated and shown to convert primary bile acids into secondary bile acids, a process termed bile acid 7α-dehydroxylation (1). We previously proposed a multistep biochemical pathway for bile acid 7α-dehydroxylation (Fig. 1). In addition, we characterized the bile acid-inducible (bai) operons from C. scindens VPI 12708 (5), C. hiranonis DSM13275 (6), and C. hylemonae DSM 15053 (7) that encode enzymes involved in the initial steps of the bile acid 7α-dehydroxylation pathway (Fig. 2). The current model of bile acid 7α-dehydroxylation suggests that free primary bile acids are actively transported into the bacterial cell by a proton-dependent bile acid transporter encoded by the baiG gene (8). Once inside, the primary bile acid is ligated to CoA in an ATP-dependent manner by the baiB gene product (9). The baiA gene encodes a bile acid, 3α-hydroxysteroid dehydrogenase, specific for primary bile acid CoA conjugates (10). We recently reported that the baiCD and baiH gene products encode stereospecific 3-dehydro-4-bile acid oxidoreductases recognizing 7α-hydroxy bile acids (CA, CDCA) and 7β-hydroxy bile acids [ursodeoxycholic acid (UDCA),; 3α, 7α-dihydro-5β-cholan-24-oic acid], respectively (11). The rate-limiting and irreversible step in this pathway is 7α-dehydration. This reaction is catalyzed by bile acid 7α-dehydratase, which is encoded by the baiE gene (12). The 3-dehydro-4,6-bile acid intermediate is then sequentially reduced and exported from the cell; however, genes in the “reductive arm” of the pathway have yet to be identified. Previously, we showed that the baiF gene product hydrolyzes bile acid CoA conjugates (13). However, others and we have hypothesized, based on amino acid sequence comparisons with known CoA transferases, that this gene product may be a bile acid CoA transferase (1, 14). Here, we present strong evidence that the baiF gene encodes a bile acid CoA transferase with broad bile acid substrate specificity. In addition, we report the discovery and characterization of a second bile acid CoA transferase encoded on what appears to be second multigene operon involved in bile acid metabolism in C. scindens.
Fig. 1.
Current model of the bile acid 7α-dehydroxylation pathway in Clostridium scindens VPI 12708.
Fig. 2.
Schematic representation of bile acid 7α-dehydroxylation operons from C. scindens VPI 12708 and C. hylemonae DSM 15053. “P” represents putative promoter regions.
MATERIALS AND METHODS
Chemicals and bacterial strains
Clostridium scindens ATCC 35704 was purchased from American Type Culture Collection. C. scindens VPI 12708 was stored as a 30% glycerol stock at −80°C prior to this study, and working stocks were cultivated in chopped meat broth. E. coli BL21 (DE3) RIL was purchased from Agilent Stratagene (Santa Clara, CA). E. coli was cultivated in Luria-Bertani medium containing appropriate antibiotics. C. scindens strains were cultivated in brain/heart infusion broth under nitrogen. Bile acids were purchased from Sigma-Aldrich (St. Louis, MO) and Steraloids (Newport, RI). ADCA was a generous gift from Dr. James P. Coleman, East Carolina University. CoA was purchased from Sigma-Aldrich.
Isolation of genomic DNA and genome-walking by PCR
Genomic DNA was isolated from C. hylemonae, C. scindens VPI 12708, and C. scindens ATCC 35704 by enzymatic treatment, followed by bead beating as described previously (7). Genome-walking DNA libraries were prepared using the Universal GenomeWalker Kit (Clontech) according to the manufacturer. The DNA extraction method used in this application and PCR conditions have been described previously (7).
RNA isolation and transcriptional analysis
Total RNA was isolated as described previously (7). cDNA for reverse transcriptase PCR was generated with the Advantage RT-for-PCR kit (Clontech). cDNA for determination of transcription initiation site was generated using the SMART-RACE cDNA synthesis kit (Clontech) according to the manufacturer, with the exception that a gene-specific primer replaced 5′ CDS primer. The gene-specific primer for determination of the baiJ gene transcription initiation site was SR12708J 5′-TTC TAT GTC CTC ATC CGT TGC CCC TG-3′. The gene-specific primer for determination of the baiJ gene transcription initiation site was DSM 15053JRace1 5′-TGC GGT AGA GCC TCC TGT GAT ACC TTG-3′. PCR was performed using the Advantage-GC 2 PCR Kit (Clontech) according to the manufacturer. An amount of 0.5 μl GC-Melt was added to each 50 μl reaction. PCR products were excised from agarose gels, GENECLEAN purified, cloned into a pCR 8 GW TOPO TA vector, and sequenced at the Virginia Commonwealth University Nucleic Acid Research Facility.
Cloning the baiF and baiK genes
The baiF gene insert was amplified with forward primer 5′-CTGCAGAGAAAAGGAGGTATAAAAACATGGCTGG-3′ (PstI site in italics) and reverse primer 5′GGATCCTTATTTTTCGAACTGCGGGTGGCTCCACTCCTCTTTCTTTCTCATATGT-3′ (BamHI site in italics). The baiK gene insert was amplified with forward primer 5′-CTG CAG GGA AAG GAG GTT ATA GGA ATG AAA GG-3′ (PstI site in italics) and reverse 5′-GGA TCC TTA TTT TTC GAA CTG CGG GTG GCT CCA TTC TTC CTC CTT TTT CCA ACG TTC-3′ (BamHI site in italics). The streptavidin tag sequence (bold) was incorporated into the C terminus of bile acid CoA transferases for one-step purification (15). PCR products were cloned into a pCR8GW TOPO TA vector (Invitrogen) and transformed by heat shock at 42°C into One Shot Chemically competent E. coli that accompanies the vector. Plasmids were isolated using the Qiaprep Miniprep Kit (Qiagen) and double-digested with PstI and BamHI (New England Biolabs). Expression vector pSport-1 was double-digested with the same restriction enzymes, and both inserts and pSport-1 linear plasmid were gel-purified using the GENECLEAN Spin Kit (MP Biomedicals). Ligation was performed using T4 DNA ligase according to the manufacturer (New England Biolabs). Ligated vector was transformed into E. coli DH5 (NEB 5α; New England Biolabs) and cultivated in LB medium containing 100 μg/ml ampicillin. pSport vectors containing insert (hereafter referred to as pSportbaiFCTSBP and pSportbaiKCTSBP) as determined by double-digestion and agarose gel electrophoresis were sequenced at the Nucleic Acid Core Facility at Virginia Commonwealth University. Expression vectors were transformed into E. coli BL21 (DE3)RIL by heat shock.
Expression and purification of bile acid CoA transferases
Colonies were selected and cultivated overnight in 1 ml LB medium containing 100 μg/ml ampicillin (LB Amp). A 5% inoculum was transferred to LB Amp at 37°C and cultivated until reaching an OD600nm 0.4, at which point gene expression was induced with 1 mM IPTG. Overexpression occurred for 3 h, at which point cells were centrifuged and washed with Buffer A (two volumes 20 mM sodium phosphate; 0.1 M sodium chloride; 10 mM 2-mercaptoethanol; 15% glycerol, pH 7.0). Cells were frozen at −20°C until further processing. Cell lysates were produced by resuspending cells in Buffer A containing 700 μg lysozyme and incubating on ice for 1 h followed by three rounds of sonication (8 s burst, 20 s pause, 2 min total). Benzonase (25 U; Novagen) was added to degrade nucleic acids. Crude lysate was centrifuged 30 min at 16,000 g. Lysate was loaded onto Streptactin Superflow Resin (IBA) equilibrated with Buffer A and washed according to the manufacturer instructions. Elution was performed with Buffer A containing 2.5 μM desthiobiotin. Protein was concentrated on Amicon Ultracel-50K centrifugal filters (Millipore, Cork, Ireland) and stored in 50% glycerol at −20°C. Protein was stable for up to 1 month without detectable loss of activity.
SDS-PAGE and Western immunoblotting
Protein samples were separated by 12% SDS-PAGE and transferred to PVDF Western blotting membranes (Roche). Expression and identity of the recombinant streptavidin-tagged proteins was verified using Strep Tag II antibody HRP conjugate (Novagen).
Synthesis and purification of bile acid CoA conjugates
CoA conjugates of ursodeoxycholic acid (UDCA), chenodeoxycholic acid (CDCA), cholic acid (CA), allocholic acid (ACA), DCA, allodeoxycholic acid (ADCA), lithocholic acid (LCA), and β-muricholic acid (β-MCA) were synthesized according to the method of Killenberg and Dukes (16), with the exception that free bile acids were extracted with ethyl acetate rather than ether. ADCA-CoA synthesized from hydrolyzed rabbit gallstones was separated from contaminating DCA-CoA by reverse-phase HPLC on an HP 1100 series equipped with a Coulter Ultrasphere semipreparative column (5 µm silica gel) (10 mm × 25 cm). Samples (100 µl) were loaded on the column (40°C) and eluted with 5-40% isopropanol in 20 mM ammonium acetate buffer (pH 7.0) over a 40 min period with a flow rate of 2 ml/min. Free CoA and bile acid CoA conjugates were monitored at 260 nm. Isopropanol from fractions containing ADCA-CoA conjugates was evaporated under nitrogen and purified by Sep Pak C-18 chromatography. Columns were washed and equilibrated with 10 ml methanol followed by 10 ml deionized water and Buffer B (3 ml 20 mM sodium phosphate buffer; 0.1 M NaCl, pH 6.0). Bile acid CoA conjugates were dissolved in Buffer B and loaded onto the column, followed by washing with 10 ml water and elution with 2 ml absolute methanol. Samples were concentrated under nitrogen and resuspended in 50% methanol. Concentrated bile acid CoA conjugates were resuspended in 0.1 M sodium acetate buffer (pH 5.5) and stored at −20°C. Concentrations were determined by UV spectrophotometry at A260 nm using a published millimolar extinction coefficient of 15.03 (16).
Bile acid CoA transferase enzyme assays
An HPLC assay was used to measure CoA transferase activity. Assays contained 0.01 μg enzyme of recombinant BaiF-SBP or 0.03 μg recombinant BaiK-SBP and 300-500 μM secondary bile acid CoA conjugate with an equal amount of free bile acid acceptor. Reactions were performed at 37°C and terminated at 1 min by addition of 75 μl 1N HCl. Samples were prepared for HPLC by Sep Pak C-18 chromatography as described above. Concentrated reaction products were resuspended in 125 μl 50% methanol. Samples (100 μl) were autoinjected onto an Agilent 1200 series HPLC equipped with an Agilent Eclipse XDB-C18 column (5 μm, 4.6 × 250 mm). CoA conjugates were separated in a buffer system of 20 mM ammonium acetate buffer (pH 6.0) with a linear gradient 0-55% isopropanol over a 35 min period. CoA conjugates were detected at 260 nm and concentrations determined by comparison of peak areas versus A260 nm against concentration curves of known quantity.
Assays testing transfer of CoA from DCA-CoA or from ADCA-CoA to CDCA required measurement of the production of [24-14C]CDCA-CoA by liquid scintillation spectrometry instead of reverse-phase HPLC separation because nearly identical hydrophobicities result in comigration of substrate and product. Following termination of reaction by HCl, free [24-14C]CDCA was extracted from the reaction mixture by organic extraction twice with two volumes ethyl acetate. Radioactive counts in the aqueous phase thus measure [24-14C]CDCA-CoA, which is not soluble in ethyl acetate. Samples were separated by reverse-phase HPLC, and fractions corresponding to peaks absorbing at 260 nm were collected and counted by liquid scintillation spectrometry. Values were normalized relative to fractions collected from reactions incubated with [24-14C] CA. One unit of bile acid CoA transferase activity is defined as the amount of enzyme required to produce 1 μmole of primary bile acid CoA conjugate formed per minute.
Sequence analysis and nucleotide accession numbers
Nucleotide and amino acid sequence alignments were performed using ClustalW (17). Amino acid sequence alignments were represented using Boxshade. Protein Mr calculations were performed using the Compute pI/MW tool on the ExPASy Proteomics Server. Open reading frames (ORF) were detected using the ORF Finder Program at the NCBI website and searched against GenBank by BLAST. GenBank accession numbers for the baiJKL operon from C. hylemonae DSM 15053 is EU675331, and baiJKL operon from C. scindens VPI 12708 is EU675330.
RESULTS
Identification of a second multigene operon containing a bile acid CoA transferase
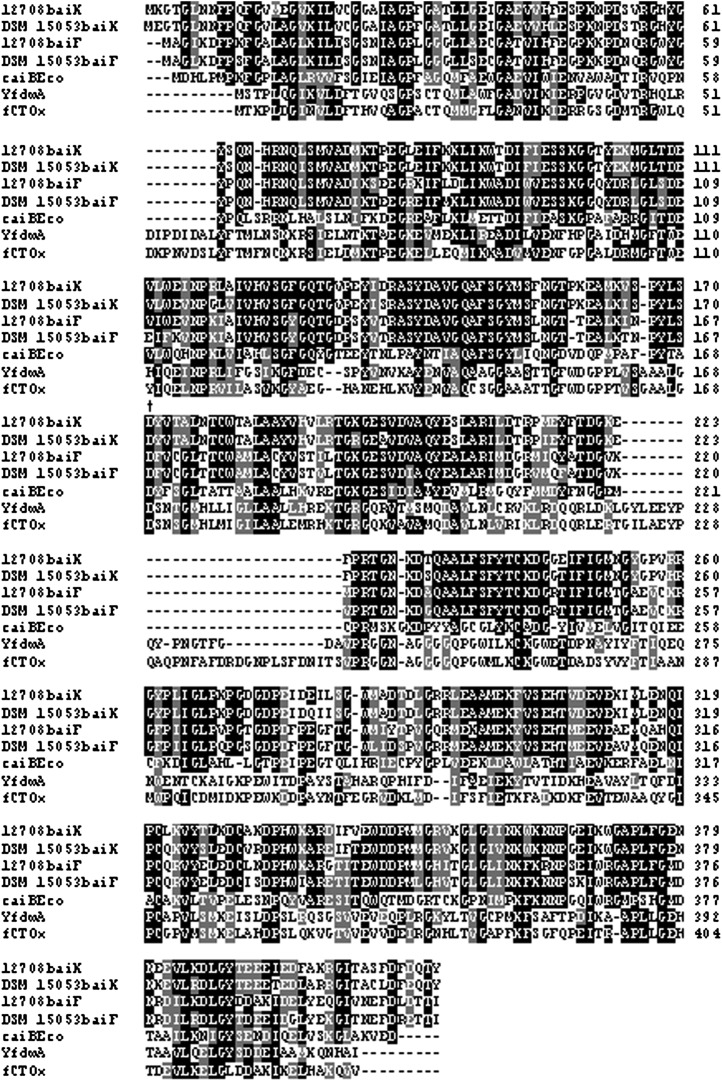
We previously reported multiple copies of the baiA gene, which encodes a primary bile acid, CoA-dependent 3α-HSDH, in the chromosome of C. scindens VPI 12708 (18). One copy of the baiA gene is located within in the large bai operon, and the other is a single cholic acid-inducible gene at a different chromosomal location (Fig. 2). More recently, we have been able to isolate and identify the baiA genes in C. hylemonae (7). Subsequent upstream DNA sequencing from the single baiA gene in this bacterium revealed a cluster of genes that appeared to be involved in bile acid metabolism by amino acid sequence analysis (Fig. 3). These same genes were also found in C. scindens VPI 12708 but not in C. scindens ATCC 35704, for which a total genome DNA sequence is available. An ORF (baiL) upstream of the baiA gene in C. hylemonae is predicted to encode a short chain pyridine nucleotide-dependent alcohol/polyol oxidoreductase. Members of this family include the baiA gene. However, these two gene products shared only 19% amino acid sequence identity and 35% similarity. A second ORF (baiK) was located upstream of the baiL gene, and amino acid sequence analysis suggests it may encode a type III CoA transferase. It has very high amino acid sequence identity with the baiF gene product from C. scindens VPI 12708 (63% identity; 77% similarity), as well as other type III CoA transferases reported in the literature (Fig. 4). Directly upstream from the hypothetical CoA transferase gene was a third ORF (baiJ) encoding a putative flavoprotein. BLAST analysis suggested this gene product encodes a flavoprotein similar to fumarate reductases and 3-ketosteroid-Δ1-dehydrogenases. The former can likely be ruled out due to the lack of four conserved N-terminal heme binding sites (CXXCH) found in fumarate reductases, such as that of Shewanella putrefaciens MR-1, whose structure has been solved (19). These three gene products share 89% amino acid sequence identity between C. scindens VPI 12708 and C. hylemonae DSM 15053.
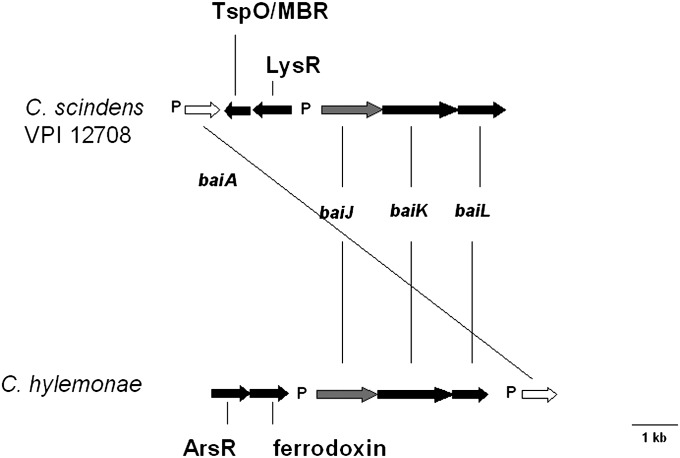
Fig. 3.
Schematic representation of baiJKL operon from Clostridium scindens VPI 12708 and Clostridium hylemonae DSM 15053. The baiA genes encode 27 kDa 3α-hydroxysteroid dehydrogenases. The baiJ genes encode a predicted 62 kDa flavoprotein similar to 3-ketosteroid-Δ1-dehydrogenases. The baiK genes encode a predicted 49 kDa type III CoA transferase homologous to the baiF gene. The baiL genes are predicted to encode a 27 kDa protein in the short chain reductase family. “P” represents conserved bai “promoter” region. TspO/MBR family acts in signal transduction as well as in transport of steroids/dicarboxylic tetrapyrrole intermediates.
Fig. 4.
Boxshade alignment of baiF and baiK gene products with members of type III CoA transferase family. fCTOx represents formyl CoA transferase from Oxalobacter formigenes (AAC45298.1), caiBEco represents carnitine CoA transferase from E. coli (BAB96607.1), and YfdwA represents the formyl CoA transferase from E. coli (YP_853505). † Active site Asp169 residue.
In addition, an ORF upstream of the baiJ gene on the antisense strand encodes a putative member of the LysR family of transcription factors. A second ORF also located on the antisense strand relative to the baiJ gene was found upstream of the putative transcription factor. This ORF was predicted to encode a polypeptide in the tryptophan-rich sensory protein (TspO)/mitochondrial benzodiazepine receptor (MBR) homolog family of signal transduction integral membrane proteins. Farther upstream, and on the same strand relative to the baiJ gene, we located the baiAl gene and promoter region that has been previously reported (18). This gene was located directly upstream of the TspO homolog. Note that the single baiA genes from both C. scindens VPI 12708 and C. hylemonae were found in proximity to this novel bai operon (Fig. 3).
Some notable features of this novel operon include the similar organization of the baiJKL genes, upstream conserved elements with the oxidative bai upstream region, and proximity of the transcription initiation site. Interestingly, the upstream ORFs are not conserved between C. scindens and C. hylemonae. Each region includes an ORF predicted to encode a transcription factor of a different family of proteins on opposite strands, which might indicate differences in transcriptional regulation. In addition, the ORF immediately upstream of the baiJ gene from C. hylemonae DSM 15053 is predicted to encode a 4Fe-4S ferrodoxin. This ORF was not located adjacent to the baiJKL genes from C. scindens. The baiJKL genes were searched against total genome sequencing data of C. scindens ATCC 35704 and were not detected. Moreover, these genes were not detected by PCR amplification of the baiK gene using cloning primers (see Materials and Methods). These results suggest that these genes are missing in this strain of bile acid 7α-dehydroxylating intestinal bacteria.
Transcriptional analysis of multigene operon in C. scindens VPI 12708
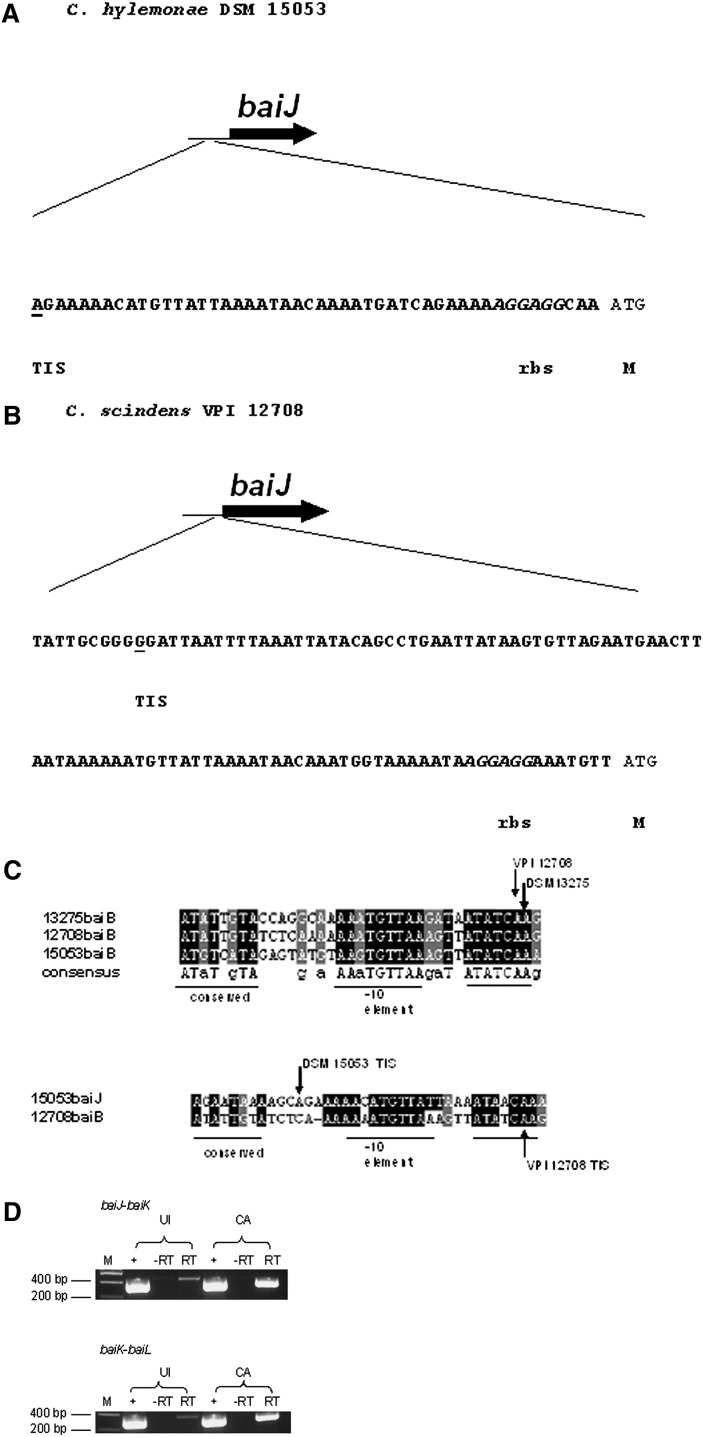
We located a sequence 24 bp upstream of the baiJ ORF in C. hylemonae with conserved elements with that of the regions upstream of the baiBCDAGHI and baiA operons from C. scindens, C. hylemonae, and C. hiranonis. 5′ SMART RACE PCR was performed on the baiJ gene, and the mRNA start site was determined at the 5′ end of the conserved bai region, suggesting the promoter is located upstream of this conserved region (Fig. 5). It is quite possible that this conserved region is a promoter region for both the baiA and baiB operons; however, this region does not appear to be a promoter for the baiJKL genes. The functional significance of this conserved region remains to be elucidated. This region was also located directly upstream of the baiJ gene in C. scindens VPI 12708 and shared a high degree of sequence identity with the putative bai promoter region found upstream of the baiJ gene of C. hylemonae. Finally, cDNA was prepared from CA versus uninduced control cells and primers were designed at the 3′ end of the baiJ gene and the 5′ end of the baiK gene and between baiK and baiL for determination of coexpression. Indeed, the baiJKL genes appear to be expressed as a polycistronic (single mRNA) operon (Fig. 5D).
Fig. 5.
Transcriptional analysis of baiJKL operons from C. hylemonae and C. scindens VPI 12708. A: Transcriptional initiation site for baiJ gene from C. hylemonae (underlined). B: Transcriptional initiation site for baiJ gene from C. scindens VPI 12708. C: Boxshade alignment of conserved putative regulatory elements upstream of the baiB between C. scindens VPI 12708, C. hylemonae DSM 15053, and C. hiranonis 13275 (above). The baiJ-conserved regulatory region from C. hylemonae DSM 15053 compared with that of the baiB upstream element (below) D: RT-PCR using primers spanning intergenic region of baiJ-baiK and baiK-baiL genes of C. scindens VPI 12708. “+” represents 10 ng C. scindens VPI 12708 genomic DNA, “−RT” represents negative control in which reverse transcriptase is not added, and “RT” represents cDNA prepared from uninduced (UI) and 50 μM cholic acid-induced (CA) cultures of C. scindens VPI 12708. M, methionine; rbs, ribosome binding site; TIS, transcription initiation site.
Cloning, expression, and purification of bile acid CoA transferases in E. coli
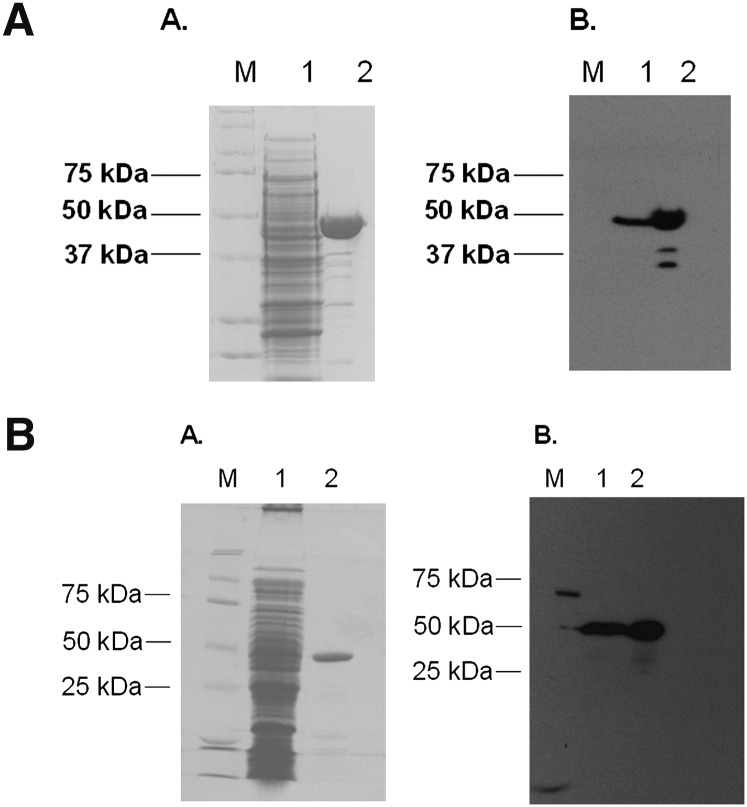
The baiF gene was PCR amplified from purified genomic DNA from Clostridium scindens ATCC 35704 and cloned into a pSport-1 expression vector. The baiF gene was overexpressed in E. coli BL21(DE3)RIL, producing a C-terminal streptavidin-tagged recombinant protein. One-step purification from 300 mg protein yielded 600 μg BaiF-Strep tag fusion protein, which matches 2% total protein expression estimated by SDS-PAGE gel (Fig. 6A). Western blot analysis using Strep Tag II antibody confirmed identity of the recombinant BaiF-Strep tag fusion protein.
Fig. 6.
Purification of the streptavidin-tagged recombinant bile acid-CoA transferases from E. coli. A. Purification of the recombinant C-terminal streptavidin tagged BaiF. B. Purification of the recombinant C-terminal streptavidin tagged BaiK. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of 30 μg crude (1) and 5g Streptactin-purified (2) fractions. Molecular weight standards are indicated to the left (M). (B) Detection of streptavidin-tagged recombinant transferases in crude (1) and purified (2) fractions using Strep-tag II antibody.
The baiK gene was also cloned into a pSport-1 expression vector and expressed as a C-terminal streptavidin-tagged recombinant protein. Expression and one-step purification from 300 mg crude extract resulted in recovery of 500 μg purified enzyme. The deduced molecular weight of the BaiK is 49.5 kDa, which was confirmed by SDS-PAGE and Western immunoblotting using Strep-Tag II antibody (Fig. 6B).
The baiF gene encodes a bile acid CoA transferase
The adenosine moiety in CoA absorbs at 260 nm, whereas bile acids do not. In addition, secondary bile acid-CoA conjugates are with one exception readily separable from primary bile acid-CoA conjugates by reverse-phase chromatography. We hypothesized that bile acid CoA transfer would proceed from secondary bile acid CoA conjugates to free primary bile acids (Fig. 1). We could thus detect the formation of primary bile acid CoA conjugates by reverse-phase HPLC by monitoring at 260 nm. Substrate saturation curves were prepared for the recombinant baiF and baiK gene products with DCA-CoA, LCA-CoA, and ADCA-CoA with CA as CoA acceptor (data not shown). Saturating substrate concentrations appeared to be inversely related to hydrophobicity. As a result, reactions were performed at concentrations of 500 μM DCA-CoA, 400 μM ADCA-CoA, and 300 μM LCA-CoA with equimolar concentrations of primary bile acid acceptors in each case. LCA-CoA did not appear to be a substrate for the recombinant baiK gene product (Table 1). The reaction was linear over a 2 min period and over protein concentrations between 0.005 μg and 0.05 μg at 37°C.
TABLE 1.
Substrate specificity of bile acid CoA transferase
| BaiFa |
BaiKa |
||||||||||
| Relative Activity |
Position and Substitutions |
Relative Activity |
|||||||||
| CoA Acceptor | CoA Donor | 3 | 5 | 6 | 7 | 12 | CoA Donor | ||||
| DCA-CoA | LCA-CoA | ADCA-CoA | DCA-CoA | LCA-CoA | ADCA-CoA | ||||||
| CA | 100 ± 15.4 | 91.6 ± 3.1 | 100 ± 9 | αOH, βH | βH | αH, βH | αOH, βH | αOH, βH | 69.6 ± 11.2 | ND | 44.5 ± 2.98 |
| ACA | 87 ± 4.3 | 100 ± 1.5 | 88.9 ± 0.9 | αOH, βH | αH | αH, βH | αOH, βH | αOH, βH | 28.1 ± 3 | ND | 100 ± 3 |
| UDCA | 51.35 ± 4.35 | 35 ± 1.2 | 44.38 ± 8.0 | αOH, βH | βH | αH, βH | αH, βOH | αH, βH | 100 ± 12 | ND | 73.61 ± 11.9 |
| CDCA | 4.1 ± 2 | 90.14 ± 6.5 | NA | αOH, βH | βH | αH, βH | αOH, βH | αH, βH | 2.15 ± 1.2 | ND | NA |
| β-MCA | 53.6 ± 2.6 | 32.87 ± 3.7 | 35 ± 2.8 | αOH, βH | βH | αH, βOH | αH, βOH | αH, βH | 11.4 ± 2.4 | ND | NA |
Reactions performed in triplicate, represented as average with standard deviation. NA, reaction not performed; ND, not detected.
Amounts of 0.02 U BaiF and 0.012 U BaiK were used per reaction. One unit of activity is defined as the amount of enzyme required to transfer CoA from 1 μmol DCA-CoA to CA min−1.
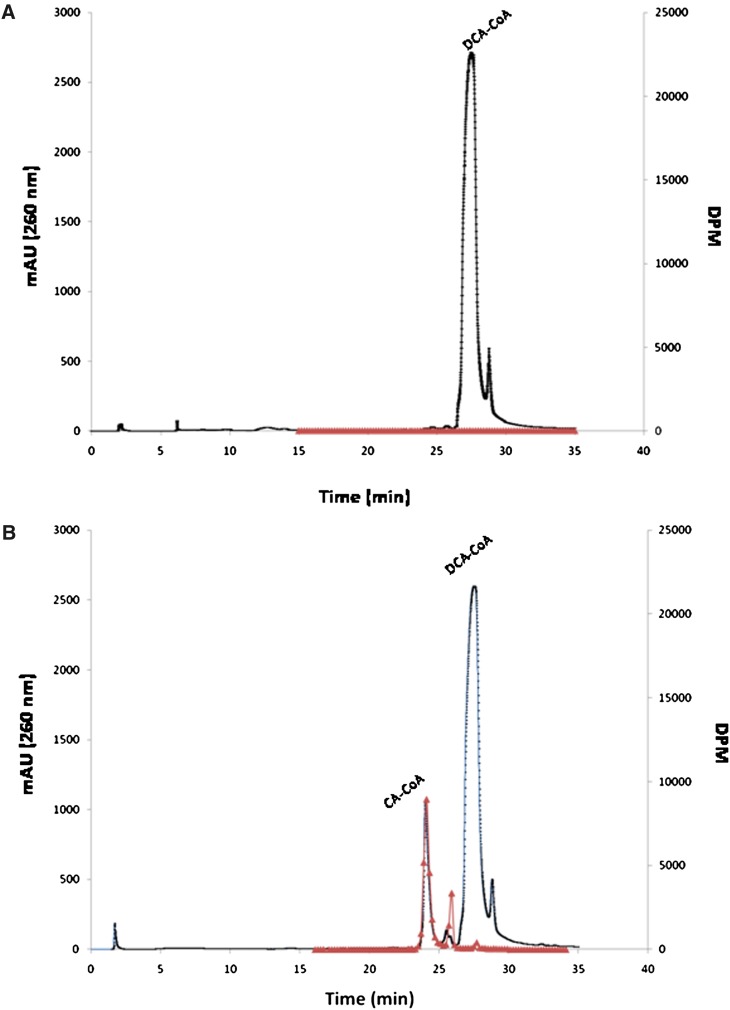
We determined that synthesized DCA-CoA eluted at 27.48 min on reverse-phase HPLC. Addition of the recombinant BaiF to a reaction mixture containing DCA-CoA and CA resulted in a product eluting at 24 min (Fig. 7). We repeated the assay with radiolabeled CA, and after termination of the reaction, free bile acids where extracted from the aqueous reaction mixture by ethyl acetate. Remaining water soluble radioactive counts represent [24-14C]CA-CoA conjugates. We collected fractions from 15 to 35 min, measured radioactivity by liquid scintillation spectrometry, and compared BaiF catalyzed reaction to the no-enzyme control. Fractions corresponding to the HPLC peak detected at 24 min also contained [24-14C] CA, whereas no such peak or radioactivity above background was detected in the no-enzyme control (Fig. 7). This data suggests that the baiF gene product transfers CoA from DCA-CoA to CA.
Fig. 7.
Reverse phase C-18 HPLC elution profile of CoA transferase reaction products catalyzed by BaiF. Reactions contained 500 μM cholic acid (17,600 DPM/nmole [24-14C] cholic acid) and 500 μM deoxycholyl-CoA A: No enzyme control elution profile. B: 0.01 μg BaiF catalyzed reaction. Reactions were acidified and extracted with ethyl acetate to remove free bile acids prior to HPLC. Radioactivity represented by triangles; absorption at 260 nm represented by dots.
Bile acid substrate specificity of recombinant bile acid CoA transferases
We then wanted to determine the substrate specificity for the recombinant baiF and baiK gene products using secondary bile acid CoA substrates and primary bile acid acceptors. The 3,7,12-trihydroxy bile acids CA and ACA were excellent acceptors, regardless of secondary bile acid CoA donor (Table 1). CDCA and CA had similar reaction rates when LCA-CoA was donor; however, transfer to CDCA was significantly diminished when DCA-CoA was donor (91% relative activity versus 4%, respectively). This was observed with both the baiF and baiK gene products. Although both the baiF and the baiK gene products were able to transfer CoA to UDCA, the baiK had the greatest reaction rate with this tertiary bile acid. Interestingly, the BaiK gene product demonstrated CoA transferase activity with DCA-CoA and ADCA-CoA but not with LCA-CoA. To determine whether lower concentrations of LCA-CoA were required, experiments were repeated with concentrations of LCA-CoA lowered from 300 μM to between 100 μM and 200 μM, with no product detected (data not shown). We tested the 3,6,7-trihydroxy bile acid β-MCA, which had a specificity similar to UDCA for the baiF gene product, and it was recognized to a lesser extent by the baiK gene product. In addition, unlike the baiF gene product, the baiK gene product appeared to prefer UDCA when DCA-CoA was donor, whereas ACA was the best acceptor when ADCA-CoA was donor (Table 1). We expected to see a broad substrate specificity as CA, CDCA, UDCA are common in human bile and the secondary bile acids LCA, DCA, and the 5α-epimer of DCA, ADCA, are produced by human bile acid 7α-dehydroyxlating bacteria.
DISCUSSION
In the current study, we cloned and expressed two genes encoding bile acid CoA transferases from C. scindens VPI 12708 in E. coli. We tested several combinations of secondary bile acid CoA donors and primary bile acid CoA acceptors and found that the baiF and baiK gene products have broad bile acid substrate specificity. However, the baiK gene product had no activity when LCA-CoA was used as the CoA donor (Table 1). Generally, the highest bile acid CoA transferase activity occurred when a homologous secondary bile acid was used as the bile acid CoA donor (i.e., cholic acid was the best acceptor when deoxcholyl-CoA was used as the CoA donor).
Bile acid 7α-dehydroxylating bacteria express a number of genes in the presence of primary bile acids (1). Upon entering the cell, bile acids are rapidly conjugated to CoA. Previously, we showed that the baiB gene product ligated CoA to primary bile acids by an ATP-dependent manner (9). We also demonstrated that the baiF gene product catalyzes the hydrolysis of cholyl-CoA (13). However, in our original experiments, we did not test for CoA transferase activity. In 2001, Heider proposed a new family of CoA transferases that, based on amino acid sequence analysis and knowledge of reaction mechanisms at the time, could not be placed into family I or II (14). This third family, involved in anaerobic metabolic pathways, includes the baiF gene and has been referred to as the “CaiB/BaiF” family. During anaerobic carnitine metabolism, (R)-carnitine is activated for degradation to butyrobetaine by ATP-dependent ligation by the caiC gene. ATP-dependent CoA ligation is thought to be required to generate a pool of CoA-thioesters that can later be usurped by an ATP-independent CoA-transferase encoded by the caiB gene product (14). In bile acid 7α-dehydroxylation, the baiB gene product would serve to generate a pool of bile acid-CoA conjugates, which could then be acted on by the bile acid CoA transferases encoded by the baiF and baiK genes (Fig. 1).
Type III CoA transferases, such as the crotonoyl-CoA (caiB) and yfdW gene products from E. coli (19–21) as well as the formyl-CoA transferase (FRC) from Oxalobacter formigenes (22), share a conserved Asp169 residue (D169 formyl CoA) involved in catalytic formation of oxo-acid mixed anhydrides as well as covalent CoA thioesters. The baiF and baiK gene products share this conserved residue (Fig. 4). It has been proposed that CoA transfer proceeds through Asp169 acylation (22–25). Mutagenesis of Asp169 (D169A, D169S, D169E) in the FRC from O. formigenes resulted in abolition of or appreciable decrease in specific activity despite no significant structural alterations (22). Recently, a catalytic mechanism has been proposed based on crystallographic data for FRC from O. formigenes (26). The proposed catalytic mechanism for FCR suggests that formyl-CoA forms an acid anhydride with Asp169, resulting in hydrolysis of CoAS–. Gln-17 and a glycine loop alternate between “open” and “closed” conformations, shielding aspartyl mixed anhydrides and aspartyl-CoA thioesters from hydrolysis between steps. -SCoA nucleophilically attacks the β-aspartyl-formylyl anhydride, releasing formate. The second substrate, oxalate, then nucleophilically attacks the aspartyl-CoA thioester, forming the second aspartyl-oxalyl anhydride, which is hydrolyzed in a final attack by CoAS−, releasing oxalyl-CoA and regenerating Asp169. This mechanism suggests that a ternary complex is not formed as observed in family II transferases. Instead, the reaction is completed before reaction products are released (26). Crystallization of CaiB yielded crystals with only CoA bound, suggesting hydrolysis occurs in the absence of the CoA-accepting substrates (21). This would provide a plausible explanation for observed CoA hydrolase activity shown for the baiF gene product in the absence of CoA substrate (13). We speculate that a similar mechanism is involved in bile acid CoA transfer by the baiF and possibly the baiK genes to that proposed for the FRC. However, the baiF and baiK gene products appear to lack the 258GGGGQ261 glycine loop; therefore, they use a different means to shield intermediates from premature hydrolysis.
We have demonstrated that the baiF and baiK gene products have bile acid CoA transferase activity with broad bile acid substrate specificity. However, in the current study, we focus only on end-product secondary bile acid donors. It is possible that intermediates following 7α/β-dehydration, including 3-dehydro-4, 3-dehydro-4,6 and 3-dehydro-intermediates of DCA, LCA, and ADCA, are the preferred substrates. Chemical synthesis of these intermediates and future kinetic studies of the baiF and baiK genes will be required to answer this question. Additionally, identification of genes in the “reductive” arm of the pathway and kinetic analysis of free bile acid versus bile acid-CoA thioesters will allow determination of the step in the pathway where CoA is transferred. Previously, we isolated a 3-dehydro-4-DCA~CoA intermediate, produced after reduction of the Δ6-bond following 7α-dehydration, from cell extracts of CA-induced cultures of C. scindens VPI 12708 (27). This suggests that CoA is transferred late in the pathway; however, additional work will be required to determine which intermediate is the preferred donor.
We have discovered a polycistronic operon that appears to be involved in bile acid metabolism in C. scindens VPI 12708 and C. hylemonae DSM 15053. The baiJ gene is similar to fumarate reductases and 3-keto steroid-Δ1-dehydrogenases, and the baiL is suggested to encode a 3α-hydroxysteroid dehydrogenase. The baiK gene located on this operon encodes a bile acid CoA transferase that may be required to enhance transferase activity when bile acids are prevalent. Coexpression of the baiK gene with genes in the “reductive” arm of the 7α/β-dehydroxylation pathway may help maintain a steady ATP-independent recycling of CoA to maximize conservation of energy in this reductive anaerobic process. Interestingly, genes within this operon have been identified in the 7α-dehydroxylating strains C. hylemonae DSM 15053, C. scindens VPI 12708, and C. hiranonas DSM 13275, but these genes were not detected in the 7α-dehydroxylating strain C. scindens ATCC 35704. Further work will be necessary to determine the function of this novel operon.
In summary, we report the first bile acid CoA transferases (baiF, baiK) found in bacteria and suggest they function to conserve an ATP in the bile acid 7-α-dehydroxylation pathway (Fig. 1).
Acknowledgments
The authors thank Patricia Cooper, Dalila Marques, and Kalyani Daita for technical assistance.
Footnotes
Abbreviations:
- ADCA
- allodeoxycholic acid
- β-MCA
- β-muricholic acid
- bai
- bile acid inducible
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- DCA
- deoxycholic acid
- FRC
- formyl-CoA transferase
- HSDH
- hydroxysteroid dehydrogenase
- LCA
- lithocholic acid
- ORF
- open reading frame
- UDCA
- ursodeoxycholic acid
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Fellowship Training Grant 2T32DK-007150-35 (J.M.R.) and National Institutes of Health Grant PO1DK-38030. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Ridlon J. M., Kang D., Hylemon P. B. 2006. Bile salt biotransformations by Human Intestinal bacteria. J. Lipid Res. 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein H., Bernstein C., Payne C. M., Dvorakova K., Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589: 47–65. [DOI] [PubMed] [Google Scholar]
- 3.Berr F., Kullak-Ublick G. A., Paumgartner G., Munzig W., Hylemon P. B. 1996. 7alpha-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology. 111: 1611–1620. [DOI] [PubMed] [Google Scholar]
- 4.Dowling R. H., Veysey M. J., Pereira S. P., Hussaini S. H., Thomas L. A., Wass J. A., Murphy G. M. 1997. Role of intestinal transit in the pathogenesis of gallbladder stones. Can. J. Gastroenterol. 11: 57–64. [DOI] [PubMed] [Google Scholar]
- 5.Mallonee D. H., White W. B., Hylemon P. B. 1990. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J. Bacteriol. 172: 7011–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells J. E., Hylemon P. B. 2000. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66: 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridlon J. M., Kang D., Hylemon P. B. 2010. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae DSM 15053. Anaerobe. 16: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mallonee D. H., Hylemon P. B. 1996. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J. Bacteriol. 178: 7053–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallonee D. H., Adams J. L., Hylemon P. B. 1992. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J. Bacteriol. 174: 2065–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallonee D. H., Lijewski M. A., Hylemon P. B. 1995. Expression in Escherichia coli and characterization of a bile acidinducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr. Microbiol. 30: 259–263. [DOI] [PubMed] [Google Scholar]
- 11.Kang D., Ridlon J. M., Moore D. R., 2nd, Barnes S., Hylemon P. B. 2008. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta. 1781: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson J. A., Mallonee D. H., Björkhem I., Hylemon P. B. 1996. Expression and characterization of a C-24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 37: 1258–1267. [PubMed] [Google Scholar]
- 13.Ye H. Q., Mallonee D. H., Wells J. E., Björkhem I., Hylemon P. B. 1999. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J. Lipid Res. 40: 17–23. [PubMed] [Google Scholar]
- 14.Heider J. 2001. A new family of CoA-transferases. FEBS Lett. 509: 345–349. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt T. G. M., Skerra A. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2: 1528–1535. [DOI] [PubMed] [Google Scholar]
- 16.Killenberg P. G., Dukes D. F. 1976. Coenzyme A derivatives of bile acids-chemical synthesis, purification, and utilization in enzymic preparation of taurine conjugates. J. Lipid Res. 17: 451–455. [PubMed] [Google Scholar]
- 17.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopal-Srivastava R., Mallonee D. H., White W. B., Hylemon P. B. 1990. Multiple copies of a bile acid-inducible gene in Eubacterium sp. strain VPI 12708. J. Bacteriol. 172: 4420–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leys D., Tsapin A. S., Nealson K. H., Meyer T. E., Cusanovich M. A., Van Beeumen J. J. 1999. Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat. Struct. Biol. 6: 1113–1117. [DOI] [PubMed] [Google Scholar]
- 20.Toyota C. G., Berthold C. L., Gruez A., Jónsson S., Lindqvist Y., Cambillau C., Richards N. G. J. 2008. Differential substrate specificity and kinetic behavior of Escherichia coli YfdW and Oxalobacter formigenes formyl coenzyme A transferase. J. Bacteriol. 190: 2556–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rangarajan E. S., Li Y., Iannuzzi P., Cygler M., Matte M. 2005. Crystal structure of Escherichia coli crotonobetainyl-CoA: carnitine CoA-transferase (CaiB) and its complexes with CoA and carnitinyl-CoA. Biochemistry. 44: 5728–5738. [DOI] [PubMed] [Google Scholar]
- 22.Jonsson S., Ricagno S., Lindqvist Y., Richards N. G. J. 2004. Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes. J. Biol. Chem. 279: 36003–36012. [DOI] [PubMed] [Google Scholar]
- 23.Ricagno S., Jonsson S., Richards N. G. J., Lindqvist Y. 2003. Formyl-CoA transferase encloses the CoA binding site at the interface of an interlocked dimer. EMBO J. 22: 3210–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gogos A., Gorman J., Shapiro L. 2004. Structure of Escherichia coli YfdW, a type III CoA transferase. Acta Crystallogr. D Biol. Crystallogr. 60: 507–511. [DOI] [PubMed] [Google Scholar]
- 25.Gruez A., Roig-Zamboni V., Valencia C., Campanacci V., Cambillau C. 2003. The crystal structure of the E. coli yfdW gene product reveals a new fold of two interlaced rings identifying a wide family of CoA transferases. J. Biol. Chem. 278: 34582–34586. [DOI] [PubMed] [Google Scholar]
- 26.Berthold C. L., Toyota C. G., Richards N. G. J., Lindqvist Y. 2008. Reinvestigation of the catalytic mechanism of formyl-CoA transferase, a class III CoA-transferase. J. Biol. Chem. 283: 6519–6529. [DOI] [PubMed] [Google Scholar]
- 27.Coleman J. P., White W. B., Egestad B., Sjövall J., Hylemon P. B. 1987. Biosynthesis of a novel bile acid nucleotide and mechanism of 7alpha-dehydroxylation by an intestinal Eubacterium species. J. Biol. Chem. 262: 4701–4707. [PubMed] [Google Scholar]