Abstract
Duplicated genes can contribute to the evolution of new functions and they are common in eukaryotic genomes. After duplication, genes can show divergence in their sequence and/or expression patterns. Qualitative complementary expression, or reciprocal expression, is when only one copy is expressed in some organ or tissue types and only the other copy is expressed in others, indicative of regulatory subfunctionalization or neofunctionalization. From analyses of two microarray data sets with 83 different organ types, developmental stages, and cell types in Arabidopsis thaliana, we determined that 30% of whole-genome duplicate pairs and 38% of tandem duplicate pairs show reciprocal expression patterns. We reconstructed the ancestral state of expression patterns to infer that considerably more cases of reciprocal expression resulted from gain of a new expression pattern (regulatory neofunctionalization) than from partitioning of ancestral expression patterns (regulatory subfunctionalization). Pollen was an especially common organ type for expression gain, resulting in contrasting expression of some duplicates in pollen. Many of the gene pairs with reciprocal expression showed asymmetric sequence rate evolution, consistent with neofunctionalization, and the more rapidly evolving copy often showed a more restricted expression pattern. A gene with reciprocal expression in pollen, involved in brassinosteroid signal transduction, has evolved more rapidly than its paralog, and it shows evidence for a new function in pollen. This study indicates the evolutionary importance of reciprocal expression patterns between gene duplicates, showing that they are common, often associated with regulatory neofunctionalization, and may be a factor allowing for retention and divergence of duplicated genes.
Keywords: gene duplication, gene expression, genome duplication, genome evolution, microarrays
Introduction
Gene duplication is one of the most important types of genetic variation that has provided the raw material for new gene functions and evolutionary innovations during eukaryotic evolution (reviewed by Dermuth and Hahn 2009; Hastings et al. 2009). Duplicated genes can be produced by various molecular mechanisms, including whole-genome (WG) duplication, segmental duplication, tandem duplication, and transposition (reviewed in Freeling 2009). WG duplications have taken place during the evolution of vertebrates, yeast, and plants, among other groups of eukaryotes. All angiosperms have undergone at least one round of ancient WG duplication during their evolutionary history, and genome sequencing projects and analyses of expressed sequence data have shown evidence for additional rounds of ancient WG duplication in some plant lineages (e.g., Blanc and Wolfe 2004b; Sterck et al. 2005; Cui et al. 2006; Barker et al. 2009; Schmutz et al. 2010; Tang et al. 2010; Jiao et al. 2011). The number of genes retained after WG duplication varies by lineage. In addition, many plants have experienced an evolutionarily recent polyploidy event, and they are cytologically polyploid (Wood et al. 2009). Tandem duplication contrasts to WG duplication in that the duplications are small scale and local, often being formed by unequal crossing over. It has been estimated that at least 14–16% of the genes in angiosperm genomes were derived from tandem duplication events (Rizzon et al. 2006).
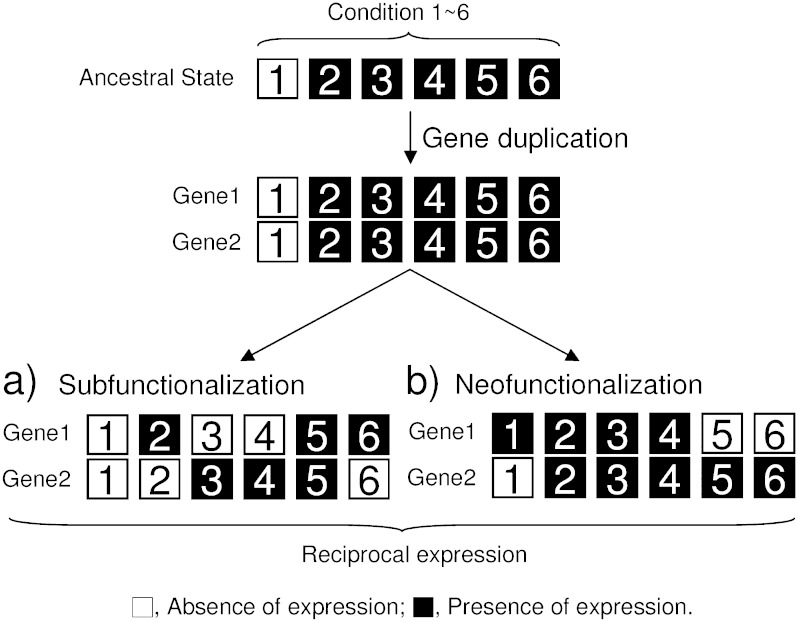
Several models for duplicate gene retention and subsequent fates have been proposed, including genetic redundancy, gene dosage balance, genetic robustness, and divergence of protein sequence and expression patterns that can lead to neofunctionalization, subfunctionalization, or subcellular relocalization (reviewed in Sémon and Wolfe 2007; Hahn 2009; Innan and Kondrashov 2010). Divergence in expression patterns and protein sequence can be responsible for duplicate gene retention or they can be a subsequent outcome after the duplicates were initially retained by other factors. Expression divergence between duplicated genes has been studied in a variety of eukaryotes. Expression divergence can be asymmetric, where one copy is always expressed at a higher level or complementary, where the duplicate with a higher expression level varies by organ or tissue type (e.g., Casneuf et al. 2006; Ganko et al. 2007). Complementary expression patterns can be quantitative, where both genes are expressed in all organ and tissue types, but the duplicate that is more highly expressed varies (Duarte et al. 2006). Complementary expression patterns also can be qualitative, here referred to as a reciprocal expression pattern, where only one copy is expressed in one or more organ or tissue types and only the other copy is expressed in others. Reciprocal expression patterns could arise by regulatory neofunctionalization, where one copy gains a new expression pattern in some organ or tissue types or regulatory subfunctionalization, where ancestral expression patterns are divided between the duplicates (Force et al. 1999; see fig. 1 herein). Reciprocal expression can be important for the retention of duplicated genes because loss of either copy would result in no expression in certain organ or tissue types and that might cause a detrimental effect or lower the fitness (e.g., Force et al. 1999). Several examples of reciprocally expressed duplicated genes have been reported in plants (e.g., Adams et al. 2003; Bottley et al. 2006; Drea et al. 2006; Chaudhary et al. 2009; Buggs et al. 2010a, 2010b; Liu and Adams 2010), suggesting that reciprocal expression can be an important factor for functional diversification of duplicated genes. Most previous studies of the evolution of duplicate gene expression on a large scale in plants used correlation methods to show considerable expression divergence between duplicated gene pairs (e.g., Blanc and Wolfe 2004a; Haberer et al. 2004; Casneuf et al. 2006; Ganko et al. 2007; Ha et al. 2007; Li et al. 2009; Throude et al. 2009), but most of those studies were not designed to detect reciprocal expression patterns. A recently published paper examined expression of genes duplicated during WG duplication in maize from each subgenome, but only the overall trends rather than details from individual gene pairs were reported as that was the question of interest (Schnable et al. 2011). Furthermore, little is known about whether reciprocal expression patterns more often result from regulatory neofunctionalization or subfunctionalization. In the only previous study designed to infer neofunctionalization or subfunctionalization in plants on a large scale, Duarte et al. (2006) used a gene family approach to study expression 280 regulatory gene pairs in six organ types and to infer the ancestral state of expression. Only a few cases of reciprocal expression were discovered in their study, probably due to the limited number of organ types and developmental stages examined.
FIG. 1.—
Schematics illustrating subfunctionalization and neofunctionalization as evolutionary causes of reciprocal expression patterns between duplicated genes. Numbers indicate different conditions such as cell types, organ types, or developmental stages. (a) Subfunctionalization showing reciprocal expression between the duplicated genes due to the partitioning of the ancestral expression pattern. (b) Neofunctionalization showing reciprocal expression due to the acquisition of a new expression pattern in gene 1 in comparison to the ancestral expression pattern.
Arabidopsis thaliana has advantages as a system for studying expression evolution of duplicated genes in plants. A large amount of microarray data is available from previous studies, including a large scale study of expression in 63 different organ and tissue types and developmental stages (Schmid et al. 2005) as well as a study of 20 different cell types and developmental stages of roots (Birnbaum et al. 2003; Brady et al. 2007), among others. The most recent WG duplication during the evolutionary history of A. thaliana occurred at or near the base of the Brassicaceae family, referred to as the alpha WG duplication (Bowers et al. 2003; Barker et al. 2009). About 2,500 pairs of genes have been retained from the alpha WG duplication (Blanc et al. 2003). In addition, about 4,000 genes have been identified as tandem duplicates in clusters of various sizes (Haberer et al. 2004; Rizzon et al. 2006).
The goal of this study was to understand the frequency, causes, and effects of reciprocal expression patterns of WG duplicates and tandem duplicates in a plant in a broad range of developmental stages, organ types, and cell types. We analyzed WG duplicates and tandem duplicates because their duplication mechanism is clear and contrasting, large scale versus small scale, whereas dispersed and transposed duplicates can arise by multiple mechanisms. We investigated the frequency of reciprocal expression patterns among 83 different organ types, developmental stages, and cell types by using ATH1 microarray data from A. thaliana (Birnbaum et al. 2003; Schmid et al. 2005; Brady et al. 2007) as well as performing additional analyses of the duplicate pairs showing reciprocal expression patterns.
Materials and Methods
Duplicated Gene Pair Selection
We obtained A. thaliana gene families from PLAZA 1.0 (Proost et al. 2009) and implemented a maximum likelihood (ML) analysis for every gene family by RAxML v.7.0.0 with an amino acid substitution matrix WAG and gamma-distributed rate variation (Stamatakis 2006). A 50% consensus tree for each gene family was obtained from 100 replicates of bootstrapping analysis. Using the 50% consensus tree topology, we pulled out all terminal gene pairs. From these pairs, pairs of WG and tandem duplicates were identified using 2,584 pairs of duplicated genes (5,168 genes) derived from the most recent WG duplication event identified by Blanc et al. (2003) and 1,826 clusters of tandemly duplicated genes (4,970 genes) identified in the current study. Identification of tandem duplicates followed the analytical procedure of Zou et al. (2009) using the following three criteria: 1) they belong to the same gene family, 2) they are located within 100 kb of each other, and 3) they are separated by ten or fewer genes that do not belong to the same gene family. The above procedures allowed us to identify gene pairs that have not experienced any subsequent duplication events. We excluded WG duplicates and tandem duplicates that are not included on the Affymetrix ATH1 microarray chip, which contains 22,746 probe sets (>80% of known Arabidopsis genes). To avoid cross-hybridization, only those genes with unique probes on the chip were selected (those that are designated with an “_at” extension and without an “s” or “x” suffix). Last, we excluded genes that were annotated as pseudogenes by TAIR (http://www.arabidopsis.org/). After these filtration steps, 1,539 WG duplicated pairs and 466 tandem duplicated pairs were subsequently used for further analyses (supplementary table S1, Supplementary Material online).
Microarray Data Analysis and Detection of Reciprocal Expression
After excluding data from mutants, raw ATH1 microarray data from 63 different organ types and developmental stages (ADA, Arabidopsis Development Atlas; Schmid et al. 2005) were obtained from the TAIR website (http://www.arabidopsis.org/). Raw ATH1 microarray data from 20 different cell types and developmental stages in roots (ARA, Arabidopsis Root Atlas; Birnbaum et al. 2003; Brady et al. 2007) were downloaded from the AREX website (http://www.arexdb.org/). Raw CEL files were processed and normalized using the MAS5.0 algorithm in Bioconductor (http://www.bioconductor.org/). Absence or presence of expression was statistically determined by using the “mas5calls” function in Bioconductor (Gautier et al. 2004; Gentleman et al. 2004). The statistical test performed the Wilcoxon signed rank–based gene expression absence/presence detection algorithm and generated a detection call (i.e., a probability value) to determine if the expression signal was significantly greater than background noise. Genes with a probability value less than 0.05 were designated as presence of expression, whereas genes with a probability value equal to or greater than 0.05 were assigned as absence of expression. Because there are three biological replicates, presence of expression was inferred when at least two of three showed presence of expression. To better visualize the reciprocal expression patterns of gene duplicates across different developmental stages, organ types, and cell types, we also generated graphs that contain the expression profiles between duplicated gene pairs (supplementary figs. S1 and S2, Supplementary Material online). Expression profile analysis and all statistical tests were implemented using the statistical package R.
After determining the absence and presence of expression using the Wilcoxon signed rank–based gene expression absence/presence detection algorithm (i.e., the mas5calls function in Bioconductor), reciprocal expression between duplicated genes was determined based on the following three Boolean criteria: 1) let yij be one of two expression status (0 and 1), where 0 stands for the absence of expression, 1 stands for the presence of expression, i = 1, 2 for gene copy 1 and gene copy 2, and j = 1, 2, … for different developmental stages, organ types, or cell types; 2) then, let min(yij) = 0 and max(yij) > 0; and 3) last, max(y1j − y2j) > 0 and min(y1j − y2j) < 0. The first criterion assigned the expression status for each organ type, developmental stage, or cell type. The second criterion filtered out genes that showed no expression across all conditions. The third criterion ensured that there is reciprocal expression between a duplicated gene pair under any given two data points.
Simulation Analysis
To examine the effects of sample numbers on detecting the frequency of reciprocal expression, we performed a simulation with a random subsampling process. Among the WG and tandem duplicates, we started at number of data points = 2 and ended at number of data points = total data points, by randomly subsampling different organ types or developmental stages in the ADA data set and in the ARA data set, and then calculated the frequency of reciprocal expression. We then repeated this procedure 1,000 times. To compare the average accumulative curve in terms of percentage of reciprocal expression to number of data points (i.e., the sample size of different organ types, developmental stages, or cell types) between the WG duplicates and the tandem duplicates from the ADA data set and the ARA data set, the Kolmogorov–Smirnov test was applied using the function “ks.test” in the statistical package R (http://www.r-project.org/).
Gene Ontology Analysis
Gene ontology (GO) annotations for A. thaliana were obtained from the website TAIR. For the GO enrichment analysis, the package topGO in Bioconductor was used (Gentleman et al. 2004). Any difference in terms of enrichment of GO categories between two different data sets was compared by using Fisher's exact test in the statistical package topGO. To correct for multiple testing, we implemented a 5% false discovery rate (FDR) adjustment algorithm using the function “p.adjust” with the method = “fdr” in the statistical package R. An FDR-adjusted P value (or Q value) smaller than 0.05 was considered to be a significant difference. We next compared the ratio of genes in each GO category between reciprocally expressed gene duplicates and all gene duplicates. At each developmental stage, organ type, or cell type, we also compared the ratio of genes in each GO category between neofunctionalized gene duplicates and all reciprocally expressed gene duplicates.
Inference of the Most Recent Common Ancestral Expression
Annotated protein sequences in A. thaliana (TAIR, v8) were downloaded from the TAIR website (http://www.arabidopsis.org/). We obtained gene families from PLAZA 1.0 (Proost et al. 2009). For the most recent common ancestral (MRCA) analysis, we followed the analytical procedure described in Zou et al. (2009) and Liu and Adams (2010). Briefly, reconstruction of the MRCA expression pattern between extant gene duplicates with reciprocal expression was conducted with a ML algorithm using the program MultiState in the package BayesTraits v.1.0 (Barker et al. 2007). To take the uncertainty of the phylogenetic tree topology into account, 100 bootstrapping trees deduced from ML analyses by RAxML v.7.0.0 with an amino acid substitution matrix WAG and gamma-distributed rate variation (Stamatakis 2006) were imported into BayesTraits, and each tree was rooted at the midpoint using the program Reroot in the package Phylip v.3.68 (Felsenstein 2009). Prior to gene family phylogeny analysis, protein sequences were aligned using the MUSCLE program with default settings (Edgar 2004). Two evolutionary transition rates comprising forward (from presence of expression to absence of expression) and reverse transition (from absence of expression to presence of expression) were used for estimating the character transition rate. Two different character states were designated: absence of expression (0) and presence of expression (1). The AddMRCA function was used to define the MRCA node of two extant duplicated genes with reciprocal expression pattern for each gene family tree (Barker et al. 2007). To take into account different tree topologies generated from 100 different bootstrapping analyses; the ancestral state probability was averaged across the 100 bootstrapping trees. If the average of ancestral state probability for absence or presence of expression was greater than 0.6, it was inferred as the ancestral expression state; this criterion is more conservative than 0.5 that was used in Zou et al. (2009).
Analysis of Synonymous Substitution Rate
To estimate the age since gene duplication, the synonymous substitution rate (Ks) between two duplicates genes was computed using a ML algorithm using the program Codeml in PAML (Yang 1997). Prior to the estimation of Ks, all pairwise alignments of amino acid sequences among the WG duplicates and the tandem duplicates were computed using the software MUSCLE with default settings (Edgar 2004) and then their protein sequence alignments were used as an alignment guide to correct for pairwise nucleotide alignments using a perl script (available upon request). The F3x4 codon frequency model was used in our analysis.
Detection of Asymmetric Sequence Evolution
After the inference of the MRCA expression pattern between extant duplicated gene pairs, we tested for asymmetric protein sequence evolution for these reciprocally expressed gene duplicates, in which one copy has accumulated more amino acid mutations than the other copy after duplication. The analytical procedure followed the description in Blanc and Wolfe (2004a). To identify the outgroup orthologous sequence, the Arabidopsis annotated protein sequences were searched against other plant annotated protein sequences from four eudicots with available genome sequences (Carica papaya, Glycine max, Populus trichocarpa, Vitis vinifera) using the BlastP program (Altschul et al. 1997). We then retrieved the best hit orthologous sequences using the reciprocal best hit method described in Hulsen et al. (2006). Two criteria were used to keep the orthologous sequences for further asymmetric sequence evolution analysis. First, we kept those sequences that shared greater than 80% identity with e values ≤ 10−5 with the Arabidopsis duplicated genes. Second, we estimated the synonymous substitution rate (Ks) for each triplet of sequences (i.e., two duplicated genes and one best hit orthologous sequence in the outgroup species) using a ML method in PAML (Yang 1997). We kept triplets that showed Ks between the Arabidopsis duplicated genes that was smaller than that between the Arabidopsis duplicated genes and the orthologous sequence in the outgroup species.
For asymmetric sequence analysis, protein sequences were aligned using the MUSCLE program with default settings (Edgar 2004). By using the Codeml program in the PAML package (Yang, 1997), we then obtained ML estimates from two different hypotheses (unconstrained rate of evolution [i.e., asymmetric sequence evolution] vs. clock-like rate of evolution [i.e., symmetric sequence evolution]) with the Jones-Taylor-Thornton substitution matrix (Jones et al. 1992) and the gamma correction to accommodate variability in substitution rates. To test if the first hypothesis fits better than the second hypothesis, a likelihood ratio test (LRT) was applied. Briefly, twice the difference of the likelihood estimate between these two hypotheses (D = −2(Ln1 − Ln2), where D indicates twice likelihood ratio, Ln1 indicates the likelihood estimate from the first hypothesis, and Ln2 indicates the likelihood estimate from the second hypothesis) was compared against a chi-square distribution with the degree of freedom (df) equal to 1. The df was obtained based on the difference of parameters used in these two different hypotheses. To correct for the issue of multiple testing, an FDR approach described previously was applied to minimize the false positives. We determined that a duplicated pair has asymmetric sequence evolution when the null hypothesis was rejected after the LRT. The branch length estimated form the nonclock model was subsequently used to calculate the relative evolutionary rate (Relrate) and asymmetric evolutionary rate (Asyrate) using the following equations: RelRate(i) = Li/(L1 + L2) and Asyrate = |L1 − L2|/(L1 + L2), where i = 1, 2 for gene copy 1 and gene copy 2, L1 indicates the branch length since gene duplication for gene copy 1, and L2 indicates the branch length since gene duplication for gene copy 2.
Detection of Asymmetric Expression Evolution
To investigate any associations between expression divergence and protein divergence, we examined expression breadth (EB) for each copy of gene duplicates and calculated an asymmetric expression index (Asy) for gene duplicates.
We defined EB by the following equation: EBi = ai/(a1 + a2 − b), where i = 1, 2 for gene copy 1 and gene copy 2, a1 indicates the number of organ types, developmental stages, and cell types with expression for copy 1, a2 indicates the number for copy 2, and b indicates the shared number for both copies.
We defined Asy using the following equation: Asy = |a1 − a2|/(a1 + a2 − b), where a1 indicates the number of organ types, developmental stages, and cell types with expression for copy 1, a2 indicates the number for copy 2, and b indicates the shared number for both copy 1 and copy 2.
Plant Materials, Nucleic Acid Extraction, and Reverse Transcription–Polymerase Chain Reaction
Total RNA was extracted from various organ types (indicated in fig. 8) from the following species: A. thaliana (ecotype Columbia), C. papaya (cultivar Sun-Up), and V. vinifera (cultivar Pinot Noir). Nucleic acid extraction and reverse transcription–polymerase chain reaction (RT-PCR) followed the description in Liu and Adams (2010). Gene-specific primers are listed in supplementary table S2 (Supplementary Material online). The partial coding sequence of CpBSL1 in C. papaya determined in this study was deposited in GenBank with the accession number JN852984.
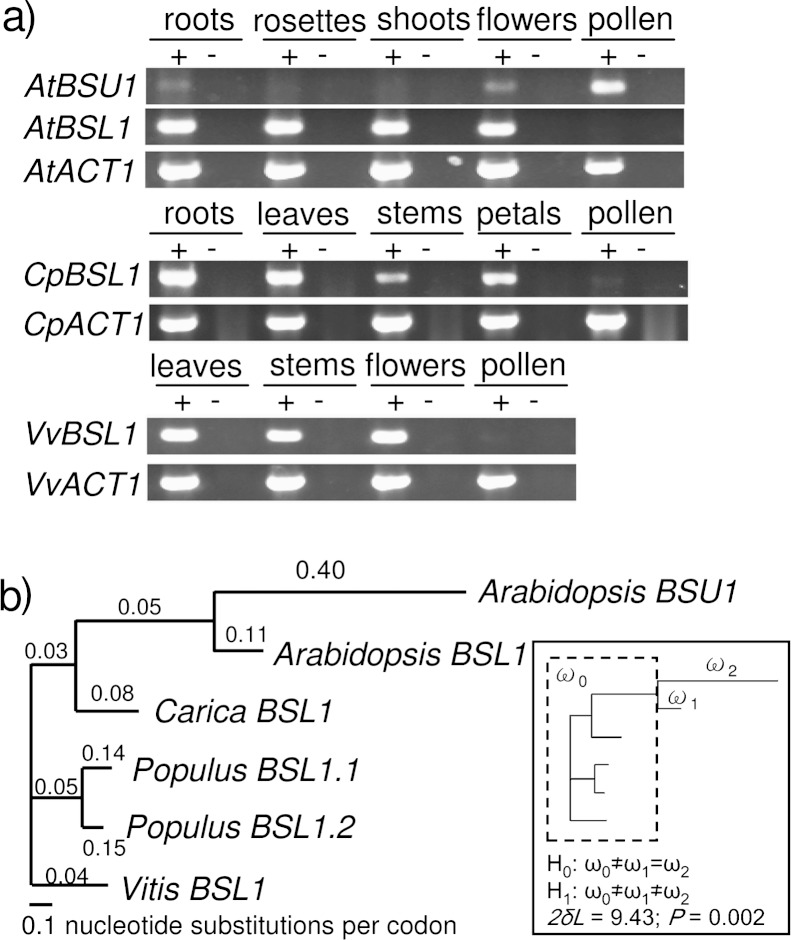
FIG. 8.—
AtBSU1 (AT1G03445) and AtBSL1 (AT4G03080) show reciprocal expression and AtBSU1 shows regulatory neofunctionalization and accelerated sequence evolution. (a) RT-PCR expression assays of the WG duplicate pair, AtBSU1 and AtBSL1, and their orthologs from outgroup species, Carica papaya (CpBSL1) and Vitis vinifera (VvBSL1). Results of RT-PCR expression assays of species-specific Actin 1 (AtACT1, CpACT1, and VvACT1) are shown in the lower panel of each set. Plus signs indicate the reactions with reverse transcriptase, whereas minus signs indicate the reactions without reverse transcriptase. (b) Sequence rate analysis of AtBSU1 and AtBSL1. Phylogenetic tree of BSU1 from Arabidopsis thaliana and BSL1 genes from A. thaliana, C. papaya, Populus trichocarpa, and V. vinifera. Branch length and branchwise Ka/Ks ratio (i.e., ω) were estimated using a free-ratio model in Codeml. A LRT between two different hypotheses (assuming the same rate [H0] and different rate [H1] between BSU1 and BSL1 in A. thaliana) indicates that BSU1 evolved about four times faster than its duplicated partner, BSL1.
Selection Analysis on BSU1
To test if there is evidence of accelerated evolution or positive selection acting on BSU1, a branch model and a branch-site model were implemented using the program Codeml in PAML (Yang 1997), following manual inspection of the MUSCLE generated alignment using BioEdit (Hall 1999). Orthologous sequences identified based on collinear analysis from C. papaya, P. trichocarpa, and V. vinifera were downloaded from the website PLAZA v.1 (Proost et al. 2009). Branchwise Ka/Ks (=ω) ratio along the phylogenetic tree was estimated using a free-ratio model. To test if the ω ratio of BSU1 and BSL1 evolved in an asymmetric fashion, two-ratio model and three-ratio models were implemented. The first model assumes that one ω ratio leads to the pro-ortholog branch and another ratio leads to the BSU1 and BSL1 branch. The second model assumes that three different ω ratios lead to the pro-ortholog branch, the BSU1 branch, and the BSL1 branch. The ω ratio values between BSU1 and BSL1 were assumed to be the same in the first model and to be different in the second model. Then, twice the difference of their likelihood log values (i.e., LRT) was compared against a chi-square distribution with the df = 1. Asymmetric sequence evolution was then determined when the second model significantly fits better than the first model. For the detection of positive selection, a branch-site test of positive selection was conducted along the BSU1 branch. Two different models (model A test 1 and model A test) were implemented (Zhang et al. 2005). The first model assumes no positive selection and the second model assumes the presence of positive selection, and the LRT was computed to compare against a chi-square distribution with 50:50 mixture of df = 0 and 1. Those codons that show posterior probability < 0.95 from a Bayes Empirical Bayes analysis are not considered as strong evidence of positively selected sites (Yang et al. 2005).
Results
Reciprocal Expression Patterns Are Common between Duplicated Genes
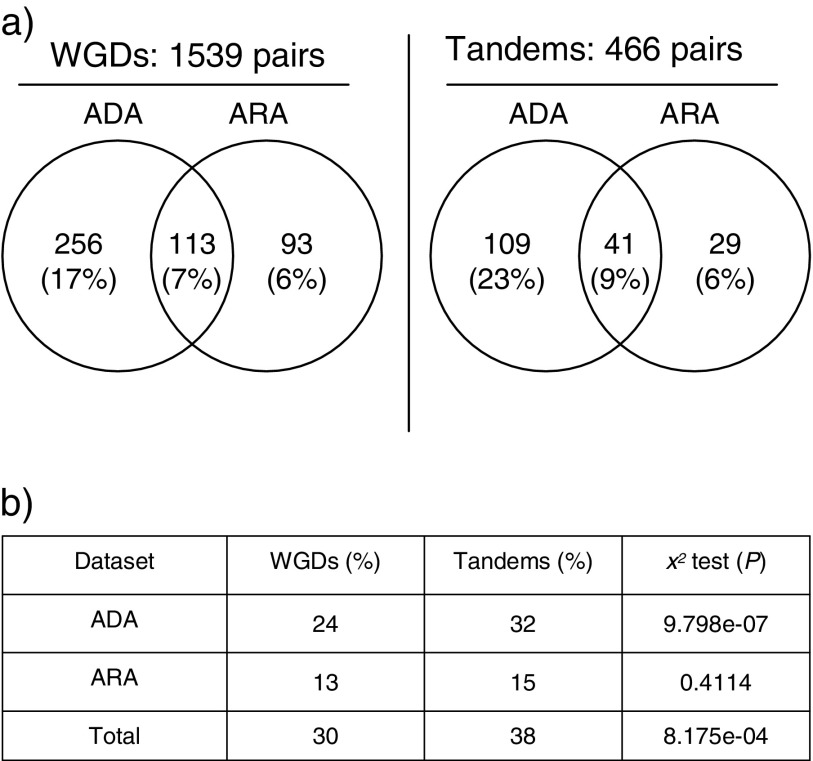
First, we identified duplicated gene pairs showing reciprocal expression patterns, where only one copy is expressed in one or more organ, tissue, or cell types and only the other copy is expressed in one or more different organ, tissue, or cell types. We analyzed expression patterns of 1,539 pairs of genes duplicated from the alpha WG duplication and 466 pairs of tandem duplicates in A. thaliana using Affymetrix ATH1 microarray data from 63 different organ types and developmental stages (ADA,; Schmid et al. 2005) and 20 different cell types and developmental stages in roots (ARA; Birnbaum et al. 2003; Brady et al. 2007). Those data sets were chosen because a large number of organs and tissues, or cell types, was assayed in a single study, and there are at least three biological replicates. The absence or presence of expression was determined using the Wilcoxon signed rank–based gene expression presence/absence detection algorithm in Bioconductor (see Materials and Methods). We found that 24% of the WG duplicates in the ADA data set and 13% in the ARA data set showed reciprocal expression patterns (fig. 2 and supplementary figs. S1 and S2 and tables S3 and S4, Supplementary Material online). Among the tandem duplicates, 32% in the ADA data set and 15% in the ARA data set show reciprocal expression patterns (fig. 2 and supplementary tables S3 and S4, Supplementary Material online). Seven percent of the WG duplicate gene pairs and 9% of the tandem duplicates showed reciprocal expression in both the ADA and the ARA data sets (fig. 2a). The tandem duplicates have a significantly higher frequency of reciprocal expression than the WG duplicates in the ADA data set (χ2, P = 9.798 × 10−7; fig. 2b) but not in the ARA data set (χ2, P = 0.4114; fig. 2b). When both data sets are considered together, there is a significantly higher frequency of reciprocal expression in the tandem duplicates (38%) than WG duplicates (30%) from the combination of the ADA data set and the ARA data set (χ2, P = 8.175 × 10−4; fig. 2b), suggestive of a higher frequency of expression diversification in tandem duplicates than WG duplicates.
FIG. 2.—
The frequency of reciprocal expression patterns in WG duplicates (WGDs) and tandem duplicates (Tandems) from the ADA and ARA data sets. (a) Venn diagram showing the frequency of reciprocal expression among WG duplicates and tandem duplicates. (b) Diagram showing a comparison of the frequency of reciprocal expression between WG duplicates and tandem duplicates using the χ2 test.
To investigate if certain types of genes more often show a reciprocal expression pattern, we conducted a GO enrichment analysis using the program topGO with the GO annotations from the TAIR website. Then, duplicated genes with reciprocal expression were compared against all duplicated genes by using Fisher's exact test. Among the WG duplicates, transcription (GO:0006350; Q = 0.0184), transcription factor activity (GO:0003700; Q = 0.0046), and DNA binding (GO:0003677; Q = 0.0199) were overrepresented in the ADA data set, whereas transferase activity (GO:0016740; Q = 0.0184), catalytic activity (GO:0003824; Q = 0.0291), and transcription factor activity (GO:0003700; Q = 0.0426) were overrepresented in the ARA data set (supplementary table S5, Supplementary Material online). These results suggested that expression pattern of transcription factor–related WG duplicates tend to diverge in a reciprocal expression fashion. Among the tandem duplicates, only catalytic activity (GO:0003824; Q = 0.0075) was detected to be overrepresented in the ADA data set (supplementary table S5, Supplementary Material online).
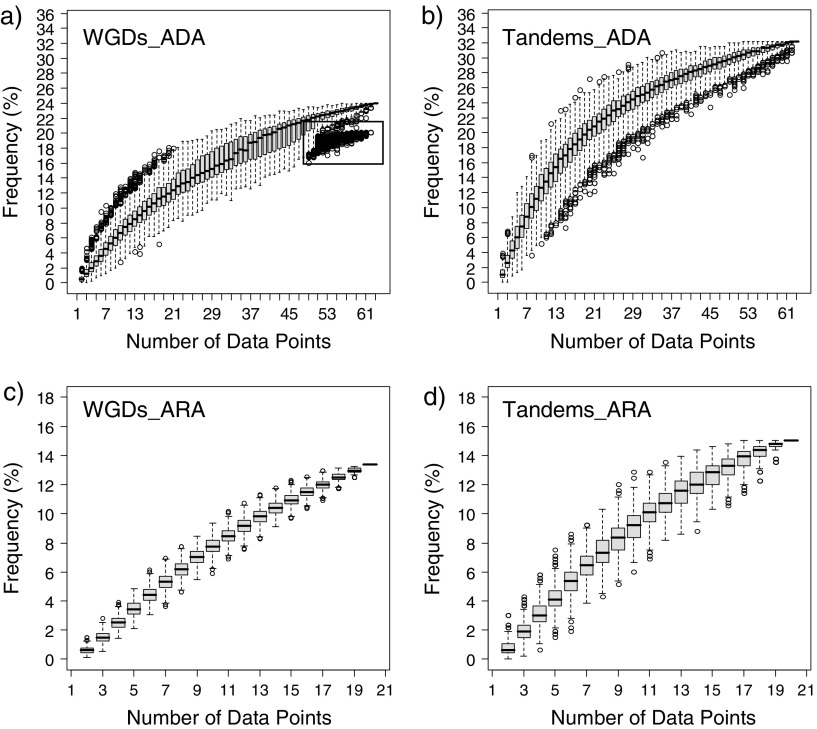
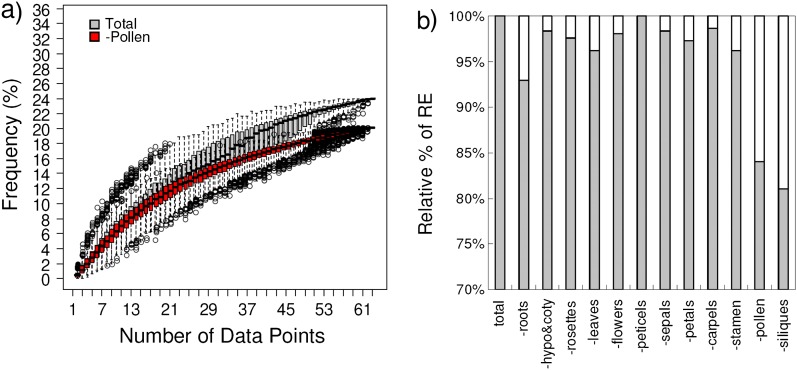
Next, we examined if the lower percentage of reciprocal expression in the ARA data set was due to the lower number of data points (20 vs. 63 in the ADA data set) or due to the less divergent structures (cell types within the root vs. a wide variety of tissues and organs in the ADA data set). We assessed the effects of the number of data points on the detection of reciprocal expression patterns between the WG duplicates and the tandem duplicates in the ADA data set and the ARA data set by performing simulation studies. For our simulations, we subsampled the number of data points randomly from all data points, starting at the number of data points = 2 and ending at the number of data points = all data points. We then repeated the simulations 1,000 times. Among the WG and tandem duplicates in the ADA and ARA data set, we found that the more data points we subsampled, the higher the percentage of duplicates with reciprocal expression we detected (fig. 3a–d). In the ADA data set, a higher percentage of reciprocal expression was found among the tandem duplicates than among WG duplicates (Kolmogorov–Smirnov test, P = 1.891 × 10−9; fig. 3a and b), consistent with our previous observations. It is noteworthy that there is a cluster of data points showing much lower percentages of reciprocal expression among the WG duplicates when more data points were subsampled (rectangular box in fig. 3a), suggesting that the inclusion of certain data points (i.e., certain organ types) might greatly contribute to the reciprocal expression patterns. Such a phenomenon was not found among the tandem duplicates (fig. 3b). In the ARA data set, no difference in terms of the percentage of reciprocal expression was observed between the WG duplicates and the tandem duplicates (Kolmogorov–Smirnov test, P = 0.8080; fig. 3c and d). We then compared the simulated curve between the ADA data set and the ARA data set. Among the tandem duplicates, a higher percentage of reciprocal expression was found in the ADA data set than in the ARA data set using the same number of data points, although there is no strong statistical support (Kolmogorov–Smirnov test, P = 0.0681; fig. 3a and c), suggesting that a higher percentage of reciprocal expression patterns among the tandem duplicates in the ADA data set is probably due to both more data points and more divergent organ types. In contrast, there is no difference in the percentage of reciprocal expression between the ADA data set and the ARA data set among the WG duplicates when using the same number of data points (Kolmogorov–Smirnov test, P = 0.9780; fig. 3b and d), suggesting that a higher percentage of reciprocal expression among the WG duplicates in the ADA data set is largely due to the larger number of data points. Overall, the number of data points (i.e., number of different organ types or developmental stages) can influence the detection of reciprocal expression. Interestingly, the higher percentage of reciprocal expression found among the tandem duplicates in the ADA data set is partly due to the more divergent organ types, suggesting that expression patterns among the tandem duplicates may diverge more across different organ types than the WG duplicates when compared with the ARA data set.
FIG. 3.—
Simulation of the effects of different sample numbers on the detection of reciprocal expression in WG duplicates (WGDs) and tandem duplicates (Tandems) from the ADA data set and ARA data set. (a–d) Box plots showing that with more different organ types or developmental stages (i.e., number of data points), the higher the frequency of reciprocal expression. (a) WGDs in the ADA data set. Black box indicates that there is a cluster of simulated data points showing a lower frequency of reciprocal expression when the sampling number approaches the maximum number in the ADA data set. (b) Tandems in the ADA data set. (c) WGDs in the ARA data set. (d) Tandems in the ARA data set.
Since we observed that a cluster of data points showed a lower percentage of reciprocal expression among the WG duplicates in the ADA data set, we further explored the possibility that some organ types might greatly contribute to the reciprocal expression patterns among the WG duplicates. We repeated our simulations but removed one data point at a time. When pollen was removed from the data pool, a significant decrease in the percentage of reciprocal expression was observed (Kolmogorov–Smirnov test, P = 0.0099; fig. 4a). This result suggested that the cluster with a lower percentage of reciprocal expression found in our previous simulations among the WG duplicates in the ADA data set is due to the removal of pollen (fig. 3a). This observation raised an interesting question as to which organ type might contribute more to the reciprocal expression patterns among the WG duplicates. To further examine the contribution of different organ types on the percentage of reciprocal expression among the WG duplicates, we scored the percentage of reciprocal expression by removing all developmental stages of one organAT1G03445 type at a time (e.g., all leaf data points) and compared the percentage of reciprocal expression before and after the removal of a particular organ type. Among different organ types (including roots, cotyledons and hypocotyls, rosettes, leaves, whole flowers, peticels, sepals, petals, carpels, stamens, pollen, and siliques), the removal of pollen and siliques significantly decreased up to 16% and 19%, respectively, the total percentage of reciprocal expression (χ2, P = 0.0117 and P = 0.0026, respectively; fig. 4b), suggesting that pollen and siliques contribute more than other organ types to the reciprocal expression patterns among the WG duplicates. Among tandem duplicates, only the removal of siliques could significantly decrease up to 20% of the total percentage of reciprocal expression (χ2, P = 0.0363; supplementary fig. S3, Supplementary Material online). Although the removal of roots can greatly decrease up to 15% of the total reciprocal expression, this observation was not strongly supported (χ2, P = 0.1148; supplementary fig. S3, Supplementary Material online). Overall, pollen and siliques are the most common structures for the occurrence of reciprocal expression patterns among the WG duplicates and siliques (and possibly roots) are the most common structures for the occurrence of reciprocal expression patterns among the tandem duplicates.
FIG. 4.—
Effects of different organ types on the frequency of reciprocal expression. (a) Simulation showing the frequency of reciprocal expression before and after removing pollen data, using different sample numbers. (b) Diagram showing that there is a significant decrease in the frequency of reciprocal expression after removing pollen data (ca. 16%; χ2, P = 0.0117) or siliques data (ca. 19%; χ2, P = 0.0026) but not after the removal of other organ types.
Reciprocal Expression Patterns Result More from Neofunctionalization than Subfunctionalization
The reciprocal expression patterns could result from regulatory subfunctionalization, where expression of each duplicate has been partitioned between organ types or from neofunctionalization, where there is gain of expression in a new organ type. Distinguishing between these possibilities requires an inference of the ancestral preduplication state of expression. The MRCA expression pattern can be inferred from other members in a gene family using a ML algorithm in a probabilistic framework, and it can be used to approximate the ancestral state of expression pattern (Gu 2004; Gu et al. 2005; Oakley et al. 2006; Fisher 2008; Zou et al. 2009; Liu and Adams 2010). The method has been applied to the inference of regulatory subfunctionalization or neofunctionalization between duplicated genes in Drosophila (Oakley et al. 2006) and Arabidopsis (Zou et al. 2009). We thus applied an integration of expression data and gene family phylogenies to infer the putative MRCA expression pattern of the reciprocally expressed gene duplicates. The phylogenetic distance and the uncertainty of the phylogenetic gene topology were taken into account, as in Pagel (1999).
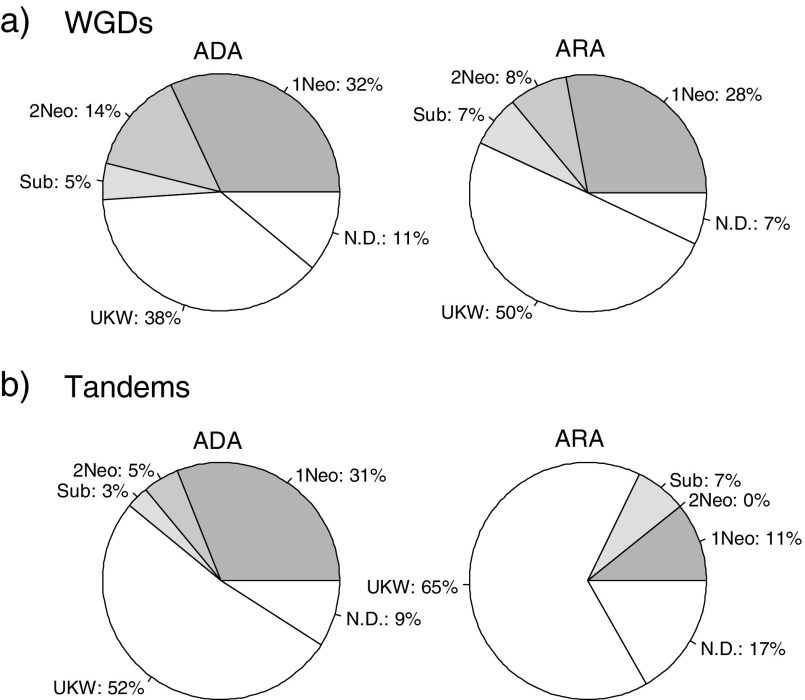
Among the WG duplicates with reciprocal expression patterns, 46% in the ADA data set and 36% in the ARA data set were inferred as neofunctionalized, whereas only 5% in the ADA data set and 7% in the ARA data set were inferred as subfunctionalized (fig. 5a and supplementary table S3, Supplementary Material online). Among the tandem duplicates, 36% in the ADA data set and 11% in the ARA data set were inferred as neofunctionalized, whereas only 3% in the ADA data set and 7% in the ARA data set were inferred as subfunctionalized (fig. 5b and supplementary table S4, Supplementary Material online). Among those neofunctionalized cases, a small percentage of them (8–14% in WG duplicates and 5% in tandem duplicates) showed gain of a new expression pattern for both copies (fig. 5). The ancestral expression state in some cases could not be assessed due to uncertainty in the phylogenetic tree topology and lack of expression data for most members (labeled as UKW in fig. 5) or lack of information such as a small gene family size with only two and three members (labeled as ND in fig. 5). The results of the ancestral expression state reconstruction suggested that reciprocal expression patterns between WG duplicates and tandem duplicates in A. thaliana result more from gain of a new expression pattern (neofunctionalization) than partitioning of the ancestral expression pattern (subfunctionalization).
FIG. 5.—
The relative frequency of subfunctionalization and neofunctionalization of expression patterns. Regulatory neofunctionalization and subfunctionalization were inferred by MRCA analysis in both WG duplicates (a) and tandem duplicates (b) from both the ADA and the ARA data sets. Abbreviations: 1Neo, neofunctionalization of one copy; 2Neo, neofunctionalization of both copies; Sub, subfunctionalization; UKW, unknown due to uncertain tree topology; and ND, not determined due to small gene family size with only two members.
Preferential Gain or Loss of Gene Expression
We next assessed if there is any preferential gain or loss of expression in particular organ types, developmental stages, and cell types among both WG duplicates and tandem duplicates in both the ADA and the ARA data sets. We compared the ratio of expression gain and expression loss at each developmental stage, organ type, and cell type by using Fisher's exact test. In the ADA data set, a significantly higher percentage of genes with expression gain than expression loss was found in pollen (Q = 0.0010; ca. 9.4% higher), the shoot apex after bolting (Q = 0.0283; ca. 6.7% higher), senescing leaf (Q = 0.0381; ca. 6.3% higher), seeds at the developmental stage 9 (Q = 0.0381; ca. 7.3% higher), and seeds at the developmental stage 10 (Q = 0.0381; ca. 7.0% higher) among the WG duplicates (supplementary fig. S4a, Supplementary Material online). Among these five organ types/developmental stages, a significantly higher percentage of expression gain than expression loss was only observed in pollen when a more stringent Bonferroni correction was applied (adjusted P = 0.0021), suggesting that pollen shows a more striking pattern in terms of expression gain after WG duplication. In contrast, there are not any particular organ types (or developmental stages) showing a significant difference between expression gain and expression loss among the tandem duplicates (supplementary fig. S4a, Supplementary Material online). We then performed GO enrichment analysis to see if there is any functional enrichment for neofunctionalized gene duplicates in pollen, seeds, shoot apex after bolting, and senescing leaf using the statistical package topGO (Alex and Rahnenführer 2009). In senescing leaf, genes that are involved in organ morphogenesis (GO:0009887; Q = 0.0291) were enriched (supplementary table S6, Supplementary Material online). In pollen, the biological process of microgametogenesis (GO:0055046; Q = 0.0014) and several molecular functions such as lipase activity (GO:0016298; Q = 0.0217), hydrolase activity (GO:0016788; Q = 0.0423) and microtubule motor activity (GO:0008574; Q = 0.0423) were enriched (supplementary table S6, Supplementary Material online). The results suggested that these neofunctionalized genes might play important roles in microgametogenesis in pollen. In the other organ structures, we did not find any significant enrichment for each GO category between the neofunctionalized duplicated genes and all reciprocally expressed duplicated genes.
In the ARA data set, we found that no particular cell types showed a significant difference between expression gain and expression loss among the WG duplicates, whereas two different cell types, phloem (Q = 0.0387) and all radial root tissues at stage 3 (Q = 0.0344), were found to show a significantly higher percentage of expression loss than expression gain among the tandem duplicates (supplementary fig. S4b, Supplementary Material online).
After assessing if particular organ types, developmental stages, and cell types showed preferential expression gain or loss, we next examined if there is any difference in expression gain or loss between WG duplicates and tandem duplicates by Fisher's exact test with 5% FDR correction for multiple tests. Among WG duplicates, a significantly higher percentage of expression gain than expression loss was observed in the ADA data set (gain: ca. 7.4% vs. loss: ca. 4.5%; Q = 3.12 × 10−32) but not in the ARA data set (gain: ca. 7.6% vs. loss: ca. 6.2%; Q = 0.3236) (supplementary fig. S4a, Supplementary Material online). In contrast, an opposite trend was found in tandem duplicates, where expression loss is significantly more common than expression gain in both the ADA data set (loss: ca. 7.4% vs. gain: ca. 5.2%; Q = 8.98e × 10−8) and the ARA data set (loss: ca. 8.1% vs. gain: ca. 1.7%; Q = 3.14 × 10−11) (supplementary fig. S4, Supplementary Materials online).
Asymmetric Sequence Evolution in Some Pairs with Neofunctionalization of Expression Patterns
Asymmetric sequence rate evolution in one member of a duplicate pair has been proposed as a likely indicator of neofunctionalization because one copy has experienced an accelerated rate of amino acid replacements in comparison to its duplicated partner (Blanc and Wolfe 2004a; Byrne and Wolfe 2007). In our asymmetric rate analysis, the best hit orthologous sequence from an outgroup species was used to polarize the evolutionary rate between gene duplicates, as done in Blanc and Wolfe (2004a). Of the WG duplicates, 43 of 267 triplets (16%) showed significant asymmetric protein sequence divergence (LTR, Q < 0.05; table 1 and supplementary table S3, Supplementary Material online). Of the tandem duplicates, 8 of 55 triplets (15%) showed significant asymmetric protein sequence divergence (LRT, Q < 0.05; table 1 and supplementary table S4, Supplementary Material online). Among them, there are 16 cases (classified as group 1) that showed both asymmetric sequence rate evolution and expression gain, inferred by the MRCA expression analysis (table 1), further supporting our inference of neofunctionalization. There were five cases (classified as group 2), where one copy showed asymmetric rate evolution and both copies were inferred as neofunctionalized by the MRCA analysis (table 1). In two cases (classified as group 3), the inference from the MRCA analysis was subfunctionalization, but there was asymmetric sequence rate analysis between the duplicates, suggesting neofunctionalization (table 1). Those two pairs might have undergone a transition stage between subfunctionalization and neofunctionalization, referred to as subneofunctionalization (He and Zhang 2005), via a combination of regulatory subfunctionalization and protein sequence neofunctionalization. The remaining cases were inferred as neofunctionalized only by asymmetric sequence rate analysis because of the lack of inference by MRCA (table 1). To test if older duplicated genes tend to show asymmetric rate evolution, we conducted a comparison of synonymous substitution rate (Ks) between pairs with symmetric evolution and asymmetric evolution. We did not see any significant difference in terms of the age of gene duplicates between the symmetric group and the asymmetric group among the WG duplicates and the tandem duplicates (t-test, P > 0.05; Wilcoxon signed-rank test, P > 0.05; supplementary fig. S5, Supplementary Material online). Tandem duplicates were on average younger than WG duplicates based on their Ks data (t-test: P = 1.659 × 10−5; Wilcoxon signed-rank test: P = 3.221 × 10−13; supplementary fig. S6, Supplementary Material online), which is consistent with previous reports (Blanc and Wolfe 2004a; Haberer et al. 2004). Overall, tandem duplicates did not show a significantly higher frequency of asymmetric rate evolution than WG duplicates (χ2, P = 0.9317).
Table 1.
List of the Putative Function/Function and the MRCA Inference of Subfunctionalization and Neofunctionalization for Reciprocally Expressed Gene Duplicates with Asymmetric Sequence Evolution
| Gene Duplicates |
Putative Function/Function | MRCA |
Asymmetric | ||
| Gene 1 | Gene 2 | ADA | ARA | ||
| WG duplicates | |||||
| AT1G07870 | AT2G28590 | Protein kinase | Neo (2) | — | Neo (2); G1 |
| AT1G55200 | AT3G13690 | Protein kinase | Unknown | — | Neo (1) |
| AT1G77280 | AT1G21590 | Protein kinase | — | Unknown | Neo (2) |
| AT4G25160 | AT5G51270 | Protein kinase | Neo (2) | — | Neo (2); G1 |
| AT5G65600 | AT5G10530 | Lectin protein kinase | Unknown | — | Neo (1) |
| AT5G03610 | AT3G09930 | GDSL-motif lipase/hydrolase | Neo (2) | — | Neo (2); G1 |
| AT5G67200 | AT3G50230 | Leucin-rich repeat transmembrane protein kinase | — | Unknown | Neo (2) |
| AT4G39860 | AT2G22270 | Unknown protein | Neo (1, 2) | — | Neo (2); G2 |
| AT3G60190 | AT2G44590 | Dynamin-related protein | Neo (2) | — | Neo (2); G1 |
| AT1G60930 | AT1G10930 | DNA helicase | Neo (1) | Unknown | Neo (1) |
| AT1G78050 | AT1G22170 | Phosphoglycerate/biphosphoglycerate mutase | Unknown | — | Neo (1) |
| AT2G02480 | AT1G14460 | DNA polymerase related | Neo (2) | — | Neo (2); G1 |
| AT2G18590 | AT4G36790 | Carbohydrate transmembrane transporter | Neo (1) | Neo (1) | Neo (1); G1 |
| AT5G44700 | AT4G20140 | Leucine-rich repeat transmembrane-type receptor kinase | Unknown | — | Neo (1) |
| AT3G59080 | AT2G42980 | Aspartyl protease | Unknown | — | Neo (2) |
| AT4G28320 | AT2G20680 | Glycosyl hydrolase | Unknown | — | Neo (1) |
| AT1G35140 | AT4G08950 | Exordium | Neo (1, 2) | Neo (1) | Neo (1); G2 |
| AT4G14760 | AT3G22790 | Kinase-interacting protein | Neo (1) | — | Neo (1); G1 |
| AT5G66390 | AT3G50990 | Peroxidase | Neo (2) | — | Neo (2); G1 |
| AT1G70510 | AT1G23380 | Class I of KN homeodomain transcription factor | Neo (1, 2) | Unknown | Neo (1); G2 |
| AT1G02460 | AT4G01890 | Glycoside hydrolase | — | Sub | Neo (2); G3 |
| AT1G53100 | AT3G15350 | Acetylglucosaminyltransferase | Unknown | — | Neo (1) |
| AT4G15430 | AT3G21620 | Unknown protein | — | Neo (1) | |
| AT1G13270 | AT3G25740 | Methionine aminopeptidase | Neo (2) | — | Neo (2); G1 |
| AT1G09350 | AT1G56600 | Galactinol synthase | Sub | Sub | Neo (1); G3 |
| AT2G34940 | AT1G30900 | Vacuolar sorting receptor | Unknown | Unknown | Neo (1) |
| AT5G57580 | AT4G25800 | Calmodulin-binding protein | Unknown | Unknown | Neo (2) |
| AT1G68540 | AT1G25460 | Oxidoreductase | — | Neo (2) | Neo (2); G1 |
| AT3G10660 | AT5G04870 | Calcium-dependent protein kinase | Neo (1) | — | Neo (1); G1 |
| AT5G14740 | AT3G01500 | Beta carbonic anhydrase | Neo (1, 2) | Unknown | Neo (1); G2 |
| AT1G02050 | AT4G00040 | Chalcone and stilbene synthase | Unknown | Neo (2) | Neo (2); G1 |
| AT1G70710 | AT1G23210 | Endo-1,4-beta-glucanase | Unknown | — | Neo (2) |
| AT4G24260 | AT5G49720 | Endo-1,4-beta-glucanase | Unknown | — | Neo (1) |
| AT4G18050 | AT5G46540 | P-glycoprotein | Neo (2) | Neo (2) | Neo (2); G1 |
| AT5G06700 | AT3G12060 | Trichome birefringence–like protein | Neo (1, 2) | — | Neo (2); G2 |
| AT3G53680 | AT2G37520 | PHD finger transcription factor | — | Unknown | Neo (1) |
| AT1G26310 | AT1G69120 | MADS-box transcription factor | Unknown | — | Neo (1) |
| AT1G10540 | AT1G60030 | Xanthine/uracil permease | Neo (1) | Neo (1) | Neo (1); G1 |
| AT2G20340 | AT4G28680 | Tyrosine decarboxylase | ND | — | Neo (2) |
| AT3G03110 | AT5G17020 | Exportin protein | ND | — | Neo (1) |
| AT2G21210 | AT4G38840 | SAUR-like auxin-responsive protein | Unknown | — | Neo (1) |
| AT4G03080 | AT1G03445 | Serine/threonine protein phosphatase | Neo (2) | — | Neo (2); G1 |
| Tandem duplicates | |||||
| AT2G44230 | AT2G44260 | Unknown protein | Unknown | Unknown | Neo (1) |
| AT5G10760 | AT5G10770 | Aspartyl protease | Unknown | — | Neo (1) |
| AT5G06720 | AT5G06730 | Peroxidase | — | Unknown | Neo (2) |
| AT3G62000 | AT3G61990 | O-methyltransferase | Unknown | Unknown | Neo (2) |
| AT4G26530 | AT4G26520 | Fructose-bisphosphate aldolase | Neo (2) | — | Neo (2); G1 |
| AT5G20940 | AT5G20950 | Glycosyl hydrolase | Unknown | — | Neo (1) |
| AT3G06460 | AT3G06470 | GNS1/SUR4 membrane protein | — | Unknown | Neo (1) |
| AT5G24900 | AT5G24910 | Cytochrome P450 | — | Unknown | Neo (1) |
Note.—MRCA, results from the most recent common ancestral expression pattern analysis; Asymmetric, asymmetric sequence rate analysis; Neo, neofunctionalization; Sub, subfunctionalization; 1, gene 1; 2, gene 2; —, no detection of reciprocal expression; unknown, unable to infer the MRCA expression due to an uncertain phylogenetic tree topology or lack of expression data for most members; ND, not determined because of the lack of enough information such as a small gene family with two or three members; G1, group 1 with both MRCA and asymmetric rate analysis suggesting neofunctionalization; G2, group 2 where MRCA inferred neofunctionalization for both copies and asymmetric rate analysis indicated neofunctionalization for one copy; and G3, group 3 where the MRCA analysis inferred subfunctionalization for both copies and asymmetric rate analysis indicated neofunctionalization for one copy.
Asymmetric Sequence Evolution Is Associated with Asymmetric Expression Divergence
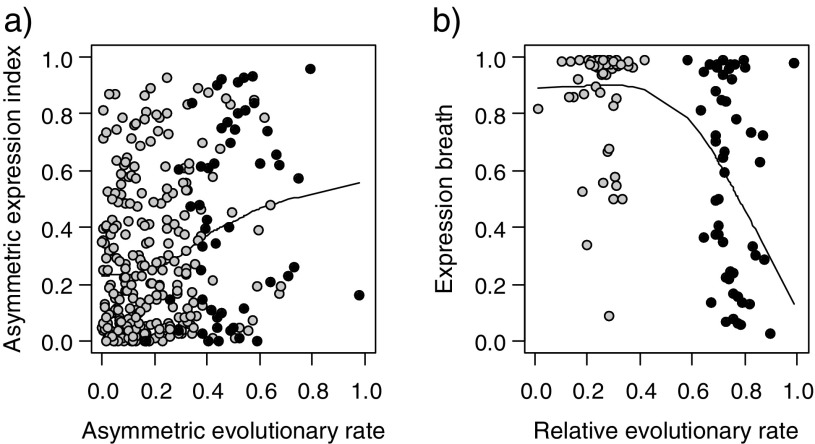
After investigating the frequency of asymmetric rate evolution for those duplicates with reciprocal expression, we conducted an analysis to see if there is any association between asymmetric sequence divergence and expression divergence. We first scored the relative evolutionary rate and asymmetric evolutionary rate among the WG duplicates and the tandem duplicates (for details, see Materials and Methods). We then scored the EB (i.e., how many conditions in which one gene is expressed) from both the ADA and the ARA data sets (for details, see Materials and Methods). We then compared the asymmetric expression index (Asy; i.e., EB difference) between gene duplicates with their asymmetric evolutionary rate as well as the EB and relative evolutionary rate between the accelerated copy and the nonaccelerated copy if gene duplicates showed asymmetric rate evolution. Due to fewer data points among the tandem duplicates, we analyzed data points from the WG duplicates and the tandem duplicates together. We observed that gene duplicates with asymmetric rate evolution have significantly higher Asy values between duplicated genes in comparison to those with symmetric rate evolution (Pearson's correlation test, r = 0.2476, P = 6.929 × 10−6; fig. 6a), suggesting that asymmetric rate evolution is often associated with asymmetric expression divergence. These results are consistent with findings in yeast, where asymmetric expression divergence of duplicated genes is associated with asymmetric protein divergence (Tirosh and Barkai 2007). When comparing EB with relative evolutionary rate, we also found that the copy with an accelerated rate of amino acid replacements (i.e., higher relative evolutionary rate) often showed a lower EB value in comparison to its nonaccelerated duplicated partner (Pearson's correlation test, r = −0.4850, P = 2.392 × 10−7; fig. 6b), suggesting that the copy with accelerated amino acid evolution tends to lose expression across multiple organ types and gain expression in a limited number of organ types.
FIG. 6.—
Asymmetric sequence evolution is associated with asymmetric expression divergence. (a) Scatter plots showing a comparison of the relationship between the asymmetric expression index and asymmetric evolutionary rate (i.e., the difference of relative branch length between two copies) among the WG duplicates and the tandem duplicates. Gray dots indicate the duplicate pairs without asymmetric sequence evolution and black dots indicate the duplicate pairs with asymmetric sequence evolution. The black line represents the local regression fit. (b) Scatter plots showing the relationship between expression breath and relative evolutionary rate (i.e., relative branch length since gene duplication) among the WG duplicates and the tandem duplicates. Gray dots indicate the nonaccelerated copy and black dots indicate the accelerated copy among duplicates with asymmetric sequence evolution. The black line represents the local regression fit using the “lowess” function in R.
Potential Cases of Neofunctionalization Involving Pollen
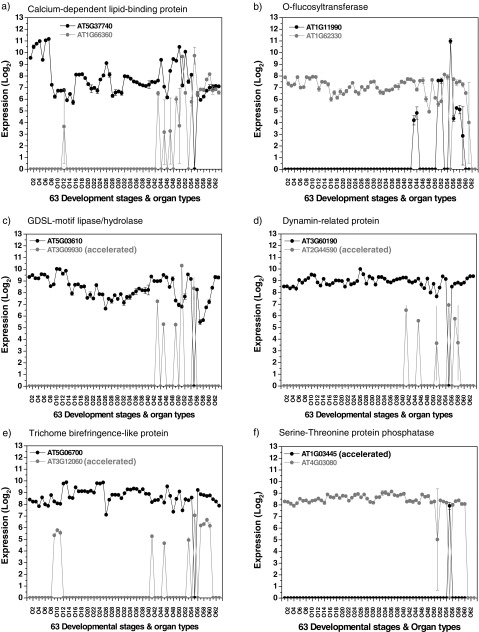
Among the reciprocally expressed WG duplicates, a significantly higher percentage of expression gain was found in pollen than in other organ types. There were 44 gene pairs that showed expression gain in pollen (supplementary table S7, Supplementary Material online). Among them, six pairs of duplicated genes showed especially contrasting reciprocal expression patterns that involved pollen (fig. 7). These six pairs of duplicated genes all showed that one copy has gained expression in pollen; in contrast, its duplicated partner has broad expression across different organ types but no expression in pollen. Four of the gene pairs also showed asymmetric sequence evolution, including GDSL-motif lipase/hydrolase genes (AT5G03610 and AT3G09930), dynamin-related genes (AT3G60190 and AT2G44590), trichome birefringence–like genes (AT5G06700 and AT3G12060), and serine–threonine protein phosphatase genes (AT1G03445 and AT4G03080) (fig. 7c–f). The serine/threonine protein phosphatase genes play important roles in the brassinosteroid signaling pathway (see details below). However, the functions for most of the other gene pairs remain uncharacterized. Dynamin-like proteins have been shown to be involved in pollen tube development (Konopka et al. 2008; Backues et al. 2010), although it is not known if the gene pairs studied here have those functions. The previously reported SSP and BSK1 gene pair (Liu and Adams 2010), which showed pollen-specific reciprocal expression and asymmetric sequence evolution, were not identified in this study because SSP has undergone a subsequent duplication and such genes were excluded from this study. Thus, there may be additional duplicated genes in the A. thaliana genome that show reciprocal expression involving pollen with regulatory neofunctionalization and asymmetric sequence rate evolution.
FIG. 7.—
Reciprocal expression involving pollen. Diagrams showing some striking reciprocally expressed gene duplicates in which one copy showed a restricted expression pattern and gain of expression in pollen (a–f) plus accelerated sequence evolution (c–f). MAS5-normalized microarray gene expression data from 63 different developmental stages and organ types. Absence or presence of expression was determined by using the mas5calls function in Bioconductor. Error bars indicate standard deviations (n = 3). The 63 different developmental stages and organ types are listed in supplementary fig. S1, Supplementary Material online.
Regulatory Neofunctionalization in Pollen of a Pair of Serine–Threonine Protein Phosphatase Genes
One example of regulatory neofunctionalization among the genes showing reciprocal expression in pollen is a pair of serine–threonine protein phosphatase genes, BSU1 (AT1G03445) and BSL1 (AT4G03080). Because the function of BSU1 has been well characterized, we have done experiments and additional analyses to further characterize the duplicated gene pair from an evolutionary perspective. BSU1 operates in the brassinosteroid signal transduction pathway by inactivating BIN2, ultimately allowing for expression of brassinosteroid target genes (Mora-García et al. 2004; Kim et al. 2009; Ryu et al. 2010). BSL1 also has been shown to interact with BIN2, albeit most functional studies were done with BSU1, suggesting that BSL1 probably plays similar role as BSU1 in the brassinosteroid signaling pathway (Kim et al. 2009).
The ADA microarray data indicated that BSU1 was only expressed in pollen, whereas BSL1 was expressed in most organ types but not in pollen (fig. 7f). To validate the expression pattern observed in the microarray data, we then performed RT-PCR, which is more sensitive. BSU1 showed strong expression in mature pollen and very weak expression in roots and whole flowers, whereas BSL1 showed expression in different organ types but not in pollen (fig. 8a). Thus, our RT-PCR assay further supports the reciprocal expression pattern between BSU1 and BSL1observed in the microarray data, indicative of their expression divergence after gene duplication. From the MRCA expression pattern analysis, we inferred that BSU1 acquired expression in pollen (table 1), indicative of regulatory neofunctionalization. To gain further support for regulatory neofunctionalization of BSU1, we conducted RT-PCR expression assays using orthologs from two outgroup species, C. papaya and V. vinifera. In both orthologs, expression was detected in multiple organ types, but no expression was detected in mature pollen (fig. 8a). This result further supports our inference from the MRCA expression analysis that BSL1 reflects the ancestral expression pattern and that BSU1 acquired expression in mature pollen.
In addition to regulatory neofunctionalization, BSU1 has considerably accelerated sequence evolution compared with BSL1 (table 1). To further examine the degree of asymmetric sequence evolution, we conducted a more detailed sequence rate analysis using multiple outgroup species. The Ka/Ks ratio analysis indicated that BSU1 evolved approximately four times faster than BSL1 (LRT, P = 0.002; fig. 8b). Moreover, a positive selection analysis using a branch-site model suggested that several codons in BSU1 underwent positive selection since gene duplication (LRT, P = 0.009; supplementary fig. S7, Supplementary Material online), further supporting the neofunctionalization model.
Collectively, the results from RT-PCR expression assays, MRCA expression analysis, and sequence rate analysis provide evidence to support the inference of neofunctionalization for BSU1 including gain of expression in pollen and accelerated sequence rate evolution compared with BSL1. BSU1 is phosphorylated by BSK1 in the brassinosteroid signal transduction cascade (Kim et al. 2009). BSK1 is not expressed in mature pollen (Liu and Adams 2010) and thus, BSU1 is not likely to be activated by BSK1 in mature pollen. Thus, it is likely that BSU1 has gained a new, as yet uncharacterized, function in pollen, perhaps playing a role in fertilization or another pollen-specific function.
Discussion
Reciprocal Expression Patterns and Regulatory Neofunctionalization Are Common among Duplicated Genes
Our study provides new insights into the evolutionary importance of reciprocal expression patterns (qualitative complementary expression) between duplicated genes in plants. First, reciprocal expression in different organ types, tissues, cell types, and developmental stages is common in both WG duplicates and tandem duplicates in A. thaliana. We have shown that 30–38% of the duplicated genes in Arabidopsis that were examined in this study are reciprocally expressed in different organ types, cell types, and developmental stages. This result contrasts to the results of Duarte et al. (2006) who found only a few cases of reciprocal expression of duplicated genes in A. thaliana among the six organ types that they examined. Considering that our study examined data from 83 different organ types, cell types, and developmental stages, it is not surprising that we found a much higher number of gene pairs with reciprocal expression patterns, as we showed in our simulations. Second, we found that transcription factors are overrepresented among the reciprocally expressed WG duplicates compared with the entire set of WG duplicates, which in itself is overrepresented with transcription factors (Blanc and Wolfe 2004a). Overrepresentation of transcription factors among WG duplicates has been explained by the gene dosage balance hypothesis (reviewed in Edger and Pires 2009; Freeling 2009). We propose that after being initially retained by gene dosage, or other reasons, many WG duplicates that are transcription factors underwent regulatory neofunctionalization that led to functional divergence and long-term preservation. Third, our results indicate that pollen and siliques are the most common structures in which reciprocal expression patterns are observed. The siliques contain both the seeds and seedpods. Had only the seeds been assayed for expression there might have been additional cases of reciprocal expression observed. Likewise, mature pollen contains both the gametophytic sperm cells and the sporophytic pollen coat; assaying only the sperm cells might reveal additional cases of reciprocal expression patterns. We discuss the implications of reciprocal expression in pollen more below.
Fourth, our results indicate that the reciprocal expression patterns of most WG and tandem duplicated gene pairs (of those that could be assessed) appear to result from regulatory neofunctionalization instead of regulatory subfunctionalization. Most previous studies of expression patterns of several hundred duplicated genes in plants were not able to infer neofunctionalization and subfunctionalization in various organ types and developmental stages because there was no attempt to infer the ancestral state of expression. Exceptions were a study by Zou et al. (2009) on stress responsiveness of duplicated genes in A. thaliana, discussed more below, and the study of Duarte et al. (2006) discussed above. Finding evidence for more regulatory neofunctionalization than subfunctionalization is consistent with recent proposals that have de-emphasized the importance of subfunctionalization as a retention mechanism for duplicated genes and instead proposed that subfunctionalization is primarily a gene divergence mechanism (Freeling 2008). Alternatively, regulatory subfunctionalization might be more important soon after formation of duplicated genes, which is not likely to be detected in the data set we analyzed considering that most of the duplicated genes in this study formed millions of years ago (Blanc and Wolfe 2004a, 2004b; Haberer et al. 2004). How common is regulatory neofunctionalization in other eukaryotes? Frequencies of regulatory subfunctionalization or neofunctionalization have been inferred in several different eukaroytes such as yeast (Tirosh and Barkai 2007), Drosophila (Oakley et al. 2006), and mammals (Farré and Albà 2010). In yeast, 45% of duplicated genes have been shown to experience regulatory neofunctionalization (Tirosh and Barkai 2007). In Drosophila, Oakley et al. (2006) inferred that regulatory neofunctionalization (ca. 28%) is more common than regulatory subfunctionalization (ca. 10%). In mammals, Farré and Albà (2010) studied the expression evolution of gene duplicates and found that 23–25% of them showed regulatory subfunctionalization and 42–52% of them were neofunctionalized, suggesting that regulatory neofunctionalization is more prevalent than regulatory subfunctionalization. Our study is consistent with these previous studies conducted in different eukaryotic kingdoms, indicating that regulatory neofunctionalization plays a more important role than regulatory subfunctionalization in the longer term evolutionary retention and divergence of duplicated genes.
Inferring ancestral expression states using ML analyses of gene expression within a gene family in a single species can be done computationally for a large number of genes, given the readily available expression data. Also, the expression data are coming from one species, allowing for unambiguous comparisons between organ types at the exact same developmental stage. However, the ancestral state reconstruction approach may overestimate the number of genes that have undergone neofunctionalization because subsequent changes in expression of other genes in the family after their common ancestor with the duplicate pair in question could lead to an incorrect inference of neofunctionalization. Overall, the inferences we made about the ancestral state of expression, and thus regulatory neofunctionalization, for the reciprocally expressed gene pairs should be regarded as testable hypotheses for the ancestral state of expression rather than a firm assessment of the ancestral state. Additional evidence for neofunctionalization of one copy after gene duplication can come from a combination of evidence for asymmetric sequence rate evolution plus information from functional studies if available. Sixteen gene pairs in this study had both asymmetric sequence rate evolution and an ancestral state expression inference of neofunctionalization (as category group 1 in table 1). In another recent study that used the ancestral state reconstruction approach to study expression patterns of duplicated genes in A. thaliana, Zou et al. (2009) found that the expression patterns in response to nine abiotic stress treatments indicated that a much higher percentage of genes lost stress responsiveness (upregulation or downregulation under stress) than gained stress responsiveness. Their results are consistent with a larger role for regulatory subfunctionalization than neofunctionalization in the evolution of stress responsiveness of duplicated genes. The results of our study contrast with their observations. However, the data sets are different in type (abiotic stresses vs. organs, developmental stages, and cell types). Another difference is that we did not analyze upregulation and downregulation of expression level (i.e., quantitative complementary expression) in this study instead focusing on reciprocal expression patterns (i.e., qualitative complementary expression).
Another factor that could influence our results is that microarrays are not as sensitive in detecting gene expression at very low levels as techniques like RT-PCR, real time PCR, and the use of GUS reporter constructs. Thus, there may be cases where the microarray data indicated that a gene was not expressed in a particular organ type, but a more sensitive detection technique might detect low levels of expression. For example, we showed very weak expression of BSU1 in roots and whole flowers, in contrast to the microarray data, although the expression in flowers could be primarily from the pollen. In addition, BSU1 has been shown to be expressed at very low levels in some organ types, expression that was not detected by RT-PCR but detected only after hybridizing RT-PCR gels with a BSU1 probe, which is even more sensitive than RT-PCR by itself (Mora-García et al. 2004). Likewise, the statistical analyses used to infer presence or absence of expression from microarray data may result in both false negatives (failure to detect expression) and false positives (incorrectly inferring expression). Despite the drawbacks of the microarray data, they provide a very useful data set for examining expression of hundreds of duplicate genes in a large number of organs, tissues, developmental stages, and cell types.
Expression Gain and Accelerated Sequence Evolution in Pollen
Among the reciprocally expressed WG duplicates, a significantly higher percentage of expression gain was found in pollen than in other organ types or developmental stages, including 44 gene pairs that showed gain of expression by one copy in pollen. Thus, pollen greatly contributes to reciprocal expression patterns of WG duplicates. The pollen transcriptome has been shown to be distinctive from the transcriptome in other structures, with many genes expressed specifically in pollen (Becker et al. 2003; Honys and Twell 2003). Expression changes after gene duplication help contribute to the distinctiveness of the pollen transcriptome. A previously reported example of expression and functional change after WG duplication, involving pollen, is the SSP and BSK1 pair (Liu and Adams 2010). SSP, the SHORT SUSPENSOR gene, is only expressed in the sperm cells of pollen, whereas BSK1 is expressed in most organ types but not pollen. Thus, SSP and BSK1 provide an example of expression change after gene duplication that has contributed to the distinctiveness of the pollen transcriptome.
We found that four pairs of duplicated genes that showed striking reciprocal expression patterns in pollen had undergone accelerated sequence evolution. Genes that are expressed in reproductive organs sometimes evolve rapidly or undergo positive evolution (reviewed by Swanson and Vacquier 2002). The rapid evolution of traits that are related to reproductive organs has been considered as an important evolutionary mechanism of speciation (Gavrilets 2000). In plants, a similar trend has been observed in several genes with pollen-specific expression (Fiebig et al. 2004; Schein et al. 2004). The accelerated evolution or positive selection of pollen-specific genes can be driven by pollen competition and sexual conflict (reviewed by Bernasconi et al. 2004). In addition, the accelerated sequence evolution and positive selection can be involved in the interaction during species recognition (Ishimizu et al. 1998) or novel phenotypic effects during pollen development (Matsuno et al. 2009) and embryogenesis (Liu and Adams 2010).
Asymmetric Sequence Rate Evolution and Neofunctionalization of the RecQ4B DNA Helicase Gene: an Example of Functional Divergence
For most of the reciprocally expressed gene pairs, the functions of both genes have not been characterized, and thus, it is not possible to show functional changes after duplication. However, the functions of both copies of a pair of reciprocally expressed DNA helicase genes (AT1G60930 and AT1G10630) have been characterized (Hartung et al. 2007). We showed that the gene pair shows asymmetric sequence rate evolution, with the RecQ4B (AT1G60930) evolving more rapidly (table 1). The products of the two duplicated genes have antagonistic functions, where RecQ4B promotes homologous recombination by stabilizing recombination intermediates, whereas RecQ4A (AT1G10630) suppresses the frequency of recombination (Hartung et al. 2007). In comparison to the functions of RecQ-like genes from other eukaryotes, homologous RecQ genes in human and yeast mainly perform the function of suppressing recombination that is similar to RecQ4A, suggesting that neofunctionalization has occurred in RecQ4B after the gene duplication event within the Brassicaceae. Hartung et al. (2007) favored the subfunctionalization model between RecQ4A and RecQ4B based on the fact that both the promotion and the suppression of recombination were observed in homologous RecQ-like genes in Escherichia coli. However, those antagonistic functions have not been found in any other eukaryote besides A. thaliana, suggesting that recent evolution of the recombination promotion function of RecQ4B occurred after gene duplication. Further supporting our inference of neofunctionalization is our finding of accelerated and asymmetric sequence evolution in RecQ4B. Thus, the function of RecQ4B in promoting recombination likely evolved during the evolution of the Brassicaceae family.
Differences in Expression Evolution between Tandem and WG Duplicates
The results from the comparison of reciprocal expression frequency between WG duplicates and tandem duplicates showed that reciprocal expression has occurred more frequently in tandem duplicates. In addition, the results of our ancestral expression pattern analysis indicate that WG duplicates showed more expression gain than expression loss, whereas tandem duplicates showed more loss than gain. Thus, expression evolution is different between these two different types of duplicates. Casneuf et al. (2006) and Ganko et al. (2007) found that gene duplicates from large scale duplication events (e.g., WG duplicates) largely have highly redundant or overlapping expression patterns and showed less expression divergence than those from small scale duplication events (e.g., tandem duplicates). One possible explanation is due to the difference of gene duplication mechanisms. Tandem duplication is often derived from unequal crossing over (Achaz et al. 2000). Duplication by unequal crossing over can disrupt the promoter and other regulatory regions, whereas that would not occur by WG duplication. Our results further support the idea that tandem duplicates tend to share less regulatory context between each other than WG duplicates, which therefore leads to a more divergent expression pattern and a higher frequency of reciprocal expression. On the other hand, more expression loss than expression gain among the tandem duplicates might be explained by the age of gene duplication. Previous studies have shown that younger duplicated genes tend to loose functions more often than older duplicated genes that often gained new functions (e.g., stress responsiveness in Zou et al. 2009). Our observations are consistent with the subneofunctionalization model, in which expression loss plays an important role at the younger stages of duplicated gene evolution, whereas expression gain plays an important role in expression divergence of older duplicated genes (i.e., the WG duplicates) (He and Zhang 2005, Rastogi and Liberles 2005).
Our study showed that a considerable number of duplicate pairs from both WG duplicates and tandem duplicates are reciprocally expressed. What are possible molecular mechanisms causing reciprocal expression between duplicates genes? One possible mechanism is divergence of cis-regulatory element regions between duplicated genes. In Arabidopsis, Haberer et al. (2004) found that both segmental duplicates and tandem duplicates showed highly similar cis-element regions even though they have high expression divergence, suggesting that minor changes in cis-element regions could lead to regulatory neofunctionalization or subfunctionalization in gene duplicates. Another possible mechanism is unequal crossing over. Because tandemly duplicated genes are often derived from unequal crossing over, it is possible that only part of a cis-element region is duplicated (Achaz et al. 2000), potentially leading to change in expression pattern after gene duplication such as reciprocal expression patterns.
One caveat of studying the evolution of the WG duplicates in A. thaliana is that it remains unknown if the most recent WG duplication in the Arabidopsis lineage (the alpha WG duplication event) originated from autopolyploidization or allopolyploidization. If it was autopolyploidy, the WG duplicates shared the same expression pattern upon the WG duplication event. If it was allopolyploidy, the WG duplicates probably originated from two different species and their expression patterns might not have been the same upon the WG duplication event. That could affect our inferences of the ancestral expression pattern. Previous studies of allopolyploids showed a few cases of reciprocal expression of duplicated genes (homeologs) in different organ and tissue types, but it tends to be at a low level in most of the studied systems (e.g., Adams et al. 2003; Buggs et al. 2010a), in contrast to biased expression of homeologs which is considerably more common.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Angel Pan and Shing Hei Zhan for helping with the simulation and statistical analyses. This study was funded by a grant from the Natural Science and Engineering Research Council (NSERC) of Canada. S.L.L. was partly supported by graduate fellowships from the University of British Columbia and G.J.B. was partly supported by a Biodiversity Research training grant from NSERC.
References
- Achaz G, Coissac E, Viari A, Netter P. Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol Biol Evol. 2000;17:1268–1275. doi: 10.1093/oxfordjournals.molbev.a026410. [DOI] [PubMed] [Google Scholar]
- Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci U S A. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues SK, Korasick DA, Heese A, Bednarek SY. The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell. 2010;22:3218–3231. doi: 10.1105/tpc.110.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20. doi: 10.1093/bioinformatics/btl558. [DOI] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi G, et al. Evolutionary ecology of the prezygotic stage. Science. 2004;303:971–975. doi: 10.1126/science.1092180. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004a;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004b;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottley A, Xia GM, Koebner RMD. Homoeologous gene silencing in hexaploid wheat. Plant J. 2006;47:897–906. doi: 10.1111/j.1365-313X.2006.02841.x. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Buggs RJ, et al. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol Ecol. 2010a;19:132–146. doi: 10.1111/j.1365-294X.2009.04469.x. [DOI] [PubMed] [Google Scholar]
- Buggs RJ, et al. Tissue-specific silencing of homeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 2010b;186:175–183. doi: 10.1111/j.1469-8137.2010.03205.x. [DOI] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. Consistent patterns of rate asymmetry and gene loss indicate widespread neofunctionalization of yeast genes after whole-genome duplication. Genetics. 2007;175:1341–1350. doi: 10.1534/genetics.106.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B, et al. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium) Genetics. 2009;182:503–517. doi: 10.1534/genetics.109.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermuth JP, Hahn MW. The life and death of gene families. Bioessays. 2009;31:23–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- Drea SC, Lao NT, Wolfe KH, Kavanagh TA. Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. Plant J. 2006;46:723–735. doi: 10.1111/j.1365-313X.2006.02741.x. [DOI] [PubMed] [Google Scholar]
- Duarte JM, et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Farré D, Albà MM. Heterogeneous patterns of gene-expression diversification in mammalian gene duplicates. Mol Biol Evol. 2010;27:325–335. doi: 10.1093/molbev/msp242. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.69. 2009. Distributed by the author. Seattle (WA): Department of Genetics, University of Washington. [Google Scholar]
- Fiebig A, Kimport R, Preuss D. Comparisons of pollen coat genes across Brassicaceae species reveal rapid evolution by repeat expansion and diversification. Proc Natl Acad Sci U S A. 2004;101:3286–3291. doi: 10.1073/pnas.0305448101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KM. Bayesian reconstruction of ancestral expression of the LEA gene families reveals propagule-derived desiccation tolerance in resurrection plants. Am J Bot. 2008;95:506–515. doi: 10.3732/ajb.95.4.506. [DOI] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 2008;4:28–40. doi: 10.1159/000126004. [DOI] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ. Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol. 2007;24:2298–2309. doi: 10.1093/molbev/msm158. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. Rapid evolution of reproductive barriers driven by sexual conflict. Nature. 2000;403:886–889. doi: 10.1038/35002564. [DOI] [PubMed] [Google Scholar]
- Gentleman L, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. Statistical framework for phylogenomic analysis of gene family expression profiles. Genetics. 2004;167:531–542. doi: 10.1534/genetics.167.1.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc Natl Acad Sci U S A. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Li WH, Chen ZJ. External factors accelerate expression divergence between duplicate genes. Trends Genet. 2007;23:162–166. doi: 10.1016/j.tig.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KF. Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 2004;136:3009–3022. doi: 10.1104/pp.104.046466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2007;104:18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 2003;132:640–652. doi: 10.1104/pp.103.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 2006;7:R31. doi: 10.1186/gb-2006-7-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, et al. Identification of regions in which positive selection may operate in S-RNase of Rosaceae: implication for S-allele-specific recognition sites in S-RNase. FEBS Lett. 1998;440:337–342. doi: 10.1016/s0014-5793(98)01470-7. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kim TW, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Backues SK, Bednarek SY. Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell. 2008;20:1363–1380. doi: 10.1105/tpc.108.059428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. Expression pattern divergence of duplicated genes in rice. BMC Bioinformatics. 2009 doi: 10.1186/1471-2105-10-S6-S8. 10(6 Suppl):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Adams KL. Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol Biol Evol. 2010;27:2817–2828. doi: 10.1093/molbev/msq169. [DOI] [PubMed] [Google Scholar]
- Matsuno M, et al. Evolution of a novel phenolic pathway for pollen development. Science. 2009;325:1688–1692. doi: 10.1126/science.1174095. [DOI] [PubMed] [Google Scholar]
- Mora-García S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley TH, Ostman B, Wilson AC. Repression and loss of gene expression outpaces activation and gain in recently duplicated fly genes. Proc Natl Acad Sci U S A. 2006;103:11637–11641. doi: 10.1073/pnas.0600750103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst Biol. 1999;48:612–622. [Google Scholar]
- Proost S, et al. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;14:28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Hwang I. Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells. 2010;29:291–296. doi: 10.1007/s10059-010-0034-y. [DOI] [PubMed] [Google Scholar]
- Schein M, Yang Z, Mitchell-Olds T, Schmid KJ. Rapid evolution of a pollen-specific oleosin-like gene family from Arabidopsis thaliana and closely related species. Mol Biol Evol. 2004;21:659–669. doi: 10.1093/molbev/msh059. [DOI] [PubMed] [Google Scholar]
- Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 2011;108:4069–4074. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17:505–512. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sterck L, et al. EST data suggest that poplar is an ancient polyploidy. New Phytol. 2005;167:165–170. doi: 10.1111/j.1469-8137.2005.01378.x. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers J, Wang X, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci U S A. 2010;107:472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throude M, et al. Structure and expression analysis of rice paleo duplications. Nucleic Acids Res. 2009;37:1248–1259. doi: 10.1093/nar/gkn1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielson R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielson R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH. Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 2009;5:e1000581. doi: 10.1371/journal.pgen.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]