Background: A schizophrenia risk DISC1 polymorphism (S704C) shows phenotypic variations in hippocampus structure and associated memory defects.
Results: DISC1 forms octamers via dimers as building blocks. S704C forms higher-order oligomers, without affecting its affinity with NDEL1.
Conclusion: Schizophrenia risk DISC1-S704C polymorphism has altered oligomeric assembly.
Significance: The improper oligomeric assembly of DISC1-S704C provides a clue in understanding the molecular basis of the known phenotype.
Keywords: Analytical Biochemistry, Biophysics, Protein Assembly, Protein-Protein Interactions, Schizophrenia, DISC1, NDEL1
Abstract
DISC1 (Disrupted in schizophrenia-1) plays essential roles in neuronal proliferation, neuronal migration and axon guidance and has been implicated in schizophrenia and related psychiatric disorders. DISC1 forms a functional complex with nuclear distribution element-like protein-1 (NDEL1), a key component that regulates microtubule organization during cell division and neuronal migration. DISC1 polymorphisms at the binding interface of DISC1-NDEL1 complex have been implicated in schizophrenia. However, it is unknown how schizophrenia risk polymorphisms perturb its interaction with NDEL1 and how they change the inherent biochemical properties of DISC1. Here, we characterize the oligomerization and binding property of DISC1 and its natural schizophrenia risk variant, S704C. Our results show that DISC1 forms octamers via dimers as building blocks and directly interacts with tetramers of NDEL1. The schizophrenia risk variant S704C affects the formation of octamers of DISC1 and exhibits higher-order self-oligomerization. However, the observed formation of new oligomeric species did not influence its binding with NDEL1. These results suggest that the improper oligomeric assembly of DISC1-S704C may underlie the observed phenotypic variation due to the polymorphism.
Introduction
DISC1 (Disrupted in schizophrenia-1) is a gene implicated in schizophrenia and bipolar disorder. Recently, DISC1 has emerged as a key molecular player involved in orchestrating several neurodevelopmental signaling pathways (1–4). Multiple studies have linked specific genetic variants such as a C-terminal truncation of DISC1 and two single-nucleotide polymorphisms (SNPs)2 of DISC1 (L607F, S704C) in schizophrenia phenotypes (5, 6). DISC1 has several interaction partners to regulate key neuronal functions such as neural progenitor proliferation, neuronal migration, and neurite outgrowth (5). Among the DISC1-interacting proteins, NDEL1 (nuclear distribution element-like protein-1) has been identified consistently as a necessary binding partner of DISC1 in migrating cortical neurons during neurogenesis (5, 7). Schizophrenic risk allelic polymorphisms are located at the NDEL1 binding region of DISC1, suggesting probable structural perturbation at the binding site (6) and hence the disturbance in neuronal migration complex assembly.
Among the three domains of DISC1 (a coiled-coil C-terminal, globular N-terminal, and a self-associating core domain (1)), the C-terminal region, which harbors a dimerization domain (8), is considered necessary for neuronal migration complex assembly. Accordingly, a transgenic mouse model expressing the construct of a dominant-negative form of DISC1 (lacking the C terminus) has been shown to exhibit characteristic schizophrenia symptoms (9).
One of the natural schizophrenia risk DISC1 polymorphisms, S704C (10, 11), shown to affect the function of prefrontal cortex, is also localized in its C-terminal domain (12, 13). This SNP has been observed to function at the molecular level, as DISC1-S704C affects the localization of key PCM1 (pericentriolar material protein-1) to the centrosome (14). Furthermore, the DISC1-S704C has been shown to preferentially bind NDE1, an NDEL1 homolog specifically expressed in earlier stages of corticogenesis in progenitor cells (15, 16). This SNP shows a great variation in phenotype; however, its effect at a biochemical level in the disease state is unclear.
In addition to the C-terminal domain, the DISC1 self-associating core domain, facilitates self-assembly and localizes DISC1 into centrosome (17). This domain has been shown to be indispensible for neuronal migration during cortical development (18). These studies emphasize the role of C-terminal and self-association domains in functional assembly of the neuronal migration complex and perturbation of this assembly in disease. However, whether DISC1 SNP alters the self-association state and modulates its binding with NDEL1 is unknown.
In this current work, we show that DISC1 self-assembles into octamers using dimers as building blocks, where as NDEL1 exists as stable tetramers. The octamers of DISC1 interact with tetramers of NDEL1, and this interaction between DISC1 and NDEL1 appears to require secondary structural elements rather than the quaternary structural assembly. The S704C allelic variant exhibits higher-order self-oligomerization, without modulating its binding to NDEL1 as compared with wild-type. This suggests that the observed difference in phenotype with the DISC1-S704C variant may be due to the formation of new oligomeric species, rather than the difference in binding to the NDEL1 protein. Based on our results, we propose models for self-association of DISC1 and its interaction.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Bacterial Expression of Target Proteins
Genes of full-length DISC1 and full-length NDEL1 were synthesized (GeneArt) for constitutive expression in E. coli. The codon-optimized DISC1 gene was further PCR-amplified and subcloned into the pMal-c4E vector containing the tobacco etch virus protease cleavage site. The DISC1-S704C construct was created using site-directed mutagenesis strategy. The NDEL1 gene was also amplified and inserted into a pET vector containing the protein G domain (GB1) and tobacco etch virus cleavage site. Fragments of DISC1 and NDEL1 were PCR-amplified and subcloned into either the pET24b or in-house GB1 fusion vectors. The constructs used for this study are listed in supplemental Table S1.
The full-length DISC1 and NDEL1 constructs were co-transformed with chaperone plasmid pTf6 (Takara) in BL21(Star) (Invitrogen) strain. The inoculated cultures were grown in the presence of 0.4% l-arabinose (for full-length DISC1/NDEL1 constructs). 0.3% of glucose was introduced in the LB culture for the maltose-binding protein (MBP) fusion constructs. The cultures were grown at 30 °C up to an OD of 0.9 and were then induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. All other constructs were transformed in the Rosetta2(DE3) strain. Cultures were grown at 37 °C before induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. After induction, all cultures were allowed to grow at 23 °C for an additional 12 h before harvesting.
Purification of Target Proteins
Harvested cells were subjected to sonication to isolate the soluble target proteins in the lysis buffer (50 mm Tris, 400 mm NaCl, 10 mm MgCl2, pH 8). Full-length MBP-DISC1 proteins (both wild-type/S704C) were fractioned from cell lysates using 65% ammonium sulfate. MBP-DISC1 was further purified by affinity using amylose resin (New England Biolabs). The fractioned MBP-DISC1 proteins were resuspended in binding buffer (50 mm Tris, 200 mm NaCl, 10 mm MgCl2, 10% glycerol, 0.2% Nonidet P-40, pH 8) and incubated with amylose resin. After 4 h of incubation at 4 °C, the resins were washed with binding buffer, wash buffer 1 (50 mm Tris, 500 mm Na2SO4, 10 mm MgCl2, pH 8) and wash buffer 2 (WB2: 50 mm Tris, 150 mm KCl, 10 mm MgCl2, 5 mm ATP, pH 8). Full-length proteins were eluted in maltose elution buffer (50 mm Tris, 100 mm NaCl, 5 mm MgCl2, 0.5 mm DTT, 25 mm maltose, pH 8.0). The eluted proteins were concentrated and diluted to a final salt concentration of 50 mm. MBP-DISC1 proteins were further purified using ion-exchange chromatography. Eluted MBP-DISC1 proteins were applied to a Resource-Q 6-ml column, washed with three column volumes of buffer A1 (25 mm Tris, 30 mm NaCl, pH 8) and eluted with buffer B1 (25 mMTris, 750 mm NaCl, pH 8 under linear gradient of 30–70% of buffer B1). Bound proteins were eluted at 400–600 mm NaCl. The eluted proteins were diluted to 100 mm NaCl, and the maltose binding protein was either cleaved off using tobacco etch virus protease (in house) or kept intact before being subjected to size exclusion chromatography using a Superdex-200 column (GE Healthcare). Full-length DISC1/MBP-DISC1 proteins were eluted in 25 mm Tris, 200 mm NaCl, 5 mm MgCl2, 0.5 mm (tris(2-carboxyethyl)phosphine (TCEP), pH 8. Full-length NDEL1 and DISC1-CTF proteins were affinity purified using nickel-nitrilotriacetic acid beads (Qiagen). Cells were lysed as described above. For sonication, 50 mm Tris-HCl, 500 mm NaCl, 5 mm MgCl2, 0.5% Nonidet P-40, 20 mm imidazole at pH 7.4 was used as a buffer. The cleared lysates were incubated with nickel-nitrilotriacetic acid beads for 3 h at 4 °C. The beads were washed with lysis buffer followed by wash buffer 1 and wash buffer 2. Proteins were eluted in nickel elution buffer (50 mm Tris-HCl, 200 mm NaCl, 500 mm imidazole, pH 8). Full-length NDEL1 and DISC1-CTF proteins were further concentrated and dialyzed in buffer A1. Proteins were purified in the Resource-Q column as described above. These proteins were eluted in 300 mm and 250 mm NaCl, respectively. Proteins were further purified by size-exclusion chromatography as described above. The concentration of the purified proteins was estimated using Nanodrop spectrophotometer (Thermoscientific).
Pulldown Assay and Size Exclusion Chromatography for Interaction Studies
Pulldown assays were performed using MBP constructs of DISC1 (DISC1/DISC1-CTF/DISC1-CTF-(680–804)/DISC1-NTF) as baits. 200 μl of cleared lysates containing ∼0.5–1 μg of DISC1 and NDEL1 (NDEL1, NDEL1-CTF) proteins were incubated with 50 μl of the amylose beads. The competition experiment was performed by competing with untagged DISC1 protein for NDEL1 protein binding. The control experiments were designed using no MBP-tagged DISC1 proteins as bait to rule-out nonspecific interactions. 0.5–1 μg of GB1/His7-tagged DISC1-CTF proteins were used as competitors. The assay volume was adjusted to 800 μl using lysis buffer. After incubating the pulldown reactions for 3 h at 4 °C, the beads were washed extensively as described above. The proteins were incubated with 100 μl of SDS-PAGE sample buffer containing 0.25% SDS at 30 °C. The samples were spun down before being loaded onto a SDS-PAGE gel. Co-purification of proteins were performed using purified DISC1 (full-length DISC1/DISC1-CTF) and purified NDEL1 as samples. DISC1 and NDEL1 were co-incubated at a 1:1 ratio before injection into precalibrated Superdex 200 column. The calibration was performed using thyroglobulin (∼700 kDa), aldolase (∼150 kDa), and ovalbumin (∼43 kDa). The elution profiles of the complex were compared with the individual proteins. The eluted peak was analyzed by SDS-PAGE to confirm the complex formation.
CD Spectroscopy
3 μm of DISC1 and 5 μm of NDEL1 samples in 20 mm phosphate, 100 mm NaCl, pH 7.4, were used for far-UV CD spectroscopy experiments using an AVIV 62DS spectropolarimeter equipped with temperature control peltier unit. All spectra were measured five times, averaged and buffer corrected. Spectra were measured from 194–245 nm under constant bandwidth of 1.5 nm slit mode with the scan step of 1 nm and averaging time of 1 s at 25 °C.
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were performed using a VP-ITC microcal calorimeter (Microcal) at 18 °C. 20 μm of wild-type DISC1 (1.4 ml) in 25 mm Tris-HCl, 200 mm NaCl, 5 mm MgCl2, 0.5 mm TCEP, pH 8, were titrated against 300 μm NDEL1 in the same buffer. Titrations were performed by adding 10-μl injections of NDEL1. Heat of dilution of NDEL1 was also measured using above experimental conditions. The isothermal titration calorimetry data were analyzed using the manufacture supplied Origin program (version 5.0).
Equilibrium Analytical Ultracentrifugation
Equilibrium sedimentation experiments were performed using a Beckman XL-I ultracentrifuge equipped with a Beckman 50-Ti rotor and six sector cells at three different concentrations. 110 μl of samples were centrifuged at 5000, 7000, and 9000 rpm for DISC1 proteins and at 5000, 10000, and 15000 rpm for NDEL1. Samples were equilibrated for 48 h in a respective speed setting at 4 deg. Scans were taken every 12 h, and data were buffer-corrected. Data were analyzed using Ultrascan. Data fitting was performed using a molecular weight distribution model and non-linear curve fitting algorithms. The statistically best-fit molecular weights were further subjected to Monte Carlo analysis to obtain the distribution of oligomers.
Surface Plasmon Resonance Experiments
Surface plasmon resonance measurements were performed on Biacore3000 (GE Healthcare). GB1-NDEL1 was covalently linked to CM5 chips (GE Healthcare) as per the manufacturer's instructions. GB1-NDEL1 was immobilized to CM5 chip at a response unit of 650. Wild-type MBP-DISC1 and MBP-DISC1-S704C were injected at the flow rate of 10 μl/min at 25 °C. To assess the binding affinity between the variants of MBP-DISC1 and GB1-NDEL1, a change in response units were compared at two different concentrations (10 μm and 2 μm) of MBP-DISC1 variants with CM5 chip immobilized GB1-NDEL1. Data were analyzed using Scrubber (BioLogic) software.
RESULTS
Full-length Recombinant DISC1 and NDEL1 Are Folded Properly with High Helical Content
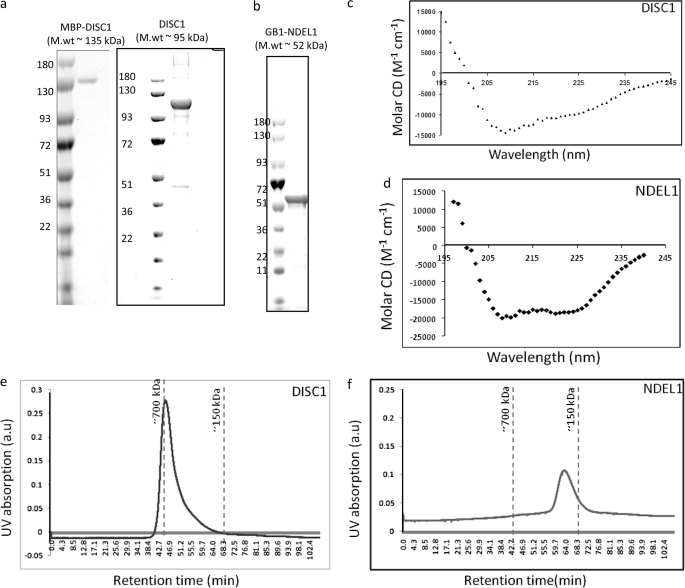
Biochemical insights into how full-length DISC1-NDEL1 complexes form and how the natural DISC1 variants affect its inherent properties are unknown. To this end, we aimed to purify the full-length DISC1 and NDEL1 proteins. To overcome the difficulties associated with the expression of full-length DISC1 (8, 19, 20), we first optimized the DISC1 and NDEL1 genes for bacterial expression by removing the mRNA secondary structural elements proximal to both the ribosomal binding and putative internal ribosomal binding sites (21). To facilitate correct folding of the expressed proteins, we co-expressed a trigger factor protein (TF16) in conjunction with our proteins of interest. These improvements dramatically increased protein production, and our optimized protocols further ensured purification to homogeneity (Fig. 1, a and b).
FIGURE 1.
Expression and purification of DISC1 and NDEL1. DISC1 and NDEL1 were co-expressed with a trigger factor in a BL21(Star) strain. Homogenously purified DISC1 with MBP (molecular mass ∼135 kDa, without MBP (molecular mass ∼95 kDa) (a) and GB1-NDEL1 (molecular mass ∼52 kDa) (b) are shown in the SDS-PAGE gel. Folding of DISC1 and NDEL1 was assessed using circular dichroism spectroscopy. Both DISC1 (c) and NDEL1 (d) show significant helical content. Size-exclusion chromatography suggests DISC1 exists as a higher-order oligomers (> hexamer) (e), whereas the NDEL1 profile suggests the protein is a tetramer (f). a.u., arbitrary unit.
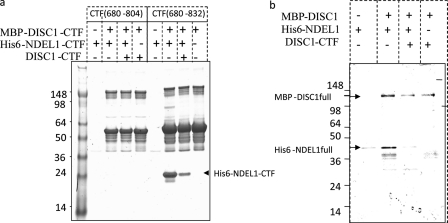
To test whether these purified proteins are folded properly, we used CD spectroscopy to analyze for secondary structural elements in DISC1 and NDEL1 (22, 23). The observed absorption minima both at 208 nm and 222 nm indicate substantial content of α helices, a characteristic of coiled-coil proteins (Fig. 1, c and d). Next, we analyzed whether these recombinant proteins could self-assemble into oligomers. Using a suitable size-exclusion column, we found that both DISC1 and NDEL1 form higher-order oligomers (Fig. 1, e and f). To investigate the direct interaction between DISC1 and NDEL1, we performed competitive pulldown experiments. For the binding studies, we used MBP and protein G-domain (GB1) as N-terminal fusion partners for DISC1 and NDEL1, respectively. The presence of MBP or GB1 at the N-terminal region neither influence self-oligomerization (supplemental Fig. S1, a and b) nor binding to NDEL1 (supplemental Fig. S1c). Secondary structure prediction algorithms have shown consistently that the NDEL1 binding site on DISC1 (DISC1 (804–834)) is helical in nature (supplemental Fig. S2). We deleted this 28-amino acid region from the C terminus of DISC1 (amino acids 804–832) similar to what was done in a yeast two-hybrid study (24). This deletion completely abolished NDEL1 binding to our recombinant DISC1, confirming this short stretch of helical region is necessary the NDEL1 binding (Fig. 2a).
FIGURE 2.
Pulldown assays further confirm that the specificity of the DISC1/NDEL1 interaction is mapped to a key helical region on the C terminus of DISC1. a, to map the NDEL1 binding region on recombinant DISC1, two DISC1 constructs (MBP-DISC1-(680–802) and MBP-DISC1-(680–832) were made based on the previous yeast two-hybrid studies (24). These expressed DISC1 proteins were subjected to pulldown assay using NDEL1-CTF (shown in an arrow). MBP-DISC1-CTF-(680–802), which lacks the crucial helical region (802–832), could not pull down NDEL1. b, proper folding of full-length DISC1 was confirmed using MBP-DISC1 as a bait to pull down full-length NDEL1. The interaction was being competed using C-terminal region of DISC1.
To demonstrate a direct interaction between full-length DISC1 and NDEL1, we performed a pulldown assay using full-length DISC1 and full-length NDEL1. We showed that DISC1 specifically interacts with NDEL1 and that this interaction can be disrupted competitively by the NDEL1 binding domain of DISC1 (Fig. 2b). These results indicate a direct interaction between full-length DISC1 and NDEL1.
DISC1 Assembles into Octamers via Dimers, Whereas NDEL1 Predominantly Exists as Tetramers
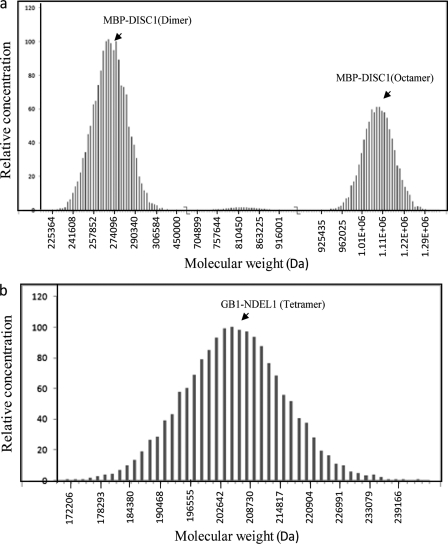
Several in vivo studies suggested that both the self-association domain and the C-terminal domain of DISC1 assist in formation of functional DISC1 oligomers (17, 18). However, how full-length DISC1self-assembles into oligomers and the nature of DISC1/NDEL1 oligomers are unknown. We used equilibrium analytical ultracentrifugation to analyze the distribution of oligomers of full-length DISC1 and NDEL1. Sedimentation equilibrium experiments have been established as an ideal method to study the protein oligomerization process under steady-state conditions in which the derived molecular mass is independent of the shape of the molecules (25). Experiments were performed at various concentrations of MBP-DISC1 and GB1-NDEL1 as well as various centrifugal speeds to extract accurate molecular weight distribution. The results of Monte Carlo fitting of the obtained data suggest that, under steady-state conditions, MBP-DISC1 (monomer molecular mass ∼ 135 kDa) exists in two populations: a dimer (molecular mass ∼270 kDa) and a higher-order oligomer (molecular mass ∼ 1.1 MDa), which corresponds to its octameric state (Fig. 3a, supplemental Fig. S3). Interestingly, GB1-NDEL1 (monomer molecular mass ∼ 50 kDa) exists predominantly as a tetramer (molecular mass ∼ 200 kDa) (Fig. 3b and supplemental Fig. S4). The results support that full-length DISC1 exist in equilibrium between octamers and dimers, whereas NDEL1 exists predominantly as a tetrameric species. The DISC1 dimer species could be seen in steady-state condition by sedimentation equilibrium, suggesting the oligomerization assembly occur via dimers.
FIGURE 3.
Distribution of oligomers in wild-type DISC1 and NDEL1. a, analytical ultracentrifugation data were fitted using the Monte Carlo simulation method using Ultrascan software. Wild-type DISC1 has been fitted to two oligomeric populations, a dimer (∼270 kDa) and an octamer (∼1.1 MDa). b, Monte Carlo data fitting of analytical ultracentrifugation data on NDEL1 predicts the existence of a unique tetramer population (∼200 kDa).
Higher-order Oligomers of DISC1 Are Dispensable for NDEL1 Interaction
The C-terminal region of DISC1 (amino acids 598–834) has been shown deleted upon chromosomal translocation in schizophrenia patients (26). This region contains a putative dimerization domain and has been studied for NDEL1 binding (8, 20, 24, 26). Here, we characterize the inherent binding affinity between the full-length DISC1 and NDEL1 using purified proteins under native conditions to explore the biochemistry of the interactions.
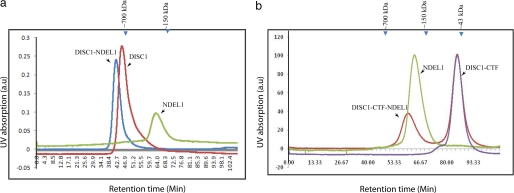
We first performed size-exclusion chromatography experiments on complexes of DISC1-NDEL1 and DISC1-CTF/NDEL1 to answer whether DISC1 or NDEL1 oligomers need to be dissociated prior to complex formation. We co-incubated full-length DISC1 and NDEL1 at a 1:1 ratio and subjected the complex to size-exclusion chromatography. The homo-oligomers of DISC1 were not dissociated by the NDEL1 interaction (Fig. 4a, supplemental Fig. S5a). Notably, due to the size of the DISC1-NDEL1 complex (∼900 kDa), the co-elution occurred at the void-volume of the column. However, a control experiment with the DISC1 (320–834)-NDEL1 complex, which forms the complex within the resolution limit, reaffirms that no dissociation of DISC1 oligomers is needed for the interaction (supplemental Fig. S5b). Likewise, the NDEL1 oligomer was not disrupted by interaction with an excess of DISC1-CTF, a binding domain of NDEL1 (Fig. 4b). These experiments confirm that the interactions between DISC1 and NDEL1 are mediated through oligomers. However, both DISC1-CTF dimers and full-length DISC1 octamers form a stable complex with NDEL1, suggesting state of oligomerization of DISC1 is dispensable for the NDEL1 complex formation. Altogether, we conclude that the secondary structural elements rather than oligomeric assembly define the NDEL1-DISC1 complex formation.
FIGURE 4.
Size-exclusion chromatography confirms the DISC1/NDEL1 oligomers did not dissociate during the interaction. Size-exclusion chromatography of full-length DISC1 and full-length NDEL1 complex characterization (left). A chromatogram of the full-length DISC1-NDEL1 complex (blue), DISC1 alone (red), and NDEL1 alone (green) reassure that the full-length DISC1 or NDEL1 and DISC1-NDEL1 form higher-order oligomers as consistent with analytical ultracentrifugation data (Fig. 3). right, DISC1-CTF and full-length NDEL1 complex characterization. Shown is a chromatogram of the DISC1-CTF-NDEL1 complex (red), DISC1-CTF alone (purple), and NDEL1 alone (green). A shift in the retention time indicates a shift toward higher molecular weight upon direct binding of DISC1/DISC1-CTF to NDEL1. No change in oligomerization of DISC1 and/or NDEL1 was observed. Purified components were used in a pre-calibrated Superdex-200 column. a.u., arbitrary unit.
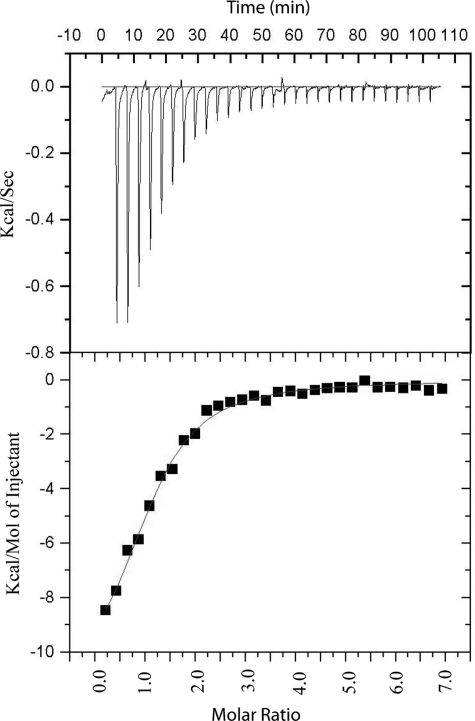
We performed isothermal titration calorimetry experiments to determine the binding affinity between full-length DISC1 and full-length NDEL1. We observed that the DISC1 octamer and NDEL1 tetramer form a 1:1 complex, with a binding constant of 2.3 μm (Fig. 5). The heat of dilution of NDEL1 did not contribute to the heat of the reaction (supplemental Fig. S6). This biophysical experiment formally proves that the DISC1 interacts directly with NDEL1 and assembles as a hetero complex.
FIGURE 5.
Isothermal titration calorimetry experiments confirm the direct binding of DISC1/NDEL1 with a binding constant of 2.3 μm.
Natural DISC1 SNP (S704C) Does Not Modulate NDEL1 Binding but Forms Higher-order Oligomers
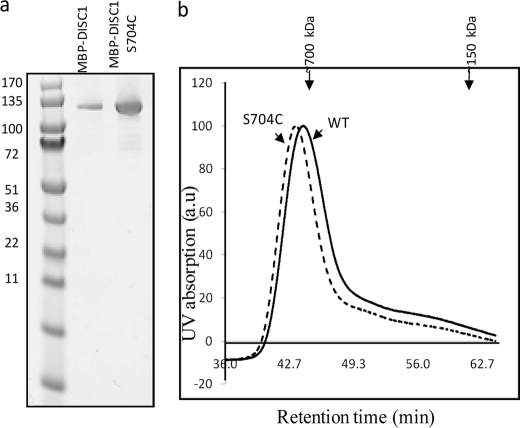
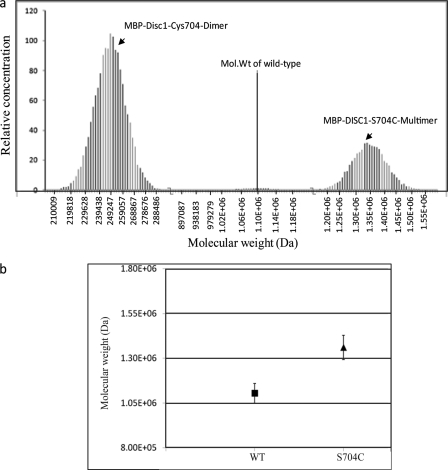
Next, we addressed, whether the natural schizophrenia risk variant DISC1-S704C affects the self-oligomerization and/or binding of DISC1 to its binding partner NDEL1. To study this, we expressed and purified full-length MBP-DISC1-S704C (Fig. 6a). Size-exclusion chromatography experiments on purified wild-type DISC1 and S704C variant (Fig. 6a) showed a subtle difference between the size of the oligomers (Fig. 6b), suggesting that these two variants differ in terms of size of the oligomers under steady-state. We further characterized the oligomerization of DISC1-S704C under equilibrium conditions using analytical ultracentrifugation. To our surprise, the SNP at the dimerization domain still did not affect the formation of the dimers (Fig. 7a and supplemental Fig. S7). However, instead of octamers, a new higher-order oligomeric species was populated corresponding to a size greater than 9-mer (Fig. 7a). The size of the higher-order oligomers of DISC1-S704C is quantitatively higher than that of wild-type protein (1.3 MDa) (Fig. 7b). This result suggests that the natural schizophrenia SNP at the dimerization domain directly affects self-assembly and populates a new higher-order oligomeric species of DISC1.
FIGURE 6.
Size-exclusion chromatography experiments of wild-type DISC1 and DISC1-S704C suggest DISC1-S704C forms a higher molecular weight oligomeric species compared with wild-type. a, SDS-PAGE gel shows homogenous purification of MBP-DISC1 and S704C variant. b, size-exclusion chromatography of wild-type DISC1 (continuous line) and DISC1S704C variant (dashed line).
FIGURE 7.
Distribution of oligomers with the MBP-DISC1-S704C variant. Analytical ultracentrifugation data of MBP-DISC1-S704C were fitted using Monte Carlo simulation method (Ultrascan software). The data have been fitted to two oligomeric populations, a dimer (∼240 kDa) and an oligomer that is higher than a nonamer (∼1.3 MDa). The observed molecular weight of wild-type DISC1 is indicated by the dotted line.
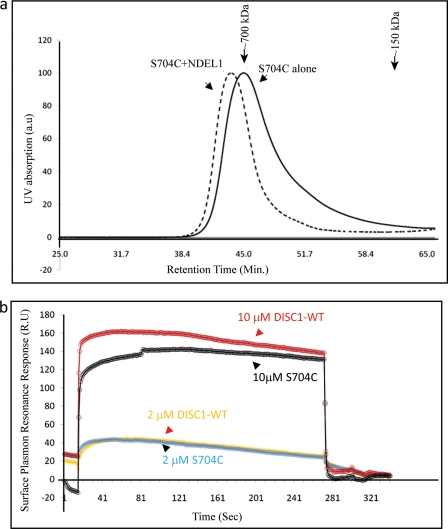
To assess whether the formation of new higher-order oligomeric species has any effect on the binding to NDEL1, we first performed size-exclusion chromatography for the equimolar complex of DISC1-S704C-NDEL1. We observed that the DISC1-S704C forms a stable complex with NDEL1, as judged by a shift in the retention time for the complex as compared with DISC1-S704C (Fig. 8a). To understand the affinity of DISC1-S704C to NDEL1, we performed surface plasmon resonance experiments. With GB1-NDEL1 immobilized on the chip, we compared the change in plasmon resonance response units upon binding either wild-type MBP-DISC1 or the S704C variant. Titrating with 2 μm DISC1 variants against immobilized NDEL1, both variants show identical binding affinity with respect to NDEL1. At a high concentration of DISC1 variants (10 μm), the S704C retains >90% of the binding affinity of the wild-type. The observed response units of the DISC1-S704C and NDEL1 interaction indicates that the variant still forms a complex with NDEL1, regardless of its oligomerization state (Fig. 8b). However, we cannot rule out effects of this SNP on other binding partners of DISC1. We conclude that the reported phenotypic changes in DISC1-S704C variant link to the formation of new higher-order oligomeric species of DISC1-S704C rather than a difference in NDEL1 binding.
FIGURE 8.
The DISC1-S704C variant shifts the molecular weight (Mol. Wt) of DISC1-S704C-NDEL1 into higher-order oligomers without affecting the binding affinity with NDEL1. a, size-exclusion chromatography profile of full-length DISC1-S704C alone (solid line) and the full-length DISC1-S704C-NDEL1 complex suggest a stable complex formation between DISC1-S704C and NDEL1. b, surface plasmon resonance experiments of the interactions between NDEL1 and wild-type DISC1 or DISC-S704C. Experiments were performed using the immobilized GB1-NDEL1 and titrated against wild-type DISC1 (red) and DISC1-S704C (green) at two different concentrations (2 and 10 μm). No major difference in NDEL1 binding between of two DISC1 variants, as both yield similar response units (RU).
DISCUSSION
Among schizophrenia candidate genes, DISC1 is associated consistently. Although its impact on disease pathogenesis is debated (10, 27), its connection to schizophrenia is credible (28), and its importance in corticogenesis is widely accepted (3, 17, 29). Likewise, evidence (30, 31) associating NDEL1 to schizophrenia biology is debated (32). However, the crucial role of NDEL1, together with DISC1 in cortical layer formation (17, 29), is well established. It is important to know what molecular mechanisms underlie the DISC1-associated schizophrenia phenotype.
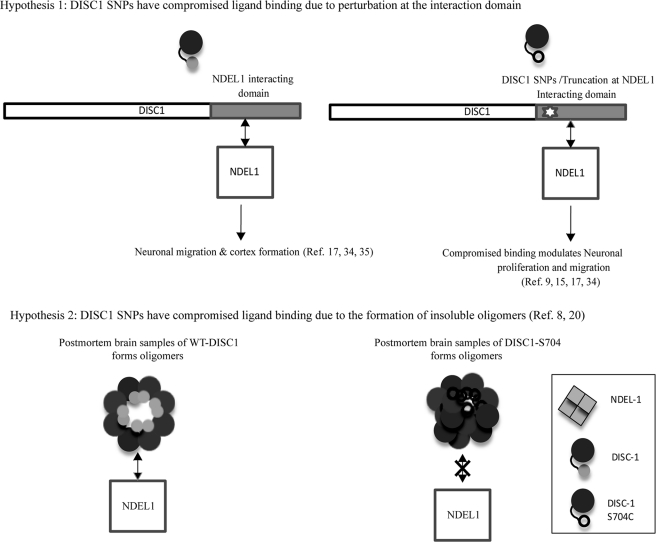
Two different models currently are proposed as molecular mechanisms of DISC1-associated schizophrenia phenotypes. The first hypothesis (Fig. 9a) proposed a compromised NDEL1 binding of schizophrenia risk DISC1 variants as a potential perturbation for cerebral cortex development (15, 17). This is supported by the following facts. 1) DISC1 schizophrenia risk SNPs are located within the NDEL1 binding region of DISC1. 2) This NDEL1 binding region on DISC1 acts as a dominant negative during corticogenesis (6, 17). These findings suggest a perturbation at the direct binding to NDEL1. Recently, DISC1 has been shown to act as a molecular switch from progenitor proliferation to migration in the developing cortex upon phosphorylation at C-terminal domain (S710) (29). DISC1-S704C shows a stronger binding affinity toward NDE1, linking the potential schizophrenia phenotype via change in DISC1-NDEL1 binding affinity (15). In summary, the hypothesis 1 suggests that the schizophrenia risk SNP/truncation leads to compromised NDEL1 binding and hence perturbs the corticogenesis. This hypothesis is further supported by a study that correlates the cortical thickness to the cognition in schizophrenia (33).
FIGURE 9.
Proposed mechanisms of DISC1-S704C-associated schizophrenia phenotype. a, hypothesis 1: schematic diagram of DISC1 primary sequence and its potential NDEL1 binding site (left). Schizophrenia risk DISC1 SNPs are located at the NDEL1 binding site, suggesting a probable perturbation at the DISC1-S704C-NDEL1 interactions (right). b, hypothesis 2: oligomerization condition of WT DISC1 extracted from the postmortem brain control sample (left) and DISC1-S704C (right) in the disease sample. Insoluble oligomerization condition of DISC1-S704C and a compromised NDEL1 binding were proposed due to the disease risk polymorphism.
The second hypothesis (Fig. 9b) suggests that insolubility of DISC1 and its multimerization is part of the molecular mechanism by which DISC1-S704C leads to schizophrenia (20). The study identified insoluble DISC1 aggregates in the post-mortem samples of schizophrenia patients (20) and further showed the formation of insoluble multimers of a truncated form of DISC1 in disease-associated polymorphism (8). In summary, this hypothesis proposed the formation of DISC1-insoluble aggregates as a potential molecular mechanism of the DISC1-associated phenotype and further proposed an effect on NDEL1 binding affinity as a probable cause for the DISC1-associated schizophrenia pathology (20). Interestingly, disease-associated DISC1 polymorphisms are located in the dimerization domain (C terminus) of DISC1 and not in the self-association domain (5, 8). It is intriguing to know how the SNPs in the DISC1 dimerization domain influence the oligomerization state of DISC1.
In the current study, we report the biochemical characterization of full-length DISC1-NDEL1 complexes, using purified components of full-length DISC1 and NDEL1. In particular, we focused on the following biochemical questions. What is the nature of oligomerization of full-length DISC1 or NDEL1 at equilibrium? How does a schizophrenia risk SNP, DISC1-S704C differ from that of wild-type DISC1 in terms of oligomerization and binding to NDEL1?
Our experimental results show that a schizophrenia risk DISC1 SNP (S704C) changes the quaternary structural assembly of DISC1 without modulating its binding to NDEL1. We therefore rule out the change in NDEL1 binding affinity with DISC1-S704C as a potential contributing factor in to the DISC1-associated phenotype. We identify a change in the oligomerization state of DISC1-S704C as a potential molecular mechanism for the observed phenotype.
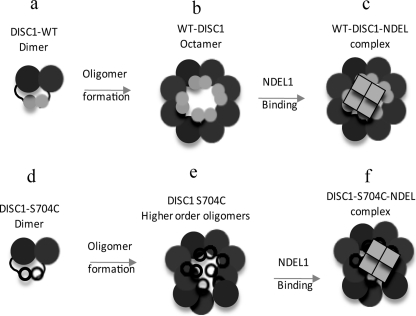
Based on our results, we propose a DISC1 oligomerization and interaction model (Fig. 10). In this biochemical model, the C-terminal dimerization domain (8) serves as a crucial building block for the formation of DISC1 oligomers (Fig 10, a and b). Any change in amino acid sequence or other disruption of this region could result in defects in the formation of proper oligomers. This is supported by our findings that the DISC1-S704C polymorphism has no influence on the formation of dimers (Fig 10d), but instead, it populates a new higher-order oligomer (Fig 10e).
FIGURE 10.
Our working model for DISC1 self-association and NDEL1 binding. Model for full-length DISC-1 self-association and NDEL1 interactions. In our model, we propose that the dimers (a) of DISC1 act as building blocks to assemble DISC1 octamers (b). The DISC1-S704C SNP near the NDEL1 binding site does not prevent the formation of DISC1-S704C dimer (d) and leads to improper oligomerization (e). Furthermore, irrespective of the nature of oligomerization, NDEL1 interacts with octamer (wild-type DISC1) (c) and higher-order oligomers (DISC1 S704C) of DISC1 (f), suggesting that the nature of oligomeric assembly is dispensable for NDEL1 binding.
Furthermore, we show here that the DISC1/NDEL1 interaction is governed by secondary structural elements in DISC1 rather than interfaces from oligomeric assembly. Regardless of the nature of the oligomerization, wild-type (Fig 10b) as well as the S704C variant (Fig 10e) and a truncated DISC1 product (Fig. 4b) form stable complexes with NDEL1 (Fig 10, c and f). Hence, our in vitro study suggests ruling out altered NDEL1 binding as a cause for the DISC1-associated phenotype and support the hypothesis linking insolubility of DISC1-S704C to the phenotype.
It would be important to know the structural mechanism beneath the formation of new oligomeric species, how these new DISC1 oligomeric species turn into insoluble DISC1 in schizophrenia pathogenesis, and also downstream consequences of the formation of improper oligomers of DISC1-NDEL1 complexes. The answers to these questions await further structural and functional studies.
In conclusion, this is the first detailed biophysical study on full-length DISC1 and NDEL1. Based on our characterization, we identify improper oligomeric assembly as a link to the observed DISC1-S704C phenotype.
Supplementary Material
Acknowledgments
We thank Professor Akira Sawa (Johns Hopkins University) for providing constructs, Dr. Tracy Young-Pearse (Brigham and Women's Hospital), Professor Dennis Selkoe (Brigham and Women's Hospital), and Dr. Jay Gopalakrishnan (Harvard Medical School) for helpful discussions, and Ms. Julia Klukowski (Brigham and Women's Hospital) and Dr. Angela Toms (Dana-Farber Cancer Institute) for technical support.
This work was supported, in whole or in part, by the National Institutes of Health Grant P01-GM047467. M. S. W. acknowledges the funding from the Foundation for Neurologic Diseases.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S7.
- SNP
- single nucleotide polymorphism
- MBP
- maltose-binding protein
- NDEL1
- nuclear distribution element-like 1
- CTF
- C-terminal fragment
- NTF
- N-terminal fragment.
REFERENCES
- 1. Brandon N. J., Millar J. K., Korth C., Sive H., Singh K. K., Sawa A. (2009) J. Neurosci. 29, 12768–12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mackie S., Millar J. K., Porteous D. J. (2007) Curr. Opin. Neurobiol. 17, 95–102 [DOI] [PubMed] [Google Scholar]
- 3. Mao Y., Ge X., Frank C. L., Madison J. M., Koehler A. N., Doud M. K., Tassa C., Berry E. M., Soda T., Singh K. K., Biechele T., Petryshen T. L., Moon R. T., Haggarty S. J., Tsai L. H. (2009) Cell 136, 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Millar J. K., Pickard B. S., Mackie S., James R., Christie S., Buchanan S. R., Malloy M. P., Chubb J. E., Huston E., Baillie G. S., Thomson P. A., Hill E. V., Brandon N. J., Rain J. C., Camargo L. M., Whiting P. J., Houslay M. D., Blackwood D. H., Muir W. J., Porteous D. J. (2005) Science 310, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 5. Ross C. A., Margolis R. L., Reading S. A., Pletnikov M., Coyle J. T. (2006) Neuron 52, 139–153 [DOI] [PubMed] [Google Scholar]
- 6. Sawa A., Snyder S. H. (2005) Science 310, 1128–1129 [DOI] [PubMed] [Google Scholar]
- 7. Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., Tsai L. H. (2004) Neuron 44, 263–277 [DOI] [PubMed] [Google Scholar]
- 8. Leliveld S. R., Hendriks P., Michel M., Sajnani G., Bader V., Trossbach S., Prikulis I., Hartmann R., Jonas E., Willbold D., Requena J. R., Korth C. (2009) Biochemistry 48, 7746–7755 [DOI] [PubMed] [Google Scholar]
- 9. Hikida T., Jaaro-Peled H., Seshadri S., Oishi K., Hookway C., Kong S., Wu D., Xue R., Andradé M., Tankou S., Mori S., Gallagher M., Ishizuka K., Pletnikov M., Kida S., Sawa A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14501–14506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hennah W., Thomson P., McQuillin A., Bass N., Loukola A., Anjorin A., Blackwood D., Curtis D., Deary I. J., Harris S. E., Isometsä E. T., Lawrence J., Lönnqvist J., Muir W., Palotie A., Partonen T., Paunio T., Pylkkö E., Robinson M., Soronen P., Suominen K., Suvisaari J., Thirumalai S., St Clair D., Gurling H., Peltonen L., Porteous D. (2009) Mol. Psychiatry 14, 865–873 [DOI] [PubMed] [Google Scholar]
- 11. Hashimoto R., Numakawa T., Ohnishi T., Kumamaru E., Yagasaki Y., Ishimoto T., Mori T., Nemoto K., Adachi N., Izumi A., Chiba S., Noguchi H., Suzuki T., Iwata N., Ozaki N., Taguchi T., Kamiya A., Kosuga A., Tatsumi M., Kamijima K., Weinberger D. R., Sawa A., Kunugi H. (2006) Hum. Mol. Genet. 15, 3024–3033 [DOI] [PubMed] [Google Scholar]
- 12. Di Giorgio A., Blasi G., Sambataro F., Rampino A., Papazacharias A., Gambi F., Romano R., Caforio G., Rizzo M., Latorre V., Popolizio T., Kolachana B., Callicott J. H., Nardini M., Weinberger D. R., Bertolino A. (2008) Eur. J. Neurosci. 28, 2129–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Callicott J. H., Straub R. E., Pezawas L., Egan M. F., Mattay V. S., Hariri A. R., Verchinski B. A., Meyer-Lindenberg A., Balkissoon R., Kolachana B., Goldberg T. E., Weinberger D. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8627–8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eastwood S. L., Walker M., Hyde T. M., Kleinman J. E., Harrison P. J. (2010) Hum. Mol. Genet. 19, 2487–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burdick K. E., Kamiya A., Hodgkinson C. A., Lencz T., DeRosse P., Ishizuka K., Elashvili S., Arai H., Goldman D., Sawa A., Malhotra A. K. (2008) Hum. Mol. Genet. 17, 2462–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Y., Walsh C. A. (2004) Neuron 44, 279–293 [DOI] [PubMed] [Google Scholar]
- 17. Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., Ross C. A., Hatten M. E., Nakajima K., Sawa A. (2005) Nat. Cell Biol. 7, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 18. Young-Pearse T. L., Suth S., Luth E. S., Sawa A., Selkoe D. J. (2010) J. Neurosci. 30, 10431–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taya S., Shinoda T., Tsuboi D., Asaki J., Nagai K., Hikita T., Kuroda S., Kuroda K., Shimizu M., Hirotsune S., Iwamatsu A., Kaibuchi K. (2007) J. Neurosci. 27, 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leliveld S. R., Bader V., Hendriks P., Prikulis I., Sajnani G., Requena J. R., Korth C. (2008) J. Neurosci. 28, 3839–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kudla G., Murray A. W., Tollervey D., Plotkin J. B. (2009) Science 324, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derewenda U., Tarricone C., Choi W. C., Cooper D. R., Lukasik S., Perrina F., Tripathy A., Kim M. H., Cafiso D. S., Musacchio A., Derewenda Z. S. (2007) Structure 15, 1467–1481 [DOI] [PubMed] [Google Scholar]
- 23. Brandon N. J., Schurov I., Camargo L. M., Handford E. J., Duran-Jimeniz B., Hunt P., Millar J. K., Porteous D. J., Shearman M. S., Whiting P. J. (2005) Mol. Cell Neurosci. 28, 613–624 [DOI] [PubMed] [Google Scholar]
- 24. Ozeki Y., Tomoda T., Kleiderlein J., Kamiya A., Bord L., Fujii K., Okawa M., Yamada N., Hatten M. E., Snyder S. H., Ross C. A., Sawa A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taylor I. A., Eccleston J. F., Rittinger K. (2004) Methods Mol. Biol. 261, 119–136 [DOI] [PubMed] [Google Scholar]
- 26. Brandon N. J., Handford E. J., Schurov I., Rain J. C., Pelling M., Duran-Jimeniz B., Camargo L. M., Oliver K. R., Beher D., Shearman M. S., Whiting P. J. (2004) Mol. Cell Neurosci. 25, 42–55 [DOI] [PubMed] [Google Scholar]
- 27. Mathieson I., Munafo M. R., Flint J. (2011) Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chubb J. E., Bradshaw N. J., Soares D. C., Porteous D. J., Millar J. K. (2008) Mol. Psychiatry 13, 36–64 [DOI] [PubMed] [Google Scholar]
- 29. Ishizuka K., Kamiya A., Oh E. C., Kanki H., Seshadri S., Robinson J. F., Murdoch H., Dunlop A. J., Kubo K., Furukori K., Huang B., Zeledon M., Hayashi-Takagi A., Okano H., Nakajima K., Houslay M. D., Katsanis N., Sawa A. (2011) Nature 473, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicodemus K. K., Callicott J. H., Higier R. G., Luna A., Nixon D. C., Lipska B. K., Vakkalanka R., Giegling I., Rujescu D., St Clair D., Muglia P., Shugart Y. Y., Weinberger D. R. (2010) Hum. Genet. 127, 441–452 [DOI] [PubMed] [Google Scholar]
- 31. Tomppo L., Hennah W., Lahermo P., Loukola A., Tuulio-Henriksson A., Suvisaari J., Partonen T., Ekelund J., Lönnqvist J., Peltonen L. (2009) Biol. Psychiatry 65, 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kähler A. K., Djurovic S., Kulle B., Jönsson E. G., Agartz I., Hall H., Opjordsmoen S., Jakobsen K. D., Hansen T., Melle I., Werge T., Steen V. M., Andreassen O. A. (2008) Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 1089–1100 [DOI] [PubMed] [Google Scholar]
- 33. Bakken T. E., Bloss C. S., Roddey J. C., Joyner A. H., Rimol L. M., Djurovic S., Melle I., Sundet K., Agartz I., Andreassen O. A., Dale A. M., Schork N. J. (2011) Arch. Gen. Psychiatry 68, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamiya A., Tomoda T., Chang J., Takaki M., Zhan C., Morita M., Cascio M. B., Elashvili S., Koizumi H., Takanezawa Y., Dickerson F., Yolken R., Arai H., Sawa A. (2006) Hum. Mol. Genet. 15, 3313–3323 [DOI] [PubMed] [Google Scholar]
- 35. Singh K. K., Ge X., Mao Y., Drane L., Meletis K., Samuels B. A., Tsai L. H. (2010) Neuron 67, 33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.