Background: Extracellular ubiquitin functions as a CXC chemokine receptor (CXCR) 4 agonist.
Results: Ubiquitin possesses distinct receptor binding and signaling sites and CXCR4 contains separate binding sites for its natural ligands.
Conclusion: Ubiquitin mimics the structure-function relationship of chemokines and interacts with CXCR4 through a ligand specific binding site on the receptor.
Significance: Agonist-selective pharmacological targeting of CXCR4 appears possible.
Keywords: Computational Biology, Cxcr4, G Protein-coupled Receptors (GPCR), Protein Domains, Ubiquitin, Extracellular Ubiquitin, RosettaDock, Ligand-Receptor Interaction Sites, Protein-Protein Docking, Stromal Cell-derived Factor-1 alpha
Abstract
Ubiquitin, a post-translational protein modifier inside the cell, functions as a CXC chemokine receptor (CXCR) 4 agonist outside the cell. However, the structural determinants of the interaction between extracellular ubiquitin and CXCR4 remain unknown. Utilizing C-terminal truncated ubiquitin and ubiquitin mutants, in which surface residues that are known to interact with ubiquitin binding domains in interacting proteins are mutated (Phe-4, Leu-8, Ile-44, Asp-58, Val-70), we provide evidence that the ubiquitin-CXCR4 interaction follows a two-site binding mechanism in which the hydrophobic surfaces surrounding Phe-4 and Val-70 are important for receptor binding, whereas the flexible C terminus facilitates receptor activation. Based on these findings and the available crystal structures, we then modeled the ubiquitin-CXCR4 interface with the RosettaDock software followed by small manual adjustments, which were guided by charge complementarity and anticipation of a conformational switch of CXCR4 upon activation. This model suggests three residues of CXCR4 (Phe-29, Phe-189, Lys-271) as potential interaction sites. Binding studies with HEK293 cells overexpressing wild type and CXCR4 after site-directed mutagenesis confirm that these residues are important for ubiquitin binding but that they do not contribute to the binding of stromal cell-derived factor 1α. Our findings suggest that the structural determinants of the CXCR4 agonist activity of ubiquitin mimic the typical structure-function relationship of chemokines. Furthermore, we provide evidence for separate and specific ligand binding sites on CXCR4. As exogenous ubiquitin has been shown to possess therapeutic potential, our findings are expected to facilitate the structure-based design of new compounds with ubiquitin-mimetic actions on CXCR4.
Introduction
Ubiquitin is a constitutively expressed protein in all eukaryotic cells and also a natural plasma protein, which is detectable in increased concentrations during various disease processes (1). Although ubiquitin functions as a post-translational protein modifier inside the cell (2), it possesses CXC chemokine receptor 4 (CXCR4)2 agonist activity when it is present outside the cell (3).
CXCR4 fulfills important biological functions during development and hematopoiesis. It also plays pleiotropic roles in the immune system and during tissue repair processes and gained particular attention as a drug target due to its role in HIV infection and metastatic diseases (4–12). Thus, identification of the molecular events leading to CXCR4 activation is of biological and pharmacological importance.
Administration of the cognate ligand of CXCR4, stromal cell-derived factor-1α (SDF-1α; chemokine (CXC motif) ligand 12) and of ubiquitin has been shown to result in anti-inflammatory and organ protective effects in various disease models (13–22). Despite these similarities, we showed previously that ubiquitin, unlike SDF-1α, does not reduce HIV-1 infectivity (23). These differential effects of the natural CXCR4 ligands led to the assumption that ubiquitin and SDF-1α may not share the same interaction sites on CXCR4. Accordingly, we provided evidence that the ubiquitin-CXCR4 interaction does not follow the typical two-site binding mechanism of chemokine-receptor interactions, including the SDF-1α-CXCR4 interaction, in which the receptor N terminus is important for ligand binding (23–30). However, the structural determinants of the ubiquitin-CXCR4 interaction remain unknown. Here, we provide evidence that ubiquitin contains separate receptor docking and activation sites, and thus, resembles the typical structure-function relationship of chemokines. Based on these data, we predicted the ubiquitin-CXCR4 interface utilizing the RosettaDock software followed by manual docking adjustments to account for charge complementarity and an anticipated conformational switch of CXCR4 during the transition from the inactive to the active state. This computational model suggests three amino acid residues as potential ubiquitin interaction sites on CXCR4. Receptor binding experiments after site-directed mutagenesis of CXCR4 confirm that these CXCR4 residues are important for the interaction with ubiquitin but that they do not contribute to the binding of SDF-1α. These data suggest that the natural ligands bind and activate CXCR4 through separate binding sites on the receptor, which may enable a selective pharmacological targeting of CXCR4.
EXPERIMENTAL PROCEDURES
Proteins and Reagents
Ubiquitin, BSA, and forskolin were purchased from Sigma. N-terminal FITC-labeled ubiquitin (FITC-ubiquitin), ubiquitin mutants in which Phe-4 (F4A), Leu-8 (L8A), Ile-44 (I44A), Asp-58 (D58A), Val-70 (V70A), or Gly-75 and Gly-76 (Ub-(1–74)AA) are mutated to Ala, a C-terminal ubiquitin deletion mutant that lacks Gly-75 and Gly-76 (Ub-(1–74)), linear ubiquitin chains of variable lengths (linear dimer-decamer, Ub2–10) and the Lys-48-linked ubiquitin dimer were obtained from Boston Biochem. Recombinant human SDF-1α was obtained from PeproTech. FITC-labeled SDF-1α (FITC-SDF-1α) was prepared using a FITC labeling kit (Pierce) according to the manufacturer's instructions.
HA-tagged CXCR4 Transfections
DNA encoding HA-tagged WT CXCR4 was as described previously (23, 31). DNA encoding HA-tagged CXCR4 in which Phe-29 (F29A), Phe-189 (F189A), or Lys-271 (K271A) are mutated to alanine was obtained from GenScript. The DNA sequences were verified by dideoxy sequencing. DNA encoding each tagged G protein-coupled receptor and empty vector (pcDNA3) was transiently transfected into HEK293 cells grown on 10-cm tissue culture dishes using TransIT-LT1 transfection reagent (Mirus Bio), according to the manufacturer's recommendation. Forty-eight hours later, cells were harvested and used for Western blotting, flow cytometry, and ubiquitin binding assays.
Ubiquitin Binding Assays
Ubiquitin binding assays were performed with HEK293 and THP-1 cells, as described (3, 23). In brief, cells were washed with ice-cold PBS, and 105 cells were suspended in 100 μl of cold (4 °C) PBS, 1% BSA, 0.01% sodium azide. FITC-ubiquitin was added and incubated for 1 min at 4 °C. Cells were washed twice, and the fluorescence intensities were measured (λexcitation/emission: 485/528 nm). Nonspecific binding was assessed as binding of FITC-ubiquitin in the presence of 300 μm native ubiquitin.
Calcium Assay
Intracellular calcium was measured using the Fluo-4 NW calcium assay kit (Molecular Probes), as described (3, 23).
cAMP Assay
Quantitative determination of cAMP levels was performed in forskolin- (5 μm, 10 min, 37 °C) treated cells using the cAMP complete enzyme immunoassay kit (Enzo Life Sciences), acetylated format, as described (3, 23).
Western Blots
Western blotting was performed as described (3, 23, 32). Mouse monoclonal anti-HA (Covance) in combination with anti-mouse IgG horseradish peroxidase- (HRP) linked whole antibody (GE Healthcare) were used for detection of HA-tagged CXCR4 WT and mutants. Mouse anti-GAPDH (Applied Biosystems) was used in combination with anti-mouse IgG HRP-linked whole antibody (GE Healthcare) as a protein loading control.
FACS Analyses
FACS was used to analyze cell surface expression of HA-tagged CXCR4 WT and mutants and to assess binding of FITC-ubiquitin and FITC-SDF-1α, as described (3, 23). For the analyses of WT and mutant HA-tagged CXCR4, cells were labeled with monoclonal mouse anti-HA in combination with anti-mouse Alexa Fluor 488 goat IgG (Invitrogen). Rabbit IgG (R&D Systems) in combination with FITC-conjugated anti-rabbit goat IgG (Abcam) was used as a negative control. The fluorescence intensities of at least 3 × 104 cells were recorded and analyzed using the FlowJo software (Tree Star).
Protein-Protein Docking
Ubiquitin (Protein Data Bank (PDB) ID: 1UBQ (33)) was docked onto the extracellular domain of CXCR4 (PDB ID: 3ODU (27)) automatically using the software RosettaDock 3.2.1 (34, 35) or was docked manually. For protein-protein docking using RosettaDock, ubiquitin was brought within 3 Å distance to the extracellular domain of CXCR4. The docking was performed by fixing CXCR4 and randomizing ubiquitin. The possible ubiquitin position was sampled 5,000–10,000 times each cycle by a 3–5 Å translation and 8° rotation. The best model from each cycle was used as the initial model for the subsequent cycle of Rosetta docking. The docking ended when the minimal change in root mean square deviation to the original coordinate of ubiquitin could achieve the lowest energy minimum. For manual docking, ubiquitin was docked onto the extracellular domain of CXCR4 using the PyMOL software. Such docking was guided by the charge complementarity of the positively charged region of ubiquitin to the negatively charged extracellular domain of CXCR4 and based on the results obtained from binding experiments with Ub-1–74, ubiquitin-F4A, and ubiquitin-V70A.
Statistics
Data are expressed as mean ± S.E. from duplicate to quadruplicate measurements of n independent experiments that were performed on different days. Data were analyzed using the GraphPad Prism 5 software. For the calculation of the Ki, the following parameters and constraints were used: concentration of labeled ligand = 1163 nm (10 μg/ml); Kd of labeled ligand equals 100 nm (3); upper plateau constant equal to 100%.
RESULTS AND DISCUSSION
Many intracellular proteins bind to ubiquitin through various ubiquitin binding domains, which interact with a number of recently identified binding loci on ubiquitin (36–38). To assess whether similar surface regions on ubiquitin are also involved in the interaction between ubiquitin and CXCR4, we tested the receptor binding and activation properties of the ubiquitin point mutants F4A, L8A, I44A, D58A, and V70A (Fig. 1A). Leu-8, Ile-44, and Val-70 are located on the hydrophobic surface patch on ubiquitin near the C-terminal end of β strand 5, the most common interaction site on ubiquitin (37, 38). Although Phe-4 represents a hydrophobic interaction area distinct from the hydrophobic patch centered around Ile-44 (39), Asp-58 represents a hydrophilic interaction surface (40).
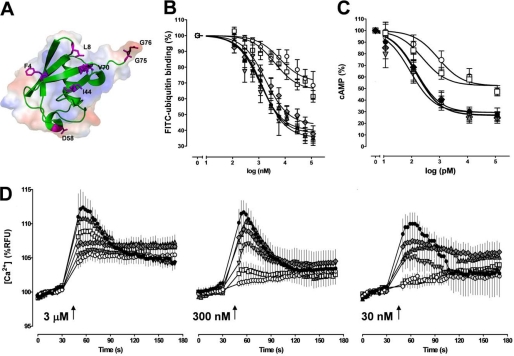
FIGURE 1.
Phe-4 and Val-70 of ubiquitin are important for receptor binding and activation. A, ribbon diagram (green) with underlying surface representation of ubiquitin. The mutated or truncated residues are highlighted in magenta. B, competition binding (1 min, 4 °C) curves for WT ubiquitin (black circle) and the mutants F4A (white square), L8A (gray upright triangle), I44A (gray inverted triangle), D58A (gray diamond), and V70A (white circle) with 1.16 μm FITC-ubiquitin (n = 4–6). FITC-ubiquitin binding is expressed as the percentage of the fluorescence signal measured in the absence of unlabeled ubiquitin (= 100%). C, effects of WT ubiquitin (black circle) and the mutants F4A (white square), L8A (gray upright triangle), I44A (gray inverted triangle), D58A (gray diamond), and V70A (white circle) on cAMP levels in forskolin-stimulated THP-1 cells (n = 7). Cells were incubated with ubiquitin WT and mutants for 15 min at 37 °C and 5% CO2. Data are expressed as the percentage of untreated cells (= 100%). D, intracellular Ca2+ flux in THP-1 cells after stimulation with WT ubiquitin (black circle) and the mutants F4A (white square), L8A (gray upright triangle), I44A (gray inverted triangle), D58A (gray diamond), and V70A (white circle). Arrows indicate the time point when ubiquitin/mutants were added at the given concentrations (n = 4–6). RFU, relative fluorescence units.
In competition binding experiments with FITC-ubiquitin, L8A, I44A, and D58A were indistinguishable from WT ubiquitin (WT, Ki, 110 ± 1 nm; bottom plateau, 35 ± 2%; L8A, Ki, 94 ± 1 nm; bottom plateau, 38 ± 2%; I44A, Ki, 87 ± 1.3 nm; bottom plateau, 39 ± 4%; D58A, Ki, 144 ± 1.2 nm; bottom plateau, 42 ± 3%). In contrast, the abilities of F4A and V70A to compete with FITC-ubiquitin for receptor binding were clearly reduced, as compared with WT ubiquitin (F4A, Ki, 318 ± 1.3 nm; bottom plateau, 65 ± 3%; V70A, Ki, 496 ± 2 nm; bottom plateau, 71 ± 4%; Fig. 1B).
To determine whether the reduced binding properties of F4A and V70A are also accompanied by attenuated activation of CXCR4, we compared the effects of WT ubiquitin and the ubiquitin mutants on intracellular cAMP levels (Fig. 1C) and Ca2+ fluxes (Fig. 1D). WT ubiquitin, L8A, I44A, and D58 resulted in a comparable reduction of cAMP levels in forskolin-stimulated THP-1 cells (EC50, 89 ± 1.5 to 144 ± 1.2 pm; bottom plateau, 27 ± 4% to 29 ± 5%; p > 0.05 versus WT ubiquitin for all mutants), whereas F4A and V70A showed diminished effects on cellular cAMP concentrations (F4A, EC50, 246 ± 1.6 pm; bottom plateau, 52 ± 4%, p < 0.01 versus WT ubiquitin; V70A, EC50, 899 ± 2 pm; bottom plateau, 52 ± 5%, p < 0.01 versus WT ubiquitin). Similar to the effects on cAMP concentrations, WT ubiquitin, L8A, I44A, and D58 promoted intracellular Ca2+ fluxes in THP-1 cells when tested in concentrations of 3 μm, 300 nm, and 30 nm. In contrast, stimulation of THP-1 cells with F4A and V70A failed to promote Ca2+ fluxes at 30 and 300 nm, respectively (Fig. 1D). These data suggest that the ubiquitin surface residues F4A and V70A are required to establish a functionally relevant interaction between ubiquitin and CXCR4.
As the C terminus of ubiquitin is essential for intracellular functions of ubiquitin (2), we next tested whether deletion or Ala mutation of the C-terminal diglycine of ubiquitin (Fig. 1A) influences receptor binding and activation properties. As shown in Fig. 2A, Ub-(1–74) and Ub-(1–74)AA were able to compete with FITC-ubiquitin binding to THP-1 cells to the same degree as WT ubiquitin. Despite the identical binding properties, Ub-(1–74)AA reduced cAMP to the same degree as WT ubiquitin, whereas Ub-(1–74) showed a 26-fold higher EC50 value (5421 ± 2 pm) than WT ubiquitin (207 ± 1 pm) and a bottom plateau of 70 ± 4.4% in dose-response experiments (bottom plateau for WT ubiquitin, 38 ± 2%, Fig. 2B). Similarly, Ub-(1–74) displayed a reduced ability to promote intracellular Ca2+ fluxes when compared with WT ubiquitin and Ub-(1–74)AA in parallel experiments (Fig. 2C). This suggests that the presence of the C-terminal tail of ubiquitin facilitates receptor activation.
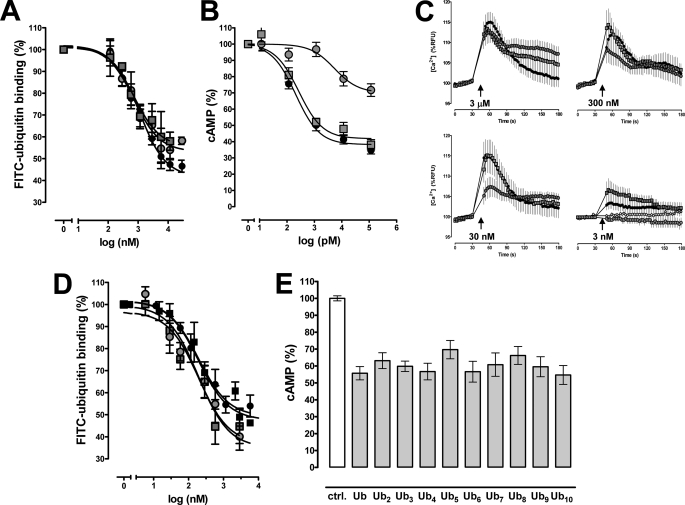
FIGURE 2.
The ubiquitin C terminus facilitates receptor activation. A, competition binding (1 min, 4 °C) curves for WT ubiquitin (black circle), ubiquitin-(1–74) (gray circle) and ubiquitin-(1–74)AA (gray square) with 1.16 μm FITC-ubiquitin (n = 5). FITC-ubiquitin binding is expressed as the percentage of the fluorescence signal measured in the absence of unlabeled ubiquitin (= 100%). B, effects of WT ubiquitin (black circle), ubiquitin-(1–74) (gray circle), and ubiquitin-(1–74)AA (gray square) on cAMP levels in forskolin-stimulated THP-1 cells (n = 7–10). Cells were incubated with ubiquitin WT and mutants for 15 min at 37 °C and 5% CO2. Data are expressed as the percentage of untreated cells (= 100%). C, intracellular Ca2+ flux in THP-1 cells after stimulation with WT ubiquitin (black circle), ubiquitin-(1–74) (gray circle), ubiquitin-(1–74)AA (gray square), and vehicle (white circle). Arrows indicate the time point when ubiquitin/mutants were added at the given concentrations (n = 4–6). RFU, relative fluorescence units. D, competition binding (1 min, 4 °C) curves for WT ubiquitin (black circle), linear diubiquitin (gray square), Lys-48-linked diubiquitin (gray circle), and linear tetraubiquitin (black square) with 1.16 μm FITC-ubiquitin (n = 5). FITC-ubiquitin binding is expressed as the percentage of the fluorescence signal measured in the absence of unlabeled ubiquitin (= 100%). E, effects of ubiquitin (Ub) and linear ubiquitin chains (Ub2–10) on cAMP levels in forskolin-stimulated THP-1 cells (n = 4). Cells were incubated with 100 nm ubiquitin/ubiquitin chains for 15 min at 37 °C and 5% CO2. Experiments with 1 and 10 nm ubiquitin/ubiquitin chains showed identical results (not shown). Data are expressed as the percentage of untreated cells (= 100%; control (ctrl.)).
Our finding that Phe-4 and Val-70 are important for receptor binding, whereas the C-terminal diglycine of ubiquitin facilitates receptor activation, is in agreement with the general two-site model of the structure-function relationship of chemokines, including SDF-1α (24, 25, 29). This model is based on the observation that chemokines have separate binding and activation sites. In analogy to SDF-1α, in which Lys-1 and Pro-2 are critical for receptor activation but not for receptor binding, the C-terminal diglycine of ubiquitin appears to mimic the function of the two N-terminal amino acid residues of SDF-1α (25).
Because ubiquitin chains are also present in the systemic circulation (41), we then tested whether diubiquitin and ubiquitin chains bind to and activate CXCR4. As shown in Fig. 2D, linear diubiquitin, diubiquitin linked via Lys-48, and linear tetraubiquitin competed with FITC-ubiquitin for receptor binding comparable with WT ubiquitin. Furthermore, WT ubiquitin and linear ubiquitin chains consisting of 2–10 ubiquitin molecules showed comparable effects on cAMP levels in forskolin-stimulated THP-1 cells when tested in equimolar concentrations. These data suggest that ubiquitin chains are able to activate CXCR4. Furthermore, our findings imply that the C-terminal ubiquitin molecule within a ubiquitin chain establishes significant contact with the receptor and that the subsequent ubiquitin units do not contribute to or interfere with the binding and activation process.
Recently, the crystal structure of CXCR4 has been solved (27). Thus, to identify candidate CXCR4 residues that could be involved in the binding of ubiquitin, we then sought to dock ubiquitin onto the extracellular domain of CXCR4 using the RosettaDock software (34). The resulting computational model of the ubiquitin-CXCR4 complex can, in part, explain how the mutations and C-terminal modifications of ubiquitin affect binding to and activation of CXCR4 (Fig. 3). In this model, Val-70 of ubiquitin is in close contact with CXCR4 (Fig. 3A). Thus, the V70A mutation should reduce the binding affinity of ubiquitin, as we have detected in competition binding experiments.
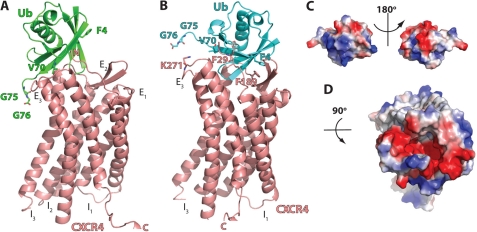
FIGURE 3.
Protein-protein docking of ubiquitin onto the extracellular domain of CXCR4. A, ribbon diagram of the ubiqutin-CXCR4 complex from the lowest energy model of Rosetta docking. Ub and CXCR4 are colored in green and salmon, respectively. The N terminus (N), extracellular loops (E1–E3), intracellular loops (I1–I3), and C terminus (C) of CXCR4 and ubiquitin residues that were identified to be important for CXCR4 binding and activation are indicated. B, ribbon diagram of the ubiquitin-CXCR4 complex after manual adjustment of the docking based on the charge complementarity and the mutational data. Ub and CXCR4 are colored in cyan and salmon, respectively. The ubiquitin and CXCR4 residues that are predicted as interaction sites are indicated. C, surface representation of ubiquitin. The orientation of ubiquitin is the same as in B. D, surface representation of the extracellular domain of CXCR4.
The Rosetta docking model further suggests that the C-terminal ubiquitin diglycine is near the extracellular loop 3, which could alter the conformation of helix 6 and induce a switch of intracellular loop 3 to activate the downstream G protein (42–44). However, this model fails to explain why the F4A mutation reduces the ability of ubiquitin to bind and activate CXCR4.
G protein-coupled receptors demonstrate a substantial conformational switch upon ligand activation and G protein binding (42–44), which is difficult to predict using computational methods. Thus, we hypothesized that the inactive conformation of CXCR4, which is depicted by the available structure, will also undergo a conformational switch upon ubiquitin binding and that a small rotation/translation would allow ubiquitin Phe-4 to make contact with CXCR4, without compromising the interactions of Val-70 and the C-terminal diglycine during CXCR4 binding. We manually moved ubiquitin Phe-4 closer to CXCR4 (Fig. 3B), which would improve charge complementarity between ubiquitin and CXCR4 (Fig. 3, C and D). In addition, Phe-4 and Val-70 of ubiquitin would make close contact with Phe-189 and Phe-29 in CXCR4, respectively. The C-terminal carboxyl group of ubiquitin would then be positioned to form a salt bridge with CXCR4 Lys-271.
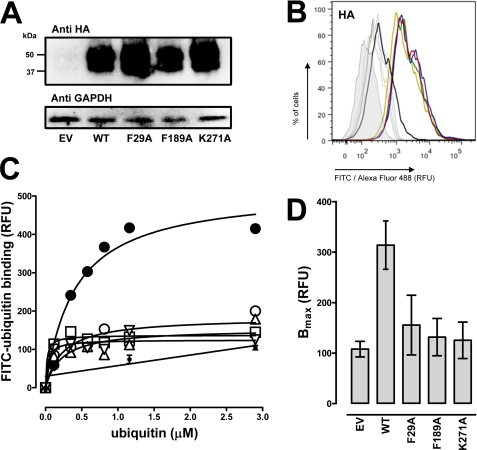
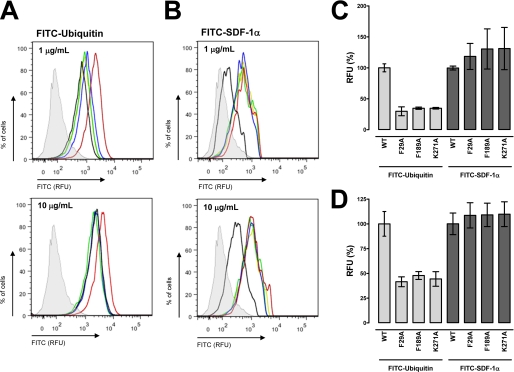
As this computational model provided a testable hypothesis, we utilized site-directed mutagenesis to assess whether the CXCR4 residues Phe-29, Phe-189, and Lys-271 interact with ubiquitin. We transfected HEK 293 cells with HA-tagged CXCR4-WT, CXCR4-F29A, CXCR4-F189A, and CXCR4-K271A expression vectors and confirmed expression by Western blotting and FACS analyses. When whole cell extracts were analyzed by Western blotting with anti-HA, we detected the expected pattern of bands for CXCR4 after transfection with the various CXCR4 cDNA clones (Fig. 4A) (3, 23). Flow cytometry analyses then documented that all HA-tagged CXCR4 expression vectors were expressed on the cell surface and that the fluorescence signals for WT CXCR4 and the CXCR4 mutants were comparable (Fig. 4B). We then tested the transfected cells for FITC-ubiquitin binding in saturation binding experiments. The results from a typical saturation binding experiment are shown in Fig. 4C, and the quantification of the Bmax values from five independent experiments is shown in Fig. 4D. As expected, FITC-ubiquitin binding was increased after transfection of the cells with WT CXCR4 (3, 23). In contrast, FITC-ubiquitin binding to cells transfected with the CXCR4 mutants was indistinguishable from binding to cells transfected with the empty vector. To confirm these findings from ubiquitin binding assays, we then analyzed FITC-ubiquitin binding to the cell surface by FACS analyses (Fig. 5A). Quantification of the mean fluorescence signals after incubation of cells with 1 and 10 μg/ml FITC-ubiquitin resulted in 70 and 60%, respectively, reduced fluorescence signals in cells transfected with the CXCR4 mutants, as compared with cells transfected with WT CXCR4 (Fig. 5, C and D). In contrast, cells transfected with WT or mutant CXCR4 showed increased binding of FITC-SDF-1α, as compared with cells transfected with empty vector (Fig. 5B). There were no differences in mean fluorescence signals between WT and mutant CXCR4 when incubated with 1 and 10 μg/ml FITC-SDF-1α (Fig. 5, C and D). These data argue in favor of the proposed model of the ubiquitin-CXCR4 interaction. Furthermore, this model would also be consistent with our previous finding, that anti-CXCR4-(1–14) does not affect the binding and activation of CXCR4 by ubiquitin, whereas anti-CXCR4-(176–293), an antibody directed against extracellular loops 2 and 3 of CXCR4, interrupts the ubiquitin-CXCR4 interaction (23).
FIGURE 4.
Phe-29, Phe-189, and Lys-271 of CXCR4 are important for ubiquitin binding. A, HA-tagged open reading frame cDNA clones of WT CXCR4, CXCR4-F29A, CXCR4-F-189A, and CXCR4-K271A were transfected into HEK293 cells followed by immunoblotting of whole cell lysates with anti-HA and anti-GAPDH. EV, empty vector. B, quantification of HA expression by flow cytometry after transfection as in A. Thick lines, cells labeled with mouse anti-HA/anti-mouse Alexa Fluor 488 goat IgG. Thin lines, control, cells labeled with rabbit IgG/anti-rabbit FITC goat IgG. Gray, unstained cells. Black, cells transfected with empty vector. Red, cells transfected with HA-tagged WT CXCR4. Blue, cells transfected with HA-tagged CXCR4-F29A. Green, cells transfected with HA-tagged CXCR4-F189A. Light green, cells transfected with HA-tagged CXCR4-K271A. RFU, relative fluorescence units. C, FITC-ubiquitin binding (1 min, 4 °C) after transfection as in A and B. White circle, empty vector. Large black circle, WT CXCR4. White square, CXCR4-F29A. White upright triangle, CXCR4-F189A. White inverted triangle, CXCR4-K271A. Small black circle, nonspecific binding was similar after transfection with empty vector and the various CXCR4 cDNA clones. D, quantification of Bmax from five independent experiments as in C.
FIGURE 5.
Phe-29, Phe-189, and Lys-271 of CXCR4 are important for ubiquitin binding but do not contribute to the binding of SDF-1α. A, FACS analyses of FITC-ubiquitin binding (1 min, 4 °C) after transfection of HEK293 cells as in Fig. 4. Top, 1 μg/ml FITC-ubiquitin. Bottom, 10 μg/ml FITC-ubiquitin. Black, cells transfected with empty vector. Red, cells transfected with HA-tagged WT CXCR4. Blue, cells transfected with HA-tagged CXCR4-F29A. Green, cells transfected with HA-tagged CXCR4-F189A. Light green, cells transfected with HA-tagged CXCR4-K271A. RFU, relative fluorescence units. B, FACS analyses of FITC-SDF-1α binding (1 min, 4 °C) after transfection of HEK293 cells as in Fig. 4. Top, 1 μg/ml FITC-SDF-1α. Bottom, 10 μg/ml FITC-SDF-1α. Same color code as in A. C and D, quantification of the mean fluorescence signals from three experiments as in A and B. C, 1 μg/ml FITC-ubiquitin/SDF-1α. D, 10 μg/ml FITC-ubiquitin/SDF-1α. Data are expressed as the percentage of binding to WT CXCR4.
Although the details of the SDF-1α-CXCR4 interface also remain to be determined, the current model suggests that the docking domain of SDF-1α interacts with the receptor N terminus, which is absent in the available structure of CXCR4 (27, 45). The flexible SDF-1α N terminus is then placed into the central ligand binding pocket to activate the receptor (27). Along with previous evidence that Phe-189 and Lys-271 of CXCR4 are not involved in SDF-1α binding (46, 47), our finding that mutations of Phe-29, Phe-189, and Lys-271 do not affect FITC-SDF-1α binding to CXCR4 is consistent with the currently available information on the structure-function relationship of the SDF-1α-CXCR4 interaction.
Conclusively, we provide initial evidence that the mechanism through which ubiquitin activates CXCR4 resembles the typical structure-function relationship of chemokines, which contain separate receptor docking and activation sites. The ubiquitin docking site for CXCR4 corresponds to surface areas that are also involved in the recognition of ubiquitin by ubiquitin binding domains of intracellular interacting proteins. The flexible ubiquitin C terminus functions as a receptor activation site and mimics the functions of the N termini of chemokines. Furthermore, we generated a computational model of the ubiquitin-CXCR4 interface, which provided testable hypotheses. Although further experimental evidence is required to accept and refine this model, it withstands initial testing by site-directed mutagenesis of CXCR4. These data provide evidence that the natural ligands bind and activate CXCR4 through unique and separate binding sites on the receptor. We have shown previously that CXCR4 activation with SDF-1α and ubiquitin stimulate similar signal transduction pathways, although the effects of both ligands on cell migration and HIV-1 infection were distinct (23). Thus, we speculate that the presence of separate CXCR4 ligand binding sites may provide the structural basis for biased agonism. However, a detailed comparison of signal transduction events and subsequent effects on cell function after CXCR4 activation with ubiquitin and SDF-1α is required to address this hypothesis.
Ubiquitin, SDF-1α, and a SDF-1α peptide analog have been shown to reduce exuberant inflammation and organ injury in various disease models (13–22, 48). Thus, our findings provide the basis for the structure-based design of novel compounds that interact with ligand-specific binding sites and thus may permit targeting of selective therapeutic effects that are mediated through CXCR4.
This research was made possible, in part, by a grant that was awarded and administered by the United States Army Medical Research and Materiel Command (USAMRMC) and the Telemedicine and Advanced Technology Research Center (TATRC) at Fort Detrick, MD, under Contract W81XWH1020122 (to M. M.). This work was also supported in part by National Institutes of Health Grants GM075159 (to A. M.) and GM081539 (to W. J. T.). Based on the results of this work, Loyola University Chicago has filed a provisional patent application. The authors are the inventors. None of the authors have received any income related to the patent or patent application.
- CXCR4
- CXC chemokine receptor 4
- SDF-1α
- stromal cell-derived factor 1α
- Ub
- ubiquitin.
REFERENCES
- 1. Majetschak M. (2011) J. Leukocyte Biol. 89, 205–219 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3. Saini V., Marchese A., Majetschak M. (2010) J. Biol. Chem. 285, 15566–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Busillo J. M., Benovic J. L. (2007) Biochim. Biophys. Acta 1768, 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagasawa T., Hirota S., Tachibana K., Takakura N., Nishikawa S., Kitamura Y., Yoshida N., Kikutani H., Kishimoto T. (1996) Nature 382, 635–638 [DOI] [PubMed] [Google Scholar]
- 6. Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., Kataoka Y., Kitamura Y., Matsushima K., Yoshida N., Nishikawa S., Kishimoto T., Nagasawa T. (1998) Nature 393, 591–594 [DOI] [PubMed] [Google Scholar]
- 7. Karin N. (2010) J. Leukocyte Biol. 88, 463–473 [DOI] [PubMed] [Google Scholar]
- 8. Zaruba M. M., Franz W. M. (2010) Expert Opin. Biol. Ther. 10, 321–335 [DOI] [PubMed] [Google Scholar]
- 9. Li M., Yu J., Li Y., Li D., Yan D., Ruan Q. (2010) Cell. Reprogram. 12, 405–415 [DOI] [PubMed] [Google Scholar]
- 10. Nagasawa T., Tachibana K., Kishimoto T. (1998) Semin. Immunol. 10, 179–185 [DOI] [PubMed] [Google Scholar]
- 11. Veldkamp C. T., Ziarek J. J., Peterson F. C., Chen Y., Volkman B. F. (2010) J. Am. Chem. Soc. 132, 7242–7243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teicher B. A., Fricker S. P. (2010) Clin. Cancer Res. 16, 2927–2931 [DOI] [PubMed] [Google Scholar]
- 13. Meiron M., Zohar Y., Anunu R., Wildbaum G., Karin N. (2008) J. Exp. Med. 205, 2643–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shyu W. C., Lin S. Z., Yen P. S., Su C. Y., Chen D. C., Wang H. J., Li H. (2008) J. Pharmacol. Exp. Ther. 324, 834–849 [DOI] [PubMed] [Google Scholar]
- 15. Hu X., Dai S., Wu W. J., Tan W., Zhu X., Mu J., Guo Y., Bolli R., Rokosh G. (2007) Circulation 116, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Majetschak M., Cohn S. M., Nelson J. A., Burton E. H., Obertacke U., Proctor K. G. (2004) Surgery 135, 536–543 [DOI] [PubMed] [Google Scholar]
- 17. Majetschak M., Cohn S. M., Obertacke U., Proctor K. G. (2004) J. Trauma 56, 991–999; discussion 999–1000 [DOI] [PubMed] [Google Scholar]
- 18. Earle S. A., Proctor K. G., Patel M. B., Majetschak M. (2005) Surgery 138, 431–438 [DOI] [PubMed] [Google Scholar]
- 19. Garcia-Covarrubias L., Manning E. W., 3rd, Sorell L. T., Pham S. M., Majetschak M. (2008) Crit. Care Med. 36, 979–982 [DOI] [PubMed] [Google Scholar]
- 20. Griebenow M., Casalis P., Woiciechowsky C., Majetschak M., Thomale U. W. (2007) J. Neurotrauma 24, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 21. Earle S. A., El-Haddad A., Patel M. B., Ruiz P., Pham S. M., Majetschak M. (2006) Transplantation 82, 1544–1546 [DOI] [PubMed] [Google Scholar]
- 22. Ahn H. C., Yoo K. Y., Hwang I. K., Cho J. H., Lee C. H., Choi J. H., Li H., Cho B. R., Kim Y. M., Won M. H. (2009) Exp. Neurol. 220, 120–132 [DOI] [PubMed] [Google Scholar]
- 23. Saini V., Staren D. M., Ziarek J. J., Nashaat Z. N., Campbell E. M., Volkman B. F., Marchese A., Majetschak M. (2011) J. Biol. Chem. 286, 33466–33477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta S. K., Pillarisetti K., Thomas R. A., Aiyar N. (2001) Immunol. Lett. 78, 29–34 [DOI] [PubMed] [Google Scholar]
- 25. Crump M. P., Gong J. H., Loetscher P., Rajarathnam K., Amara A., Arenzana-Seisdedos F., Virelizier J. L., Baggiolini M., Sykes B. D., Clark-Lewis I. (1997) EMBO J. 16, 6996–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doranz B. J., Orsini M. J., Turner J. D., Hoffman T. L., Berson J. F., Hoxie J. A., Peiper S. C., Brass L. F., Doms R. W. (1999) J. Virol. 73, 2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veldkamp C. T., Seibert C., Peterson F. C., Sakmar T. P., Volkman B. F. (2006) J. Mol. Biol. 359, 1400–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kofuku Y., Yoshiura C., Ueda T., Terasawa H., Hirai T., Tominaga S., Hirose M., Maeda Y., Takahashi H., Terashima Y., Matsushima K., Shimada I. (2009) J. Biol. Chem. 284, 35240–35250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gozansky E. K., Louis J. M., Caffrey M., Clore G. M. (2005) J. Mol. Biol. 345, 651–658 [DOI] [PubMed] [Google Scholar]
- 31. Malik R., Marchese A. (2010) Mol. Biol. Cell 21, 2529–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Majetschak M., Krehmeier U., Bardenheuer M., Denz C., Quintel M., Voggenreiter G., Obertacke U. (2003) Blood 101, 1882–1890 [DOI] [PubMed] [Google Scholar]
- 33. Vijay-Kumar S., Bugg C. E., Cook W. J. (1987) J. Mol. Biol. 194, 531–544 [DOI] [PubMed] [Google Scholar]
- 34. Wang C., Bradley P., Baker D. (2007) J. Mol. Biol. 373, 503–519 [DOI] [PubMed] [Google Scholar]
- 35. Lyskov S., Gray J. J. (2008) Nucleic Acids Res. 36, W233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 38. Winget J. M., Mayor T. (2010) Mol. Cell 38, 627–635 [DOI] [PubMed] [Google Scholar]
- 39. Sloper-Mould K. E., Jemc J. C., Pickart C. M., Hicke L. (2001) J. Biol. Chem. 276, 30483–30489 [DOI] [PubMed] [Google Scholar]
- 40. Alam S. L., Sundquist W. I. (2006) Nat. Struct. Mol. Biol. 13, 186–188 [DOI] [PubMed] [Google Scholar]
- 41. Takada K., Nasu H., Hibi N., Tsukada Y., Shibasaki T., Fujise K., Fujimuro M., Sawada H., Yokosawa H., Ohkawa K. (1997) Clin. Chem. 43, 1188–1195 [PubMed] [Google Scholar]
- 42. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. (2011) Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. (2011) Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Standfuss J., Edwards P. C., D'Antona A., Fransen M., Xie G., Oprian D. D., Schertler G. F. (2011) Nature 471, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Veldkamp C. T., Seibert C., Peterson F. C., De la Cruz N. B., Haugner J. C., 3rd, Basnet H., Sakmar T. P., Volkman B. F. (2008) Sci. Signal. 1, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brelot A., Heveker N., Montes M., Alizon M. (2000) J. Biol. Chem. 275, 23736–23744 [DOI] [PubMed] [Google Scholar]
- 47. Choi W. T., Tian S., Dong C. Z., Kumar S., Liu D., Madani N., An J., Sodroski J. G., Huang Z. (2005) J. Virol. 79, 15398–15404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fan H., Wong D., Ashton S. H., Borg K. T., Halushka P. V., Cook J. A. (2011) Inflammation, in press [DOI] [PMC free article] [PubMed] [Google Scholar]