Background: Ufd2 is a U-box-containing ubiquitin-protein ligase.
Results: Mps1 turnover is regulated by Ufd2 in yeast and mammalian cells.
Conclusion: Our study leads to novel insights into the cell cycle control and physiological significance of the Ufd2 pathway.
Significance: Understanding the functions of Ufd2 will elucidate a poorly characterized pathway in proteolysis that may be crucial to unravel the mechanisms underlying human diseases.
Keywords: Cancer, Cell Cycle, Protein Degradation, Protein Turnover, Ubiquitin Ligase, Ubiquitination
Abstract
Ufd2 is a U-box-containing ubiquitylation enzyme that promotes ubiquitin chain assembly on substrates. The physiological function of Ufd2 remains poorly understood. Here, we show that ubiquitylation and degradation of the cell cycle kinase Mps1, a known target of the anaphase-promoting complex E3, require Ufd2 enzyme. Yeast cells lacking UFD2 exhibit altered chromosome stability and several spindle-related phenotypes, expanding the biological function of Ufd2. We demonstrate that Ufd2-mediated Mps1 degradation is conserved in humans. Our results underscore the significance of Ufd2 in proteolysis and further suggest that Ufd2-like enzymes regulate far more substrates than previously envisioned.
Introduction
Substrate ubiquitylation is carried out through the concerted actions of several enzymes, including a ubiquitin (Ub)2-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub-protein ligase (E3) (1, 2). For substrates targeted for proteasome-mediated degradation, successive Ub molecules are added to form a Ub chain on the substrates facilitated by the same E3 (2) or, in some cases, an E4 enzyme that is specifically involved in Ub chain elongation (3). Among the few known E4 enzymes, the best characterized and also first to be identified is Ufd2, which was demonstrated to elongate only an existing, short oligo-Ub chain on model UFD substrates assembled by the Ufd4 E3 in vitro and in vivo (3, 4). The physiological function of Ufd2 remains poorly understood because only a handful of Ufd2 targets have been identified (3).
Here, we identified the kinase Mps1 as a target of the Ufd2 enzyme. Mps1 is a conserved protein kinase critical for spindle pole body (SPB; the functional equivalent of the centrosome) duplication, spindle checkpoint, kinetochore biorientation, and chromosome-microtubule attachment (5, 6). The biological function of Mps1 is likely fulfilled through its kinase activity by phosphorylating SPB components (e.g. Spc98 and Cdc31) and other spindle checkpoint proteins (e.g. Dam1 and Ndc80 in yeast and BubR1 and Borealin in humans) (5–7). Mps1 contains three D-box motifs and was shown to be regulated throughout the cell cycle by the anaphase-promoting complex (APC) E3 (8), which is one of two major E3 enzymes employed to propel the cells cycling through different stages via highly coordinated ubiquitylation and destruction of various cell cycle regulators (9–11). A major challenge for APC is then how to destruct these diverse targets timely and efficiently to ensure proper cell cycle progression (9, 10). The link between Ufd2 and Mps1 suggests that some APC targets may be regulated by other Ub ligases. Consistent with the critical roles of Mps1 in spindle checkpoint and SPB duplication, UFD2 deletion leads to several Mps1-related cell cycle phenotypes. Furthermore, Ufd2-mediated Mps1 degradation is conserved in humans and is important for mitotic progression. Our study leads to novel insights into the cell cycle control and physiological significance of the Ufd2 pathway and further suggests that the involvement of Ufd2 in proteolysis in vivo may be more prevalent than previously anticipated.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
The synthetic genetic array compatible S288c strain Y8835 (MATa ura3::natR can1::STE2pr-Sp_his5 lyp1 his3 1 leu2 0 met15 0) and isogenic ufd2 (ufd2::natR) cells were used for the genome-wide screen (12). Haploid strains bearing rpn10Δ, mad1Δ, Mps1-tandem affinity purification tag (TAP), Ase1-TAP, Spc110-TAP, Spc98-TAP, or Ufd2-TAP in the BY4741 background were obtained from Open Biosystems (Huntsville, AL). The UFD2 gene was replaced with KanMX4 using PCR-mediated homologous recombination to construct strain YHR213 (Mps1-TAP UFD2::KanMX4). Strains EJY130 (UFD2::LEU2) and ufd1-1 were obtained from Dr. Alex Varshavsky (California Institute of Technology). The cdc16-1 strain was provided by Dr. David Morgan (University of California, San Francisco). The plasmid pWP2 (PGAL-MPS1-myc) was obtained from Dr. Andrew Murray (Harvard University). The yeast strain YHR207 (UFD2U-boxΔ-TAP in the BY4741 background) was generated by replacing the Ufd2 sequences after amino acid 856 with a TAP-HIS3 cassette in BY4741 by homologous recombination. The replacement was confirmed by PCR and immunoblotting. UFD2 was deleted from the sectoring assay strain YPH278 (MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 CFIII (CEN3L.YPH278) URA3 SUP11), yielding YHR230. YPH278 and YPH277 were obtained from Dr. P. Hieter for the sectoring assay. Double mutants for Fig. 3B were created by crossing UFD2::natR to bub1::kanMX4, bub3::kanMX4, tid3-1::kanMX4, and spc110-221::kanMX (all in the BY4741 background), respectively. Colonies derived from at least three independent spores, carrying both markers, were tested for suppression phenotypes.
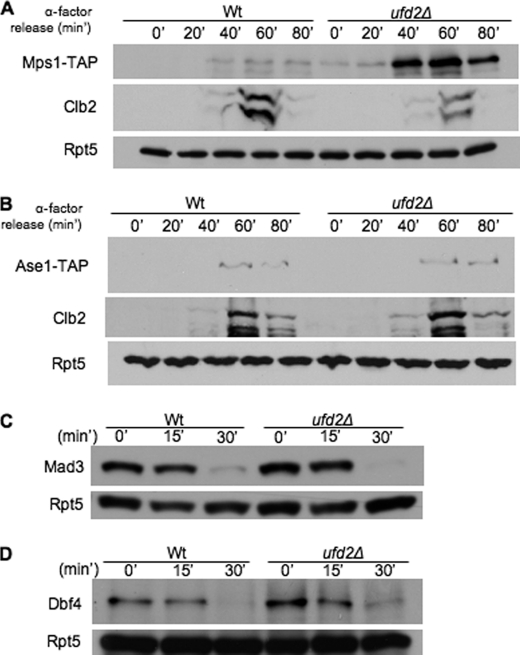
FIGURE 3.
Ufd2 is not required for degradation of all APC substrates. A and B, involvement of Ufd2 in the degradation of APC substrates. Shown are the results from immunoblot analysis of endogenously expressed, C-terminally TAP-tagged Mps1, Ase1, or Clb2 in wild-type and ufd2Δ cultures synchronized with α-factor. Shown is the time after removal of α-factor in wild-type and ufd2Δ cultures. C and D, degradation of two APC substrates (Mad3 and Dbf4) in wild-type or ufd2Δ cells. Yeast cells containing GAL1-regulated Mad3 or Dbf4 tagged with HA and the IgG-binding site from protein A were synchronized in G1 by α-factor arrest and induced with galactose for 2 h. Induction was terminated by the addition of 2% glucose and 100 μg/ml cycloheximide. Samples were withdrawn at intervals and processed for immunoblotting with anti-HA antibody.
Cultures were grown in rich (yeast/peptone/dextrose (YPD)) or synthetic medium containing standard ingredients and 2% glucose (SD medium), 2% raffinose (SR medium), 2% galactose (SG medium), or 2% raffinose + 2% galactose (SRG medium). For the synthetic dosage lethal screens, haploids were selected on synthetic dextrose medium (2% glucose, 1.7 g/liter yeast nitrogen base without ammonium sulfate and amino acids, 1 g/liter monosodium glutamic acid, and 2 g/liter amino acid dropout mixture lacking uracil, arginine, lysine, and histidine) supplemented with the following chemicals: 100 mg/liter clonNAT (WERNER BioAgents), 200 mg/liter Geneticin (Invitrogen), 50 mg/liter l-canavanine (Sigma), and 50 mg/liter S-(2-aminoethyl)-l-cysteine hydrochloride (Sigma).
Genome-wide Synthetic Dosage Lethal Screen
The URA3-marked overexpression library and the screening procedures used in this study were described previously (12). Overexpressed genes that uniquely caused a reduction in colony size of >20% in the ufd2Δ background were considered for downstream analysis.
Expression Shut-off Assay
Yeast cells carrying plasmids expressing His6- and GST-tagged proteins from the GAL1 promoter were grown at 30 °C to A600 ∼ 1 in SR Ura− medium with auxotrophic supplements and 2% raffinose as the carbon source. Protein expression was induced with 2% galactose for 1.5 h and then repressed by the addition of 2% glucose. Cycloheximide (100 μg/ml) was also added to stop translation. Samples were withdrawn at the indicated time points and harvested by centrifugation. Proteins were extracted by glass bead lysis of cells, processed for immunoprecipitation with glutathione-Sepharose beads (GE Healthcare), and resolved by 7% SDS-PAGE. Immunoblots were probed with anti-His6 antibody (Abcam), followed by detection with goat anti-mouse HRP conjugate using ECL reagents (GE Healthcare). The stable protein Rpt5 was used as a loading control in the expression shut-off experiments.
Detection of Ubiquitylated Substrates
Yeast cells expressing GAL1-regulated substrate Myc-Mps1 and HA-tagged Ub were grown to log phase in SR medium. 2% galactose was added to induce protein expression for 3 h. MG132 was added as described (12) for 1.5 h to detected ubiquitylated species. Cells were lysed with glass beads and immunoprecipitated with glutathione-Sepharose or Myc beads for 2 h at 4 °C. The immunoprecipitates were resolved by SDS-PAGE, transferred to PVDF membrane, and immunoblotted with anti-HA antibody (Covance), followed by anti-mouse HRP conjugates and ECL reagents. Non-ubiquitylated Mps1 was detected by anti-His6 or anti-Myc antibody.
FACS Analysis
Yeast cells were grown in raffinose-containing medium to early log phase and then arrested in G2/M phase using 30 μl/ml nocodazole for 4 h, during which 2% galactose was added to overexpress Mps1 for 2 h. Samples at time 0 were collected after nocodazole was removed by three washes. Samples were collected at the indicated time points after release from the G2/M arrest and stained with propidium iodide for flow cytometry analysis on a BD FACSCalibur flow cytometer using CellQuest software.
Sectoring Assay
Yeast YPH278 cells contained the defective ade2-101 ochre mutation, which can be suppressed by the SUP11 gene, carried on a nonessential 90-kb chromosome fragment (13). YPH278 cells that maintained this chromosome fragment produced white colonies; the loss of it resulted in a red color or sectors. The UFD2 gene was deleted in the YPH278 strain; the resulting mutant was compared with YPH278. As described above, cells were grown first in the uracil-lacking medium that retained the SUP11-containing chromosome, followed by growth in rich YPD medium and plating on YPD plates.
Mammalian Cell Line and Reagents
The plasmid expressing FLAG- or HA-tagged UFD2a was obtained from Dr. Keiichi Nakayama. UFD2a-deficient cell line NB-1 was provided by Dr. Akira Nakagawara (Chiba University, Chiba, Japan). Plasmids were transfected using Lipofectamine 2000 (Invitrogen).
Mammalian cells were cultured using DMEM supplemented with 10% FBS. Cells were kept in 37 °C incubation with 5% CO2. RNAi knockdown was performed following the manufacturer's protocol (Dharmacon) using HiPerFect transfection reagent (Qiagen). Antibodies for UFD2a and Cdc27 were purchased from BD Biosciences and Sigma, respectively.
Analysis of Mitotic Progression in Mammalian Cells
To assess the mitotic delay, cells grown on coverslips coated with fibronectin-like polymer (Sigma) were incubated with 100 μm monastrol (Enzo Chemicals) for 4 h and washed with fresh medium lacking monastrol. At 0, 15, 30, 45, or 60 min after release from monastrol, cells were fixed for 10 min in PBS containing 4% formaldehyde (Ted Pella) and 0.2% Triton X-100 (Sigma) and stained with Hoechst and antibodies against α- and γ-tubulin, and at least 100 mitotic cells were characterized in each of three replicates. This experiment was repeated to score HA-positive mitotic cells after transfection of NB-1 cells with HA-tagged UFD2a using Lipofectamine 2000. All images were acquired at ambient temperature using an Olympus IX-81 microscope with a ×100 Plan Apo oil immersion objective (1.4 numerical aperture) and a QCam Retiga EXi Fast 1394 camera using the SlideBook software package (Intelligent Imaging Innovations, Denver, CO).
RESULTS
Mps1 Turnover Requires Ufd2 and Rad23/Dsk2
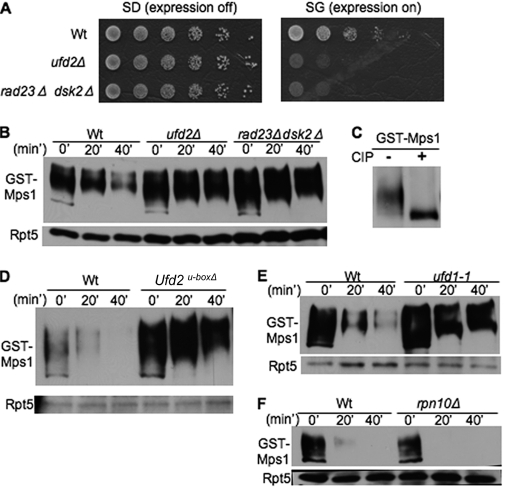
To understand the biological function of Ufd2, we employed a genome-wide screen to isolate the cellular targets of Ufd2 (supplemental Fig. S1A) (12). We found that MPS1 overexpression induced severe toxicity in ufd2Δ cells, linking Mps1 to the Ufd2 pathway (Fig. 1A). We then used an expression shut-off assay to determine the stability of Mps1 in wild-type or ufd2Δ cells. Mps1, a phosphorylated protein (Fig. 1C) (5), was degraded in wild-type cells but significantly stabilized in ufd2Δ cells (Fig. 1B), suggesting that Mps1 turnover is regulated by Ufd2. Ufd2 contains a conserved U-box motif essential for its ubiquitylation activity (14). Interestingly, Ufd2 can also promote substrate degradation without its enzymatic activity in a U-box-independent manner (15). We found that Mps1 was stabilized in ufd2U-boxΔ cells (Fig. 1D), suggesting that its enzymatic activity is essential for Mps1 degradation.
FIGURE 1.
Identification of Mps1 as substrate of Ufd2-Rad23/Dsk2 pathway. A, overexpression of Mps1 leads to slower growth of ufd2Δ or rad23Δ dsk2Δ double mutant cells. GST-His6-Mps1 isolated from the genome-wide screen was transformed into the wild type or the indicated mutants. These cells were grown to similar densities, and 5-fold serial dilutions were spotted onto SD or SG medium. B, efficient degradation of Mps1 requires Ufd2, Rad23, and Dsk2. Wild-type and ufd2Δ cells containing GAL1 promoter-driven GST-His6-Mps1 were first grown in raffinose-containing medium. Expression of Mps1 was induced by the addition of galactose. Samples were taken after promoter shut-off at the time points indicated and analyzed by anti-His6 Western blotting. Equal amounts of protein extracts were used and confirmed by blotting with anti-Rpt5 antibody in all of the expression shut-off experiments (lower panel). Proteins are identified on the left. C, Mps1 is phosphorylated. GST-His6-Mps1 was expressed in wild-type cells and recovered by immunoprecipitation. Mps1 immunoprecipitates were incubated with or without alkaline phosphatase and visualized by immunoblotting. CIP, calf intestinal phosphatase. D, the U-box motif is critical for Mps1 degradation. GST-tagged Mps1 was transformed into the wild type and ufd2U-boxΔ mutants. Mps1 degradation was assayed as described for B. E and F, Mps1 degradation requires Ufd1 but not Rpn10. GST-His6-Mps1 stabilities in ufd1-1 and rpn10Δ were determined.
Downstream of Ufd2, Ub-binding proteins (e.g. Dsk2, Rad23, Rpn10, and the Ufd1-Cdc48-Npl4 complex) are involved in targeting proteins to the proteasome (1, 16). These Ub receptors have different substrates and sometimes perform overlapping functions (14, 17). Previously, we demonstrated that Ufd2 works with the Ub-binding proteins Dsk2 and Rad23 (18). Consistent with this notion, the overexpression of Mps1 led to growth retardation in cells lacking DSK2 and RAD23, and Mps1 was degraded in a Dsk2/Rad23-dependent manner (Fig. 1, A and B). Interestingly, DSK2 was first identified as a suppressor of a kar1 mutant 14 years ago (19). Kar1 is a key component of SPB crucial for SPB duplication, but the underlying cause of this genetic interaction between DSK2 and KAR1 remained elusive. Our results reveal Mps1, a known SPB regulator (5, 6), as the first protein that links Dsk2 to SPB duplication. We also found that Mps1 was stabilized in ufd1-1 mutant cells but efficiently degraded in rpn10Δ cells (Fig. 1, E and F), suggesting that Mps1 is a substrate of the Ufd1-Cdc48-Npl4 complex. Because the Cdc48 complex is characterized mainly by its proteolytic role in endoplasmic reticulum-associated protein degradation, the identification of Mps1 as its substrate provides another tool to study the function of the Cdc48 pathway in nuclear protein degradation (17, 20). Interestingly, whereas Ufd2 substrates (e.g. Pex29 and Mps1) require Cdc48 and Rad23, not all Cdc48 or Rad23 substrates are regulated by Ufd2 (e.g. ricin A chain and Ubc6*) (12, 21).
Link between Ufd2 and Various APC Substrates
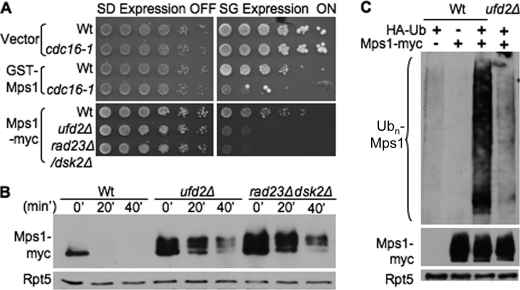
The E3 enzyme for Mps1 degradation was previously demonstrated to be the APC/cyclosome (8). Because the Mps1 allele we used is N-terminally fused to the GST and His6 tags, we examined whether GST-His6-Mps1 is regulated by the APC E3 as well. We found that GST-His6-Mps1 overexpression was toxic in yeast cells harboring mutant cdc16, an essential subunit of APC (Fig. 2A), suggesting that GST-His6-Mps1 is likely regulated by APC. To address this issue more directly, we obtained the C-terminally Myc-tagged version of Mps1 previously used to demonstrate the involvement of APC (8). We found that overexpression of Mps1-Myc caused growth retardation in ufd2Δ and rad23Δ dsk2Δ cells (Fig. 2A), and its degradation was also compromised in ufd2Δ and rad23Δ dsk2Δ cells (Fig. 2B), indicating that Mps1 with either the GST-His6 or Myc tag is subject to regulation by both APC and Ufd2. Furthermore, Mps1 ubiquitylation was significantly impaired in ufd2Δ cells (Fig. 2C). We evaluated the Ub chain linkages assembled onto Mps1 using the Ub mutants defective for Ub chain synthesis. Similar to its effect on Pex29 ubiquitylation (12), mutation at Lys-48 significantly reduced Mps1 ubiquitylation (supplemental Fig. S1B).
FIGURE 2.
Involvement of Ufd2 and APC pathway in Mps1 degradation. A, overexpression of various Mps1 alleles causes toxicity in cdc16-1 and ufd2Δ mutants. The plasmid bearing GST-His6-Mps1 or Mps1-Myc or the empty vector was transformed into the strains indicated. 5-Fold serial dilutions were plated as described in the legend to Fig. 1A. B, Mps1-Myc degradation involves the Ufd2-Rad23/Dsk2 pathway. Mps1-Myc stability in the indicated strains was determined as described in the legend to Fig. 1B except that immunoprecipitation was done with Myc beads, followed by immunoblotting with anti-Myc antibody. C, Ufd2 regulates Mps1 ubiquitylation. Myc-tagged Mps1 was transformed with HA-tagged Ub into wild-type or ufd2Δ cells. Because Mps1-Myc ubiquitylation was difficult to detect under normal conditions, we treated wild-type or ufd2Δ cells with the proteasome inhibitor MG132, followed by immunoprecipitation with Myc beads and blotting with anti-HA antibody for ubiquitylated Mps1 species (upper panel). The amounts of Mps1 and Rpt5 in the extracts were also determined by immunoblotting (middle and lower panels).
One concern regarding our results is that the GAL1 promoter-driven Mps1 was overexpressed and may not need Ufd2 for its normal degradation. We next investigated the influence of Ufd2 on the concentration of endogenously expressed Mps1. We first determined steady-state levels of Mps1 in unsynchronized wild-type and ufd2Δ cells. Mps1 levels were (∼2.4-fold) higher in ufd2Δ cells than in wild-type cells (supplemental Fig. S1C), suggesting that endogenous Mps1 is regulated by Ufd2. We also monitored Mps1 levels by immunoblot analysis of samples obtained at various time points using synchronized cultures of wild-type and ufd2Δ cells. In wild-type cultures, cells in the G1 phase contained little Mps1, which appeared as cells entered the G2/M phase (∼40 min after release). Interestingly, ufd2Δ cells contained Mps1 throughout the cell cycle and at levels that exceeded those in wild-type cells (Fig. 3A), supporting that endogenous Mps1 is under the control of Ufd2. In contrast, two other APC substrates, Clb2 and Ase1, exhibited similar expression profiles in wild-type and ufd2Δ cells (Fig. 3, A and B), suggesting that Ufd2-dependent regulation is specific for Mps1. Furthermore, the overexpression of Dbf4 or Mad3, two additional APC substrates, did not cause growth retardation of ufd2Δ cells, and their degradation was not significantly altered in ufd2Δ cells (Fig. 3, C and D). Our data indicate that not all APC substrates require Ufd2 for their degradation.
Cell Cycle and Spindle-related Phenotypes in ufd2Δ Cells
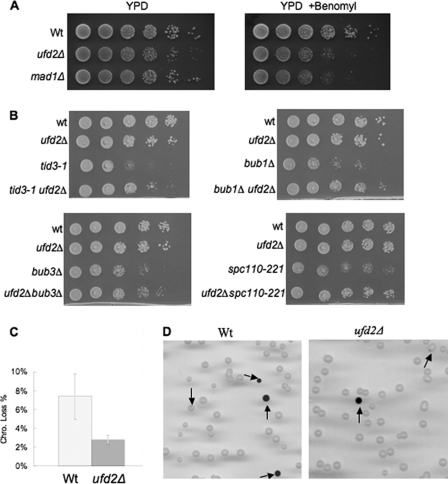
The Mps1 kinase is critical for spindle checkpoint and SPB duplication (5, 7, 22). We therefore examined whether increased Mps1 levels in ufd2Δ cells lead to spindle-related defects, which are often manifested as an increased sensitivity to the antimicrotubule drug benomyl. Interestingly, like mad1Δ mutants that are defective in the spindle assembly checkpoint, ufd2Δ cells showed reduced fitness relative to wild-type cells on benomyl-containing medium (Fig. 4A), supporting the involvement of Ufd2 in spindle regulation.
FIGURE 4.
Cell cycle and spindle-related phenotypes associated with ufd2Δ cells. A, the ufd2Δ mutant strains are sensitive to the microtubule-destabilizing drug benomyl. Yeast strains were grown to similar densities, and 7-fold serial dilutions were spotted onto YPD plates with or without benomyl (15 μg/ml). B, deletion of UFD2 suppresses slow growth of bub1Δ, tid3-1, bub3Δ, and spc110-221 mutant cells. Yeast cells with the indicated genotypes were spotted onto synthetic complete plates in serial 10-fold dilutions and incubated at 30 °C for 2 days. C, enhanced chromosome stability in yeast cells lacking UFD2. Sectoring assay for the chromosome stability was carried out as described previously (13). The experiments were done five times, and the means ± S.D. are shown. D, representative images from the sectoring assay for the chromosome loss using the SUP11-containing YPH278 and ufd2Δ mutants. Colonies containing red color or sectors are marked with arrows.
Proper mitotic spindle regulation could prevent chromosome loss by ensuring accurate chromosome segregation (7, 22). We then wondered whether ufd2Δ cells, in which higher Mps1 levels are maintained (Fig. 3A and supplemental Fig. S1C), may exhibit altered chromosome stability. A colony sectoring assay was employed to assess the chromosome stability in wild-type and ufd2Δ cells (Fig. 4, C and D) (13). These cells harbor a nonessential chromosome fragment, the loss of which leads to the accumulation of red pigment. Colonies exhibiting red color or red sectors were much less frequent (∼2.6-fold) in ufd2Δ cells than in wild-type cells, indicating enhanced chromosome stability in ufd2Δ cells, a phenotype that few yeast mutants exhibit. Yeast cells carrying an extra chromosome IV or X were recently shown to exhibit increased chromosome stability but different responses to benomyl (23). Interestingly, increased chromosome maintenance was also observed in wild-type cells that express moderately higher level of Mps1 regulated by its endogenous promoter on a yeast high copy plasmid (supplemental Fig. S1D). The data also suggest that higher levels of Mps1 promote genome stability.
Quantitative genetic data derived from an ongoing large-scale synthetic genetic array analysis (24)3 suggested possible genetic interactions between UFD2 and components of the spindle checkpoint pathway. Indeed, deletion of UFD2 suppressed the slow growth phenotypes of bub1Δ and bub3Δ mutants, as well as a key kinetochore component mutant that is defective in spindle checkpoint and chromosome segregation, tid3-1 (i.e. Ndc80) (Fig. 4B). The latter has been shown to be a kinase target of Mps1 (25). Furthermore, deletion of UFD2 rescued the growth defect of yeast cells containing a mutant allele of SPB component Spc110 (Fig. 4B), which is known to be phosphorylated by Mps1 kinase (5–7). Higher Mps1 levels alone did not suppress bub1Δ or bub3Δ mutants (data not shown), suggesting that other factors may be involved in the genetic suppression. It would be of interest to isolate high copy suppressors of bub1Δ or tid3-1 and then evaluate whether they are Ufd2 targets. Nevertheless, our data establish a suppressing role for the UFD2 deletion in mutations of other spindle regulators. Because the functions of these proteins are largely conserved from yeast to humans, it is possible that defects in spindle regulation and/or chromosome maintenance associated with mutations in corresponding checkpoint genes or centrosome components could also be suppressed by a Ufd2 mutation in human cell lines.
Phenotypes Associated with Mps1 Overexpression
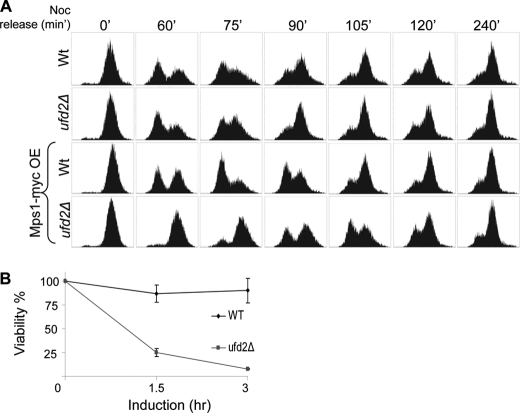
To further examine toxicity induced by the accumulation of GAL1-induced high levels of Mps1 in ufd2Δ cells (Fig. 1A), we monitored cell cycle progression by FACS analysis. In the absence of Mps1 overexpression, no striking differences were detected between wild-type and ufd2Δ cells with respect to the kinetics of cell cycle progression (Fig. 5A). In contrast, upon release from nocodazole-induced G2/M arrest, ufd2Δ cells overexpressing Mps1 exhibited a significant delay in entering the G1 phase of the next cycle (Fig. 5A, lower panels). Interestingly, transient overexpression of Mps1 was highly toxic to ufd2Δ cells. Roughly 92% of ufd2Δ cells could not survive after 3 h of Mps1 induction (Fig. 5B), suggesting that significant damage was inflicted by the accumulated excessive levels of Mps1. Together, these data underscore the biological significance of Ufd2-mediated Mps1 degradation.
FIGURE 5.
Mps1 overexpression-induced phenotypes. A, accumulation of large amounts of Mps1 in ufd2Δ cells delays cell cycle progression. Wild-type or ufd2Δ cells were arrested in G2/M by nocodazole (Noc) for 4 h. Cells were released from the arrest after the removal of nocodazole, and samples taken at various time points after the release were subject to FACScan analysis of DNA content. In the two lower panels, Mps1-Myc expression was induced for 2 h with the addition of galactose and was repressed with glucose as cells were released from the G2/M block. Representative FACS results are shown. OE, overexpression. B, plating efficiency of wild-type or ufd2Δ mutant cells that transiently overexpressed Mps1. Mid-log phase yeast cells were split in two sets: one set was kept unperturbed, the second set was treated with galactose for 1.5 or 3 h to transiently induce Mps1 expression. Equal numbers of cells were plated on glucose-containing medium to repress Mps1 overexpression. The percentage of yeast cells that formed colonies after Mps1 induction was determined. Values shown are derived from three independent experiments.
The molecular mechanism underlying Mps1-related phenotypes in ufd2Δ cells remains unclear and requires a comprehensive phosphorylation profile of various Mps1 targets in wild-type and ufd2Δ cells and careful analysis of the phosphorylation-defective mutations in Mps1 substrates implicated. To gain molecular insights into the phenotypes of ufd2Δ cells, it will be beneficial to look for suppressors (e.g. mutations or overexpression) of these phenotypes by genetic screens.
Ufd2-mediated Mps1 Degradation Is Conserved from Yeast to Humans
Thus far, the known functions of Mps1, APC, and Ufd2 are largely conserved from yeast to humans (3, 5, 7, 11, 26, 27). We examined whether UFD2a, one human counterpart of Ufd2, is also involved in MPS1 turnover in mammalian cells. UFD2a has been linked previously to proteolysis (27). Analysis of the neuroblastoma cell line NB-1 (28) revealed that UFD2a is among the six genes that were deleted in this cell line. Further studies also support that UFD2a is a candidate neuroblastoma tumor suppressor because it is localized in a region closely associated with neuroblastoma and subject to mutations in tumors (29).
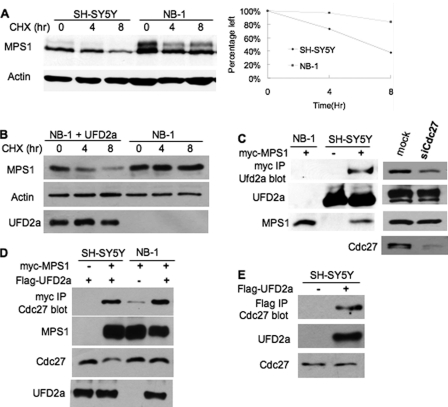
RNAi knockdown of UFD2a was inefficient (data not shown). To assess the role of UFD2a in proteolysis, we employed the wild-type UFD2a-expressing SH-SY5Y and UFD2a-deficient NB-1 (29) neuroblastoma cell lines (Fig. 6C). Interestingly, degradation of human MPS1 was impaired in NB-1 cells (Fig. 6A). Importantly, UFD2a restored Mps1 turnover in NB-1 cells (Fig. 6B). Although it is difficult to detect the interaction between Mps1 and APC or Ufd2 in yeast (for unknown reasons), we found that human MPS1 interacted with endogenous UFD2a by co-immunoprecipitation (Fig. 6C, left panel). Interestingly, the MPS1-UFD2a interaction was reduced in Cdc27 knockdown cells containing compromised APC activity (Fig. 6C, right panel). APC was recently shown to be involved in human MPS1 degradation (30). MPS1 also bound Cdc27, a subunit of APC (Fig. 6D). The MPS1-Cdc27 association was decreased in NB-1 cells deficient in UFD2a (Fig. 6D). We also detected UFD2a-Cdc27 binding (Fig. 6E), suggesting that UFD2a and APC work together in promoting MPS1 ubiquitylation and degradation.
FIGURE 6.
UFD2a regulates human MPS1 degradation. A, human MPS1 degradation is compromised in UFD2a-deficient cells. Wild-type UFD2a-expressing SH-SY5Y cells and UFD2a-deficient NB-1 cells were transiently transfected with Myc-tagged MPS1. 24 h after transfection, cells were split onto a 6-well plate. After 48 h of transfection, cells were treated with 100 μg/ml cycloheximide (CHX) to start the chase. Samples were taken at the indicated time points after protein synthesis shut-off and analyzed by Western blotting with anti-Myc antibody. B, UFD2a expression restores MPS1 turnover in NB-1 cells. The plasmid expressing FLAG-tagged UFD2a or the vector plasmid was transfected into NB-1 cells bearing Myc-tagged MPS1. MPS1 degradation was assessed as described for A. UFD2a expression was ascertained by Western blotting with anti-FLAG antibody. C, MPS1 binds endogenous UFD2a by co-immunoprecipitation. UFD2a-deficient NB-1 cells and UFD2a-containing SH-SY5Y cells were transiently transfected with Myc-tagged human MPS1 or vector as indicated. After 48 h of transfection, cell extracts were prepared and subjected to immunoprecipitation (IP) with anti-Myc antibody, followed by immunoblotting with anti-UFD2a antibody. In the lower panels, 10% input lysates were analyzed by immunoblotting to ascertain the expression of UFD2a and Myc-MPS1. In the right panels, the MPS1-UFD2a interaction was determined in Cdc27 siRNA (siCdc27)- or mock-treated SH-SY5Y cells. Cdc27 knockdown efficiency is shown in the lower right panel. D, UFD2a affects MPS1-Cdc27 association. The binding experiments were carried out as described for C using FLAG-tagged UFD2a and Myc-tagged MPS1 in SH-SY5Y or NB-1 cells. E, UFD2a interacts with Cdc27. The immunoprecipitations were done as described for C. Endogenous Cdc27 was detected using anti-Cdc27 antibody.
We evaluated whether mitotic progression is altered in NB-1 cells. Monastrol leads to the production of monopolar spindles through its inhibition of the Eg5 kinesin, thereby promoting aberrant microtubule-kinetochore attachments whose correction requires the activity of MPS1 (5, 7, 22). Compared with SH-SY5Y cells, NB-1 cells exhibited a delay in the recovery from monastrol treatment (supplemental Fig. S2), suggesting that the UFD2a-mediated degradation of MPS1 contributes to mitotic progression in human cells. Combined, these results suggest that the Mps1-Ufd2 connection is highly conserved.
DISCUSSION
Our study leads to novel insights into the physiological significance of the Ufd2 pathway. Without the help of Ufd2, inefficient ubiquitylation and degradation of substrates such as Mps1 lead to altered cellular regulation. Yeast cells lacking UFD2 exhibit several phenotypes, including increased chromosome stability, enhanced sensitivity to the microtubule-destabilizing drug benomyl, and genetic suppression of yeast mutants defective in spindle checkpoint (Fig. 4). MPS1 overexpression in ufd2Δ cells impedes cell cycle progression and induces cellular toxicity (Fig. 5). Impaired degradation of human MPS1 associated with UFD2a deficiency in NB-1 neuroblastoma cells (Fig. 6 and supplemental Fig. S2) may contribute to tumorigenesis of neuroblastomas. Understanding the functions and mechanisms of Ufd2 will elucidate a poorly characterized pathway in proteolysis that may be crucial to unravel the mechanisms underlying human diseases.
Our understanding of Ufd2-like U-box-containing Ub ligases remains primitive (3, 12). Our results reveal a novel link between Ufd2 and APC, which has a long and still growing list of substrates critical for cell cycle progression and checkpoint control (10, 31, 32). Several mechanisms have been employed to ensure timely ubiquitylation of multiple APC substrates (10, 32, 33, 34). For example, multiple E2 enzymes have been found to cooperate with APC. Interestingly, Ubc4 and Ubc1 perform sequential monoubiquitylation and multiubiquitylation (34).
APC seems to possess potent multiubiquitylation activity in vitro (33, 34). Hence, the involvement of Ufd2 in Mps1 degradation is surprising and further suggests that the efficient multiubiquitylation ability of E3 enzymes does not preclude the employment of another Ub ligase for their substrate ubiquitylation in vivo. However, how Ufd2-like enzymes participate in substrate ubiquitylation remains elusive. We plan to set up an in vitro ubiquitylation assay to further delineate the roles of Ufd2 and APC in Mps1 ubiquitylation. As discussed previously (12), there is no single determinant that confers the Ufd2 specificity, such as a particular E3, degradation signal sequence, oligomerization status, or cellular localization. We favor the model that the combination of these factors may be influential to the selection of Ufd2-like Ub ligases, the support for which would require additional structural and functional studies.
Supplementary Material
Acknowledgments
We are grateful to A. Murray, M. Rose, D. Morgan, K. Nakayama, and S. Jentsch for strains and plasmids. We thank Dr. I. Kim and G. H. Baek for discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
C. Liu, D. van Dyk, V. Choe, J. Yan, S. Majumder, M. Costanzo, X. Bao, C. Boone, K. Huo, M. Winey, H. Fisk, B. Andrews, and H. Rao, unpublished data.
- Ub
- ubiquitin
- SPB
- spindle pole body
- APC
- anaphase-promoting complex
- TAP
- tandem affinity purification tag.
REFERENCES
- 1. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hochstrasser M. (2009) Chem. Rev. 109, 1479–1480 [DOI] [PubMed] [Google Scholar]
- 3. Hoppe T. (2005) Trends Biochem. Sci. 30, 183–187 [DOI] [PubMed] [Google Scholar]
- 4. Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., Jentsch S. (1999) Cell 96, 635–644 [DOI] [PubMed] [Google Scholar]
- 5. Fisk H. A., Mattison C. P., Winey M. (2004) Cell Cycle 3, 439–442 [PubMed] [Google Scholar]
- 6. Lan W., Cleveland D. W. (2010) J. Cell Biol. 190, 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zich J., Hardwick K. G. (2010) Trends Biochem. Sci. 35, 18–27 [DOI] [PubMed] [Google Scholar]
- 8. Palframan W. J., Meehl J. B., Jaspersen S. L., Winey M., Murray A. W. (2006) Science 313, 680–684 [DOI] [PubMed] [Google Scholar]
- 9. Skaar J. R., Pagano M. (2009) Curr. Opin. Cell Biol. 21, 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matyskiela M. E., Rodrigo-Brenni M. C., Morgan D. O. (2009) J. Biol. 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pines J. (2011) Nat. Rev. Mol. Cell Biol. 12, 427–438 [DOI] [PubMed] [Google Scholar]
- 12. Liu C., van Dyk D., Xu P., Choe V., Pan H., Peng J., Andrews B., Rao H. (2010) J. Biol. Chem. 285, 10265–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spencer F., Gerring S. L., Connelly C., Hieter P. (1990) Genetics 124, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005) Cell 120, 73–84 [DOI] [PubMed] [Google Scholar]
- 15. Hosoda M., Ozaki T., Miyazaki K., Hayashi S., Furuya K., Watanabe K., Nakagawa T., Hanamoto T., Todo S., Nakagawara A. (2005) Oncogene 24, 7156–7169 [DOI] [PubMed] [Google Scholar]
- 16. Dikic I., Wakatsuki S., Walters K. J. (2009) Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 18. Kim I., Mi K., Rao H. (2004) Mol. Biol. Cell 15, 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biggins S., Ivanovska I., Rose M. D. (1996) J. Cell Biol. 133, 1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Musacchio A., Salmon E. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 23. Sheltzer J. M., Blank H. M., Pfau S. J., Tange Y., George B. M., Humpton T. J., Brito I. L., Hiraoka Y., Niwa O., Amon A. (2011) Science 333, 1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. (2009) EMBO J. 28, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoppe T., Cassata G., Barral J. M., Springer W., Hutagalung A. H., Epstein H. F., Baumeister R. (2004) Cell 118, 337–349 [DOI] [PubMed] [Google Scholar]
- 27. Matsumoto M., Yada M., Hatakeyama S., Ishimoto H., Tanimura T., Tsuji S., Kakizuka A., Kitagawa M., Nakayama K. I. (2004) EMBO J. 23, 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohira M., Kageyama H., Mihara M., Furuta S., Machida T., Shishikura T., Takayasu H., Islam A., Nakamura Y., Takahashi M., Tomioka N., Sakiyama S., Kaneko Y., Toyoda A., Hattori M., Sakaki Y., Ohki M., Horii A., Soeda E., Inazawa J., Seki N., Kuma H., Nozawa I., Nakagawara A. (2000) Oncogene 19, 4302–4307 [DOI] [PubMed] [Google Scholar]
- 29. Carén H., Holmstrand A., Sjöberg R. M., Martinsson T. (2006) Eur. J. Cancer 42, 381–387 [DOI] [PubMed] [Google Scholar]
- 30. Cui Y., Cheng X., Zhang C., Zhang Y., Li S., Wang C., Guadagno T. M. (2010) J. Biol. Chem. 285, 32988–32998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song L., Rape M. (2010) Mol. Cell 38, 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pines J. (2009) Mol. Cell 34, 135–136 [DOI] [PubMed] [Google Scholar]
- 33. Rape M., Reddy S. K., Kirschner M. W. (2006) Cell 124, 89–103 [DOI] [PubMed] [Google Scholar]
- 34. Rodrigo-Brenni M. C., Morgan D. O. (2007) Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.