Background: The membrane proteins, TatA, TatB, and TatC, enable the transmembrane passage of folded precursor proteins.
Results: The transmembrane helix of TatA makes contacts with TatC and precursors recognized by the TatBC complex.
Conclusion: Following its insertion into the TatBC receptor complex, a Tat signal sequence contacts a nearby monomer of TatA.
Significance: TatA interacts with twin-arginine precursors prior to the actual translocation event.
Keywords: Escherichia coli, Membrane Proteins, Membrane Trafficking, Protein Export, Protein Sorting, TatA, Twin-arginine Translocation
Abstract
Twin-arginine translocation (Tat) is a unique protein transport pathway in bacteria, archaea, and plastids. It mediates the transmembrane transport of fully folded proteins, which harbor a consensus twin-arginine motif in their signal sequences. In Gram-negative bacteria and plant chloroplasts, three membrane proteins, named TatA, TatB, and TatC, are required to enable Tat translocation. Available data suggest that TatA assembles into oligomeric pore-like structures that might function as the protein conduit across the lipid bilayer. Using site-specific photo-cross-linking, we have investigated the molecular environment of TatA under resting and translocating conditions. We find that monomeric TatA is an early interacting partner of functionally targeted Tat substrates. This interaction with TatA likely precedes translocation of Tat substrates and is influenced by the proton-motive force. It strictly depends on the presence of TatB and TatC, the latter of which is shown to make contacts with the transmembrane helix of TatA.
Introduction
Twin-arginine (Tat)2-specific protein translocation machineries serve to transport fully folded proteins across cellular membranes of bacteria, archaea, and plant chloroplasts. Tat client proteins are distinguished from other preproteins by a conserved SRRXFLK sequence motif in their N-terminal signal sequences (RR signal peptides). Most Tat translocases comprise three essential membrane proteins, termed TatA, TatB, and TatC in bacteria and Tha4, Hcf106, and cpTatC in chloroplasts. Despite sharing 20% sequence identity, TatA and TatB are functionally distinct proteins each spanning the membrane by a single transmembrane helix. On the contrary, TatC is a polytopic membrane protein with six predicted transmembrane spans.
The three Tat subunits assemble into various complexes. TatB (1–4) and TatC (1, 5, 6) each have the propensity to individually homo-oligomerize. Together they form multimers of functional hetero-dimeric core units (1, 4, 7–9), which when purified from bacteria often also contain some TatA (1, 10, 11). Likewise, homo-oligomeric assemblies of TatA have been described both in vitro (10, 12–15) and in vivo (4, 16, 17).
The prevailing model for how Tat-specific translocation occurs suggests that RR precursors target the membrane via the TatBC complex (8, 9, 13, 14, 18–21) and that the RR consensus motif is initially recognized by TatC (19, 22–24). Some authors have proposed that an RR signal sequence might first interact with membrane lipids (25, 26) before being handed over to the TatBC receptor complex. Although unfolded polypeptide chains can be translocated by the Tat apparatus (27–29), folding of a Tat substrate generally is a prerequisite for its transport (30–34). Tat translocases were shown to discriminate against improperly folded substrates (30, 35, 36), but it is not understood how this is mechanistically achieved. Some bacterial Tat-dependent redox proteins require dedicated chaperones that guarantee membrane targeting only after folding and co-factor incorporation (reviewed in Refs. 37 and 38). Following a productive binding of a Tat substrate to TatBC, TatA is recruited (15, 19, 39) to initiate the transmembrane passage of the RR precursor protein. Functionally assembled Tat translocases seem to have multiple precursor-binding sites to allow for the simultaneous transport of several precursor proteins (8, 40).
Recent solution (41) and solid state (42) NMR structures of a Bacillus subtilis TatA orthologue confirmed the previously predicted single transmembrane and single amphipathic helix of TatA, which are separated by a hinge region and arranged in an L-shaped manner that is stabilized by extensive contacts between both helices. When reconstituted into planar bicelles, the N-terminal transmembrane domain was found deeply inserted in the lipid bilayer with the hinge region and the proximal part of the amphipathic helix both being immersed in the membrane (42). An Nout orientation of the transmembrane helices of TatA is backed by numerous findings, but inverse topologies of TatA have also been suggested (43, 44). In Escherichia coli, TatA is expressed at ∼25- and 50-fold higher levels than TatB and TatC, respectively. Because of its relative abundance and the finding that it assembles into oligomeric, pore-like structures (12, 45), the idea was borne that TatA functions as a form-fitting transmembrane conduit for folded protein substrates. Some findings, however, seem to be at odds with the pore model (28, 46), and alternative modes of how TatA might achieve the transmembrane passage of Tat client proteins have been suggested (15, 47). Tat-specific translocation requires the proton-motive force (PMF), but how the proton gradient energizes the transport process is not understood in detail. Experimental evidence has been provided that the recruitment of TatA to a TatBC-precursor complex is a PMF-dependent step (39). Whether TatA is recruited only from membrane-integrated TatA protomers or also from a soluble pool of TatA is also still an issue. A special situation prevails in Gram-positive bacteria, which naturally express only TatA and TatC orthologues, suggesting that in these organisms, TatA plays the functional roles of both TatA and TatB. This in fact could experimentally be verified by the isolation of bifunctional tatA alleles suppressing a TatB deficiency in E. coli (48).
To learn more about the functions and properties of TatA, we have generated multiple E. coli TatA variants carrying a site-specific photo-activated cross-linker within both its helices and its unstructured C terminus. This approach allowed characterizing intermolecular contacts between TatA and Tat substrates as well as between TatA and the other subunits of the Tat translocase.
EXPERIMENTAL PROCEDURES
Plasmids
TorA-mCherry was expressed from plasmid pPJ3 (pET22b+/TorA-mCherry), which is a derivative of plasmid pPJ1 (pET22b+/TorA-MalE335). To construct pPJ1, TorA-MalE335 was amplified by PCR using Pfu polymerase (Fermentas), plasmid pTorA-MalE (49) as template, and the oligonucleotide primers 5-NdeI TorA-MalE and 3-XhoI TorAMalE335 (supplemental table). The PCR fragment and the recipient vector pET22b+ were digested with NdeI and XhoI and ligated. To obtain plasmid pPJ3, the MalE335 fragment of pPJ1 was excised using EcoRI and XhoI and replaced by an mCherry fragment that had been amplified by PCR using plasmid pRSET-mCherry (50) as template and the primers 5-EcoRI-mCherry and 3-XhoI-mCherry (supplemental table) introducing flanking EcoRI and XhoI restriction sites. To generate plasmid pPJ5 encoding the KK variant of TorA-mCherry, plasmid pPJ3 was site-specifically mutagenized according to the QuikChange site-directed mutagenesis kit protocol (Stratagene) using the primers 5-TorA KK for and 3-TorA KK rev (supplemental table).
Plasmid pKSMSufI-RR (19) was used for the expression of pSufI and plasmid pKSMSufI-KK (19) to synthesize the KK variant of SufI. Plasmid p8737 (51) containing the tatABCD operon under T7 promoter control served as template to introduce the various stop codons into tatA as described (18) using the pairs of forward and reverse primers listed in the supplemental table. Plasmid pSup-BpaRS-6TRN(D86R) (52) encodes a Bpa-tRNA synthetase and a Bpa-specific amber suppressor tRNA.
To construct plasmid pQE60_TatAhis-stopL10, the tatA gene sequence was amplified from plasmid pQE60-TatABC (22) using the primers TatA ncoI bglII for and ncoI bglII rev (supplemental table). Plasmid pQE60 encoding the His tag and the PCR fragment coding for a stop codon-less tatA were restricted with NcoI and BglII and ligated. The amber stop codon at position Leu-10 of TatA was engineered as detailed above.
In Vitro Reactions
Cell extracts used for in vitro synthesis of precursor proteins were obtained from E. coli strains SL119 (53) and Top10 (Invitrogen). Cells were grown and S-135 cell extracts were prepared as described (54). Coupled transcription/translation of the different precursor proteins from plasmid DNAs was performed as described (54).
Membrane vesicles were added 10 min after starting the synthesis reaction and incubated for 25 min at 37 °C. To assay protein translocation into inside-out inner membrane vesicles (INV), 15 μl of each reaction were treated with 15 μl of 10% (w/v) trichloroacetic acid (TCA), and 30 μl were incubated with proteinase K according to Ref. 54. Carbonyl cyanide m-chlorophenyl-hydrazone (CCCP) was added at the indicated concentrations together with INV. For cross-linking, 50-μl reactions were placed in Eppendorf tubes on ice, irradiated with UV light (365 nm, 80 Joule) for 10–20 min, and then mixed with 50 μl of 10% (w/v) TCA. Control experiments had shown that in these conditions, cross-linking reached its maximum within 7 min. ATP depletion using glucose and hexokinase was performed as described previously (18).
Sodium carbonate extraction was performed by treating a 50-μl reaction aliquot with 50 μl of freshly prepared, ice-cold 0.4 m Na2CO3. After 30 min at 4 °C, samples were centrifuged for 30 min at 70,000 rpm in a Beckman TLA100 ultracentrifuge (Beckman-Coulter) using a TLA 100.3 rotor. Supernatants were withdrawn, precipitated with 100 μl of 10% (w/v) TCA, and resuspended in SDS-PAGE loading buffer. The pellets were directly dissolved in 30 μl of SDS-PAGE loading buffer.
Membrane Vesicles
Tat+-INV were prepared as described previously (54) from E. coli strain BL21(DE3) transformed with plasmids p8737. Tat+-INV containing TatA amber mutants were prepared from E. coli strain BL21(DE3) transformed with plasmid pSup-BpaRS-6TRN(D86R) and p8737 carrying the individual tatA mutations. Expression of tat genes was induced with 1 mm isopropyl β-d-thiogalactopyranoside when cell cultures had reached an optical density at 600 nm (A600) of 0.5, and growth was continued up to an A600 of 2.0 or for a total of 3 h. To suppress the amber stop codons, 1 mm p-benzoyl-phenylalanine (Bpa) was added at the same time as isopropyl β-d-thiogalactopyranoside from a 1 m Bpa stock solution prepared in 1 m NaOH.
Miscellaneous
For Western blotting, 4 μl of INV (A280 ∼40 units/ml) were each dissolved in 96 μl of SDS-PAGE loading buffer. Of this solution, 5 μl were used for SDS-PAGE and Western transfer, when blots were decorated with anti-TatA antibodies, and 20 μl were used when using anti-TatB and anti-TatC antibodies. Polyclonal antibodies against TatA, TatB, and TatC were raised in rabbits as described (19, 51). The second antibody was goat anti-rabbit IgG coupled to alkaline phosphatase (Sigma). Detection was performed using nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate stock solution (Roche Applied Science) following the manufacturer's instructions. SDS-electrophoresis using 15% polyacrylamide gels was performed as described previously (54). For UV irradiation of membrane vesicles, 4 μl of INV were each diluted with 96 μl of INV Puffer (54) in 96-well microtiter plates, irradiated on ice for 20 min, transferred to an Eppendorf tube, and precipitated with 100 μl of 10% TCA. The supernatant obtained after centrifugation for 10 min at 13,000 rpm was discarded, and each pellet was dissolved in 100 μl of SDS-PAGE loading buffer.
RESULTS
Upon Binding to the Tat Translocase, RR Precursors Make Multiple Contacts with TatA
To probe for contacts between RR precursors and TatA, we individually exchanged numerous amino acids of the transmembrane, the amphipathic helix, and the C-terminal part of E. coli TatA against the photo-activatable cross-linker Bpa (Fig. 1A). To this end, amber stop codons were introduced by mutagenizing PCR into the selected positions within the tatA gene (cf. Fig. 1A) contained in the tatABCD operon of plasmid p8737. This plasmid allows T7 promoter-dependent high expression of TatABC, which is required to obtain inner membrane vesicles displaying sufficient Tat-specific translocation activity (51, 55). These amber stop codon variants of tatA were transformed into E. coli BL21(DE3) cells expressing an amber suppressor tRNA and a cognate Bpa-tRNA synthetase. When transformants were grown in the presence of Bpa, suppression of the individual amber stop codons by the incorporation of the cross-linker Bpa occurred. This is shown by the Bpa-mediated increase in full-size TatA over the level of chromosomally encoded wild-type TatA obtained in the absence of Bpa (Fig. 1E). INV containing the Bpa-TatA variants were then prepared from the Bpa-grown cells. Radiolabeled RR precursor proteins synthesized by cell-free transcription/translation were allowed to bind to, and translocate into, these vesicles, and cross-links that formed upon UV irradiation of the samples were analyzed by SDS-PAGE and phosphorimaging.
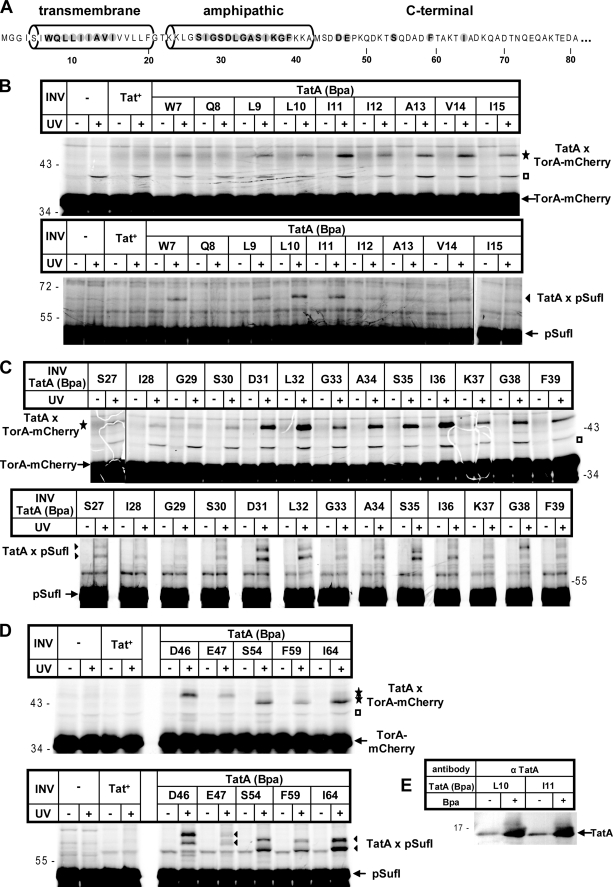
FIGURE 1.
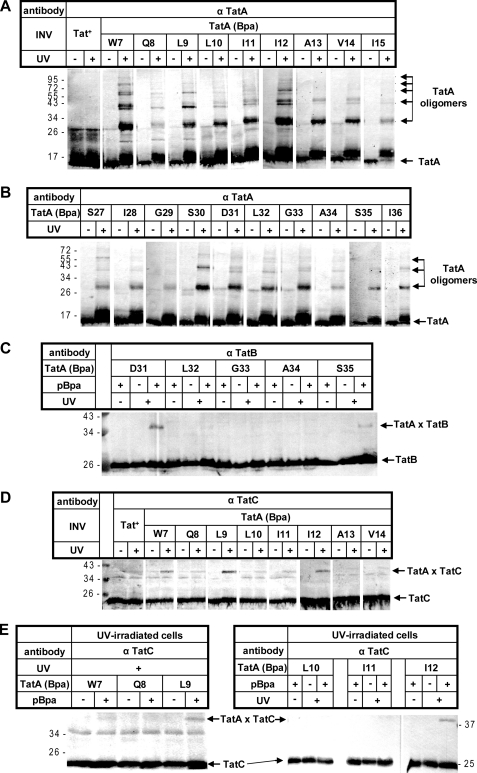
Extensive contacts between large parts of TatA and membrane-bound RR precursor proteins. A, amino acid sequence of E. coli TatA (shown are 81 of the 89 residues) with its predicted transmembrane and amphipathic helices according to Ref. 16. Amino acids exchanged against Bpa by amber stop codon suppression are highlighted. B–D, the RR precursor proteins TorA-mCherry and pSufI were in vitro synthesized and radiolabeled by cell-free transcription/translation. Where indicated, E. coli INV were added 10 min after starting the synthesis. INV were prepared from TatABC-overproducing E. coli strains expressing either wild-type TatA (Tat+) or one of the indicated TatA (Bpa) variants. Following an incubation period of 25 min, reactions were stopped directly with TCA or first UV-irradiated on ice (+UV). Radiolabeled proteins were separated by SDS-PAGE and visualized by phosphorimaging. UV-dependent adducts to TorA-mCherry and pSufI are labeled with stars and triangles, respectively. Open squares, TatA-independent cross-linking product. Numbers to the left and right indicate the molecular masses of marker proteins. E, INV prepared from strains that expressed the indicated TatA variants and that were grown in the absence or presence of Bpa were immunoblotted against anti-TatA antiserum (α TatA).
Fig. 1, B–D, shows the results for the model Tat substrate TorA-mCherry as well as the natural RR precursor SufI (FtsP). TorA-mCherry is translocated into INV to a comparable extent as pSufI (cf. supplemental figure, panel E). Virtually all membrane vesicles that carried Bpa at one position of a consecutive stretch from Trp-7 up to Ile-15 within the transmembrane helix of TatA yielded an ∼47-kDa adduct to TorA-mCherry (Fig. 1B, star). These adducts were obtained only upon irradiation with UV light to initiate cross-linking to Bpa (compare −/+ UV). The radiolabeled ∼39-kDa band (open square) is an unspecific UV-induced product that does not result from cross-linking to TatA because it also appeared in samples that did not contain Bpa variants of TatA (Fig. 1B, lanes no INV and Tat+). TatA(Q8Bpa)3 was the only TatA variant that did not show a clear UV-dependent cross-link that exceeded the background signal of an unspecific band seen with INV expressing only wild-type TatA (Tat+-INV). As expected for an interaction between TorA-mCherry and the vesicle-borne TatA, a Bpa-specific adduct was missing in samples that did not contain INV. Essentially identical results were obtained for the natural RR precursor pSufI (Fig. 1B, triangle), although in this case, cross-links were less well discernable from the background noise of labeled bands.
Similarly, Bpa incorporated at positions Ser-27 up to Phe-39 within the amphipathic helix of TatA yielded UV-dependent and Bpa-specific cross-links both with TorA-mCherry and with pSufI (Fig. 1C, star and triangles). In contrast to Bpa mutants of the transmembrane helix of TatA, most TatA variants of the amphipathic helix gave rise to two adducts to pSufI (triangles), which differed in their apparent molecular masses by ∼4 kDa. The ratio between the intensities of both adducts varied with the cross-linker position. For instance, the lower species dominated for TatA(K37Bpa), whereas the upper species dominated for TatA(G38Bpa). The origin of the double cross-links, however, is not known. A mutant precursor of SufI with a non-cleavable signal peptide gave the same double cross-links (not shown), ruling out that the pair of adducts resulted from individual cross-links to the precursor and mature forms of SufI. Similar double cross-links had been obtained for pSufI and the amphipathic helix of TatB (18) and conceivably might reflect two conformers of in vitro synthesized pSufI that result in differently branched adducts. Cross-links to TorA-mCherry and pSufI were also observed for Bpa positions at Asp-46, Glu-47, Ser-54, Phe-59, and Ile-64, all located in the C-terminal part of TatA following the amphipathic helix (Fig. 1D). Why some of these adducts to the C terminus of TatA displayed smaller apparent molecular masses than others is not known.
The results depicted in Fig. 1 indicate that RR precursor proteins upon contact with TatABC-containing membrane vesicles come into close proximity to large parts of TatA encompassing both its helices. All vesicles carrying Bpa mutants of TatA were tested for their translocation activity toward TorA-mCherry and pSufI. All of them yielded decreased transport efficiencies (supplemental figure). There was, however, no clear correlation between the translocation proficiency of individual TatA variants and the intensity with which they cross-linked to the RR precursors (compare Fig. 1, B–D, with the supplemental figure). Furthermore, the fact that the varying intensities of cross-linking to both helices (cf. Fig. 1, B and C) did not follow a clear helical pattern suggests that the interacting TatA molecules must be rather flexible at this stage of contact. Moreover, quite different from the binding of RR precursors to TatB (18), no higher molecular mass adducts between the RR precursors and TatA were obtained. Such adducts would be expected to form if an oligomeric TatA structure would allow more than one TatA monomer to bind to a single precursor molecule.
Contacts of RR Precursors to TatA Require an Intact RR Signal and the Presence of TatB and TatC
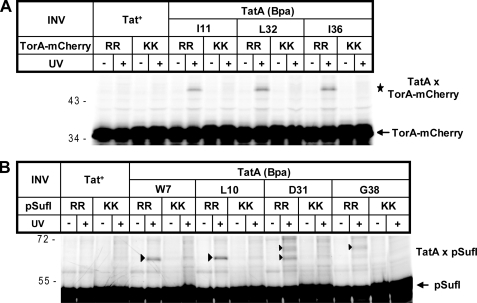
To examine the functional relevance of the precursor-TatA contacts visualized in Fig. 1, we performed the same cross-linking experiments, now using pSufI and TorA-mCherry variants, in which the double arginine had been replaced by the transport-defective twin-lysine pair. As can be seen in Fig. 2, cross-linking to sites in both helices of TatA was drastically reduced in the case of the KK precursors. This suggests that the observed TatA cross-links reflect a functional stage in twin-arginine translocation that requires a previous quality control of the RR signal peptide by TatBC.
FIGURE 2.
Cross-linking to TatA depends on an intact twin-arginine motif. Experimental details are as in the legend for Fig. 1. A, comparison between the wild-type precursor TorA-mCherry (RR) and a mutant derivative, in which the RR pair had been exchanged by two lysines (KK). B, as in A except that the wild-type RR precursor of pSufI is compared with its KK mutant. In both panels, UV-dependent adducts to TorA-mCherry and pSufI are labeled with stars and triangles, respectively.
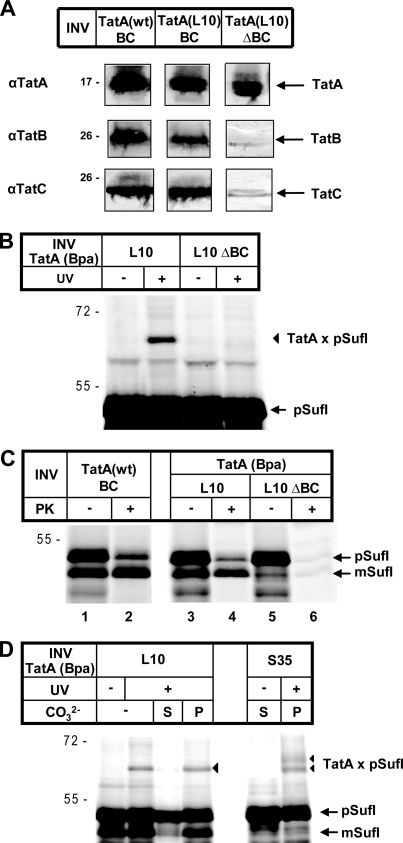
Likewise, the cross-link between pSufI and TatA(L10Bpa) was completely lost upon deprivation of TatB and TatC from INV (Fig. 3, A–C). As mentioned above, all TatA variants were overexpressed in E. coli strains from a plasmid-borne copy of the tatABC operon. Fig. 3A demonstrates that wild-type TatA and TatA(L10Bpa) were recovered to the same levels from equivalent amounts of the respective INV (left and middle columns). Both INV populations also contained comparable levels of TatB and TatC. The TatA(L10Bpa) mutant vesicles strongly cross-linking to pSufI (Fig. 3B) showed translocation activity, although less than INV carrying TatA(wt) (see also the supplemental figure). Translocation activity is indicated by the amounts of pSufI and mature SufI that each remained undigested by proteinase K (Fig. 3C, lanes 2 and 4). In contrast, INV (TatA(L10)ΔBC) obtained from a strain overexpressing solely TatA(L10Bpa) and therefore containing only the chromosomally encoded low amounts of TatB and TatC (Fig. 3A, right column) were largely inactive in translocating pSufI (Fig. 3C, lane 6). Despite the comparably high amounts of TatA present in the TatA(L10)ΔBC vesicles, an adduct to pSufI was not generated (Fig. 3B), indicating that a complete TatABC translocase was required for the observed contacts between RR precursors and TatA to form.
FIGURE 3.
Cross-linking of an RR precursor to translocase-embedded TatA. A, immunoblot analysis of the TatABC contents of INV that had been prepared from cells overproducing, besides TatBC, wild-type TatA (Tat+-INV, here denominated TatA(wt)BC) or the L10Bpa variant of TatA (TatA(L10)BC)). TatA(L10)ΔBC INV were obtained from a strain overexpressing selectively the TatA variant and therefore containing only the low amounts of chromosomally expressed TatBC. The amounts of vesicles applied to SDS-PAGE and immunoblot analysis were normalized with respect to their adsorption at 280 nm (54). B, no cross-linking to TatA in the presence of substoichiometric amounts of TatBC. For experimental details, see the legend for Fig. 1. UV-dependent adducts to pSufI are labeled with triangles. C, pSufI was synthesized in vitro in the presence of the indicated INV. To assay translocation of pSufI into these vesicles, one aliquot of each reaction was treated with proteinase K (PK) to visualize the vesicle-protected, i.e. translocated fractions of the precursor and mature form of SufI (pSufI and mSufI, respectively). In the absence of high amounts of TatBC, translocation is negligible. D, pSufI was synthesized in vitro in the presence of the indicated INV. Samples were UV-irradiated as indicated and either directly TCA-precipitated or first treated with Na2CO3 and separated into soluble (S) and pelletable (P) material.
Further support for the observed substrate-TatA cross-links reflecting an authentic interaction between an RR precursor and the intact Tat translocase comes from the results of Fig. 3D demonstrating a firm anchorage of the cross-links in the lipid bilayer of the membrane vesicles. After cross-linking pSufI to TatA(L10Bpa) and TatA(S35Bpa) membrane vesicles, reactions were treated with sodium carbonate and centrifuged to separate peripheral from integral membrane proteins. The TatA-pSufI adducts (Fig. 3D, triangles) were entirely recovered from the pellet fraction, indicating that pSufI was bound to membrane-embedded TatA. Incidentally, the TatA(S35Bpa) INV used in this experiment did not show translocation of pSufI as judged from the missing band of mature SufI (mSufI) when compared with the TatA(L10Bpa) variant. The two adducts obtained for the Ser-35 variant of TatA (Fig. 3D, triangles) can therefore not result from one cross-link to the precursor and one to the mature form of SufI as discussed above.
Contacts between RR Precursors and TatA Are Sensitive toward Dissipation of the Proton-motive Force and Kinetically Not Linked to Translocation
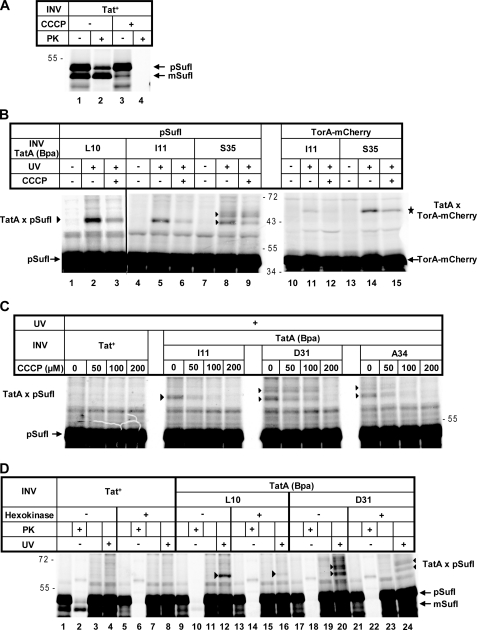
It is not known which step of the Tat-specific translocation pathway is dependent on the PMF. A common view is that following binding of an RR precursor to TatB and TatC, TatA protomers are recruited to this complex in a PMF-dependent manner. A more recent analysis suggested that the electrochemical potential of the PMF was required for an early as well as a late translocation event (56). We therefore were interested in finding out whether or not the observed precursor-TatA contacts were influenced by the PMF. To dissipate the H+ gradient of the membrane vesicles, we used the protonophore CCCP. As illustrated in Fig. 4A, CCCP at 100 μm concentration inhibited translocation of pSufI into INV, as indicated by the complete disappearance of proteinase K-resistant material in the presence of CCCP (compare lanes 2 and 4). The same concentration of CCCP also markedly lowered cross-linking between residue Leu-10 and Ile-11 within the transmembrane helix of TatA and TorA-mCherry and pSufI (Fig. 4B, lanes 2, 3, 5, and 6, triangle; lanes 11 and 12, star). In addition, CCCP negatively affected cross-linking of both precursors to Ser-35 within the amphipathic helix of TatA, although the effect seemed to be less pronounced than for the cross-link to the transmembrane helix (Fig. 4B, lanes 8 and 9, triangles; lanes 14 and 15, star). Fig. 4C, however, illustrates that with increasing concentrations of CCCP, the TatA-pSufI adducts (triangles) to the amphipathic helix of TatA (Asp-31 and Ala-34) were suppressed to the same extent as the cross-link to Ile-11 in the transmembrane helix of TatA.
FIGURE 4.
A collapse of the proton-motive force at the membrane vesicles interferes with cross-linking of precursor and TatA. A, pSufI was synthesized in vitro in the presence of Tat+-INV. CCCP at a final concentration of 100 μm or alternatively its solvent dimethyl sulfoxide (DMSO) was added during the incubation with INV as indicated. Translocation of pSufI was analyzed as described in the legend for Fig. 3C. B, cross-linking of pSufI and TorA-mCherry to the indicated Bpa variants of TatA in the absence or presence of 100 μm CCCP. UV-dependent adducts to TorA-mCherry and pSufI are labeled with stars and triangles, respectively. C, as in B except that CCCP was titrated from 0 to 200 μm. D, dissipation of the PMF by the combined addition of glucose and hexokinase 10 min before reactions synthesizing pSufI were supplemented with the indicated INV. Samples were either UV-irradiated to obtain adducts to TatA (triangles) or treated with proteinase K (PK) to monitor translocation.
To confirm the PMF sensitivity of the precursor-TatA interactions, we also employed ATP depletion to prevent H+ pumping by the F1-ATPase. In the experiment depicted in Fig. 4D, INV were added only after the in vitro synthesis of pSufI was stopped with puromycin. As observed before (51) and for still unknown reasons, the posttranslational addition of INV usually leads to a decrease in Tat-specific translocation efficiency (Fig. 4, panels A and D, compare lanes 1 and 2). Translocation was, however, completely abolished when glucose and hexokinase had been added to remove the ATP from the in vitro reactions prior to the addition of INV (Fig. 4D, lanes 5 and 6). Cross-linking to both Leu-10 in the transmembrane helix (compare lanes 12 and 16, triangles) and Asp-31 in the amphipathic helix of TatA (compare lanes 20 and 24, triangles) was clearly diminished in these conditions. Thus, cross-linking of RR precursors to TatA, as observed under our experimental conditions, is negatively affected by dissipation of the PMF of the vesicles.
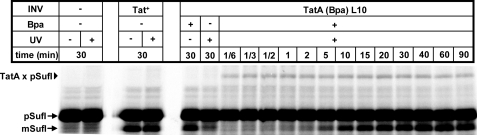
To further characterize this precursor-TatA interaction, we recorded the kinetics of cross-linking of pSufI to Leu-10 in the transmembrane helix of TatA (Fig. 5). The adduct (triangle) appeared as early as 10 s after initiating the reaction by adding membrane vesicles and obviously reached its maximum already after 20 s. Thus, the steady-state level of the SufI-TatA adducts was reached markedly before that of the translocation of SufI. Provided that the TatA cross-linked species represents a true intermediate of SufI on its way across the membrane, binding to TatA would precede the actual translocation event.
FIGURE 5.
Cross-linking of an RR precursor to the transmembrane helix of TatA occurs with faster kinetics than accumulation of translocated material. pSufI was synthesized in the absence or presence of the indicated INV. UV irradiation yielding the pSufI-TatA cross-link (triangle) was started at the indicated times following the addition of INV. The appearance of the signal sequence-less mSufI by the INV-associated signal peptidase indicates the time-dependent translocation into the vesicles. For most of the assays shown, TatA(Bpa)L10 INV had been prepared from cells grown on Bpa (+ Bpa). Cells grown in the absence of Bpa (-Bpa) could not suppress the stop codon at position Leu-10 of TatA. INV derived from these cells therefore failed to cross-link to pSufI.
Even in the Absence of Tat Substrates, the Transmembrane Helix of TatA Is Juxtaposed to TatC
The contacts between an RR precursor and the transmembrane helix of TatA that we picked up by our cross-linking approach appeared as soon as membranes were added. Moreover, all cross-links obtained account in size for 1:1 complexes between precursor and TatA, although the placement of the cross-linker into TatA would have allowed us to detect interactions of a single precursor molecule with more than one TatA monomer. These findings would argue against the precursor-TatA adducts representing translocation intermediates that were trapped on their way across a cylindrical structure made of multiple TatA transmembrane helices. Conversely, if Tat substrates come into contact with TatA, notably its transmembrane helix, as soon as they target the TatBC receptor complex, TatA should be located in close proximity to TatBC. We therefore irradiated INV containing individual Bpa variants of TatA with UV light in the absence of in vitro synthesized RR precursors. The irradiated samples were then probed by immunoblots using TatA, TatB, and TatC antibodies for nearest neighbors of TatA in the absence of ongoing Tat translocation. The results are summarized in Fig. 6. For almost every TatA variant of the transmembrane (Fig. 6A) and the amphipathic helix (Fig. 6B), multiple oligomeric complexes of TatA became visible when the Western blots were decorated with anti-TatA antiserum. This is consistent with previous data (15, 16, 45, 57, 58) suggesting oligomerization of TatA via both helices. When the cross-linked INV were probed with anti-TatB antiserum, clear TatB cross-reactive adducts were obtained for Asp-31 and Ser-35 in the amphipathic helix of TatA (Fig. 6C). By size, these adducts correspond to 1:1 TatA-TatB complexes, which, however, were obtained only when the cross-linker was positioned in the amphipathic helix of TatA.
FIGURE 6.
Both in inner membrane vesicles as well as in intact cells, the transmembrane helix of TatA is in close proximity to TatC. A and B, the indicated vesicles were irradiated with UV light to initiate cross-linking to the Bpa variants of TatA. Homo-oligomeric TatA adducts that were revealed by immunoblots using anti-TatA antibodies (α TatA) are marked. C, as in A and B except that the blot is decorated with anti-TatB antibodies. TatA adducts recognized by anti-TatB antibodies are indicated. D, as in C except that TatA adducts recognized by anti-TatC antibodies are highlighted. E, as in D except that whole cells had been irradiated with UV light.
In marked contrast, probing the irradiated INV with anti-TatC antiserum revealed numerous 1:1 TatA-TatC adducts (Fig. 6D). They were exclusively found for cross-linker sites in the transmembrane helix of TatA and were most prominent for the positions Trp-7, Leu-9, and Ile-12. To exclude that these intersubunit contacts of the Tat translocase were artifacts of the isolated membrane vesicles, we also irradiated whole cells expressing the Bpa variants of TatA. As shown in Fig. 6E, cross-links between the positions Leu-9 and Ile-12 in the transmembrane helix of TatA and TatC could clearly be detected. These results strongly suggest that even in the idling state of the Tat translocase, the transmembrane part of TatA is juxtaposed to TatC. This situation could well explain why an RR precursor upon membrane interaction also comes into contact with one TatA monomer. Whether the PMF dependence of these contacts to TatA reflects the recruitment of the TatA monomer to TatBC or a necessary conformational switch in any of the interacting partners required for proper positioning remains to be elucidated.
DISCUSSION
Although in chloroplasts the three components of the Tat translocase clearly partition into two separable complexes, a hetero-oligomeric TatBC complex and a homo-oligomeric TatA complex (14), TatBC complexes when purified from E. coli were often found to also contain TatA (10, 11). In addition, TatA was reported to stabilize the TatBC complex (59). In agreement with those findings, the studies presented here have revealed functional and structural vicinity between TatA and the TatBC receptor complex, both in the presence and in the bona fide absence of a Tat substrate protein. Thus, by Bpa-scanning analysis of the transmembrane helix of TatA, we identified 1:1 adducts between TatA and TatC, indicating intramembrane proximity between both Tat subunits. This is consistent with a previous study employing bimolecular fluorescence complementation (4). Our photo-cross-linking analysis also revealed that TatA provides multiple contact sites for RR precursors probably already at the stage of membrane targeting, which is known to occur at the TatBC subunits (8, 9, 13, 14, 19–21, 40). The formation of precursor-TatA adducts reached saturation markedly before translocation into membrane vesicles did. Cross-links between RR precursors and TatA were not obtained in the absence of TatBC, suggesting a functional hierarchy of precursor contacts with the Tat translocase subunits such that interaction with TatA required a previous recognition of the RR precursor by the TatBC receptor complex. In complete agreement with an obligate identification of the RR consensus motif prior to establishing contacts with TatA, KK mutant precursors failed to cross-link to TatA, whereas they have repeatedly been found to still physically associate with the TatBC complex (13, 36, 60). In contrast to the primary membrane targeting of RR precursors at TatB and TatC (51, 61), contacts with TatA turned out to be sensitive toward dissipation of the PMF. This means that the observed interaction of RR precursors with TatA does not directly coincide with their binding to TatBC but rather reflects a subsequent targeting event that is distinguished by its requirement for the PMF.
Cross-links between RR precursors and TatA were obtained all along a stretch encompassing the first 64 residues of the 89-amino acid-long E. coli TatA. Contact sites include the transmembrane and the amphipathic helix of TatA as well as its unstructured C terminus. A previous cross-linking analysis of the interaction of membrane-targeted, folded RR precursor proteins with the TatA homologue TatB revealed extended contacts with the amphipathic helix of TatB (18). We were therefore not surprised to also obtain cross-links between RR precursors and the amphipathic helix of TatA, conceivably reflecting extramembrane contacts with a surface-exposed precursor protein. On the contrary, the observed cross-links to residues of the transmembrane helix of TatA as N-proximal as Trp-7 strongly suggest contacts occurring in the lipid bilayer of the plasma membrane.
Consistent with the proposed pore function of TatA, those intramembrane contacts between a Tat substrate and TatA could in principle be snapshots of the transmembrane passage of the precursor. Several of our findings, however, contradict such an assumption and rather suggest that the observed precursor-TatA contacts precede the actual transmembrane translocation process. First of all, the experimentally verified ring-like arrangement of the transmembrane helices of TatA would predict that a folded Tat substrate protein on its way across such a cylindrical TatA structure makes contacts with numerous TatA protomers. All precursor-TatA cross-links obtained in this study were, however, 1:1 adducts, i.e. complexes consisting of one precursor molecule and one TatA monomer only. This is in complete contrast to a previous study on larger complexes between RR precursors and TatB oligomers that were interpreted to suggest encapsulation of a membrane-targeted RR precursor by multiple amphipathic helices of TatB (18). Furthermore, we would have expected that contacts of a translocating Tat substrate with a ring of transmembrane helices of TatA were restricted to the inner surface of the transmembrane helices that face the lumen of the pore. In contrast, our cross-linking data did not disclose any preferred binding face on the transmembrane helices of TatA, indicating that at the stage where our precursor-TatA adducts were formed, TatA hardly could have been trapped in a rigid oligomeric structure. Moreover, recent EPR spectroscopy of detergent-solubilized E. coli TatA suggested that residues Ile-12 and Val-14 of the transmembrane part define an interface between adjacent TatA subunits (45), whereas in our experimental setup, both residues were free to contact RR precursors. Notably, the fact that we could not observe cross-linking of RR precursors to more than one TatA molecule was not the result of a defective oligomerization of TatA caused by the incorporation of Bpa because the Bpa variants of TatA were all able to form higher-order homo-oligomers, as illustrated in Fig. 6. Finally, as mentioned earlier, Bpa variants of TatA cross-linked to the RR precursors, even if the Bpa mutation impaired their translocation activity, for which reason the observed interaction between precursor and TatA cannot encompass a translocation event.
For the aforementioned reasons, it seems unlikely that the contacts between RR precursors and the transmembrane helix of TatA visualized here by site-directed photo-cross-linking reflect an intermediary stage of the transmembrane passage. The question then arises as to what pretranslocational event could lead to an interaction between a membrane-targeted RR precursor and the trans-sided end of the transmembrane helix of TatA? One likely scenario would be that upon targeting, the signal peptide with its N terminus fixed in the cytoplasm (62) loops into the plane of the membrane to reach out toward the N terminus of TatA. Experimental evidence has been provided indicating that TatB and TatC cooperate in accommodating and recognizing Tat signal sequences. Thus, the findings that RR precursors cross-link to Leu-9 in the N-proximal part of the transmembrane part of TatB (18) and that suppressor mutants restoring transport of a defective precursor, whose RR motif had been changed to KQ, mapped to the adjacent Glu-8 residue of TatB (21) point to a deep insertion of a Tat signal sequence prior to the translocation of the passenger protein. Such a transmembrane topography of a Tat signal sequence mediated by TatB and TatC could satisfactorily explain why it can contact the N-proximal part of the transmembrane helix of TatA. Cross-linking of a membrane-bound precursor to the transmembrane part of TatA would then indicate that at some point, TatA joins with TatC and TatB in forming a signal sequence-binding pocket. In fact, a direct interaction of a Tat signal sequence with TatA was previously demonstrated by placing a photo-cross-linker into the signal sequence itself (19). Notably, these cross-links to TatA were abolished by dissipating the PMF of membrane vesicles (19). That an RR signal peptide is able to recruit TatA to the TatBC receptor in a PMF-dependent manner has been shown by the K. Cline group (39) for the Tat system of plant chloroplasts.
As outlined above, our cross-linking data would be consistent with a twin-arginine signal peptide recruiting a TatA monomer to the TatBC-precursor complex. If so, the signal sequence-bound TatA monomer could later on serve as the nucleation point for the formation of the proposed oligomeric TatA pore structure in line with previous results demonstrating the oligomerization of the chloroplast TatA orthologue by a Tat signal peptide (57). We do, however, not want to exclude the possibility that the signal peptide initially recruits dimers or tetramers of TatA. Nevertheless using the same experimental approach that previously allowed detection of two TatB molecules cross-linking to a single Tat substrate (18), here we did not obtain precursor-TatA adducts that contained more than a monomer of TatA.
We would like to interpret our data such that a twin-arginine signal upon recognition by TatC and its transmembrane accommodation in a concerted TatBC binding pocket recruits TatA monomers in a PMF-sensitive manner. Whether the association itself is driven by the PMF or whether the PMF affects the conformation of any of the binding partners in the TatBC precursor complex so as to enable capturing of TatA remains to be deciphered.
Supplementary Material
Acknowledgments
We gratefully acknowledge Stefan Zoufaly for the help constructing plasmid pPJ5, Gottfried Eisner for plasmid pQE_TatAhis, Tobias Flecken for plasmid pQE60_TatAhis-stopL10, and Michael Moser and Sascha Panahandeh for technical advice.
This work was supported by Sonderforschungsbereich 746 and Forschergruppe 929 of the Deutsche Forschungsgemeinschaft.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental table and figure.
Throughout this study, designations indicate TatA variants using the following style: TatA(Q8Bpa) indicates a TatA variant with the natural glutamine residue at position 8 being exchanged against p-benzoyl-phenylalanine, TatA(L10Bpa) indicates a TatA variant with the natural leucine residue at position 10 being exchanged against p-benzoyl-phenylalanine, and so forth.
- Tat
- twin-arginine translocation
- Bpa
- p-benzoyl-phenylalanine
- CCCP
- carbonyl cyanide m-chlorophenyl-hydrazone
- INV
- inside-out inner membrane vesicle(s)
- KK
- twin-lysine
- RR
- twin-arginine
- PMF
- proton-motive force.
REFERENCES
- 1. Orriss G. L., Tarry M. J., Ize B., Sargent F., Lea S. M., Palmer T., Berks B. C. (2007) FEBS Lett. 581, 4091–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Behrendt J., Lindenstrauss U., Brüser T. (2007) FEBS Lett. 581, 4085–4090 [DOI] [PubMed] [Google Scholar]
- 3. Lee P. A., Orriss G. L., Buchanan G., Greene N. P., Bond P. J., Punginelli C., Jack R. L., Sansom M. S., Berks B. C., Palmer T. (2006) J. Biol. Chem. 281, 34072–34085 [DOI] [PubMed] [Google Scholar]
- 4. Kostecki J. S., Li H., Turner R. J., DeLisa M. P. (2010) PLoS One 5, e9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Punginelli C., Maldonado B., Grahl S., Jack R., Alami M., Schröder J., Berks B. C., Palmer T. (2007) J. Bacteriol. 189, 5482–5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maldonado B., Kneuper H., Buchanan G., Hatzixanthis K., Sargent F., Berks B. C., Palmer T. (2011) FEBS Lett. 585, 478–484 [DOI] [PubMed] [Google Scholar]
- 7. Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., Robinson C. (2001) J. Biol. Chem. 276, 20213–20219 [DOI] [PubMed] [Google Scholar]
- 8. Tarry M. J., Schäfer E., Chen S., Buchanan G., Greene N. P., Lea S. M., Palmer T., Saibil H. R., Berks B. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13284–13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richter S., Brüser T. (2005) J. Biol. Chem. 280, 42723–42730 [DOI] [PubMed] [Google Scholar]
- 10. Oates J., Barrett C. M., Barnett J. P., Byrne K. G., Bolhuis A., Robinson C. (2005) J. Mol. Biol. 346, 295–305 [DOI] [PubMed] [Google Scholar]
- 11. McDevitt C. A., Hicks M. G., Palmer T., Berks B. C. (2005) Biochem. Biophys. Res. Commun. 329, 693–698 [DOI] [PubMed] [Google Scholar]
- 12. Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., Berks B. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McDevitt C. A., Buchanan G., Sargent F., Palmer T., Berks B. C. (2006) FEBS J. 273, 5656–5668 [DOI] [PubMed] [Google Scholar]
- 14. Cline K., Mori H. (2001) J. Cell Biol. 154, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dabney-Smith C., Mori H., Cline K. (2006) J. Biol. Chem. 281, 5476–5483 [DOI] [PubMed] [Google Scholar]
- 16. Greene N. P., Porcelli I., Buchanan G., Hicks M. G., Schermann S. M., Palmer T., Berks B. C. (2007) J. Biol. Chem. 282, 23937–23945 [DOI] [PubMed] [Google Scholar]
- 17. Leake M. C., Greene N. P., Godun R. M., Granjon T., Buchanan G., Chen S., Berry R. M., Palmer T., Berks B. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15376–15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maurer C., Panahandeh S., Jungkamp A. C., Moser M., Müller M. (2010) Mol. Biol. Cell 21, 4151–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. (2003) Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 20. de Leeuw E., Granjon T., Porcelli I., Alami M., Carr S. B., Müller M., Sargent F., Palmer T., Berks B. C. (2002) J. Mol. Biol. 322, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 21. Kreutzenbeck P., Kröger C., Lausberg F., Blaudeck N., Sprenger G. A., Freudl R. (2007) J. Biol. Chem. 282, 7903–7911 [DOI] [PubMed] [Google Scholar]
- 22. Holzapfel E., Eisner G., Alami M., Barrett C. M., Buchanan G., Lüke I., Betton J. M., Robinson C., Palmer T., Moser M., Müller M. (2007) Biochemistry 46, 2892–2898 [DOI] [PubMed] [Google Scholar]
- 23. Marrichi M., Camacho L., Russell D. G., DeLisa M. P. (2008) J. Biol. Chem. 283, 35223–35235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gérard F., Cline K. (2006) J. Biol. Chem. 281, 6130–6135 [DOI] [PubMed] [Google Scholar]
- 25. Schlesier R., Klösgen R. B. (2010) Biol. Chem. 391, 1411–1417 [DOI] [PubMed] [Google Scholar]
- 26. Bageshwar U. K., Whitaker N., Liang F. C., Musser S. M. (2009) Mol. Microbiol. 74, 209–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hynds P. J., Robinson D., Robinson C. (1998) J. Biol. Chem. 273, 34868–34874 [DOI] [PubMed] [Google Scholar]
- 28. Cline K., McCaffery M. (2007) EMBO J. 26, 3039–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richter S., Lindenstrauss U., Lücke C., Bayliss R., Brüser T. (2007) J. Biol. Chem. 282, 33257–33264 [DOI] [PubMed] [Google Scholar]
- 30. DeLisa M. P., Tullman D., Georgiou G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6115–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanders C., Wethkamp N., Lill H. (2001) Mol. Microbiol. 41, 241–246 [DOI] [PubMed] [Google Scholar]
- 32. Musser S. M., Theg S. M. (2000) Eur. J. Biochem. 267, 2588–2598 [DOI] [PubMed] [Google Scholar]
- 33. Santini C. L., Ize B., Chanal A., Müller M., Giordano G., Wu L. F. (1998) EMBO J. 17, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maurer C., Panahandeh S., Moser M., Müller M. (2009) FEBS Lett. 583, 2849–2853 [DOI] [PubMed] [Google Scholar]
- 35. Matos C. F., Robinson C., Di Cola A. (2008) EMBO J. 27, 2055–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Panahandeh S., Maurer C., Moser M., DeLisa M. P., Müller M. (2008) J. Biol. Chem. 283, 33267–33275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sargent F. (2007) Microbiology 153, 633–651 [DOI] [PubMed] [Google Scholar]
- 38. Turner R. J., Papish A. L., Sargent F. (2004) Can. J. Microbiol. 50, 225–238 [DOI] [PubMed] [Google Scholar]
- 39. Mori H., Cline K. (2002) J. Cell Biol. 157, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma X., Cline K. (2010) EMBO J. 29, 1477–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu Y., Zhao E., Li H., Xia B., Jin C. (2010) J. Am. Chem. Soc. 132, 15942–15944 [DOI] [PubMed] [Google Scholar]
- 42. Walther T. H., Grage S. L., Roth N., Ulrich A. S. (2010) J. Am. Chem. Soc. 132, 15945–15956 [DOI] [PubMed] [Google Scholar]
- 43. Chan C. S., Zlomislic M. R., Tieleman D. P., Turner R. J. (2007) Biochemistry 46, 7396–7404 [DOI] [PubMed] [Google Scholar]
- 44. Gouffi K., Gérard F., Santini C. L., Wu L. F. (2004) J. Biol. Chem. 279, 11608–11615 [DOI] [PubMed] [Google Scholar]
- 45. White G. F., Schermann S. M., Bradley J., Roberts A., Greene N. P., Berks B. C., Thomson A. J. (2010) J. Biol. Chem. 285, 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jakob M., Kaiser S., Gutensohn M., Hanner P., Klösgen R. B. (2009) Biochim. Biophys. Acta 1793, 388–394 [DOI] [PubMed] [Google Scholar]
- 47. Brüser T., Sanders C. (2003) Microbiol. Res. 158, 7–17 [DOI] [PubMed] [Google Scholar]
- 48. Blaudeck N., Kreutzenbeck P., Müller M., Sprenger G. A., Freudl R. (2005) J. Biol. Chem. 280, 3426–3432 [DOI] [PubMed] [Google Scholar]
- 49. Blaudeck N., Kreutzenbeck P., Freudl R., Sprenger G. A. (2003) J. Bacteriol. 185, 2811–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 51. Alami M., Trescher D., Wu L. F., Müller M. (2002) J. Biol. Chem. 277, 20499–20503 [DOI] [PubMed] [Google Scholar]
- 52. Ryu Y., Schultz P. G. (2006) Nat. Methods 3, 263–265 [DOI] [PubMed] [Google Scholar]
- 53. Lesley S. A., Brow M. A., Burgess R. R. (1991) J. Biol. Chem. 266, 2632–2638 [PubMed] [Google Scholar]
- 54. Moser M., Panahandeh S., Holzapfel E., Müller M. (2007) Methods Mol. Biol. 390, 63–79 [DOI] [PubMed] [Google Scholar]
- 55. Yahr T. L., Wickner W. T. (2001) EMBO J. 20, 2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bageshwar U. K., Musser S. M. (2007) J. Cell Biol. 179, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dabney-Smith C., Cline K. (2009) Mol. Biol. Cell 20, 2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Warren G., Oates J., Robinson C., Dixon A. M. (2009) J. Mol. Biol. 388, 122–132 [DOI] [PubMed] [Google Scholar]
- 59. Mangels D., Mathers J., Bolhuis A., Robinson C. (2005) J. Mol. Biol. 345, 415–423 [DOI] [PubMed] [Google Scholar]
- 60. Alder N. N., Theg S. M. (2003) FEBS Lett. 540, 96–100 [DOI] [PubMed] [Google Scholar]
- 61. Ma X., Cline K. (2000) J. Biol. Chem. 275, 10016–10022 [DOI] [PubMed] [Google Scholar]
- 62. Fincher V., McCaffery M., Cline K. (1998) FEBS Lett. 423, 66–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.