Background: The Kunitz-STI family is a paradigm of protease-inhibitor interaction in particular and protein-protein recognition in general.
Results: PPI is a versatile protease inhibitor that targets several subfamilies of serine proteases.
Conclusion: The β-trefoil fold constitutes an evolutionary platform for protease inhibition and molecular recognition.
Significance: Fold plasticity influences protein evolution toward multiple function and binding promiscuity.
Keywords: Protease, Protease Inhibitor, Protein Evolution, Protein Structure, Protein-Protein Interactions, Serine Protease, Structural Biology, Surface Plasmon Resonance (SPR), X-ray Crystallography, X-ray Scattering
Abstract
Proteases carry out a number of crucial functions inside and outside the cell. To protect the cells against the potentially lethal activities of these enzymes, specific inhibitors are produced to tightly regulate the protease activity. Independent reports suggest that the Kunitz-soybean trypsin inhibitor (STI) family has the potential to inhibit proteases with different specificities. In this study, we use a combination of biophysical methods to define the structural basis of the interaction of papaya protease inhibitor (PPI) with serine proteases. We show that PPI is a multiple-headed inhibitor; a single PPI molecule can bind two trypsin units at the same time. Based on sequence and structural analysis, we hypothesize that the inherent plasticity of the β-trefoil fold is paramount in the functional evolution of this family toward multiple protease inhibition.
Introduction
Protease inhibitors are nature's instruments for the regulation of the activity of their target proteases (1). They act by blocking them in emergency cases and as switches in many signaling pathways. In plants, many protease inhibitors serve in defensive mechanisms, and their expression levels are increased in injured tissues (2). The activation of serine protease inhibitors in tobacco is also known to increase the mortality rate of herbivores, especially neonate larvae (3).
In plants, serine protease inhibitors are widespread. They are classified into a number of families: serpins, Kunitz-STI,3 Bowman-Birk, potato-type I and II, squash, and thaumatin-like inhibitors (2, 4). For several decades, the Kunitz-STI superfamily has served as one of the model systems for the study of protein structure and protease-inhibitor recognition. Many principles have been discovered first for members of this family and were later confirmed for other families of inhibitors. The Kunitz-STI family belongs to the β-trefoil fold superfamily, which displays an extremely high plasticity regarding their interacting partners (5). The latter involves protein, DNA, and carbohydrate recognition, and some of them are even enzymes. Variations in the lengths, sequences, and conformations of the many loops that cover the surface of the protein define the specificity of the interaction. Given the high variability of the loop regions, these proteins have a wide variety of targets and could in principle engage multiple binders at the same time (5).
Kunitz-type protease inhibitors act by inserting a protruding loop into the active site of their target protease(s) (6). It is commonly assumed that most members of this family have only a single reactive site loop, which for the archetypical soybean trypsin inhibitor (STI) is located between residues Ser-60 and Phe-66. However, several cases of inhibitors possessing two reactive sites, and thus binding two target molecules simultaneously, have been reported (7–10). These have been dubbed “double-headed” or “Janus-type” inhibitors.
The past three decades have provided a wealth of structural and thermodynamic data that shed light on how proteinaceous inhibitors counteract the proteolytic action of serine proteases (1, 6, 11). However, despite the abundance of data obtained from x-ray crystallography for free inhibitors and complexes with serine proteases (7–9, 12–14), very little is known on the specifics of multiple target recognition by these Janus-type proteins with only one structure in the Protein Data Bank of a Kunitz-STI inhibitor bound to two protease molecules (7).
Papaya protease inhibitor (PPI) is a double-headed Kunitz-type serine protease inhibitor isolated from the latex of Carica papaya. It has a high stability and is particularly resistant to proteolysis by proteases from different families (15). It likely serves as defense protein as it is induced by wounding and is inactive against endogenous proteases from C. papaya (15). Here we report the crystal structure of PPI in two crystal forms, together with the solution structure of its complexes with one and two trypsin units, as determined by small angle x-ray scattering (SAXS) and perform a detailed kinetic study of the protease-inhibitor interaction probing the serine protease superfamily in a systematic manner. We also discuss how the potential of the β-trefoil fold to accept surface mutations influences protein evolution toward multiple function and multiple target recognition.
EXPERIMENTAL PROCEDURES
Proteins
PPI was purified from commercially available papaya latex, as a spray-dried powder, kindly provided by Enzymase International S.A. as described before (15). For details on the different proteases used in this work, see the supplemental material.
Preparation and Purification of PPI-Trypsin and PPI-Chymotrypsin Complexes
For the preparation of the 1:1 (PPI:protease) complexes, equimolar amounts of PPI and the corresponding protease were incubated for 30 min at 37 °C in 50 mm Tris-HCl buffer, 20 mm Ca2+, pH 8.0, for trypsin or in 50 mm Tris-HCl, 60 mm Ca2+, pH 8.0, for α-chymotrypsin. The 1:2 (PPI:protease) complexes were prepared under the same experimental conditions as for the 1:1 complexes by mixing 2 moles of protease per mole of PPI. The four resulting complexes were concentrated on a 5000 molecular weight cut-off Vivaspin 15R concentrator (Sartorius) to a volume of 3 ml and loaded onto a Sephadex G-75 column (40 × 2.6-cm inner diameter) pre-equilibrated with the same degassed buffer. Fractions of 4.2 ml were collected at a flow rate of 63 ml/h. The fractions corresponding to each complex were pooled, concentrated to a volume of 3 ml, and reloaded onto the same column under the same conditions. Finally, the fractions corresponding to the respective complexes were pooled and concentrated up to 30 mg/ml. Bovine serum albumin (66.2 kDa), hen egg white ovalbumin (45.0 kDa), bovine carbonic anhydrase (31.0 kDa), and horse heart cytochrome c (12.4 kDa) were used as protein molecular mass standards.
Crystallization and Structure Determination
The crystallization of PPI in two crystal forms has been described before (15, 16). Crystals of form I were flash-cooled directly in the x-ray beam after soaking for ∼1 min in a cryoprotectant solution consisting of 1.4 m ammonium sulfate, 0.1 m MES, 0.01 m CoCl2, pH 6.5, and 20% glycerol. Data for crystal form II were collected after the addition of a suitable cryoprotectant or mounted in glass capillaries for data collection at room temperature. X-ray data were collected at European Molecular Biology Laboratory (EMBL) stations BW7A and X13 of the Deutsches Elektronen Synchrotron (DESY) synchrotron (Hamburg, Germany) using an MAR CCD detector and at station ID14-1 of the European Synchrotron Radiation Facility (ESRF) synchrotron (Grenoble, France) using an Area Detector Systems Corp. Quantum Q4 CCD detector. All data were indexed and processed with the HKL2000 suite (17). Intensities were converted to structure factor amplitudes using the CCP4 program TRUNCATE (30).
The structure of crystal form I was determined by molecular replacement using the structure of STI (Protein Data Bank (PDB) number 1avx:B) as a search model. The initial molecular replacement model resulting from PHASER was used as starting model in Arp/wArp, which was able to automatically build around 90% of the structure. The final model was obtained after alternating cycles of refinement with phenix.refine and manual build using Coot and has an Rfree of 19.7% and Rwork of 18.3% with excellent statistics (see Table 1).
TABLE 1.
Data collection and refinement statistics
| Crystal form I | Crystal form II | |
|---|---|---|
| Data collection | ||
| Detector | MAR CCD | MAR CCD |
| Beamline | ID14–1 | BW7A |
| Data collection temperature (K) | 100 | 100 |
| Wavelength (˚) | 0.8073 | 0.9150 |
| Unit cell parameters (˚) | a = 44.26, b = 81.99, c = 140.89 | a = b = 74.70, c = 78.97 |
| Space group | P212121 | P31 |
| Resolution range (˚) | 20–1.7 (1.76–1.7) | 15.0–2.6 (2.69–2.6) |
| No. of observed reflections | 318,867 (22,384) | 178,588 (20,260) |
| No. of unique reflections | 51,816 (5596) | 15,065 (1809) |
| Rmerge | 0.065 (0.165) | 0.110 (0.763) |
| Completeness | 90.4 (82.2) | 100 (100) |
| I/σ(I) | 17.3 (6.1) | 24.4 (2.3) |
| Redundancy | 6.15 (4.00) | 11.88 (11.20) |
| Refinement | ||
| Resolution (˚) | 1.7 | 2.6 |
| Rwork/Rfree | 18.3/19.7 | 17.3/22.8 |
| Bond lengths (˚) | 0.017 | 0.007 |
| Bond angles (°) | 1.93 | 1.08 |
| Ramachandran profile | ||
| Core (%) | 97.4 | 94.1 |
| Allowed regions (%) | 2.6 | 5.9 |
| Outliers (%) | 0.0 | 0.0 |
The structure of crystal form II was determined by molecular replacement with PHASER. The coordinates from the refined form I were used as search model. These PPI crystals are typically merohedral twins. For structure determination and refinement, we selected the dataset with the lowest twining fraction (0.26). Accordingly, the structure was refined against the twined data as implemented in phenix.refine, using the twin operator h,-h-k,-l. The final model (Rwork = 17.7%, Rfree = 22.8%) was validated with MolProbity (18). The statistics of the refinement are shown in Table 1.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were carried out on a Biacore3000 system (GE Healthcare) at 25 °C in 20 mm Tris, pH 7.5, 150 mm NaCl, 20 mm CaCl2, 0.005% Tween 20 and with a flow rate of 30 μl/min. To qualitatively test the specificity of binding of PPI for proteases of different families, the inhibitor was immobilized on a CM5 sensor chip via amine coupling in 10 mm sodium acetate buffer, pH 4.0, and different serine proteases were passed over the chip. In a second series of experiments, we used a sandwich arrangement to validate the presence of multiple binding sites in PPI. In our design, the protease was coupled to a CM5 chip, and then PPI was passed over. Taking advantage of the slow dissociation rate of PPI, we could probe other reaction sites by injecting a new protease.
We selected bovine trypsin, chymotrypsin, and elastase (the tight binders) for kinetic experiments. In every case, a short pulse of guanidinium hydrochloride (6 m) was used to regenerate the chip. All the binding data were analyzed with the BIAevaluation 4.1 software. (Please refer to the supplemental material for further details on the methods and data analysis.)
Small Angle X-ray Scattering
SAXS data for characterizing the different PPI-trypsin complexes and examine their shapes and dimensions were collected at the synchrotron beamlines X33 (DESY Hamburg, Germany) and SWING (Soleil Paris, France). The radius of gyration (RG) of the different particles calculated from the Guinier analysis, together with other SAXS-derived parameters, are shown in the supplemental Table S2. For all samples, PPI, trypsin, PPI:trypsin, and trypsin:PPI:trypsin, Guinier plots of the data show a very good fit to linearity, indicating the absence of aggregation. The indirect Fourier transform package GNOM (19) was used to compute the distance distribution P(r) functions from the scattering curve and calculate the maximum dimension of the particles (Dmax). CRYSOL (20) was used to compare the experimental data with the scattering curve computed from all the different models derived from the crystal structures of PPI and trypsin. For rigid body modeling of the different PPI-trypsin complexes, we used SASREF (21) combined with distance restrains obtained from the docking experiments.
In Silico Docking
The inhibitory potential of all PPI surface loops was tested by docking a PPI monomer into the trypsin active site, using the docking program HADDOCK 2.0. In each docking run, the active site of trypsin was targeted by defining residues 40, 171, 172, 175, 192, 204, and 206 as active residues and residues 42, 43, 140, 174, 194, and 196 as passive residues. Additionally, the surface loops 20–25, 40–45, 76–82, 124–132, and 190–198 were defined as fully flexible segments to enable the optimization of additional trypsin-PPI contacts in the wider region surrounding the active site. With this receptor definition, distinct docking runs were set up by defining different regions of the inhibitors STI and PPI as the interaction partners of the constant target region. In each run, surface loops surrounding the prospective binding loop were defined as flexible regions to optimize secondary interactions between the proteins. Solutions were scored based on the final energy of the docked complex, the total buried surface area in the complex, and the predicted interactions with the active site and specificity pocket of trypsin.
RESULTS
Overall Structure of PPI
A BLAST search shows that PPI is a member of the miraculin family of taste-modifying proteins, which are active against serine proteases. The structure of PPI was determined in two crystal forms at high resolution (Table 1). Both forms contain two monomers in their asymmetric unit. All four monomers have well defined electron density for residues 2–183 and show visible electron density for the GlcNAcβ[(1-4)GlcNAcβ(1-4)Manβ(1-3)Man]β(1-3)Fuc attached to Asn-84 and the first GlcNAc residue attached to Asn-90. The molecule is exceptionally rigid, with root mean square deviation values between the four independently determined molecules ranging between 0.20 and 0.45 Å for all Cα atoms and between 0.27 and 0.63 Å for all heavy (nitrogen, carbon, oxygen, and sulfur) atoms (Fig. 1A). This structural rigidity is confirmed by the small angle x-ray scattering profile of PPI with a Kratky plot typical of a compact globular protein (supplemental Fig. S1) and likely reflects its resistance toward proteolysis and the harshness of its natural environment, the papaya latex (15).
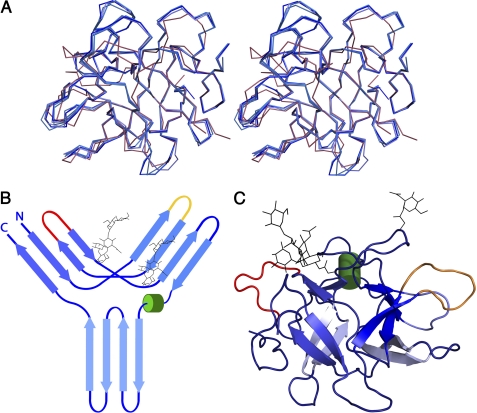
FIGURE 1.
Crystal structure of PPI and schematic representation of the β-trefoil fold. A, stereo view of the superposition of the Cα trace of all four PPI monomers onto the Cα trace of STI. B, topology diagram of PPI, showing the typical β-trefoil fold of the Kunitz-STI family. Cylinders represent helices, and arrows represent strands. C, graphic representation of the crystal structure of PPI as observed in the crystal structure. The two known glycosylation sites, Asn-84 and Asn-90, are represented as black lines, and the reactive loops β2-β3 and β4-β5 are in yellow and red, respectively.
The overall structure consists of six two-stranded hairpins that adopt the β-trefoil fold typical of the Kunitz-STI family (13). Three of these hairpins form a barrel structure, and the other three are in a triangular array that caps the barrel and gives the molecule a pseudo-three-fold axis (Fig. 1, B and C). PPI and STI (the canonical representative of this family of protease inhibitors) share 26% sequence identity, and their structures superpose with an overall root mean square deviation of 1.45 Å for 144 common Cα atoms (Fig. 1A).
The crystal structure of PPI does not lead directly to insights into its mechanism of action. The canonical loop located between strands β4 and β5 (residues 64–71) contains a 2-amino acid insertion, which makes it very unlikely to fit into the active site of trypsin in the same manner as the corresponding loop in STI. Also, other loop conformations from related inhibitors such as BASI and API-1 (7, 9) that have been observed to bind to the active site of trypsin are not observed in PPI, leaving its mode of inhibition unexplained.
PPI Is a Versatile Protease Inhibitor
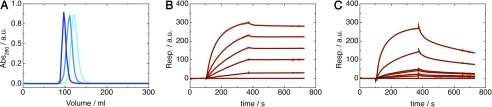
Previous measurements indicated that PPI inhibits trypsin and chymotrypsin (15). Analytical gel filtration experiments show that a single, monomeric PPI molecule is able to interact simultaneously with two trypsin molecules to form a hetero-trimeric complex. PPI elutes at an apparent molecular mass of 23 kDa, in close agreement with its theoretical monomeric mass of 23,490 Da. Upon the addition of substoichiometric amounts of trypsin, a new species appears with an apparent molecular mass of 47 kDa suggestive of a hetero-dimeric PPI:trypsin complex. At a 1:1 ratio of PPI to trypsin, only this complex is observed, indicating a tight interaction between both proteins. Further addition of trypsin leads to the appearance of a novel, faster migrating species at the expense of the hetero-dimeric complex. This novel species has an apparent molecular mass of 70 kDa and likely consists of one PPI sandwiched between two trypsin molecules (Fig. 2A).
FIGURE 2.
PPI-protease interactions. A, size exclusion chromatography profiles of PPI (in cyan) and PPI-trypsin complexes with different stoichiometries (PPI-trypsin in light blue and PPI-(trypsin)2 in dark blue). a.u., arbitrary units. B and C, binding of PPI to trypsin (B) and chymotrypsin (C) monitored with SPR. Black lines represent the best fit of the model function (which assumes two independent PPI-binding sites) to the experimental data (red lines).
The total absence of the hetero-trimeric complex at a 1:1 ratio indicates that the two binding sites on PPI differ significantly in their affinities for trypsin. Indeed, if two sites of equal affinity would be present, at a 1:1 ratio, one would observe an equilibrium between free PPI, PPI:trypsin, and trypsin:PPI:trypsin in a 1:2:1 ratio. Similar observations were made for the interaction of PPI with chymotrypsin, where a hetero-dimeric and a hetero-trimeric species are also observed (data not shown).
Binding Specificity of PPI to Serine Proteases
To further investigate the specificity and stoichiometry of the interaction between PPI and proteases, we used SPR to probe a set of 11 commercially available serine proteases, three cysteine proteases purified from papaya latex, and one commercially available metalloprotease (supplemental Table S1). Using PPI immobilized onto the flow cell surface, the binding constants and kinetics of association (kon) and dissociation (koff) were determined (Table 2). The typical tight-binding interaction described for other members of the Kunitz-STI family was detected only for members of the S1 family of serine proteases, especially for trypsin, chymotrypsin, and elastase (Fig. 2, B and C, supplemental Table S1). Additionally, PPI was also capable of binding to subtilisin and proteinase K, which are members of the S8B family. The rest of the proteases showed no appreciable binding to PPI.
TABLE 2.
PPI/protease binding statistics
| Species | kon1 | koff1 | kon2 | koff2 | KD1 | KD2 | χ2 |
|---|---|---|---|---|---|---|---|
| 1/m | 1/s | 1/m | 1/s | nm | nm | ||
| PPI-trypsin | 5.9 105 | 2.0 10−7 | 6.2 105 | 5.1 10−2 | 0.00034 | 82.2 | 1.9 |
| PPI-chymotrypsin | 2.6 105 | 1.3 10−3 | 0.6 105 | 4.3 10−2 | 5.0 | 716.7 | 2.6 |
| PPI-elastase | 5.4 103 | 4.4 10−4 | 82.7 | 0.9 |
The observed association rates (kon) showed that PPI responded slightly faster to trypsin than chymotrypsin (by a factor of ∼2 for site H and a factor of ∼10 for site L, Table 2), which suggests that indeed trypsin-like proteases are the preferred interaction partners of PPI. This is also in agreement with the measured koff values, which are significantly slower for trypsin, reflecting the fact that breaking specific short range interactions between the two proteins required for the dissociation is more difficult in the case of trypsin.
Trypsin and chymotrypsin bind to PPI with a 2:1 stoichiometry in contrast to elastase or subtilisin. The dissociation constants from each binding site differ by several orders of magnitude. To relate the two binding sites for trypsin to those for chymotrypsin, we designed a sandwich-SPR experiment where we coupled the protease to a CM5 chip via amine coupling, and then a saturating amount of PPI was injected. Given the strength of the interaction between PPI and the trypsin/chymotrypsin, the off-rate of this complex is sufficiently slow to allow accurate measurements of a second binding event on the exposed second site (supplemental Fig. S2). This experiment allowed the independent determination of the kinetic parameters of a second binding site for trypsin and chymotrypsin. This alternative interaction turns out to be slightly weaker for both enzymes, suggesting that PPI contains a high affinity (site H) site and a low affinity (site L) site for trypsin and chymotrypsin.
In Silico Docking Suggests Candidates for the Two Active Site Loops of PPI
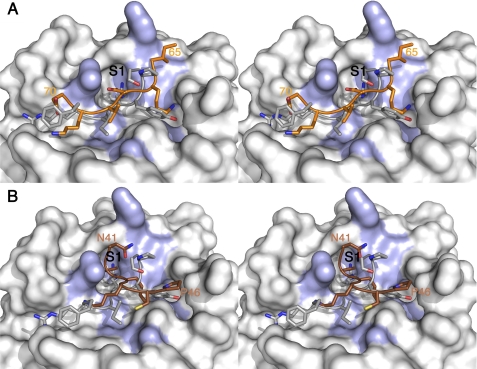
As attempts to crystallize the complexes of PPI with trypsin or chymotrypsin were unsuccessful, we looked for alternative methods to obtain structural information of the interaction between PPI and trypsin. Initially, we focused on flexible in silico docking using each of 11 loops present in PPI. As a positive control, the β4-β5 loop (residues 60–66) of the STI was targeted to the trypsin active site, resulting in a top solution with energy of −161 kcal/mol and a buried surface area of 2088 Å2 (supplemental Fig. S3). The position and conformation of the STI loop in the docked trypsin complex are a close match to those in the known crystal structure of this complex (PDB entry 1AVW). The detailed conformation of the STI side chains near the trypsin active site also correspond very well to the known structure (supplemental Fig. S3), with Arg-63 occupying the specificity pocket (S1) and Tyr-62 covering the side chain of the catalytic Ser-192.
The docking runs equally target loop β4-β5 (residues 64–71) of PPI into the active site of trypsin. The top cluster has an energy of −126 kcal/mol and a buried surface area of 1829 Å2. In this cluster, the orientation of the β4-β5 loop differs from what is seen in the crystal structure between trypsin and STI, which is not unexpected given its insertion of two amino acids, which precludes the classic interaction mode. In our model, the side chain of Asn-67 occupies the S1 pocket of trypsin, and the catalytic Ser-195 is covered by residue Val-68 of PPI (Fig. 3A). This is a recurrent theme in most miraculin-like Kunitz-STI trypsin inhibitors (22) and suggests that the S1 Lys/Asn substitution involves additional changes in the inhibitory mechanism. In the PPI-trypsin complex, the residues expected to enter the active site, loop away from the protease. This model suggests that PPI works by occluding the active site of the protease rather than using the more traditional “Laskowski-like” mechanism (6).
FIGURE 3.
Details of the interactions between PPI and trypsin based on the docking models. A and B, stereo view of the interactions involving the reactive loops β2-β3 (A) and β4-β5 (B) in orange in comparison with the canonical loop from STI (in silver). The main residues on the surface of trypsin, interacting with PPI are colored in blue, and the S1 pocket is labeled in black.
The docking run for PPI loop β2-β3 (residues Pro-39 to Pro-46 of PPI) also resulted in a top cluster targeted to the active site. The corresponding interaction energy of −113 kcal/mol together with the buried surface area of 1712 Å2 suggest a lower affinity interaction when compared with the canonical loop. In all solutions, Lys-43 occupies the specificity pocket in a manner that is very similar to the mode of binding of Arg-63 in the STI complex, whereas Lys-42 very effectively covers the catalytic serine (Fig. 3B).
The remaining loops dock in a more scattered fashion without systematically recurring interactions to trypsin in their conformational ensemble. Although several of them show significant interaction energies, the specificity pocket of trypsin remains unoccupied or clearly suboptimal interactions are observed for the lowest energy solution. Loop β5-β6 is glycosylated, and the attached glycans would likely hamper this loop in its interaction with a protease. In addition, PPI contains several Lys and Arg residues that could potentially interact with the active site of trypsin. Lys-82 from loop β5-β6 is too close to one of the glycosylation sites, and Lys-114 from loop β7-β8 and Arg-149 from β9-β10 are located close to β4-β5, making it unlikely that trypsin or chymotrypsin would be able to bind to them without severe steric clashes with a trypsin bound to β4-β5. Arg-146 is in a scaffolding role that shapes loop β9-β10. The rest of the Lys/Arg residues are located in loops β2-β3 and β4-β5 or in β-strands.
The docking results are a strong indication that loops β2-β3 and β4-β5 are the reactive centers of PPI toward serine proteases. Therefore we based the modeling of the PPI-serine protease interactions on these results and used this information for the SAXS shape reconstruction of the complexes.
Solution Structure of the PPI-Trypsin Complexes
We tried to crystallize the different trypsin-PPI complexes to obtain a molecular description of their structure. We obtained good quality crystals under several different conditions, but they invariably contained only trypsin. We resorted to a different strategy based on SAXS measurements to obtain the overall shapes of the complexes and determine the relative orientations of both trypsin-binding sites on the surface of PPI.
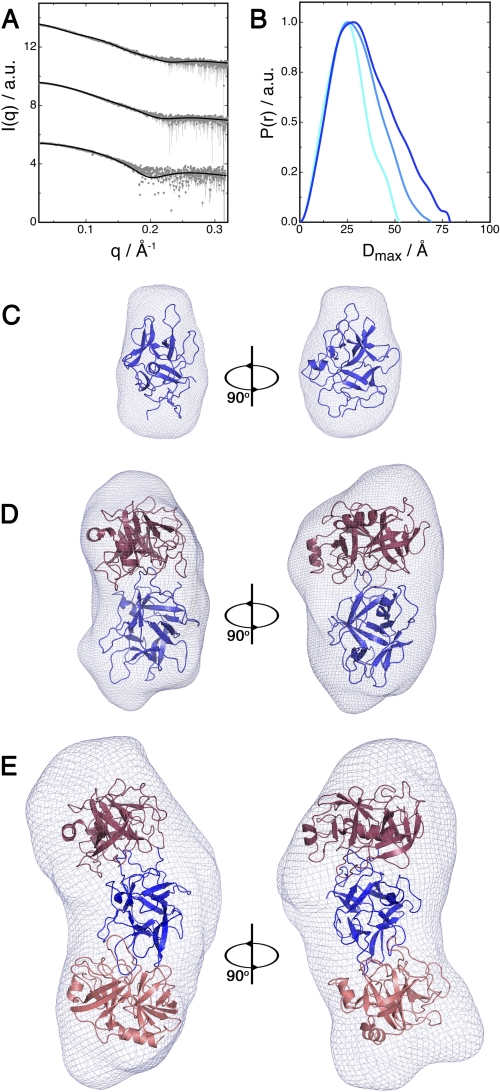
The structural parameters of PPI and the PPI-trypsin complexes calculated from the experimental scattering curves (Fig. 4A) are shown in supplemental Table S2. The estimated molecular weight of all particles agrees well with those predicted from the sequence and observed by gel filtration. Kratky plots are consistent in every case with a properly folded, homogeneous, and well structured species (supplemental Fig. S4, A and B). The distance distributions P(r) computed from the experimental data of free PPI and of the binary complex show a bell-shaped curve typical for globular particles (Fig. 4B). The P(r) function of the ternary complex, on the other hand, has a skewed profile pointing toward an elongated particle with a length of about 3 nm.
FIGURE 4.
Solution structure of PPI and the PPI-trypsin complexes. A, experimental SAXS scattering curves for free PPI and the PPI-trypsin and PPI-(trypsin)2 complexes (from top to bottom). a.u., arbitrary units. B, distance distribution function of free PPI, PPI-trypsin, and PPI-(trypsin)2 in cyan, light blue, and dark blue, respectively. C–E, SAXS-reconstructed pseudo-atomic structures of PPI (C), PPI-trypsin complex (D), and PPI-(trypsin)2 complex (E). The rigid body models are superposed onto ab initio reconstructed shapes.
To obtain pseudo-atomic models of the binary and ternary complexes, we used the simulated annealing refinement protocol implemented in SASREF for the rigid body modeling of the different PPI-trypsin complexes (21). The resulting models were further compared with ab initio shapes reconstructed using DAMMIF (see “Experimental Procedures” for details). The best model for the trypsin-PPI hetero-dimer places the β4-β5 loop of PPI into the trypsin active site. A systematic search rotating the β4-β5 loop within the trypsin active site confirmed the solution obtained by the docking study as the one giving the best agreement between the experimental SAXS scattering curve and the theoretical curves calculated from the pseudo-atomic model. In a similar approach, we determined the structure of the ternary complex (see “Experimental Procedures”), which revealed the second trypsin molecule binding to the loop β2-β3. The results from the SAXS-based modeling are in agreement with the docking results. Therefore we used a hybrid approach combining SAXS-based rigid body modeling restrained with the information from the docking experiment to build the refined models of the complexes. This approach is expected to significantly improve the resolution limit for the consensus models, thus allowing for meaningful analysis of protein-protein interactions in the complex beyond the nominal resolution of SAXS. Overall, the ab initio and rigid body models calculated from the SAXS data are consistent with each other and give us an idea of the shape of the binary and ternary PPI-trypsin complexes (Fig. 4, C–E).
Loop Versatility and Promiscuity in the β-Trefoil Fold
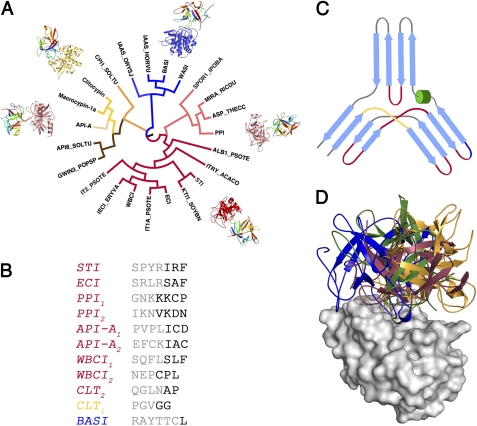
To investigate the function/evolution relationship between PPI and other proteins within the Kunitz-STI family, we calculated a structure-based phylogenetic tree using the structures of known inhibitors with different protease specificities. The analysis of the tree shows that as the proteins diverge from the STI-like core, so does the inhibitory mechanism, and for more distant homologues, even the protease specificity is completely lost (Fig. 5, A–C). PPI belongs to the subgroup of trypsin/chymotrypsin inhibitors closest to the canonical STI-like group; however, the conformation of the PPI canonical loop (β4-β5) is different from the one observed for the canonical STI-like group, as is typically observed for miraculin-like inhibitors (22). More distantly related are the BASI/WASI-like proteins, which are subtilisin inhibitors and also active against α-amylases (23).
FIGURE 5.
Loop versatility and binding modes of Kunitz-STI inhibitors. A, structure-based phylogenetic analysis of the Kunitz-STI family. Colors represent different specificity for the target protease/enzyme: blue, α-amylase/subtilisin; orange, aspartic proteases; yellow, cysteine/serine proteases; brown, serine protease inhibition outside the canonical loop; red, serine protease canonical inhibition; pink, serine proteases inhibition by the miraculin family. B, sequence of the reactive loop from the different subfamilies of Kunitz-STI inhibitors, colored according to A. C, topological position of the different reactive loops mapped on the β-trefoil fold and colored according to A. D, orientation of different Kunitz-STI inhibitors in relation to the active site of trypsin. The inhibitors are colored according to A, and trypsin is colored in light gray.
The most distant inhibitors involve proteins such as macrocypins and clitocypins, from organisms even outside the plant kingdom (10). This group of proteins typically inhibits cysteine proteases, but some of them are aspartic protease inhibitors (API-8), and others such as API-A and API-B are capable of binding two trypsin units at the same time (7). Not surprisingly, for API-A, one of these trypsin-inhibiting loops uses a novel mechanism, whereas the other adopts a canonical conformation, yet it is located between β-strands β9 and β10, as the opposite of β4-β5 for the canonical loop of STI (13). Moreover, its conformation is restrained by two disulfide bridges, which suggests the presence of convergent evolution at play.
A remarkable consequence from this prolific molecular recognition display is that outside the interaction on P1 and P1′, the relative orientation protease/inhibitor differs significantly (Fig. 5D). This is closely related to the mechanism by which these inhibitors resist proteolysis and is crucial for defining the specificity and strength of the interaction.
DISCUSSION
PPI Is a Broad Spectrum Serine Protease Inhibitor
The overall structure of PPI is similar to other Kunitz-type protease inhibitors from plants. Despite low sequence similarity, the β-trefoil fold is present in many superfamilies of proteins including soybean Kunitz family inhibitors, cytokines, agglutinins, ricin B-like lectins, fibroblast growth factors, interleukins, tetanus, and botulinum neurotoxins (5, 6).
Kunitz-STI inhibitors are known to interact with multiple proteases and other non-proteolytic enzymes with different activities (6). In this sense, PPI stands out for its ability to interact with several subfamilies of serine proteases and its remarkably high resistance to proteolytic degradation. This is largely due to the unique suitability of the β-trefoil fold for molecular recognition and its tolerance to point mutations, given the fact that most of the protein surface consists of loop regions.
The structure of the PPI-trypsin ternary complex is in contrast with that of the API-A-trypsin complex, the other member of the family for which a ternary complex with trypsin has been determined (7). API-A engages trypsin through loops β5-β6 and β9-β10, whereas PPI uses β2-β3 and β4-β5; consequently, the overall shape of both ternary complexes differs significantly.
The SPR and gel filtration experiments showed that PPI binds strongly serine proteases from the trypsin/chymotrypsin clan and also interacts with members of the subtilisin clan (Table 2). Taken together this supports a possible function of PPI as a broad spectrum inhibitor.
The PPI Reactive Loops
The canonical loop of serine protease inhibitors interacts with proteinases predominantly via a lock-and-key mechanism. In the Kunitz-STI family, this loop possesses a substrate-like protruding shape that allows the P1 side chain to stay hyper-solvent accessible while keeping the carbonyl oxygen atoms of P2 and P1′ residues projecting toward the concave side of the loop (1, 6). The Kunitz-STI inhibitor was the first one of a very large number of protease inhibitors, described to be involved in the “standard mechanism” or “Laskowski mechanism” of serine protease inhibition (6). They act by binding tightly to the active sites of their targets, as a substrate would, but are resistant to proteolysis. Other protease inhibitors bind to the substrate binding cleft, projecting the inhibitory loop away from the active site residues, and in this way, they evade proteolysis (1, 6).
PPI strongly inhibits trypsin and α-chymotrypsin via a slow, tight-binding mechanism. Combining structural information from x-ray crystallography, small angle x-ray scattering, and docking, we assigned the PPI inhibitory activity to loops β2-β3 and β4-β5. The first reactive loop of the β-trefoil fold has been already described to be involved in the inhibition of cysteine proteases by macrocypins and clitocypins (10). In the case of PPI, this loop is involved in the inhibition of serine proteases. The binding energies and surface area buried upon binding obtained from the docking experiments suggest that it is a slightly weaker trypsin binder when compared with the other PPI reactive loop. However, the interaction is still very strong, and the proteins form a stable ternary complex at a 2:1 ratio. In contrast to the second reactive loop of PPI, β2-β3 is constrained by the disulfide bridge between Cys-45 and Cys-89 and also by the presence of Pro-39 and Pro-46, which limits significantly the conformational space that the loop can explore.
The second reactive loop of PPI is located in the canonical position (identified based on sequence similarity within the Kunitz-STI superfamily) and encompasses residues from Lys-66 to Ile-72. However, the structure of this loop is far from the canonical arrangement observed in other members of the family (13). An insertion of three residues between the P1 and P2 sites of PPI reactive loop disrupts its conformation dramatically. Hence the loop can no longer bind the target in the canonical way due to several steric clashes with residues from the protease.
The peculiarities of the PPI reactive site also extend to the scaffolding residues that tether the loop in the most favorable binding conformation. Structural studies on Kunitz-STI inhibitors (24–27) revealed that a conserved Asn maintains the canonical conformation of the reactive site through a network of hydrogen bonds. Moreover, this dense hydrogen-bonding network that supports the reactive loop is one of the paradigms of serine protease inhibitors. This feature combines with an acyl enzyme and the correct orientation of the religating amide to arrest proteolysis. Interestingly, this network is completely absent in PPI. Tyr-16 replaces the conserved Asn, and consequently, the reactive site conformation holds mainly through Van der Waals interactions provided by the hydroxy-phenyl group of this tyrosine with the backbone and the side chains of Ala-64 and Val-68.
β-Trefoil Fold, an Evolutionary Platform for Protein-Protein Interactions
This fold is common to several protein families, all of which are involved in recognition functions. The Kunitz-STI family has been found to inhibit proteases and α-amylases, cytokines such as fibroblast growth factors, and interleukin-1-mediated immune response, and it is also involved in carbohydrate binding in plant and animal lectins (1, 5, 28). More surprisingly, in the CSL family of Notch-type receptors, a β-trefoil domain contributes to DNA binding and harbors the site of mutually exclusive interactions that switches from Notch repression to activation (29).
The remarkable functional plasticity of this fold relies on the fact that its sequence constraints are weak and that the surface of the protein is formed mainly from loops that differ in sequence, length, and conformation (5). Such a display holds enormous possibilities for the creation of potential binding sites. Moreover, the native state in the energy landscape of the β-trefoil family is accessible from multiple routes, and therefore these proteins would fold even if the most easily traversed paths were blocked by small changes (31, 32). In such a scenario, one can picture this fold as highly plastic and receptive to point mutations in its many surface loops, which can accumulate and become active upon a certain selective pressure.
All these features, together with an internal pseudo-three-fold symmetry, bestow proteins from this superfamily with the ability to interact with multiple partners at the same time. Kunitz-STI inhibitors are a particularly good example. From the analysis of the topology of the fold, a single molecule could potentially engage other enzymes units through 11 different loops. Indeed, the structures of PPI in complex with two trypsin units, BASI/WASI in complex with α-amylase and subtilisin (9, 23), API-A in complex with trypsin (7), and clitocypin in complex with serine/cysteine proteases (10), show that a single inhibitor molecule is capable of interacting with two enzymes. Moreover, the position of the reactive loops varies within the families, with occurrences in loops β2-β3, β4-β5, β5-β6, β6-β7, and β10-β11, and the relative specificities are also interchangeable with position β2-β3 being used for the inhibition of serine and cysteine proteases.
Supplementary Material
Acknowledgments
We acknowledge the use of beamtime at the EMBL beamlines at the DESY (Hamburg, Germany), ESRF (Grenoble, France), and SWING (Paris, France) synchrotrons. Enzymase International is gratefully acknowledged for having generously supplied C. papaya latex as a spray-dried powder.
This work was supported by grants from the Vlaams Interuniversitair Instituut voor Biotechnologie (VIB), the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO), and the Onderzoeksraad of the Vrije Universiteit Brussel.

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods, Tables S1 and S2, and Figs. S1–S4.
The atomic coordinates and structure factors (codes 3S8K and 3S8J) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- STI
- soybean trypsin inhibitor
- PPI
- papaya protease inhibitor
- API
- aspartic protease inhibitor
- SAXS
- small angle x-ray scattering
- BASI
- barley α-amylase/subtilisin inhibitor
- WASI
- wheat α-amylase/subtilisin inhibitor.
REFERENCES
- 1. Bode W., Huber R. (1992) Eur. J. Biochem. 204, 433–451 [DOI] [PubMed] [Google Scholar]
- 2. Ryan C. A. (1990) Annu. Rev. Phytopathol. 28, 425–449 [Google Scholar]
- 3. Zavala J. A., Patankar A. G., Gase K., Hui D., Baldwin I. T. (2004) Plant Physiol. 134, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Leo F., Volpicella M., Licciulli F., Liuni S., Gallerani R., Ceci L. R. (2002) Nucleic Acids Research 30, 347–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murzin A. G., Lesk A. M., Chothia C. (1992) J. Mol. Biol. 223, 531–543 [DOI] [PubMed] [Google Scholar]
- 6. Laskowski M., Jr., Kato I. (1980) Annu. Rev. Biochem. 49, 593–626 [DOI] [PubMed] [Google Scholar]
- 7. Bao R., Zhou C. Z., Jiang C., Lin S. X., Chi C. W., Chen Y. (2009) J. Biol. Chem. 284, 26676–26684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dattagupta J. K., Podder A., Chakrabarti C., Sen U., Mukhopadhyay D., Dutta S. K., Singh M. (1999) Proteins Struct. Funct. Genet. 35, 321–331 [DOI] [PubMed] [Google Scholar]
- 9. Micheelsen P. O., Vévodová J., De Maria L., Ostergaard P. R., Friis E. P., Wilson K., Skjøt M. (2008) J. Mol. Biol. 380, 681–690 [DOI] [PubMed] [Google Scholar]
- 10. Renko M., Sabotic J., Mihelic M., Brzin J., Kos J., Turk D. (2010) J. Biol. Chem. 285, 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bode W., Huber R. (2000) Biochim. Biophys. Acta 1477, 241–252 [DOI] [PubMed] [Google Scholar]
- 12. Chen C., Hsu C. H., Su N. Y., Lin Y. C., Chiou S. H., Wu S. H. (2001) J. Biol. Chem. 276, 45079–45087 [DOI] [PubMed] [Google Scholar]
- 13. Song H. K., Suh S. W. (1998) J. Mol. Biol. 275, 347–363 [DOI] [PubMed] [Google Scholar]
- 14. McCoy A. J., Kortt A. A. (1997) J. Mol. Biol. 269, 881–891 [DOI] [PubMed] [Google Scholar]
- 15. Azarkan M., Dibiani R., Goormaghtigh E., Raussens V., Baeyens-Volant D. (2006) Biochim. Biophys. Acta 1764, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 16. Azarkan M., Garcia-Pino A., Dibiani R., Wyns L., Loris R., Baeyens-Volant D. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1239–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konarev P. V., Petoukhov M. V., Volkovb V. V., Svergun D. I. (2006) J. Appl. Crystallogr. 39, 277–286 [Google Scholar]
- 20. Svergun D. I., Barberato C., Koch M. H. J. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 21. Petoukhov M. V., Svergun D. I. (2005) Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gahloth D., Selvakumar P., Shee C., Kumar P., Sharma A. K. (2010) Arch. Biochem. Biophys. 494, 15–22 [DOI] [PubMed] [Google Scholar]
- 23. Vallée F., Kadziola A., Bourne Y., Juy M., Rodenburg K. W., Svensson B., Haser R. (1998) Structure 6, 649–659 [DOI] [PubMed] [Google Scholar]
- 24. Dasgupta J., Khamrui S., Dattagupta J. K., Sen U. (2006) Biochemistry 45, 6783–6792 [DOI] [PubMed] [Google Scholar]
- 25. De Meester P., Brick P., Lloyd L. F., Blow D. M., Onesti S. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 589–597 [DOI] [PubMed] [Google Scholar]
- 26. Ravichandran S., Dasgupta J., Chakrabarti C., Ghosh S., Singh M., Dattagupta J. K. (2001) Protein Eng. 14, 349–357 [DOI] [PubMed] [Google Scholar]
- 27. Dattagupta J. K., Podder A., Chakrabarti C., Sen U., Dutta S. K., Singh M. (1996) Acta Crystallogr. D. Biol. Crystallogr. 52, 521–528 [DOI] [PubMed] [Google Scholar]
- 28. Loris R. (2002) Biochim. Biophys. Acta 1572, 198–208 [DOI] [PubMed] [Google Scholar]
- 29. Kovall R. A., Hendrickson W. A. (2004) EMBO J. 23, 3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project Number 4 (1994) TRUNCATE, Daresbury Laboratory, Daresbury, Warrington, UK [Google Scholar]
- 31. Capraro D. T., Roy M., Onuchic J. N., Jennings P. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14844–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chavez L. L., Gosavi S., Jennings P. A., Onuchic J. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10254–10258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.