Background: How Wzz proteins control O antigen chain length is not well understood.
Results: Residue 321, located within the second coiled-coil region, of the Wzz2 protein influences the chain length produced.
Conclusion: Position 321 affected the chain length regulating activity and oligomer stability of Wzz2.
Significance: Prior studies focusing on Wzz and Wzy interactions in regulating chain length do not fully explain Wzz activity.
Keywords: Carbohydrate Biosynthesis, Lipopolysaccharide (LPS), Membrane Proteins, Polysaccharide, Pseudomonas aeruginosa
Abstract
The production of preferred lipopolysaccharide O antigen chain lengths is important for the survival of pathogenic Gram-negative bacteria in different environments, yet how Wzz proteins regulate these lengths is not well understood. The Wzz2 proteins from two different serotype O11 Pseudomonas aeruginosa strains are responsible for the expression of different very long chain lengths despite high sequence homology. Site-directed mutagenesis was performed to determine whether a specific amino acid was responsible for this difference in chain length; the residue present in position 321 within the second predicted coiled-coil region was able to determine which chain length was produced. A panel of site-directed mutants introducing different amino acids at this position implicated that the charge of the amino acid affected chain length, with positively charged residues associated with shorter chain lengths. Expression data also suggested this site was important for overall stability of the protein because mutants predicted to disrupt proper folding of the α helix led to lower protein levels. Cross-linking studies found that Wzz2 proteins producing shorter chain lengths had more stable higher-order oligomers. Mapping residue 321 onto the solved Escherichia coli Wzz FepE crystal structure predicted it to be located within α helix 8, which participates in intermonomeric interactions. These data further support the observation that Wzz oligomerization is necessary for chain length regulating activity but also provide evidence that differences in complex stability or changes in the conformation of the oligomer can lead to shifts in the length of the O antigen side chain.
Introduction
Lipopolysaccharide (LPS) is a major component of the outer membrane of a Gram-negative bacterium. It is composed of lipid A, the core oligosaccharide, and the O antigen side chain. The O antigen side chain, which consists of repeating sugar subunits, is important for the virulence of pathogenic Gram-negative bacteria because it can be involved in the attachment to host cells, protection from the innate immune system, and proper localization of surface virulence factors (1). Different Gram-negative bacteria produce side chains of different lengths, and even within a species the length can vary depending on strain. Several species produce O antigen side chains with two different preferred lengths; there is evidence these chain lengths have been optimized to allow for better survival within the host during an infection (2, 3).
The Wzz proteins, which are members of the polysaccharide co-polymerase family of proteins (4), are responsible for determining the number of O antigen subunits that are linked together to form the side chain. In strains producing two different preferred chain lengths, different Wzz proteins are used to regulate each chain length (5–7). Although the sequence similarity among Wzz proteins from different species is relatively low, they all contain two transmembrane domains at the amino- and carboxyl-terminal ends of the protein that anchors them in the inner membrane. The middle portion is exposed in the periplasm; structural analyses indicate it is largely α-helical (8, 9). Because O antigen side chain synthesis occurs in the periplasm, this allows for the Wzz protein to interact with either the Wzy polymerase or the growing chain of O antigen subunits to determine the number of subunits linked together before ligation to lipid A+core.
Despite the importance of achieving proper O antigen side chain lengths for the survival of the bacteria during infections, how the Wzz proteins regulate this activity is not well understood. Mutational analyses have determined that single amino acid changes over the entire length of the protein could lead to shifts in preferred chain length or completely abolish the ability of Wzz to regulate any chain length (10, 11). Because these studies were unable to define a specific active site associated with regulating O antigen chain length, it is now assumed that Wzz does not possess any inherent enzymatic activity for regulating chain length but instead acts as a structural scaffold for the other O antigen assembling components.
Cross-linking studies indicate Wzz proteins form homo-oligomers (11), which has been confirmed via several structural analyses (9, 12, 13). This ability to homo-oligomerize is tied to their length-regulating activity; mutants that no longer oligomerize also fail to regulate chain length (11). Oligomers with sizes consistent of octamers have been observed via Western blot (6). The crystal structures to the periplasmic region of three Wzz proteins have been solved, and all three crystallized as oligomers with one, Wzz FepE, existing as a nonamer (9). The crystal structures suggested a general shape for Wzz oligomers, with the monomers coming together to form an open barrel structure that extends ∼100 Å into the periplasmic space. Because there is a rough correlation between the number of Wzz proteins associated together in an oligomer and the length of the O antigen side chain produced, a model has been proposed where the bell-shaped Wzz oligomer serves as a scaffold around which the rest of the O antigen assembly proteins organize. According to this theory, an increased number of Wzz monomers in the oligomer could aggregrate a larger number of the Wzy polymerase, leading to increased chain lengths (9).
Pseudomonas aeruginosa encodes two Wzz proteins, Wzz1 and Wzz2 (6). Daniels et al. (6) demonstrated that Wzz2 is highly homologous across different serotypes of P. aeruginosa, and we have taken advantage of this similarity to perform our own site-directed mutagenesis studies in an attempt better understand Wzz activity. Previous studies have attempted to alter chain length with mutations designed to disrupt the conformation of the Wzz protein, whereas we were able to compare variations in wild-type sequences of two Wzz2 proteins from two strains of the same serotype that produce different very long chain lengths. Our results found a major role for the second coiled-coil region in determining the chain length produced. This location appears to be involved in monomer-monomer interactions and raises intriguing possibilities about the role of oligomer stability for the chain length regulating activity of Wzz.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Strains were grown at 37 °C in LB either rotating or shaking at 200 rpm for liquid cultures or on l-agar or trypticase soy agar for plate cultures. Escherichia coli Top10 was used for all cloning experiments. Media were supplemented with gentamicin (250 μg/ml for P. aeruginosa, 15 μg/ml for E. coli), or ampicillin (100 μg/ml for E. coli).
DNA Manipulations
Chromosomal P. aeruginosa DNA was isolated using the Wizard Genome Prep kit (Promega). PCRs were performed using Roche Applied Science HiFi polymerase, and amplicons were purified using the Qiagen PCR purification kit following the manufacturer's instructions. Plasmid DNA was isolated using the Qiagen Miniprep kit. Restriction enzymes were purchased from New England Biolabs and used following the manufacturer's instructions. Ligations were performed with the Roche Applied Science Rapid DNA ligation kit.
Cloning of wzz2 Genes and Site-directed Mutagenesis
The wzz2 genes were initially amplified from genomic DNA preps of the different P. aeruginosa strains using the primers wzz2 Fd Eco (CAT GAA TTC ATG CCT TCC TCA CAG CTT CCG) and wzz2 HA Rv Hind (CAT AAG CTT CTA GGC ATA GTC TGG GAC GTC ATA TGG ATA GGT CCC GGA AAG GCT CCG), which include restriction sites and a C-terminal HA tag. The PCR products were cloned through TOPO2.1 (Invitrogen) before being digested out and inserted into pHERD30T (14). Site-directed mutagenesis was performed using the QuikChange II site-directed mutagenesis kit according to the manufacturer's instructions (Agilent Technologies), and all mutations were confirmed by sequencing.
LPS Preparation
Strains were grown by diluting 150 μl of an overnight culture into 3 ml of LB supplemented with gentamicin. Cultures were grown for two hours with rotation at 37 °C. Wzz2 expression was induced with arabinose, and the cultures were allowed to grow for another 2 h. Cultures were standardized to an A600 = 0.5 and 1 ml pelleted in an Eppendorf tube. The pellet was resuspended in 2× SDS buffer (0.1 m Tris-HCl, pH 6.8, 4% β-mercaptoethanol, 4% SDS, 20% glycerol) and boiled for 10 min. Proteinase K was added at 10 μg/ml, and samples were incubated for 3 h at 60 °C. Samples were preserved at −20 °C until they could be analyzed by SDS-PAGE.
Protein Expression and Cross-linking
Cultures were grown either as above or by diluting 1:50 from an overnight culture into 10 ml of LB with gentamicin followed by shaking at 200 rpm at 37 °C. After induction with 2% arabinose for 2 h, samples were diluted to A600 = 0.5 and pelleted. For whole cell lysates, 200 μl of 2× SDS buffer was added, and the samples were boiled for 10 min. For cross-linking analysis, pellets were washed with 1 ml of PBS and then resuspended in 1 ml of PBS. 0.5% Formaldehyde was added, and samples were allowed to incubate at room temperature for 1 h. Samples were then pelleted, resuspended in 200 μl of 2× SDS, and boiled. To follow cross-linking over time, samples were grown as above in 10 ml of LB. After induction, samples were pelleted and washed with LB before being resuspended in 10 ml of LB with gentamicin and 1% glucose to repress gene expression. Whole cell lysates and cross-linking samples were treated as above when removed at indicated time points.
SDS-PAGE and Western Blotting
Samples were analyzed on either 8 or 12% polyacrylamide gels. Following transfer to nitrocellulose membranes, LPS was detected using a polyclonal serogroup O11 antiserum (Accurate Chemical & Scientific) followed by a goat anti-rabbit IgG coupled to horseradish peroxidase (Sigma-Aldrich). The HA tag was detected using an anti-HA clone HA-7 antibody (Sigma) with a goat anti-mouse IgG coupled to horseradish peroxidase secondary (Sigma).
RESULTS
P. aeruginosa Serotype O11 Strains Produce Different Very Long Chain Lengths
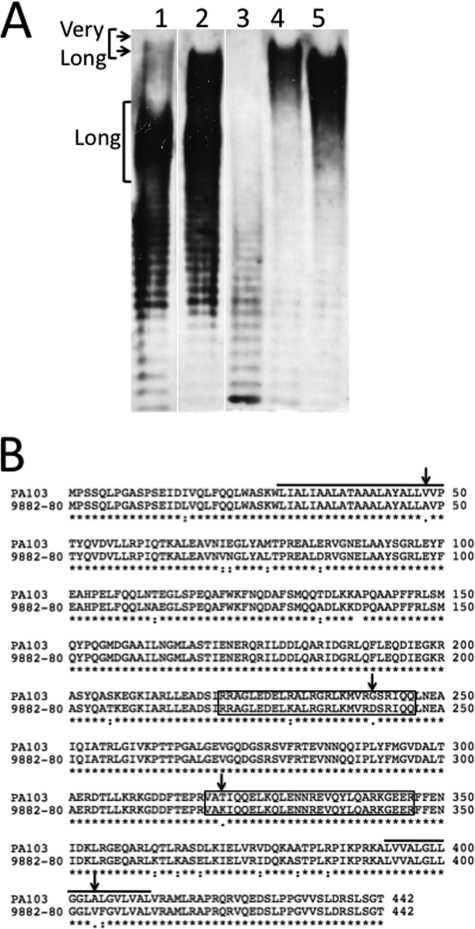
Previous evidence suggested that the wzz2 sequence was highly conserved among the 20 serotypes of P. aeruginosa (6). However, when the LPS was isolated from two serotype O11 strains, PA103 and 9882-80, a slight difference in the size of very long chain length was evident; PA103 produces a longer very long chain length compared with 9882-80 (Fig. 1A, lanes 1 and 2). Because this difference would not be explained by differences in the O antigen subunit or other O antigen assembly proteins, we hypothesized it would be due to differences in the Wzz2 proteins between the two strains. To test this, the wzz2 genes from each strain were cloned into an expression vector and used to complement a PA103 wzz double deletion mutant (DΔM) (3), which produces an unregulated distribution of chain lengths (Fig. 1A, lane 3). Although expression of both proteins led to the production of a very long chain length, the differences in length remained; complementation with the PA103 wzz2 gene produces a longer very long chain length compared with the 9882-80 wzz2 gene (Fig. 1A, lanes 3 and 4). We believed that comparing chain length differences between two such similar Wzz2 proteins from the same serotype might provide better insight into Wzz activity compared with previous studies.
FIGURE 1.
Wzz2 proteins produce different very long O antigen chain lengths. A, Western blot of LPS isolated from the wild-type P. aeruginosa serotype O11 strains and the DΔM complemented with the two wild-type Wzz2 proteins. The top arrow indicates the longer “very long” chain length produced by PA103, whereas the bottom arrow marks the shorter very long chain length produced by 9882-80. Lanes are as follows: lane 1, PA103; lane 2, 9882-80; lane 3, DΔM+pHERD30T; lane 4, DΔM+pPA103 wzz2; lane 5, DΔM+p9882-80 wzz2. B, TCOFFEE alignment between PA103 Wzz2 and 9882-80 Wzz2. Transmembrane domains are indicated by the black bars, and the coiled-coil regions (predicted using the COILS program) are outlined in boxes. Sites chosen for site-directed mutagenesis are marked with arrows.
An alignment was performed between the two Wzz2 proteins to identify regions of non-homology that might explain why each produces a different very long chain length. Overall, the proteins were very similar to each other, although there were a few positions where non-conserved amino acids existed between the two proteins. Fig. 1B shows the sequences of the Wzz2 proteins; the solid black lines indicate transmembrane domains, and the predicted coiled-coil regions are outlined in boxes. It was interesting to note that these differences fell within the transmembrane domains or the predicted coiled-coil regions in the periplasm because these have been implicated to be important for Wzz activity and oligomerization in other studies (11, 15, 16). Four amino acids were chosen, shown with arrows, for site-directed mutagenesis to determine whether these sites were responsible for the shift in very long chain length seen between PA103 and 9882-80.
Residue in Second Coiled-coil Region Determines Chain Length Produced
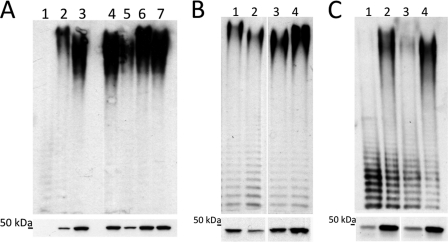
To determine whether any of the four amino acids chosen based on the alignment had an effect on chain length, each amino acid was singly changed in the 9882-80 wzz2 gene to reflect the amino acid present in that position in the PA103 protein. These four mutated 9882-80 wzz2 genes were used to complement the DΔM and LPS from the strains visualized. Three of the mutations in the 9882-80 Wzz2 produced an O antigen chain length similar to the strain complemented with wild-type 9882-80 wzz2, demonstrating that the differences in sequence seen in the transmembrane domains or the first coiled-coil region are not responsible for the shift in very long chain lengths between PA103 and 9882-80 (Fig. 2A, lanes 4, 5, and 7). However, one mutation (K321T), located within the second coiled-coil region, did produce an altered chain length. When the lysine in position 321 was changed to a threonine, the 9882-80 K321T Wzz2 protein produced a longer very long chain length that more resembled that made by the PA103 protein than the wild-type 9882-80 protein (Fig. 2A, lane 6).
FIGURE 2.
Amino acid 321 within second coiled-coil region determines chain length. A, effect of site-directed mutants in 9882-80 Wzz2 protein. For each panel, the top panel shows LPS profile of strains, whereas the bottom panel depicts expression levels of Wzz2 proteins. The lanes depict the DΔM containing the vectors as follows: lane 1, pHERD30T; lane 2, pPA103 wzz2; lane 3, p9882-80 wzz2; lane 4, p9882-80 A48V; lane 5, p9882-80 D241G; lane 6, p9882-80 K321T; lane 7, p9882-80 V404A. B, changing Wzz2 protein at position 321 affects chain length. The lanes depict the DΔM containing the vectors as follows: lane 1, pPA103 wzz2; lane 2, pPA103 T321K; lane 3, p9882-80 wzz2; lane 4, p9882-80 K321T. C, amount of Wzz2 protein does not affect chain length produced. Lanes 1 and 2 are the DΔM expressing pPA103 wzz2, and lanes 3 and 4 indicate pPA103 T321K. Amounts of arabinose shown are 0.2% (lanes 1 and 3) versus 2.0% (lanes 2 and 4).
To further confirm the importance of this position when determining chain length, the reverse mutation was made in the PA103 wzz2 gene. When the threonine was changed to a lysine (T321K), a decrease in the chain length resembling that produced by 9882-80 Wzz2 was evident (Fig. 2B, lane 2). These results indicate that the amino acid present in position 321 of the Wzz2 protein has an important role for Wzz2 activity when producing very long chain length. However, a slight difference in the amount of Wzz2 protein produced from the different strains was noted, with lower protein amounts present for the cloned genes producing the shorter very long chain length (Fig. 2B, lower panel, lanes 2 and 3 versus lanes 1 and 4). Differential expression of the PA103 wild-type Wzz2 and the PA103 T321K Wzz2 proteins was performed to ensure that it is residue 321 determining chain length and not overall amount of protein. Fig. 2C illustrates that the difference in longer and shorter very long chain length still exists even when the proteins are present in different quantities. The amount of the chain lengths correlates with the amount of Wzz2, but the length of the side chain is dependent on the amino acid present in position 321.
Effect of Panel of Site-directed Mutants on Chain Length
Threonine is a polar amino acid, whereas lysine is positively charged, suggesting that the charge of the amino acid at position 321 could explain why different chain lengths are produced. A panel of site-directed mutants was generated for position 321 in the PA103 wzz2 gene to see the effect on chain length for amino acids of different charges and different sizes. When residue 321 was switched to an arginine, which is similarly charged to lysine, the chain length was reduced in a manner similar to that seen for PA103 T321K and the 9882-80 wild-type Wzz2 proteins (Fig. 3A, lane 4). However, the histidine mutant did not exhibit this decrease in chain length despite being positively charged (Fig. 3A, lane 8). A majority of the mutants did not noticeably affect the length of the very long side chain, but several did impact the amount of the very long chain length produced. Most notably, the glycine mutant produced a reduced amount of very long side chain (Fig. 3A, lane 7). The most dramatic phenotype of any mutant was that seen when residue 321 was deleted. This protein lost the ability to regulate chain length altogether so that no very long chain length was detected, resulting in a phenotype similar to strain containing empty vector (Fig. 3A, compare lane 11 versus lane 1).
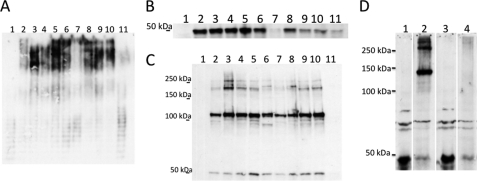
FIGURE 3.
Effect of site-directed mutations at amino acid 321 of PA103 Wzz2 protein. A, LPS of all DΔM strains complemented with various site 321 mutations of pPA103 wzz2 after 2 h of induction. B, whole cell lysates from all strains to demonstrate expression levels using αHA antibody. C, formaldehyde cross-linking to determine oligomerization of strains. Strains represent the DΔM containing the plasmids as follows: lane 1, pHERD30T; lane 2, pPA103 wzz2; lane 3, pPA103 T321K; lane 4, pPA103 T321R; lane 5, pPA103 T321A; lane 6, pPA103 T321D; lane 7, pPA103 T321G; lane 8, pPA103 T321H; lane 9, pPA103 T321Y; lane 10, pPA103 T321W; lane 11, pPA103 Δ321. D, cross-linking of the Δ321 mutant Wzz2 protein compared with wild-type PA103 Wzz2. Wild-type PA103 Wzz2 was induced with 0.2% arabinose, whereas Δ321 Wzz2 was induced with 2.0% arabinose to produce similar levels of protein. Lanes 1 and 3 are boiled lysates that demonstrate the Wzz2 monomers produced. Lanes 2 and 4 are the cross-linked samples; PA103 Wzz2 exhibits cross-linking, whereas Δ321 Wzz2 does not.
Protein Expression and Cross-linking Profiles of Site-directed Mutants
Given the effects seen during differential expression of the PA103 wild-type and PA103 T321K Wzz2 proteins, where the amount of protein correlated with the amount of very long chain length, it is possible that poor protein expression could explain some of the results seen using the panel of 321 mutants to complement very long chain length production in the DΔM. To determine whether this was the case, the different PA103 wzz2 genes were induced similarly, and expression was determined by detection of the C-terminal HA tag. Fig. 3B shows the expression levels of all the mutated Wzz2 proteins after 2 h of induction. Most of the proteins express to levels similar to the wild-type PA103 Wzz2, but the PA103 T321G and Δ321 Wzz2 proteins exhibit reduced amounts (Fig. 3B, lanes 7 and 11). Because glycine is a known helix breaker, and deletion of residue 321 would interrupt the proper folding of the predicted coiled-coil region, it suggests that the α helices present in this region are important for Wzz2 stability.
Formaldehyde cross-linking has been used to detect the homo-oligomerization of Wzz proteins (11). Cross-linking of all of the PA103 mutated Wzz2 proteins was performed to determine whether there were any differences among the different Wzz2 proteins. All of the proteins except the Δ321 protein exhibit cross-linking profiles similar to the wild-type PA103 Wzz2, indicating these mutations do not affect the number of Wzz monomers coming together (Fig. 3C). Using the exposure presented to detect distinct cross-linked Wzz2 bands, the monomer of Δ321 could not be detected in Fig. 3C (lane 11). The cross-linking experiment was repeated, adjusting the arabinose levels for wild-type PA103 Wzz2 to 0.2% to make expression comparable to the unstable Δ321 protein. Even with protein levels adjusted, the Δ321 Wzz2 protein did not cross-link at all (Fig. 3D, lane 4); only the monomer was detected. Membrane preparations of this strain demonstrated that the Δ321 Wzz2 protein did insert into the inner membrane (data not shown) but is presumably misfolded to an extent that it is incapable of forming monomer-monomer interactions. Because the Δ321 wzz2 gene did not result in production of a very long chain length, this illustrates the importance of Wzz oligomerization for its chain length regulating activity.
Stability of Threonine and Lysine Versions of Wzz2 Oligomers
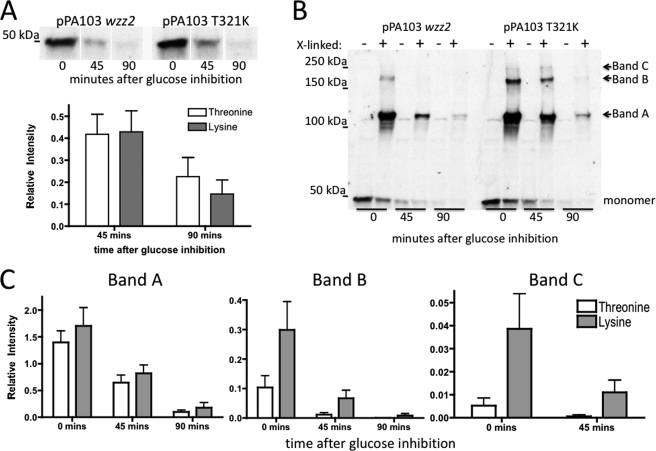
Our data indicate that different amino acids in position 321 can affect the stability of the Wzz2 protein. When comparing the cross-linking profile of PA103 wild-type Wzz2 to the T321K protein, an increase in the amount of higher-order oligomers was evident for T321K Wzz2 (Fig. 3C, lanes 2 and 3). This suggested that there was a difference in stability of the oligomer between threonine and lysine versions of the Wzz2 proteins. We took advantage of the fact that the different wzz2 genes are expressed from the pHERD30T vector, which contains a PBAD promoter, allowing us to repress gene expression by the addition of glucose. Wild-type PA103 Wzz2, PA103 T321K, wild-type 9882-80 Wzz2, and 9882-80 K321T were induced for 2 h, and then protein production inhibited by the addition of glucose; protein degradation and oligomer stability were tracked using the C-terminal HA tag. The experiment was repeated three times, resulting in six different samples each for threonine (PA103 Wzz2 and 9882-80 K321T) versus lysine (9882-80 Wzz2 and PA103 T321K) versions of the Wzz2 proteins. The results were quantified by determining the intensity of each band and standardizing it to the amount of protein present in the whole cell lysate at the beginning of the assay. Fig. 4 depicts representative examples of the results showing the PA103 Wzz2 and PA103 T321K Wzz2 proteins. When comparing the total Wzz2 amount in the whole cell lysate at each time point, no difference in Wzz2 degradation between the different proteins was apparent (Fig. 4A). However, when looking at cross-linked samples taken at different time points, it appeared that more of the protein existed as higher-order oligomers (bands A, B, and C) for the lysine versions of the protein (Fig. 4B). When quantifying the results, the lysine Wzz2 proteins had higher amounts of protein existing as oligomers compared with threonine versions of the protein (Fig. 4C). Although the results did not achieve statistical significance, the difference did become more apparent for higher-order oligomers (Fig. 4C, bands B and C) compared with dimers (assumed to be band A in Fig. 4B). This suggests that lysine versions of the Wzz2 protein hold together as an oligomer more stably than threonine versions.
FIGURE 4.
Stability of Wzz2 oligomers. A, expression levels from whole cell lysates were compared before addition of glucose (time 0) and at 45 and 90 min after inhibition. The experiment was repeated three times with PA103 Wzz2, PA103 T321K Wzz2, 9882-80 Wzz2, and 9882-80 K321T Wzz2. Densitometry was performed with ImageJ software, and relative intensity was normalized to the amount of protein present in the time 0 sample. The top panels show protein expression from a representative experiment, and the bottom panel illustrates densitometry results for whole cell lysates. The white bars indicate threonine versions (PA103 Wzz2 and 9882-80 K321T Wzz2), whereas gray bars indicate lysine versions (PA103 T321K Wzz2 and 9882-80 Wzz2) of the proteins. B, samples were cross-linked for 1 h and then boiled. The blot is representative of an example. Relative intensity of each band was normalized to the intensity of the protein amount present in the whole cell lysate at time 0. −, whole cell lysate samples; +, samples subjected to formaldehyde cross-linking. C, graph of relative intensities of different oligomer bands (indicated in margin of B) at different time points. One-way analysis of variance analysis was used within the GraphPad Prism program to determine statistics.
Predicted Position of Residue 321 in Wzz2 Structure
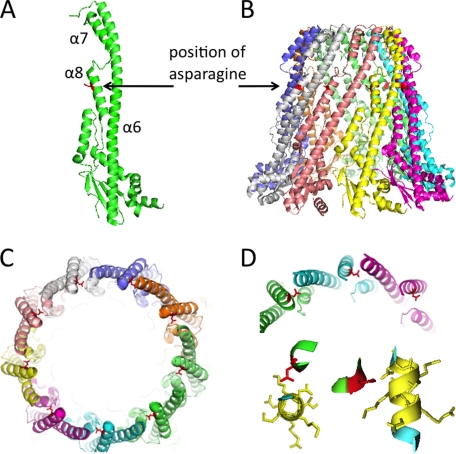
Crystal structures of the periplasmic region of three different Wzz proteins have been solved (9). We used the available structures to predict the location of residue 321 along the Wzz2 protein. A Phyre alignment was performed (Phyre webserver), which found homology ranging from 11–13% between Wzz2 and the secondary structure of all three available Wzz crystal structures. Because FepE (from E. coli O157:H7, 377 amino acids full length) is most similar to Wzz2 (442 amino acids) in regard to protein size and the length of the O antigen side chain produced, it was chosen to model the location of residue 321. An asparagine at position 300 within the FepE sequence aligned with position 321 of Wzz2. Locating the aligning asparagine within the FepE crystal structure demonstrated that this residue was located within α helix 8 and occurs about two-thirds of the way up of the periplasmic barrel formed by the Wzz oligomer (Fig. 5, A and B). It exists closer to the inner surface of the barrel structure but is turned toward the monomer alongside (Fig. 5C). The Phyre alignment indicated that the aligning residue for WzzB(ST) (from Salmonella typhimurium LT2) and WzzE (from E. coli O157:H7) also occurred within α helix 8 (data not shown). It was noted that residues in α helix 8 might be involved with monomer-monomer stability by interacting with the α helix 6 of the neighboring monomer (9); Fig. 5D illustrates the potential interactions with the amino acids occurring along α helix 6 with the FepE asparagine. This region of α helix 6 in the FepE protein did not align well with our Wzz2 sequence, so we were unable to identify the residues along α helix 6 that might interact with position 321 in Wzz2.
FIGURE 5.
Location of amino acid 321 mapped onto the E. coli Wzz FepE crystal structure (Protein Data Bank code 3B8N). A, the asparagine (red) aligning with residue 321 in Wzz2 is located in α helix 8 of the FepE monomer. B, position of aligning asparagine within barrel structure formed by an oligomer. C, aerial view of inner cavity of the FepE oligomer, showing asparagine (red) along the inner surface and pointing to the neighboring monomer. D, close-up of aligning asparagine demonstrating its potential interaction with α helix 6. The top is a close-up view of C. The bottom panel gives an aerial view (bottom left panel) and a side view (bottom right panel) of the amino acids present in α helix 6 that may potentially interact with residue 321 to aid in monomer-monomer stability. All images were generated in PyMOL.
DISCUSSION
In this study we were able to take advantage of the high sequence similarity between two serotype O11 Wzz2 proteins to investigate O antigen chain length regulation. Because the Wzz2 proteins from the two serotype O11 strains produce two different very long O antigen side chains, we were able to use complementation of our DΔM to identify the amino acid responsible for this shift in chain length. The amino acid in position 321, located within the second predicted coiled-coil region, was able to define which chain length was produced. Our use of a panel of site-directed mutants indicated the charge of this amino acid plays a role in determining which chain length is produced and also hinted at differences in protein stability depending on the residue present at position 321. Our cross-linking time course studies suggest the stability of higher order oligomers plays a role in determining chain length. Mapping the location of residue 321 onto the FepE crystal structure indicated it occurred within α helix 8. α Helix 8 interacts with the α helix 6 on neighboring monomers, providing further evidence for this region being involved in monomer-monomer stability.
The most current theory of Wzz activity focuses on the amount of Wzy associated with the Wzz oligomer; larger oligomers can associate with an increased amount of Wzy, leading to more rounds of polymerization and longer chain lengths (9). When trying to explain our results based on this theory of Wzz activity, we found many aspects inconsistent with our data. For example, the evidence presented in favor of the above model suggested that higher levels of Wzz protein leads to shorter chain lengths (9). The idea that Wzz levels influence the chain length produced do not agree with our expression data, where differential expression of the Wzz2 proteins led only to differences in the amount of the preferred chain length. Additionally, in the mutagenesis study recently performed by Papadapolous and Morona (17) on Shigella flexneri WzzSF, it was found that expression levels of their various mutants did not correlate with the resulting chain length; several Wzz proteins were undetectable via Western blotting yet still produced a regulated chain length. These results indicate that chain length is not correlated with the amount of Wzz protein present, as might be expected if Wzz activity is dependent on its interaction with Wzy.
It has been noted that single amino acid changes over the entire length of the Wzz protein can cause slight shifts in the chain length produced (10, 11). The current model, with its emphasis on the complex created between Wzz oligomers and the Wzy polymerase, does not offer any explanation for these observations. The only difference found between threonine and lysine versions of the Wzz2 proteins shown here that might explain the difference in chain lengths produced was the amount of oligomerization; lysine versions of Wzz2 had more of the available protein existing as higher-order oligomers, and these complexes were more stable over the time course experiment. Papadopalous and Morona (17) also noted that chain length was related to the stability of Wzz protein interactions; they described a positive correlation between dimer stability and the production of longer chain lengths. This initially seems to contradict our results, where the Wzz2 proteins that produced shorter very long chain lengths were found to have more stable oligomers, but several key differences in our studies may explain the discrepancy. For example, the introduction of a five-amino acid linker, as performed in their study (17), potentially has a much more dramatic effect on Wzz conformation opposed to the site-directed mutants used in our studies, which were naturally occurring variants from wild-type P. aeruginosa strains. Additionally, higher-order oligomers are much more readily detectable for the P. aeruginosa Wzz2 protein compared with the S. flexneri WzzSF protein, allowing us to track stability of the complex over time more easily.
In addition to the Papadopalous and Morona (17) study mentioned above, another recent study was performed to investigate Wzz activity, and both studies reached conclusions that support our own. One of the mutations identified by Papadopalous and Morona (17) mapped to the same area on α helix 8 on the Wzz FepE crystal structures as the location of our residue 321. This mutated wzzsf gene did not express well compared with other mutations found, demonstrating the importance of this region in protein stability. Kalynych et al. created chimeras between Salmonella enterica WzzST and S. flexneri WzzSF to identify which regions of the proteins were responsible for determining chain length (18). The authors determined that regions spanning from amino acids 200 to 274 in the WzzST and WzzSF proteins dictated which chain length the chimeric Wzz protein would produce. The aligning amino acid in the WzzST crystal structure with our residue 321 is a glutamine in position 256, contained within this region. They also found that this region was important for the stability of the oligomers formed (18). The results of both these studies support our data that α helix 8 plays an important role in Wzz chain length regulating activity, protein expression, and oligomer stability.
One class of mutants in the Papadopalous and Morona study (17) produced a chain length longer than that made by the wild-type protein. Both mutants mapped to the base of the Wzz monomer and were directed toward the center of the cavity within the oligomer (17). They hypothesized that the addition of the five-amino acid linker in this region may have affected the width of the cavity, making it larger and leading to an increase in the number of subunits linked together. Although the interface between monomers is largely hydrophobic, Tocilj et al. (9) noted that the area contained several charged residues that may be important for monomer-monomer interactions (9). In the case of our Wzz2 proteins, the charged lysine may lead to a tighter association between monomers, and the stability of the lysine Wzz2 proteins we observed is in fact evidence for a more compact barrel structure. In this case, a more compact periplasmic domain would lead to a shorter chain length as the Wzz2 “ruler” would be shortened physically. Studies are under way to determine whether there is any significant difference in the conformation of the oligomer between threonine and lysine versions of Wzz2.
We propose a model where Wzz proteins have a preferred oligomerization state and that when they exist in this conformation, they will produce the chain length inherent to their sequence. This would explain how undetectable levels of protein still regulate chain length and the lack of correlation between Wzz amount and the length of the side chain (17). Because there is evidence for an interaction between Wzz and O antigen, the length of the side chain may be determined by physically interacting with the barrel of the oligomer (9). With the barrel acting as a ruler in the periplasm, different conformations of the complex would lead to the production of different side chains. In this scenario, a more compact barrel would produce a shorter chain length because the area along which the O antigen subunit and the oligomer potentially interact would be reduced. The observed correlation between oligomer dimensions and chain length produced would therefore be the result of larger periplasmic barrel structures and not necessarily the amount of associated Wzy.
Our studies have again illustrated the connection between O antigen chain length and the ability of Wzz to form oligomers but provide further evidence that the chain length produced may be dependent on either the stability or the shape of the oligomer formed. Although the interaction between Wzz and Wzy is obviously important in the synthesis of the O antigen side chain, current models may place too much emphasis on the ratio of these proteins. The periplasmic barrel may play an important role in determining the length of the side chain, possibly by acting as way to physically measure the length. Further studies into the structure of this region, especially between two similar proteins producing two different lengths as were used in these studies, may yield important insights into how Wzz proteins regulate chain length.
Footnotes
This work was supported in part by National Institutes of Health Grants 1 R01 AI068112 and 1 R21 AI53842 (to J. B. G.).
REFERENCES
- 1. Kintz E., Goldberg J. B. (2008) Future Microbiol. 3, 191–203 [DOI] [PubMed] [Google Scholar]
- 2. Morona R., Daniels C., Van Den Bosch L. (2003) Microbiology 149, 925–939 [DOI] [PubMed] [Google Scholar]
- 3. Ivanov I. E., Kintz E. N., Porter L. A., Goldberg J. B., Burnham N. A., Camesano T. A. (2011) J. Bacteriol. 193, 1259–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morona R., Purins L., Tocilj A., Matte A., Cygler M. (2009) Trends Biochem. Sci. 34, 78–84 [DOI] [PubMed] [Google Scholar]
- 5. Stevenson G., Kessler A., Reeves P. R. (1995) FEMS Microbiol. Lett. 125, 23–30 [DOI] [PubMed] [Google Scholar]
- 6. Daniels C., Griffiths C., Cowles B., Lam J. S. (2002) Environ. Microbiol. 4, 883–897 [DOI] [PubMed] [Google Scholar]
- 7. Murray G. L., Attridge S. R., Morona R. (2003) Mol. Microbiol. 47, 1395–1406 [DOI] [PubMed] [Google Scholar]
- 8. Guo H., Lokko K., Zhang Y., Yi W., Wu Z., Wang P. G. (2006) Protein Expr. Purif. 48, 49–55 [DOI] [PubMed] [Google Scholar]
- 9. Tocilj A., Munger C., Proteau A., Morona R., Purins L., Ajamian E., Wagner J., Papadopoulos M., Van Den Bosch L., Rubinstein J. L., Féthière J., Matte A., Cygler M. (2008) Nat. Struct. Mol. Biol. 15, 130–138 [DOI] [PubMed] [Google Scholar]
- 10. Franco A. V., Liu D., Reeves P. R. (1998) J. Bacteriol. 180, 2670–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daniels C., Morona R. (1999) Mol. Microbiol. 34, 181–194 [DOI] [PubMed] [Google Scholar]
- 12. Tang K. H., Guo H., Yi W., Tsai M. D., Wang P. G. (2007) Biochemistry 46, 11744–11752 [DOI] [PubMed] [Google Scholar]
- 13. Larue K., Kimber M. S., Ford R., Whitfield C. (2009) J. Biol. Chem. 284, 7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiu D., Damron F. H., Mima T., Schweizer H. P., Yu H. D. (2008) Appl. Environ. Microbiol. 74, 7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marolda C. L., Haggerty E. R., Lung M., Valvano M. A. (2008) J. Bacteriol. 190, 2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Purins L., Van Den Bosch L., Richardson V., Morona R. (2008) Microbiology 154, 1104–1116 [DOI] [PubMed] [Google Scholar]
- 17. Papadopoulos M., Morona R. (2010) J. Bacteriol. 192, 3385–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalynych S., Ruan X., Valvano M. A., Cygler M. (2011) J. Bacteriol. 193, 3710–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]